Abstract
Herpesviruses have evolved a series of abilities involved in the process of host infection that are conducive to virus survival and adaptation to the host, such as immune escape, latent infection, and induction of programmed cell death for sustainable infection. The herpesvirus gene UL24 encodes a highly conserved core protein that plays an important role in effective viral infection. The UL24 protein can inhibit the innate immune response of the host by acting on multiple immune signaling pathways during virus infection, and it also plays a key role in the proliferation and pathogenicity of the virus in the later stage of infection. This article reviews the mechanism by which the UL24 protein mediates herpesvirus immune escape and its effects on viral proliferation and virulence by influencing syncytial formation, DNA damage and the cell cycle. Reviewing these studies will enhance our understanding of the pathogenesis of herpesvirus infection and provide evidence for new strategies to combat against viral infection.
Keywords: herpesvirus, UL24, immune escape, cGAS-STING, DNA damage response, pathogenicity, virulence
Introduction
Herpesviruses are a group of enveloped, double-stranded DNA viruses with similar biological characteristics that are classified within the Herpesviridae family. To date, more than 100 species have been identified, which are often divided into α, β, and γ subfamilies in addition to unclassified herpesviruses (McGeoch et al., 1995; Boyne and Whitehouse, 2006; Ilouze et al., 2006; Santos, 2016; Rathbun and Szpara, 2021). Herpesvirus possesses a double-stranded DNA genome arranged linearly, enclosed within an icosahedral capsid. Encircling the capsid are tegument proteins, while the outermost layer of the virion consists of a lipid bilayer adorned with proteins and glycoproteins (Figure 1A; Deng et al., 2020; Draganova et al., 2020; Gatherer et al., 2021). Herpesviruses infect the skin, mucous membranes and nervous tissue of a wide range of hosts, seriously affecting the health of humans and other animals (Gupta et al., 2007; Zaravinos et al., 2009; Crimi et al., 2019). Among these, the viruses that often infect humans include herpes simplex virus type 1 and type 2 (HSV-1, HSV-2), varicella zoster virus (VZV), Epstein–Barr virus (EBV), human cytomegalovirus (HCMV), Kaposi’s sarcoma herpes virus (KSHV), and human roseoloviruses, which comprise three different species, human herpesviruses 6A, 6B, and 7 (HHV-6A, HHV-6B, HHV-7), and are genetically related to human cytomegalovirus (Zerboni et al., 2014; Agut et al., 2016; Damania et al., 2022; Ijezie et al., 2023; Martin de Frémont et al., 2023). In addition, horse herpes virus (EHV), pseudorabies virus (PRV), Marek’s disease virus (MDV) and duck plague virus (DPV) infect animals (Pomeranz et al., 2005; Fritsche and Borchers, 2011; Ruan et al., 2022; Zheng et al., 2023). Throughout the host infection process, viral proteins have evolved diverse functions that contribute to the virus’s enhanced survival.
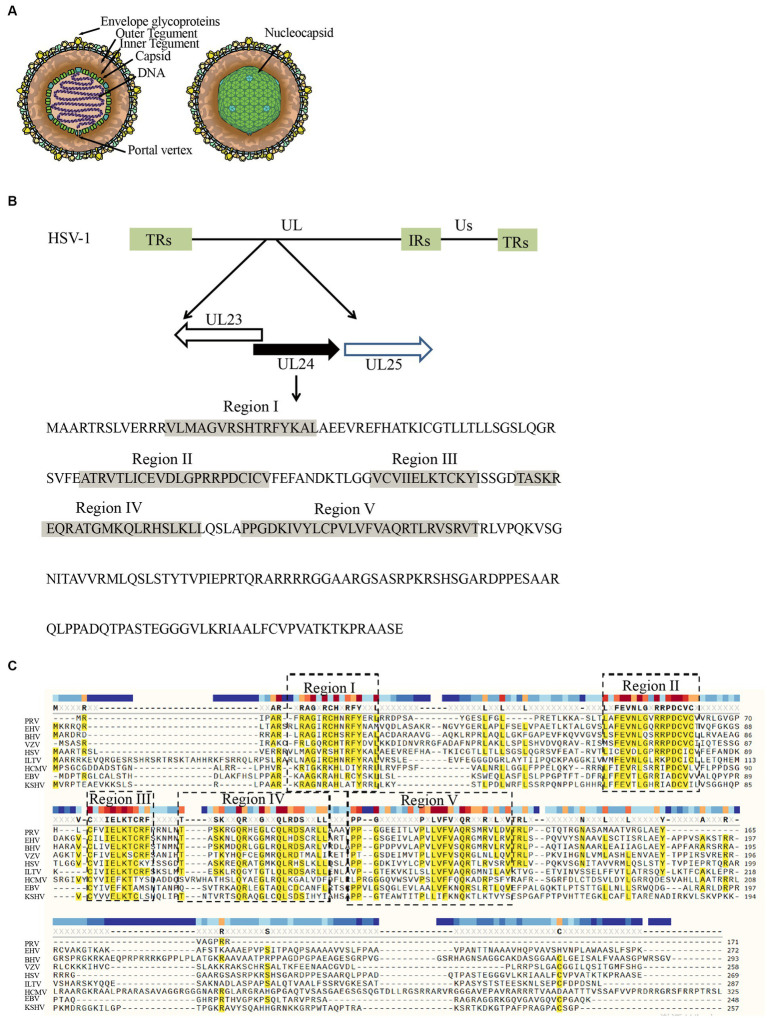
Figure 1.
Structure of the HSV-1 genome and the region encoding the UL24 gene. (A) Structure of the HSV-1 (Hulo et al., 2011). (B) The diagram demonstrates the locations of the UL23, 24, and 25 ORFs and the direction of transcription (arrows) for the parental strain (Pearson and Coen, 2002). Indicated by shading are the locations of domains (Regions I–V) conserved among various herpesvirus homologs of UL24 (Jacobson et al., 1998). (C) Multiple alignment of UL24 homologs revealed that they have five conserved functional domains (Dezélée et al., 1996).
The coding gene of the herpesvirus UL24 protein family is located in a unique long region, and the similarity of amino acids is very high. Except for channel catfish herpesvirus, UL24 protein is conserved in the whole herpesvirus family (Davison, 1992; Bertrand et al., 2010). The proteins encoded by the HCMV UL76, VZV ORF35, KSHV ORF20 and EHV-1 ORF37 genes also belong to the herpesvirus UL24 protein family (Bateman et al., 2004; Ito et al., 2005; Kasem et al., 2010; Hoffman et al., 2021). Except for the HCMV UL76 gene, the UL24 genes and TK genes of other herpesviruses are arranged in a head-to-head manner at the 5′ end (Figure 1B; Jacobson et al., 1989; Dezélée et al., 1996; Shimojima et al., 1997; Pearson and Coen, 2002; Ito et al., 2005; Li et al., 2006).
UL24 is currently considered a core gene of herpesviruses and is present in both mammalian and avian herpesviruses (Nunberg et al., 1989; Lymberopoulos and Pearson, 2007; Jia et al., 2009; Bertrand et al., 2010; Carvalho et al., 2012; Mahmoudian et al., 2012). The transcription of herpesvirus genes presents as a continuous cascade pattern, which is divided into immediate early genes (α), early genes (β), and late genes (γ) according to the chronological order of expression (Sacks et al., 1985; Liu et al., 2015; Zhou et al., 2023). After viral DNA synthesis, UL24 gene products appear in cumulative form late in infection, indicating that UL24 is a late gene (Hong-Yan et al., 2001; Pearson and Coen, 2002; Pearson et al., 2004; Mahmoudian et al., 2012). In herpesviruses, the UL24 protein consists of five highly conserved functional domains that determine most of its functions and are important for the life cycle of the virus (Figure 1C; Jacobson et al., 1989; Shimojima et al., 1997; Knizewski et al., 2006; Nascimento et al., 2009). As herpesvirus research advances, the role of UL24 is becoming increasingly understood. In this review, we examine the function of the UL24 protein by introducing the role of the herpes virus UL24 protein in immune escape, pathogenicity, and the cell cycle.
UL24 participates in immune escape
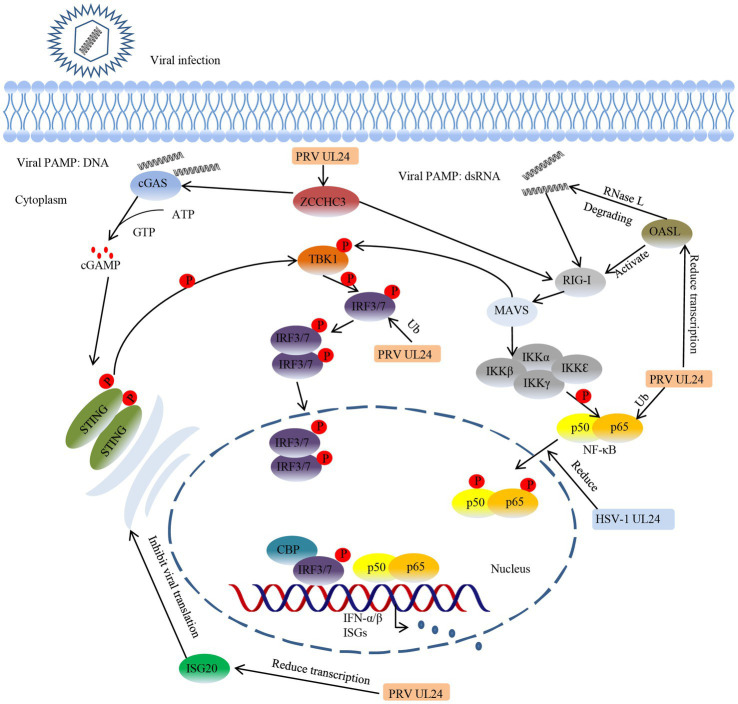
Innate immune responses are the first line of host defense against pathogens, and host cells recognize pathogens through a series of pattern recognition receptors (PRRs) that trigger the production of type I interferons (IFNs), including IFN-α and IFN-β (Akira et al., 2006; Kawai and Akira, 2010; Liu C. H. et al., 2017; Thoresen et al., 2021; Liu et al., 2022). After binding to the receptors on the cell membrane, IFNs can interact with a series of cellular proteins, eventually leading to the expression of numerous antiviral proteins, thus playing a role in resisting infection and eliminating the virus. To break through this innate immune response and proliferate effectively in host cells, herpesviruses have evolved numerous ways to resist the innate immune response of the host. One of the most important strategies is immune escape (Kolb et al., 2016; Li et al., 2020; Zhu and Zheng, 2020; He et al., 2022; Kong et al., 2022). UL24, as a viral tegument protein, has been shown to act on multiple immune signaling pathways to participate in immune escape from the host antiviral response (Table 1). For example, the HSV-1 UL24 protein interacts with the p65 and p50 subunits of NF-κB and reduces their frequency of nuclear translocation, thereby impeding immune pathway signaling (Xu et al., 2017). The PRV UL24 protein not only blocks the activation of NF-κB induced by tumor necrosis factor-α (TNF-α) by degrading p65 (Wang et al., 2020), but also degrades interferon regulatory factor 7 (IRF7) through the protease pathway to inhibit the cGAS/STING immune pathway and ultimately downregulate the host innate immune response (Liu et al., 2021). Host antiviral factors such as oligoadenylate synthetase-like (OASL), interferon-induced protein 20 (ISG20), and zinc finger CCHC-type containing protein 3 (ZCCHC3) can inhibit the proliferation of herpesvirus (Lian et al., 2018a,b). However, PRV UL24 protein can damage the RIG-I signaling pathway and inhibit the transcription of OASL, interferon-stimulated genes (ISGs) and ZCCHZ3, thus antagonizing the antiviral effects of OASL, ISG20 and ZCCHZ3 (Chen et al., 2021a,b, 2022). Interleukin-8 (IL-8) is a key component of some viruses that infect cells. This cytokine can inhibit the activity of IFN-α and regulate virus transmission and replication (Murayama et al., 1994; Craigen et al., 1997; Khabar et al., 1997). It has been confirmed that HCMV UL76 upregulates IL-8 production (Costa et al., 2013). Therefore, we speculate that HCMV UL76 can upregulate IL-8 and thereby inhibit IFN-α activity, which also has a positive effect on virus resistance to the host immune response. In addition to its role in mammalian herpesviruses, DPV UL24 protein has also been found in avian herpesvirus research to inhibit the activity of IFN-β and participate in immune escape, but the specific mechanism is not clear (Gao et al., 2022). In general, UL24 functions in multiple immune signaling pathways and plays an active role in viral resistance to host immune responses (Figure 2).
Table 1.
The mechanism by which herpesvirus UL24 participates in immune escape.
| Virus | Signaling pathway | Target protein | Mechanism |
|---|---|---|---|
| HSV-1 | RIG-I | P65, p50 | UL24 inhibits p65 and p50 localization into the nucleus (Xu et al., 2017) |
| PRV | RIG-I | P65 | UL24 induces p65 degradation (ubiquitination) (Wang et al., 2020) |
| PRV | cGAS-STING | IRF7 | UL24 induces IRF7 degradation (ubiquitination) (Liu et al., 2021) |
| PRV | RIG-I | OASL | UL24 inhibits OASL transcription (Chen et al., 2021a) |
| PRV | RIG-I | ISG20 | UL24 inhibits ISG20 transcription (Chen et al., 2021b) |
| PRV | RIG-I | ZCCHZ3 | UL24 inhibits ZCCHZ3 transcription (Chen et al., 2022) |
| HCMV | No data | IL-8 | UL76 can upregulate IL-8 (Costa et al., 2013) |
| DPV | cGAS-STING | IFN-β | UL24 inhibits the activity of IFN-β (Gao et al., 2022) |
Figure 2.
HSV-1 UL24 and PRV UL24 evade innate immunity by inhibiting the cGAS/STING and RIG-I signaling pathways. The host innate immune system can recognize pathogen-associated molecular patterns (PAMPs) through pattern recognition receptors (PRRs), thereby initiating innate immune responses and subsequent adaptive immune responses. Viral PAMPs containing herpesvirus DNA and dsRNA, PRV UL24 and HSV-1 UL24 can inhibit the innate immunity induced by viral PAMPs. PRV UL24 downregulates the expression of the antiviral factors ISG20, OASL and ZCCHZ3 and promotes the degradation of IRF7 and p65 to inhibit the host immune response. HSV-1 UL24 can reduce the entry of NF-κB subunits p50 and p65 into the nucleus to block the signal transmission of the immune pathway.
UL24 affects virus pathogenicity
Viral pathogenicity is usually determined by two factors: the virus itself and host factors. Regarding the herpesvirus, the viral proteins that determine the pathogenicity of the virus are mostly the envelope protein and the tegument protein, which play important roles in the pathogenesis of the herpesvirus (Tang et al., 2017; Shibazaki et al., 2020; Ning et al., 2022; Shen et al., 2023). In a study on the influence of the UL24 protein on virus pathogenicity, it was shown that HSV-1 UL24, especially its conserved domain that influences viral transmission to the host, is important for the virus to cause disease in the host (Leiva-Torres et al., 2010). When mice were infected with a UL24-knockout virus, the transmission of the virus to the trigeminal ganglion was blocked, which greatly reduced the virus titer in the trigeminal ganglion. The mice did not show clinical symptoms, and the latent infection and reactivation of the virus in the trigeminal ganglion were also greatly reduced (Jacobson et al., 1998; Rochette et al., 2015). Reduced pathogenicity was also observed in UL24 mutants of other herpesviruses, such as HSV-2 (Blakeney et al., 2005; Visalli et al., 2014). EHV-1 did not produce any neurotoxicity or lethal effects on mice after deletion of ORF37 (Kasem et al., 2010). The deletion of ORF35 also reduced the pathogenicity of VZV (Ito et al., 2005). In summary, the virulence of herpesviruses was significantly reduced in the existing studies following the deletion of the UL24 gene when compared to the wild-type virus. This indicates that the UL24 protein acts as a virulence factor for the herpesvirus and plays a crucial role in its pathogenicity.
The role of UL24 in viral replication
A major reason for the reduced virulence of the herpesvirus after the deletion of the UL24 protein is that the proliferation and transmission of the virus are greatly reduced, especially in neurons; this means that UL24 has a regulatory effect on the replication and proliferation of the virus (Rochette et al., 2015). Studies have shown that the UL24 protein is not necessary for the growth of the virus but plays an important role in the replication process of the virus (Ito et al., 2005; Leiva-Torres et al., 2010). For example, HSV-1 replication is downregulated in vitro after deletion of UL24 (Sanabria-Solano et al., 2016). During viral infection, OASL usually functions as an antiviral protein that inhibits viral replication and proliferation (Schoggins et al., 2015). However, the expression of OASL during KSHV infection is beneficial to viral replication. Following the deletion of ORF20, the production of new virions and the replication of viral DNA in KSHV-infected cells were significantly reduced compared with those in cells infected with wild-type viruses, suggesting that ORF20 plays a key role in regulating the replication of KSHV (Hoffman et al., 2021). The mechanism involves ORF20 interacting with OASL, leading to an increase in OASL expression and subsequently promoting the replication of KSHV (Bussey et al., 2018). This study is the first to report that KSHV ORF20 can bind with OASL to promote viral replication and proliferation. In addition, zinc finger proteins in host cells play unique biological functions in RNA metabolism, DNA repair and protein processing (Schmitges et al., 2016; Cassandri et al., 2017). Among these, ZCCHZ3 can significantly inhibit the replication of herpesviruses, while the expression of ZCCHZ3 is inhibited when UL24 is overexpressed, which in turn promotes the replication of the virus (Lian et al., 2018a; Chen et al., 2022).
The UL24 protein not only uses host proteins to promote viral replication but also interacts with other viral proteins to participate in the regulation of viral replication. The HSV-1 UL24 protein is also a potential PD-(D/E)XK endonuclease that can interact with a PD-(D/E)XK exonuclease encoded by the UL12 gene to promote the cleavage of redundant viral nucleic acids, which is important for viral replication and provides evidence for the involvement of the UL24 protein in viral replication (Bujnicki and Rychlewski, 2001; Knizewski et al., 2006). Due to the large size of the UL24 family of proteins, the specific role of UL24 homologous proteins in viral replication in individual herpesviruses remains to be confirmed. Dunn W et al. first proposed that HCMV UL76 is a viral replication enhancement gene (Dunn et al., 2003; Yu et al., 2003), while Wang S K et al. showed that the HCMV UL76 gene encodes a protein that inhibits HCMV replication (Wang et al., 2004). Later, Isomura H et al. confirmed that UL76 is involved in the regulation of UL77 gene expression. Since UL77 is important for viral replication, the author speculated that UL76 may be important for HCMV replication (Isomura et al., 2010). Although there are different views on the effect of UL76 on viral replication, the UL24 gene is highly conserved in herpesviruses, so we speculate that HCMV UL76 is an important gene for the promotion of viral replication. In conclusion, the UL24 protein not only interacts with viral proteins to promote viral replication but also regulates host proteins to provide favorable conditions for viral replication.
UL24 induces nucleolin (C23) and nucleophosmin (B23) distribution
Nucleolar proteins are required for effective infection by herpesviruses, and several nucleolar proteins are repositioned during infection (Callé et al., 2008; Sagou et al., 2010; Strang et al., 2010; Greco et al., 2012; Atari et al., 2022). C23, B23 and fibrillarin are multifunctional nucleolar proteins. They can regulate the transcription of RNA polymerase I and contribute to rRNA maturation and ribosome biogenesis (Boisvert et al., 2007; Mongelard and Bouvet, 2007; Rickards et al., 2007; Cong et al., 2012). During herpesvirus infection, some viral proteins play an important role in the distribution of nucleolin and nucleophosmin (Bertrand and Pearson, 2008; López et al., 2008; Lymberopoulos et al., 2011). Late in HSV-1 infection, nucleolins are dispersed throughout the nucleus in a manner dependent on UL24 protein expression (Lymberopoulos and Pearson, 2007). Ectopic expression of the UL24 protein can also specifically induce the distribution of nucleolin, which confirms that its functional region is a conserved N-terminal region of UL24. In addition, the endonuclease encoded by the UL24 gene is very important due to its function in inducing nucleolar protein dispersal (Lymberopoulos and Pearson, 2007; Bertrand and Pearson, 2008; Bertrand et al., 2010).
UL24 inhibits cell fusion
Cell fusion is an important biological process that plays an important role in the development, growth and immune responses of organisms (Lu and Kang, 2009; Iosilevskii and Podbilewicz, 2021). Herpesvirus entry and exit from host cells is a complex multistep process, and cell fusion is an important method of entry. Virus-induced cell fusion can be promoted or inhibited by different viral proteins (Manservigi et al., 1977; Connolly et al., 2011, 2021). Vesicles from the Golgi apparatus participate in the further assembly of virions and induce membrane fusion to release newly synthesized virions (Wisner and Johnson, 2004; Mingo et al., 2012; Sucharita et al., 2022). Herpesvirus UL24 protein can be localized to the Golgi apparatus, and its C-terminus is necessary for localization. Therefore, some scholars speculate that the C-terminal domain of the UL24 protein is involved in the regulation of membrane fusion in the late stage of viral infection (Bertrand and Pearson, 2008). The conjecture that the UL24 protein regulates cell fusion was confirmed by a study in which it was found that the UL24-knockout mutant of HSV-1 (UL24X) could not express UL24 protein and therefore lost the function of inhibiting cell fusion (Ben Abdeljelil et al., 2013). The UL24 protein regulates the cell fusion process by interacting with the gB, gD, gK and UL20 proteins (Bzik et al., 1984; Baines et al., 1991; Avitabile et al., 2004; Pataki et al., 2022b). When viruses invade cells, glycoproteins gB, gD, gH and gL form complexes to modify cell membrane proteins and induce the formation of syncytia, the classic manifestation of herpesvirus infection (Atanasiu et al., 2010, 2016; Böhm et al., 2016; Pataki et al., 2022a). As a fusion protein, the gB protein promotes the formation of syncytia, while UL24, gK and UL20 inhibit cell fusion (Atanasiu et al., 2010, 2013; Fan et al., 2023). UL24 can change the localization of gB, gD and F-actin to inhibit cell fusion and cause syncytial plaques in infected cells (Avitabile et al., 2004; Bertrand et al., 2010). Although the formation of syncytia contributes to the spread of the virus between cells, why viral proteins such as UL24 and gK inhibit cell fusion and their specific roles in inhibiting cell fusion remain to be explored. We believe that the UL24 protein’s inhibition of cell fusion might serve as a way to partially shield the virus from being eliminated by the host.
UL24 induces DNA damage in host cells
Herpesvirus infection can specifically induce chromosome damage in host cells (Fortunato et al., 2000; Fortunato and Spector, 2003; Bencherit et al., 2017). For example, HCMV infection of fibroblasts can induce DNA breakage between DFNA7 and DFNA49 on chromosome 1q23.3, and this damage is associated with hearing impairment (Nystad et al., 2008). Therefore, it is very important to explore the mechanism of DNA damage induced by herpesviruses. Studies have shown that the UL24 homologous protein encoded by the HCMV UL76 gene can cause double-strand breaks in host DNA and increase the amount of cH2AX phosphorylation, followed by the appearance of abnormal chromosomes such as micronuclei (Siew et al., 2009). After DNA damage is induced by the UL76 protein, the expression of IL-8 is upregulated, thus promoting viral replication, and this process also facilitates the effective transmission of the virus through neutrophils (Murayama et al., 1994; Craigen et al., 1997; Costa et al., 2013). While the function of the UL24 protein is typically associated with its five conserved functional domains, the region where the HCMV UL76 protein induces DNA damage is located in its nonconserved C-terminus (Zhang et al., 2015). UL76 interacts with the S5a protein of the ubiquitin protease system and exists in the form of aggregates, and their binding promotes the induction of DNA damage by UL76 (Lin et al., 2013). The S5a protein itself can interact with the DNA damage repair proteins hHR23a, hHR23b and XPC to form complexes (Sugasawa et al., 1997; Hiyama et al., 1999; Fujiwara et al., 2004). Whether UL76 can damage the function of the DNA damage repair complex through S5a and thus inhibit the DNA damage repair process remains to be further studied. In conclusion, herpesvirus UL76 protein induces DNA damage in host cells, which is beneficial to its own survival. Although this conclusion is based on HCMV U76, the UL76 protein belongs to the highly conserved UL24 protein family of herpesviruses. Therefore, we speculate that these results may translate to other herpesviruses.
UL24 causes cell cycle arrest and induces apoptosis
The cell cycle is a biological clock that controls the phases of life of a cell. The cell cycle is a precise regulatory process of intracellular and extracellular signal interactions. The signaling molecules controlling its operation are cyclin and cyclin-dependent protein kinase (Morris and Divita, 1999; Gutiérrez-Escribano and Nurse, 2015; Swaffer et al., 2016). At different stages of the cell cycle, different cyclin-CDK complexes drive the stable operation of the cell cycle (Gavet and Pines, 2010; Basu et al., 2022). To date, there has been some progress in the study of herpesvirus regulation of the cell cycle, and relevant studies have shown that the viral UL24 protein can cause cell cycle arrest in G2/M phase and cause apoptosis (Ehmann et al., 2000; Song et al., 2000). The cyclin B complex is an important mediator controlling cell cycle transition from G2 phase to mitosis (Ducommun et al., 1991; Smith and Proud, 2008). The expression of the UL24 proteins of HSV-1, MHV-68, HCMV and KSHV in host cells can hyperphosphorylate the Cdc2 protein and increase the expression of cyclin B, thereby downregulating the activity of the Cdc2/cyclin B complex and eventually causing cell cycle arrest at the G2/M phase (Nascimento and Parkhouse, 2007; Nascimento et al., 2009; Paladino et al., 2014). It is an important characteristic of viruses to adapt to the environment of the cell; herpesviruses affect the regulatory proteins of the cell cycle and thereby control the cell division cycle (Paladino et al., 2014; Trapp-Fragnet et al., 2014; Zhao et al., 2019; Bogdanow et al., 2021; Yockteng-Melgar et al., 2022). According to reports, UL24 proteins of α, β and γ herpesviruses can induce cell cycle arrest, which provides favorable conditions for the virus to actively adapt to the environment of the cell.
ICP27 and TK contribute to virulence by regulating UL24
The expression of proteins is affected by many factors such as interactions between viral proteins form a complex network and can affect the expression or function of other viral proteins. The proteins can combine into complexes to serve the entire life cycle of the virus (Reynolds et al., 2002; Ryckman and Roller, 2004; Liu et al., 2014; Takeshima et al., 2019; Deng et al., 2022). ICP27 is a conserved immediate early protein of herpesviruses that is involved in gene regulation at different stages of the virus. Concurrently, it can terminate host gene expression at the middle stage of viral infection. Its main mechanism is to inhibit mRNA splicing at the posttranscriptional level and to promote nuclear export of transcription products (Koffa et al., 2001; Fontaine-Rodriguez and Knipe, 2008; Tang et al., 2016). The transcription of UL24 is very complex, and the process produces six transcripts. The expression of UL24 protein is mainly related to the expression of transcripts produced by the first transcription initiation site (5.6 kb, 1.4 kb) (Pearson and Coen, 2002). Studies have shown that ICP27 can regulate the production of UL24 transcripts (Pearson et al., 2004). It has been shown that the UL24 protein interacts with ICP27 (Gao et al., 2017). Some scholars have found that ICP27 expression has no effect on the accumulation of UL24 1.4 kb short fragment transcripts but can regulate the transcription level of 5.6 kb long fragment transcripts (Hann et al., 1998). The expression of UL24 protein was reduced by 70% when ICP27-knockout virus was used to infect cells compared with wild-type virus (Pearson et al., 2004). ICP27 not only regulates the expression of UL24 protein but also regulates its cellular localization. It has been found that ICP27 can promote the transport of UL24 from the nucleus to the cytoplasm during viral infection (Gao et al., 2017). In addition, UL23 is also involved in the regulation of UL24. In the early stage of HSV-1 infection, the decrease in thymidine kinase expression promotes the accumulation of UL24 mRNA, especially the 1.4 kb transcription product, which indicates that the attenuation regulation of UL24 mRNA accumulation requires the participation of thymidine kinase (Cook and Coen, 1996; Cook et al., 1996).
Summary and prospects
In herpesviruses, the UL24 protein, as a component of the tegument, plays a vital role in viral infection of the host. Recent studies have shown that UL24 can induce nucleolar protein redistribution, inhibit cell fusion, induce host cell DNA damage and block progression of the cell cycle, all of which are undoubtedly infectious strategies that have been evolved by viruses for improved survival. In addition, in the process of fighting against the immune response of the host, UL24 also provides great help for the virus to evade the immune response. It can interact with a variety of immune regulatory proteins and antiviral factors to downregulate their expression or inhibit their function and ultimately inhibit the host antiviral response.
The synthesis of new virions in cells is a complex process. There are many studies on the function of the UL24 protein (Leuzinger et al., 2005; Mettenleiter et al., 2006; Sugimoto et al., 2008; Fan et al., 2020), but the specific role of pUL24 in the virus life cycle needs to be further explored.
During primary infection, herpesviruses can establish a lifelong latent infection in the trigeminal ganglion and the pharyngeal tonsil (Doll et al., 2019; Toomer et al., 2022). Although no studies have reported the direct relationship between UL24 protein and latent infection, deletion of UL24 protein can reduce the transmission efficiency of the virus in vivo and in vitro, especially transmission to the trigeminal ganglion, which may lead to impairment of the establishment and activation of latent viral infection. At present, the research and development of live vaccines and DNA vaccines that use gene deletion is in a rapid development stage, and research on herpesvirus-related vaccines such as PRV, MDV and DPV is relatively mature (Yu et al., 2012; Liu S. A. et al., 2017; Ruan et al., 2022; Sun A. et al., 2022; Sun Y. et al., 2022; Wu et al., 2022; Jiang et al., 2023). Deletion of the UL24 protein can reduce the virulence of the virus, so it is also important to further explore whether UL24-knockout strains can be used as gene deletion candidate vaccines.
The deepening of the understanding of viral proteins will inject new vitality into the treatment of herpesviruses and the development of new vaccines.
Author contributions
PR: Data curation, Writing – original draft, Writing – review & editing. MW: Conceptualization, Writing – review & editing. AC: Funding acquisition, Project administration, Writing – review & editing. XZ: Writing – review & editing. QY: Writing – review & editing. YW: Writing – review & editing. SZ: Writing – review & editing. BT: Writing – review & editing. JH: Writing – review & editing. XO: Writing – review & editing. QG: Writing – review & editing. DS: Writing – review & editing. YH: Writing – review & editing. ZW: Writing – review & editing. DZ: Writing – review & editing. RJ: Writing – review & editing. SC: Writing – review & editing. ML: Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by China Agriculture Research System of MOF and MARA (CARS-42-17) and the Program Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Agut H., Bonnafous P., Gautheret-Dejean A. (2016). Human herpesviruses 6A, 6B, and 7. Microbiol. Spectr. 4:7. doi: 10.1128/microbiolspec.DMIH2-0007-2015, PMID: [DOI] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. (2006). Pathogen recognition and innate immunity. Cells 124, 783–801. doi: 10.1016/j.cell.2006.02.015, PMID: [DOI] [PubMed] [Google Scholar]
- Atanasiu D., Saw W. T., Cohen G. H., Eisenberg R. J. (2010). Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J. Virol. 84, 12292–12299. doi: 10.1128/JVI.01700-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu D., Saw W. T., Eisenberg R. J., Cohen G. H. (2016). Regulation of herpes simplex virus glycoprotein-induced Cascade of events governing cell-cell fusion. J. Virol. 90, 10535–10544. doi: 10.1128/JVI.01501-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasiu D., Saw W. T., Gallagher J. R., Hannah B. P., Matsuda Z., Whitbeck J. C., et al. (2013). Dual split protein-based fusion assay reveals that mutations to herpes simplex virus (HSV) glycoprotein gB alter the kinetics of cell-cell fusion induced by HSV entry glycoproteins. J. Virol. 87, 11332–11345. doi: 10.1128/JVI.01700-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atari N., Rajan K. S., Chikne V., Cohen-Chalamish S., Doniger T., Orbaum O., et al. (2022). Lytic reactivation of the Kaposi's sarcoma-associated herpesvirus (KSHV) is accompanied by major nucleolar alterations. Viruses 14:1720. doi: 10.3390/v14081720, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avitabile E., Lombardi G., Gianni T., Capri M., Campadelli-Fiume G. (2004). Coexpression of UL20p and gK inhibits cell-cell fusion mediated by herpes simplex virus glycoproteins gD, gH-gL, and wild-type gB or an endocytosis-defective gB mutant and downmodulates their cell surface expression. J. Virol. 78, 8015–8025. doi: 10.1128/JVI.78.15.8015-8025.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines J. D., Ward P. L., Campadelli-Fiume G., Roizman B. (1991). The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65, 6414–6424. doi: 10.1128/jvi.65.12.6414-6424.1991, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S., Greenwood J., Jones A. W., Nurse P. (2022). Core control principles of the eukaryotic cell cycle. Nature 607, 381–386. doi: 10.1038/s41586-022-04798-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Coin L., Durbin R., Finn R. D., Hollich V., Griffiths-Jones S., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32, D138–D141. doi: 10.1093/nar/gkh121, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Abdeljelil N., Rochette P. A., Pearson A. (2013). The UL24 protein of herpes simplex virus 1 affects the sub-cellular distribution of viral glycoproteins involved in fusion. Virology 444, 263–273. doi: 10.1016/j.virol.2013.06.021, PMID: [DOI] [PubMed] [Google Scholar]
- Bencherit D., Remy S., Le Vern Y., Vychodil T., Bertzbach L. D., Kaufer B. B., et al. (2017). Induction of DNA damages upon Marek's disease virus infection: implication in viral replication and pathogenesis. J. Virol. 91:e01658. doi: 10.1128/JVI.01658-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand L., Leiva-Torres G. A., Hyjazie H., Pearson A. (2010). Conserved residues in the UL24 protein of herpes simplex virus 1 are important for dispersal of the nucleolar protein nucleolin. J. Virol. 84, 109–118. doi: 10.1128/JVI.01428-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand L., Pearson A. (2008). The conserved N-terminal domain of herpes simplex virus 1 UL24 protein is sufficient to induce the spatial redistribution of nucleolin. J. Gen. Virol. 89, 1142–1151. doi: 10.1099/vir.0.83573-0, PMID: [DOI] [PubMed] [Google Scholar]
- Blakeney S., Kowalski J., Tummolo D., DeStefano J., Cooper D., Guo M., et al. (2005). Herpes simplex virus type 2 UL24 gene is a virulence determinant in murine and guinea pig disease models. J. Virol. 79, 10498–10506. doi: 10.1128/JVI.79.16.10498-10506.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanow B., Phan Q. V., Wiebusch L. (2021). Emerging mechanisms of G(1)/S cell cycle control by human and mouse cytomegaloviruses. MBio 12:e0293421. doi: 10.1128/mBio.02934-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm S. W., Backovic M., Klupp B. G., Rey F. A., Mettenleiter T. C., Fuchs W. (2016). Functional characterization of glycoprotein H chimeras composed of conserved domains of the pseudorabies virus and herpes simplex virus 1 homologs. J. Virol. 90, 421–432. doi: 10.1128/JVI.01985-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F. M., van Koningsbruggen S., Navascués J., Lamond A. I. (2007). The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585. doi: 10.1038/nrm2184, PMID: [DOI] [PubMed] [Google Scholar]
- Boyne J. R., Whitehouse A. (2006). gamma-2 Herpes virus post-transcriptional gene regulation. Clin. Microbiol. Infect. 12, 110–117. doi: 10.1111/j.1469-0691.2005.01317.x, PMID: [DOI] [PubMed] [Google Scholar]
- Bujnicki J. M., Rychlewski L. (2001). The herpesvirus alkaline exonuclease belongs to the restriction endonuclease PD-(D/E)XK superfamily: insight from molecular modeling and phylogenetic analysis. Virus Genes 22, 219–230. doi: 10.1023/A:1008131810233, PMID: [DOI] [PubMed] [Google Scholar]
- Bussey K. A., Lau U., Schumann S., Gallo A., Osbelt L., Stempel M., et al. (2018). The interferon-stimulated gene product oligoadenylate synthetase-like protein enhances replication of Kaposi's sarcoma-associated herpesvirus (KSHV) and interacts with the KSHV ORF20 protein. PLoS Pathog. 14:e1006937. doi: 10.1371/journal.ppat.1006937, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzik D. J., Fox B. A., DeLuca N. A., Person S. (1984). Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137, 185–190. doi: 10.1016/0042-6822(84)90022-9, PMID: [DOI] [PubMed] [Google Scholar]
- Callé A., Ugrinova I., Epstein A. L., Bouvet P., Diaz J. J., Greco A. (2008). Nucleolin is required for an efficient herpes simplex virus type 1 infection. J. Virol. 82, 4762–4773. doi: 10.1128/JVI.00077-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho R. F., Spilki F. R., Cunha E. M., Stocco R. C., Arns C. W. (2012). Molecular data of UL24 homolog gene (ORF37) from Brazilian isolates of equine herpesvirus type 1. Res. Vet. Sci. 93, 494–497. doi: 10.1016/j.rvsc.2011.05.019, PMID: [DOI] [PubMed] [Google Scholar]
- Cassandri M., Smirnov A., Novelli F., Pitolli C., Agostini M., Malewicz M., et al. (2017). Zinc-finger proteins in health and disease. Cell Death Discov. 3:17071. doi: 10.1038/cddiscovery.2017.71, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Kong N., Xu J., Wang J., Zhang M., Ruan K., et al. (2021a). Pseudorabies virus UL24 antagonizes OASL-mediated antiviral effect. Virus Res. 295:198276. doi: 10.1016/j.virusres.2020.198276 [DOI] [PubMed] [Google Scholar]
- Chen X., Shan T., Sun D., Zhai H., Dong S., Kong N., et al. (2022). Host zinc-finger CCHC-type containing protein 3 inhibits pseudorabies virus proliferation by regulating type I interferon signaling. Gene 827:146480. doi: 10.1016/j.gene.2022.146480, PMID: [DOI] [PubMed] [Google Scholar]
- Chen X., Sun D., Dong S., Zhai H., Kong N., Zheng H., et al. (2021b). Host interferon-stimulated gene 20 inhibits pseudorabies virus proliferation. Virol. Sin. 36, 1027–1035. doi: 10.1007/s12250-021-00380-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong R., Das S., Ugrinova I., Kumar S., Mongelard F., Wong J., et al. (2012). Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res. 40, 9441–9454. doi: 10.1093/nar/gks720, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly S. A., Jackson J. O., Jardetzky T. S., Longnecker R. (2011). Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 9, 369–381. doi: 10.1038/nrmicro2548, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly S. A., Jardetzky T. S., Longnecker R. (2021). The structural basis of herpesvirus entry. Nat. Rev. Microbiol. 19, 110–121. doi: 10.1038/s41579-020-00448-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook W. J., Coen D. M. (1996). Temporal regulation of herpes simplex virus type 1 UL24 mRNA expression via differential polyadenylation. Virology 218, 204–213. doi: 10.1006/viro.1996.0180, PMID: [DOI] [PubMed] [Google Scholar]
- Cook W. J., Wobbe K. K., Böni J., Coen D. M. (1996). Regulation of neighboring gene expression by the herpes simplex virus type 1 thymidine kinase gene. Virology 218, 193–203. doi: 10.1006/viro.1996.0179, PMID: [DOI] [PubMed] [Google Scholar]
- Costa H., Nascimento R., Sinclair J., Parkhouse R. M. (2013). Human cytomegalovirus gene UL76 induces IL-8 expression through activation of the DNA damage response. PLoS Pathog. 9:e1003609. doi: 10.1371/journal.ppat.1003609, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen J. L., Yong K. L., Jordan N. J., MacCormac L. P., Westwick J., Akbar A. N., et al. (1997). Human cytomegalovirus infection up-regulates interleukin-8 gene expression and stimulates neutrophil transendothelial migration. Immunology 92, 138–145. doi: 10.1046/j.1365-2567.1997.00310.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimi S., Fiorillo L., Bianchi A., D'Amico C., Amoroso G., Gorassini F., et al. (2019). Herpes virus, Oral clinical signs and QoL: systematic review of recent data. Viruses 11:463. doi: 10.3390/v11050463, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damania B., Kenney S. C., Raab-Traub N. (2022). Epstein-Barr virus: biology and clinical disease. Cells 185, 3652–3670. doi: 10.1016/j.cell.2022.08.026, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J. (1992). Channel catfish virus: a new type of herpesvirus. Virology 186, 9–14. doi: 10.1016/0042-6822(92)90056-U, PMID: [DOI] [PubMed] [Google Scholar]
- Deng L., Wan J., Cheng A., Wang M., Tian B., Wu Y., et al. (2022). Duck plague virus US3 protein kinase phosphorylates UL47 and regulates the subcellular localization of UL47. Front. Microbiol. 13:876820. doi: 10.3389/fmicb.2022.876820, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Wang M., Cheng A., Yang Q., Wu Y., Jia R., et al. (2020). The pivotal roles of US3 protein in cell-to-cell spread and virion nuclear egress of duck plague virus. Sci. Rep. 10:7181. doi: 10.1038/s41598-020-64190-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezélée S., Bras F., Vende P., Simonet B., Nguyen X., Flamand A., et al. (1996). The BamHI fragment 9 of pseudorabies virus contains genes homologous to the UL24, UL25, UL26, and UL 26.5 genes of herpes simplex virus type 1. Virus Res. 42, 27–39. doi: 10.1016/0168-1702(96)01293-2, PMID: [DOI] [PubMed] [Google Scholar]
- Doll J. R., Thompson R. L., Sawtell N. M. (2019). Infectious herpes simplex virus in the brain stem is correlated with reactivation in the trigeminal ganglia. J. Virol. 93:e02209. doi: 10.1128/JVI.02209-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draganova E. B., Zhang J., Zhou Z. H., Heldwein E. E. (2020). Structural basis for capsid recruitment and coat formation during HSV-1 nuclear egress. Elife 9:e56627. doi: 10.7554/eLife.56627, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducommun B., Brambilla P., Félix M. A., Franza B. R., Karsenti E., Draetta G. (1991). cdc2 phosphorylation is required for its interaction with cyclin. EMBO J. 10, 3311–3319. doi: 10.1002/j.1460-2075.1991.tb04895.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W., Chou C., Li H., Hai R., Patterson D., Stolc V., et al. (2003). Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100, 14223–14228. doi: 10.1073/pnas.2334032100, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmann G. L., McLean T. I., Bachenheimer S. L. (2000). Herpes simplex virus type 1 infection imposes a G(1)/S block in asynchronously growing cells and prevents G(1) entry in quiescent cells. Virology 267, 335–349. doi: 10.1006/viro.1999.0147, PMID: [DOI] [PubMed] [Google Scholar]
- Fan D., Wang M., Cheng A., Jia R., Yang Q., Wu Y., et al. (2020). The role of VP16 in the life cycle of alphaherpesviruses. Front. Microbiol. 11:1910. doi: 10.3389/fmicb.2020.01910, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q., Hippler D. P., Yang Y., Longnecker R., Connolly S. A. (2023). Multiple sites on glycoprotein H (gH) functionally interact with the gB fusion protein to promote fusion during herpes simplex virus (HSV) entry. mBio 14:e0336822. doi: 10.1128/mbio.03368-22, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine-Rodriguez E. C., Knipe D. M. (2008). Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J. Virol. 82, 3538–3545. doi: 10.1128/JVI.02395-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato E. A., Dell’Aquila M. L., Spector D. H. (2000). Specific chromosome 1 breaks induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 97, 853–858. doi: 10.1073/pnas.97.2.853, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunato E. A., Spector D. H. (2003). Viral induction of site-specific chromosome damage. Rev. Med. Virol. 13, 21–37. doi: 10.1002/rmv.368, PMID: [DOI] [PubMed] [Google Scholar]
- Fritsche A. K., Borchers K. (2011). Detection of neuropathogenic strains of equid herpesvirus 1 (EHV-1) associated with abortions in Germany. Vet. Microbiol. 147, 176–180. doi: 10.1016/j.vetmic.2010.06.014, PMID: [DOI] [PubMed] [Google Scholar]
- Fujiwara K., Tenno T., Sugasawa K., Jee J. G., Ohki I., Kojima C., et al. (2004). Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitin-like domain of HR23B. J. Biol. Chem. 279, 4760–4767. doi: 10.1074/jbc.M309448200, PMID: [DOI] [PubMed] [Google Scholar]
- Gao L., Liu R., Yang F., Li X., Liu C., Qi X., et al. (2022). Duck enteritis virus inhibits the cGAS-STING DNA-sensing pathway to evade the innate immune response. J. Virol. 96:e0157822. doi: 10.1128/jvi.01578-22, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Jia R., Wang M., Yang Q., Chen S., Liu M., et al. (2017). Duck enteritis virus (DEV) UL54 protein, a novel partner, interacts with DEV UL24 protein. Virol. J. 14:166. doi: 10.1186/s12985-017-0830-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatherer D., Depledge D. P., Hartley C. A., Szpara M. L., Vaz P. K., Benkő M., et al. (2021). ICTV virus taxonomy profile: Herpesviridae 2021. J. Gen. Virol. 102:001673. doi: 10.1099/jgv.0.001673, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O., Pines J. (2010). Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J. Cell Biol. 189, 247–259. doi: 10.1083/jcb.200909144, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco A., Arata L., Soler E., Gaume X., Couté Y., Hacot S., et al. (2012). Nucleolin interacts with US11 protein of herpes simplex virus 1 and is involved in its trafficking. J. Virol. 86, 1449–1457. doi: 10.1128/JVI.06194-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Warren T., Wald A. (2007). Genital herpes. Lancet 370, 2127–2137. doi: 10.1016/S0140-6736(07)61908-4, PMID: [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Escribano P., Nurse P. (2015). A single cyclin-CDK complex is sufficient for both mitotic and meiotic progression in fission yeast. Nat. Commun. 6:6871. doi: 10.1038/ncomms7871, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann L. E., Cook W. J., Uprichard S. L., Knipe D. M., Coen D. M. (1998). The role of herpes simplex virus ICP27 in the regulation of UL24 gene expression by differential polyadenylation. J. Virol. 72, 7709–7714. doi: 10.1128/JVI.72.10.7709-7714.1998, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T., Wang M., Cheng A., Yang Q., Wu Y., Jia R., et al. (2022). Duck plague virus UL41 protein inhibits RIG-I/MDA5-mediated duck IFN-β production via mRNA degradation activity. Vet. Res. 53:22. doi: 10.1186/s13567-022-01043-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama H., Yokoi M., Masutani C., Sugasawa K., Maekawa T., Tanaka K., et al. (1999). Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J. Biol. Chem. 274, 28019–28025. doi: 10.1074/jbc.274.39.28019, PMID: [DOI] [PubMed] [Google Scholar]
- Hoffman D., Rodriguez W., Macveigh-Fierro D., Miles J., Muller M. (2021). The KSHV ORF20 protein interacts with the viral processivity factor ORF59 and promotes viral reactivation. Microbiol. Spectr. 9:e0014521. doi: 10.1128/Spectrum.00145-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong-Yan Z., Murata T., Goshima F., Takakuwa H., Koshizuka T., Yamauchi Y., et al. (2001). Identification and characterization of the UL24 gene product of herpes simplex virus type 2. Virus Genes 22, 321–327. doi: 10.1023/A:1011118424474, PMID: [DOI] [PubMed] [Google Scholar]
- Hulo C., de Castro E., Masson P., Bougueleret L., Bairoch A., Xenarios I., et al. (2011). ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res. 39, D576–D582. doi: 10.1093/nar/gkq901, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijezie E. C., O'Dowd J. M., Kuan M. I., Faeth A. R., Fortunato E. A. (2023). HCMV infection reduces Nidogen-1 expression, contributing to impaired neural rosette development in brain organoids. J. Virol. 97:e0171822. doi: 10.1128/jvi.01718-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilouze M., Dishon A., Kahan T., Kotler M. (2006). Cyprinid herpes virus-3 (CyHV-3) bears genes of genetically distant large DNA viruses. FEBS Lett. 580, 4473–4478. doi: 10.1016/j.febslet.2006.07.013, PMID: [DOI] [PubMed] [Google Scholar]
- Iosilevskii Y., Podbilewicz B. (2021). Programmed cell fusion in development and homeostasis. Curr. Top. Dev. Biol. 144, 215–244. doi: 10.1016/bs.ctdb.2020.12.013, PMID: [DOI] [PubMed] [Google Scholar]
- Isomura H., Stinski M. F., Murata T., Nakayama S., Chiba S., Akatsuka Y., et al. (2010). The human cytomegalovirus UL76 gene regulates the level of expression of the UL77 gene. PLoS One 5:e11901. doi: 10.1371/journal.pone.0011901, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Sommer M. H., Zerboni L., Baiker A., Sato B., Liang R., et al. (2005). Role of the varicella-zoster virus gene product encoded by open reading frame 35 in viral replication in vitro and in differentiated human skin and T cells in vivo. J. Virol. 79, 4819–4827. doi: 10.1128/JVI.79.8.4819-4827.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson J. G., Chen S. H., Cook W. J., Kramer M. F., Coen D. M. (1998). Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242, 161–169. doi: 10.1006/viro.1997.9012, PMID: [DOI] [PubMed] [Google Scholar]
- Jacobson J. G., Martin S. L., Coen D. M. (1989). A conserved open reading frame that overlaps the herpes simplex virus thymidine kinase gene is important for viral growth in cell culture. J. Virol. 63, 1839–1843. doi: 10.1128/jvi.63.4.1839-1843.1989, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R., Cheng A., Wang M., Zhu D., Ge H., Xin H., et al. (2009). Cloning, expression, purification and characterization of UL24 partial protein of duck enteritis virus. Intervirology 52, 326–334. doi: 10.1159/000242354, PMID: [DOI] [PubMed] [Google Scholar]
- Jiang C., Ma Z., Bai J., Sun Y., Cao M., Wang X., et al. (2023). Comparison of the protective efficacy between the candidate vaccine ZJ01R carrying gE/gI/TK deletion and three commercial vaccines against an emerging pseudorabies virus variant. Vet. Microbiol. 276:109623. doi: 10.1016/j.vetmic.2022.109623, PMID: [DOI] [PubMed] [Google Scholar]
- Kasem S., Yu M. H., Yamada S., Kodaira A., Matsumura T., Tsujimura K., et al. (2010). The ORF37 (UL24) is a neuropathogenicity determinant of equine herpesvirus 1 (EHV-1) in the mouse encephalitis model. Virology 400, 259–270. doi: 10.1016/j.virol.2010.02.012, PMID: [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. (2010). The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat. Immunol. 11, 373–384. doi: 10.1038/ni.1863, PMID: [DOI] [PubMed] [Google Scholar]
- Khabar K. S., Al-Zoghaibi F., Al-Ahdal M. N., Murayama T., Dhalla M., Mukaida N., et al. (1997). The alpha chemokine, interleukin 8, inhibits the antiviral action of interferon alpha. J. Exp. Med. 186, 1077–1085. doi: 10.1084/jem.186.7.1077, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knizewski L., Kinch L., Grishin N. V., Rychlewski L., Ginalski K. (2006). Human herpesvirus 1 UL24 gene encodes a potential PD-(D/E)XK endonuclease. J. Virol. 80, 2575–2577. doi: 10.1128/JVI.80.5.2575-2577.2006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffa M. D., Clements J. B., Izaurralde E., Wadd S., Wilson S. A., Mattaj I. W., et al. (2001). Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20, 5769–5778. doi: 10.1093/emboj/20.20.5769, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A. W., Lee K., Larsen I., Craven M., Brandt C. R. (2016). Quantitative trait locus based virulence determinant mapping of the HSV-1 genome in murine ocular infection: genes involved in viral regulatory and innate immune networks contribute to virulence. PLoS Pathog. 12:e1005499. doi: 10.1371/journal.ppat.1005499, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Z., Yin H., Wang F., Liu Z., Luan X., Sun L., et al. (2022). Pseudorabies virus tegument protein UL13 recruits RNF5 to inhibit STING-mediated antiviral immunity. PLoS Pathog. 18:e1010544. doi: 10.1371/journal.ppat.1010544, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiva-Torres G. A., Rochette P. A., Pearson A. (2010). Differential importance of highly conserved residues in UL24 for herpes simplex virus 1 replication in vivo and reactivation. J. Gen. Virol. 91, 1109–1116. doi: 10.1099/vir.0.017921-0, PMID: [DOI] [PubMed] [Google Scholar]
- Leuzinger H., Ziegler U., Schraner E. M., Fraefel C., Glauser D. L., Heid I., et al. (2005). Herpes simplex virus 1 envelopment follows two diverse pathways. J. Virol. 79, 13047–13059. doi: 10.1128/JVI.79.20.13047-13059.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu S., Kong X. (2006). Characterization of the genes encoding UL24, TK and gH proteins from duck enteritis virus (DEV): a proof for the classification of DEV. Virus Genes 33, 221–227. doi: 10.1007/s11262-005-0060-6, PMID: [DOI] [PubMed] [Google Scholar]
- Li Y., Wang M., Cheng A., Jia R., Yang Q., Chen S., et al. (2020). Duck enteritis virus VP16 antagonizes IFN-β-mediated antiviral innate immunity. J Immunol Res 2020:9630452. doi: 10.1155/2020/9630452, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H., Wei J., Zang R., Ye W., Yang Q., Zhang X. N., et al. (2018a). ZCCHC3 is a co-sensor of cGAS for dsDNA recognition in innate immune response. Nat. Commun. 9:3349. doi: 10.1038/s41467-018-05559-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H., Zang R., Wei J., Ye W., Hu M. M., Chen Y. D., et al. (2018b). The zinc-finger protein ZCCHC3 binds RNA and facilitates viral RNA sensing and activation of the RIG-I-like receptors. Immunity 49, 438–448.e435. doi: 10.1016/j.immuni.2018.08.014, PMID: [DOI] [PubMed] [Google Scholar]
- Lin S. R., Jiang M. J., Wang H. H., Hu C. H., Hsu M. S., Hsi E., et al. (2013). Human cytomegalovirus UL76 elicits novel aggresome formation via interaction with S5a of the ubiquitin proteasome system. J. Virol. 87, 11562–11578. doi: 10.1128/JVI.01568-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Cheng A., Wang M., Chen S., Jia R., Zhu D., et al. (2015). Duck enteritis virus UL54 is an IE protein primarily located in the nucleus. Virol. J. 12:198. doi: 10.1186/s12985-015-0424-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. H., Liu H., Ge B. (2017). Innate immunity in tuberculosis: host defense vs pathogen evasion. Cell. Mol. Immunol. 14, 963–975. doi: 10.1038/cmi.2017.88, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang F., Cao Y., Dang Y., Ge B. (2022). The multifaceted functions of cGAS. J. Mol. Cell Biol. 14:mjac031. doi: 10.1093/jmcb/mjac031, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. A., Stanfield B. A., Chouljenko V. N., Naidu S., Langohr I., Del Piero F., et al. (2017). Intramuscular immunization of mice with the live-attenuated herpes simplex virus 1 vaccine strain VC2 expressing equine herpesvirus 1 (EHV-1) glycoprotein D generates anti-EHV-1 immune responses in mice. J. Virol. 91:e02445. doi: 10.1128/jvi.02445-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhang M., Ye C., Ruan K., Xu A., Gao F., et al. (2021). Inhibition of the DNA-sensing pathway by pseudorabies virus UL24 protein via degradation of interferon regulatory factor 7. Vet. Microbiol. 255:109023. doi: 10.1016/j.vetmic.2021.109023, PMID: [DOI] [PubMed] [Google Scholar]
- Liu Z., Kato A., Shindo K., Noda T., Sagara H., Kawaoka Y., et al. (2014). Herpes simplex virus 1 UL47 interacts with viral nuclear egress factors UL31, UL34, and Us3 and regulates viral nuclear egress. J. Virol. 88, 4657–4667. doi: 10.1128/JVI.00137-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- López M. R., Schlegel E. F., Wintersteller S., Blaho J. A. (2008). The major tegument structural protein VP22 targets areas of dispersed nucleolin and marginalized chromatin during productive herpes simplex virus 1 infection. Virus Res. 136, 175–188. doi: 10.1016/j.virusres.2008.05.010, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Kang Y. (2009). Cell fusion as a hidden force in tumor progression. Cancer Res. 69, 8536–8539. doi: 10.1158/0008-5472.CAN-09-2159, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lymberopoulos M. H., Bourget A., Ben Abdeljelil N., Pearson A. (2011). Involvement of the UL24 protein in herpes simplex virus 1-induced dispersal of B23 and in nuclear egress. Virology 412, 341–348. doi: 10.1016/j.virol.2011.01.016, PMID: [DOI] [PubMed] [Google Scholar]
- Lymberopoulos M. H., Pearson A. (2007). Involvement of UL24 in herpes-simplex-virus-1-induced dispersal of nucleolin. Virology 363, 397–409. doi: 10.1016/j.virol.2007.01.028, PMID: [DOI] [PubMed] [Google Scholar]
- Mahmoudian A., Markham P. F., Noormohammadi A. H., Browning G. F. (2012). Kinetics of transcription of infectious laryngotracheitis virus genes. Comp. Immunol. Microbiol. Infect. Dis. 35, 103–115. doi: 10.1016/j.cimid.2011.11.001, PMID: [DOI] [PubMed] [Google Scholar]
- Manservigi R., Spear P. G., Buchan A. (1977). Cell fusion induced by herpes simplex virus is promoted and suppressed by different viral glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 74, 3913–3917. doi: 10.1073/pnas.74.9.3913, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin de Frémont G., Vanjak A., Sbihi Z., Knapp S., Garzaro M., Chbihi M., et al. (2023). Characteristics of circulating KSHV-infected viroblasts during active KSHV+ multicentric Castleman disease. Blood Adv. 7, 1682–1691. doi: 10.1182/bloodadvances.2022008456, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Cook S., Dolan A., Jamieson F. E., Telford E. A. (1995). Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247, 443–458. doi: 10.1006/jmbi.1995.0152, PMID: [DOI] [PubMed] [Google Scholar]
- Mettenleiter T. C., Klupp B. G., Granzow H. (2006). Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9, 423–429. doi: 10.1016/j.mib.2006.06.013, PMID: [DOI] [PubMed] [Google Scholar]
- Mingo R. M., Han J., Newcomb W. W., Brown J. C. (2012). Replication of herpes simplex virus: egress of progeny virus at specialized cell membrane sites. J. Virol. 86, 7084–7097. doi: 10.1128/JVI.00463-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongelard F., Bouvet P. (2007). Nucleolin: a multiFACeTed protein. Trends Cell Biol. 17, 80–86. doi: 10.1016/j.tcb.2006.11.010, PMID: [DOI] [PubMed] [Google Scholar]
- Morris M. C., Divita G. (1999). Characterization of the interactions between human cdc25C, cdks, cyclins and cdk-cyclin complexes. J. Mol. Biol. 286, 475–487. doi: 10.1006/jmbi.1998.2475, PMID: [DOI] [PubMed] [Google Scholar]
- Murayama T., Kuno K., Jisaki F., Obuchi M., Sakamuro D., Furukawa T., et al. (1994). Enhancement human cytomegalovirus replication in a human lung fibroblast cell line by interleukin-8. J. Virol. 68, 7582–7585. doi: 10.1128/jvi.68.11.7582-7585.1994, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento R., Dias J. D., Parkhouse R. M. (2009). The conserved UL24 family of human alpha, beta and gamma herpesviruses induces cell cycle arrest and inactivation of the cyclinB/cdc2 complex. Arch. Virol. 154, 1143–1149. doi: 10.1007/s00705-009-0420-y, PMID: [DOI] [PubMed] [Google Scholar]
- Nascimento R., Parkhouse R. M. E. (2007). Murine gammaherpesvirus 68 ORF20 induces cell-cycle arrest in G2 by inhibiting the Cdc2-cyclin B complex. J. Gen. Virol. 88, 1446–1453. doi: 10.1099/vir.0.82589-0, PMID: [DOI] [PubMed] [Google Scholar]
- Ning Y., Huang Y., Wang M., Cheng A., Jia R., Liu M., et al. (2022). Evaluation of the safety and immunogenicity of duck-plague virus gE mutants. Front. Immunol. 13:882796. doi: 10.3389/fimmu.2022.882796, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunberg J. H., Wright D. K., Cole G. E., Petrovskis E. A., Post L. E., Compton T., et al. (1989). Identification of the thymidine kinase gene of feline herpesvirus: use of degenerate oligonucleotides in the polymerase chain reaction to isolate herpesvirus gene homologs. J. Virol. 63, 3240–3249. doi: 10.1128/jvi.63.8.3240-3249.1989, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystad M., Fagerheim T., Brox V., Fortunato E. A., Nilssen Ø. (2008). Human cytomegalovirus (HCMV) and hearing impairment: infection of fibroblast cells with HCMV induces chromosome breaks at 1q23.3, between loci DFNA7 and DFNA49 -- both involved in dominantly inherited, sensorineural, hearing impairment. Mutat. Res. 637, 56–65. doi: 10.1016/j.mrfmmm.2007.07.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladino P., Marcon E., Greenblatt J., Frappier L. (2014). Identification of herpesvirus proteins that contribute to G1/S arrest. J. Virol. 88, 4480–4492. doi: 10.1128/JVI.00059-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataki Z., Rebolledo Viveros A., Heldwein E. E. (2022a). Herpes simplex virus 1 entry glycoproteins form complexes before and during membrane fusion. MBio 13:e0203922. doi: 10.1128/mbio.02039-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataki Z., Sanders E. K., Heldwein E. E. (2022b). A surface pocket in the cytoplasmic domain of the herpes simplex virus fusogen gB controls membrane fusion. PLoS Pathog. 18:e1010435. doi: 10.1371/journal.ppat.1010435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson A., Coen D. M. (2002). Identification, localization, and regulation of expression of the UL24 protein of herpes simplex virus type 1. J. Virol. 76, 10821–10828. doi: 10.1128/JVI.76.21.10821-10828.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson A., Knipe D. M., Coen D. M. (2004). ICP27 selectively regulates the cytoplasmic localization of a subset of viral transcripts in herpes simplex virus type 1-infected cells. J. Virol. 78, 23–32. doi: 10.1128/JVI.78.1.23-32.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz L. E., Reynolds A. E., Hengartner C. J. (2005). Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 69, 462–500. doi: 10.1128/MMBR.69.3.462-500.2005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathbun M. M., Szpara M. L. (2021). A holistic perspective on herpes simplex virus (HSV) ecology and evolution. Adv. Virus Res. 110, 27–57. doi: 10.1016/bs.aivir.2021.05.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. E., Wills E. G., Roller R. J., Ryckman B. J., Baines J. D. (2002). Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76, 8939–8952. doi: 10.1128/JVI.76.17.8939-8952.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickards B., Flint S. J., Cole M. D., LeRoy G. (2007). Nucleolin is required for RNA polymerase I transcription in vivo. Mol. Cell. Biol. 27, 937–948. doi: 10.1128/MCB.01584-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette P. A., Bourget A., Sanabria-Solano C., Lahmidi S., Lavallée G. O., Pearson A. (2015). Mutation of UL24 impedes the dissemination of acute herpes simplex virus 1 infection from the cornea to neurons of trigeminal ganglia. J. Gen. Virol. 96, 2794–2805. doi: 10.1099/vir.0.000189, PMID: [DOI] [PubMed] [Google Scholar]
- Ruan P., Feng X., Cheng A., Wang M., Zhang W., Wu Y., et al. (2022). Evaluation of safety and immunogenicity of duck-plague virus gC/gE double gene deletion. Front. Immunol. 13:963009. doi: 10.3389/fimmu.2022.963009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckman B. J., Roller R. J. (2004). Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78, 399–412. doi: 10.1128/JVI.78.1.399-412.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks W. R., Greene C. C., Aschman D. P., Schaffer P. A. (1985). Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55, 796–805. doi: 10.1128/jvi.55.3.796-805.1985, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagou K., Uema M., Kawaguchi Y. (2010). Nucleolin is required for efficient nuclear egress of herpes simplex virus type 1 nucleocapsids. J. Virol. 84, 2110–2121. doi: 10.1128/JVI.02007-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanabria-Solano C., Gonzalez C. E., Richerioux N., Bertrand L., Dridi S., Griffiths A., et al. (2016). Regulation of viral gene expression by the herpes simplex virus 1UL24 protein (HSV-1UL24 inhibits accumulation of viral transcripts). Virology 495, 148–160. doi: 10.1016/j.virol.2016.05.006, PMID: [DOI] [PubMed] [Google Scholar]
- Santos C. A. (2016). Cytomegalovirus and other β-herpesviruses. Semin. Nephrol. 36, 351–361. doi: 10.1016/j.semnephrol.2016.05.012, PMID: [DOI] [PubMed] [Google Scholar]
- Schmitges F. W., Radovani E., Najafabadi H. S., Barazandeh M., Campitelli L. F., Yin Y., et al. (2016). Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Res. 26, 1742–1752. doi: 10.1101/gr.209643.116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J. W., Wilson S. J., Panis M., Murphy M. Y., Jones C. T., Bieniasz P., et al. (2015). Corrigendum: a diverse range of gene products are effectors of the type I interferon antiviral response. Nature 525:144. doi: 10.1038/nature14554, PMID: [DOI] [PubMed] [Google Scholar]
- Shen B., Ruan P., Cheng A., Wang M., Zhang W., Wu Y., et al. (2023). Characterization of a unique novel LORF3 protein of duck plague virus and its potential pathogenesis. J. Virol. 97:e0157722. doi: 10.1128/jvi.01577-22, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibazaki M., Kato A., Takeshima K., Ito J., Suganami M., Koyanagi N., et al. (2020). Phosphoregulation of a conserved herpesvirus tegument protein by a virally encoded protein kinase in viral pathogenicity and potential linkage between its evolution and viral phylogeny. J. Virol. 94:e01055. doi: 10.1128/JVI.01055-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima Y., Jang H. K., Ono M., Maeda K., Tohya Y., Mikami T. (1997). Identification and DNA sequence analysis of the Marek's disease virus serotype 2 genes homologous to the thymidine kinase and UL24 genes of herpes simplex virus type 1. Virus Genes 14, 81–87. doi: 10.1023/a:1007943624997, PMID: [DOI] [PubMed] [Google Scholar]
- Siew V. K., Duh C. Y., Wang S. K. (2009). Human cytomegalovirus UL76 induces chromosome aberrations. J. Biomed. Sci. 16:107. doi: 10.1186/1423-0127-16-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. M., Proud C. G. (2008). cdc2-cyclin B regulates eEF2 kinase activity in a cell cycle- and amino acid-dependent manner. EMBO J. 27, 1005–1016. doi: 10.1038/emboj.2008.39, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Liu J. J., Yeh K. C., Knipe D. M. (2000). Herpes simplex virus infection blocks events in the G1 phase of the cell cycle. Virology 267, 326–334. doi: 10.1006/viro.1999.0146, PMID: [DOI] [PubMed] [Google Scholar]
- Strang B. L., Boulant S., Coen D. M. (2010). Nucleolin associates with the human cytomegalovirus DNA polymerase accessory subunit UL44 and is necessary for efficient viral replication. J. Virol. 84, 1771–1784. doi: 10.1128/JVI.01510-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucharita S., Tikoo S., Van Drunen Littel-van den Hurk S. (2022). Bovine Herpesvirus-1 glycoprotein M mediates the translocation to the Golgi apparatus and packaging of VP8. Viruses 14:1985. doi: 10.3390/v14091985, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Ng J. M., Masutani C., Maekawa T., Uchida A., van der Spek P. J., et al. (1997). Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol. Cell. Biol. 17, 6924–6931. doi: 10.1128/MCB.17.12.6924, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Uema M., Sagara H., Tanaka M., Sata T., Hashimoto Y., et al. (2008). Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 82, 5198–5211. doi: 10.1128/JVI.02681-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun A., Zhao X., Zhu X., Kong Z., Liao Y., Teng M., et al. (2022). Fully attenuated meq and pp38 double gene deletion mutant virus confers superior immunological protection against highly virulent Marek's disease virus infection. Microbiol. Spectr. 10:e0287122. doi: 10.1128/spectrum.02871-22, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zhao L., Fu Z. F. (2022). Effective cross-protection of a lyophilized live gE/gI/TK-deleted pseudorabies virus (PRV) vaccine against classical and variant PRV challenges. Vet. Microbiol. 267:109387. doi: 10.1016/j.vetmic.2022.109387, PMID: [DOI] [PubMed] [Google Scholar]
- Swaffer M. P., Jones A. W., Flynn H. R., Snijders A. P., Nurse P. (2016). CDK substrate phosphorylation and ordering the cell cycle. Cells 167, 1750–1761.e1716. doi: 10.1016/j.cell.2016.11.034, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima K., Arii J., Maruzuru Y., Koyanagi N., Kato A., Kawaguchi Y. (2019). Identification of the capsid binding site in the herpes simplex virus 1 nuclear egress complex and its role in viral primary envelopment and replication. J. Virol. 93:e01290. doi: 10.1128/JVI.01290-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Patel A., Krause P. R. (2016). Herpes simplex virus ICP27 regulates alternative pre-mRNA polyadenylation and splicing in a sequence-dependent manner. Proc. Natl. Acad. Sci. U. S. A. 113, 12256–12261. doi: 10.1073/pnas.1609695113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. D., Liu J. T., Wang T. Y., Sun M. X., Tian Z. J., Cai X. H. (2017). Comparison of pathogenicity-related genes in the current pseudorabies virus outbreak in China. Sci. Rep. 7:7783. doi: 10.1038/s41598-017-08269-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen D., Wang W., Galls D., Guo R., Xu L., Pyle A. M. (2021). The molecular mechanism of RIG-I activation and signaling. Immunol. Rev. 304, 154–168. doi: 10.1111/imr.13022, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomer G., Workman A., Harrison K. S., Stayton E., Hoyt P. R., Jones C. (2022). Stress triggers expression of bovine herpesvirus 1 infected cell protein 4 (bICP4) RNA during early stages of reactivation from latency in pharyngeal tonsil. J. Virol.:e0101022. doi: 10.1128/jvi.01010-22, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp-Fragnet L., Bencherit D., Chabanne-Vautherot D., Le Vern Y., Remy S., Boutet-Robinet E., et al. (2014). Cell cycle modulation by Marek's disease virus: the tegument protein VP22 triggers S-phase arrest and DNA damage in proliferating cells. PLoS One 9:e100004. doi: 10.1371/journal.pone.0100004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visalli R. J., Natuk R. J., Kowalski J., Guo M., Blakeney S., Gangolli S., et al. (2014). Vaccination with a HSV-2 UL24 mutant induces a protective immune response in murine and guinea pig vaginal infection models. Vaccine 32, 1398–1406. doi: 10.1016/j.vaccine.2013.10.079, PMID: [DOI] [PubMed] [Google Scholar]
- Wang S. K., Duh C. Y., Wu C. W. (2004). Human cytomegalovirus UL76 encodes a novel virion-associated protein that is able to inhibit viral replication. J. Virol. 78, 9750–9762. doi: 10.1128/JVI.78.18.9750-9762.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. Y., Yang Y. L., Feng C., Sun M. X., Peng J. M., Tian Z. J., et al. (2020). Pseudorabies virus UL24 abrogates tumor necrosis factor alpha-induced NF-κB activation by degrading P65. Viruses 12:51. doi: 10.3390/v12010051, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisner T. W., Johnson D. C. (2004). Redistribution of cellular and herpes simplex virus proteins from the trans-golgi network to cell junctions without enveloped capsids. J. Virol. 78, 11519–11535. doi: 10.1128/JVI.78.21.11519-11535.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Tan S., He Q., Wang M., Chen S., Jia R., et al. (2022). Deletion of double copies of the US1 gene reduces the infectivity of recombinant duck plague virus in vitro and in vivo. Microbiol. Spectr. 10:e0114022. doi: 10.1128/spectrum.01140-22, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Su C., Pearson A., Mody C. H., Zheng C. (2017). Herpes simplex virus 1 UL24 abrogates the DNA sensing signal pathway by inhibiting NF-κB activation. J. Virol. 91:e00025. doi: 10.1128/JVI.00025-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yockteng-Melgar J., Shire K., Cheng A. Z., Malik-Soni N., Harris R. S., Frappier L. (2022). G(1)/S cell cycle induction by Epstein-Barr virus BORF2 is mediated by P53 and APOBEC3B. J. Virol. 96:e0066022. doi: 10.1128/jvi.00660-22, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Silva M. C., Shenk T. (2003). Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100, 12396–12401. doi: 10.1073/pnas.1635160100, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Jia R., Huang J., Shu B., Zhu D., Liu Q., et al. (2012). Attenuated Salmonella Typhimurium delivering DNA vaccine encoding duck enteritis virus UL24 induced systemic and mucosal immune responses and conferred good protection against challenge. Vet. Res. 43:56. doi: 10.1186/1297-9716-43-56, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaravinos A., Bizakis J., Spandidos D. A. (2009). Prevalence of human papilloma virus and human herpes virus types 1-7 in human nasal polyposis. J. Med. Virol. 81, 1613–1619. doi: 10.1002/jmv.21534, PMID: [DOI] [PubMed] [Google Scholar]
- Zerboni L., Sen N., Oliver S. L., Arvin A. M. (2014). Molecular mechanisms of varicella zoster virus pathogenesis. Nat. Rev. Microbiol. 12, 197–210. doi: 10.1038/nrmicro3215, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Yao Y., Chen J., Wang M. (2015). Unconserved C terminal of human cytomegalovirus tegument protein pUL76 elicits nuclear aggresome formation and induces DNA damage in transfected cells. J. Biomed. Sci. 22:95. doi: 10.1186/s12929-015-0205-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Wang M., Cheng A., Yang Q., Wu Y., Jia R., et al. (2019). Duck plague virus promotes DEF cell apoptosis by activating caspases, increasing intracellular ROS levels and inducing cell cycle S-phase arrest. Viruses 11:196. doi: 10.3390/v11020196, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Liang Z., Lin Q., Chen M., Chang C., Zhou J., et al. (2023). Pathology, viremia, apoptosis during MDV latency in vaccinated chickens. Virology 579, 169–177. doi: 10.1016/j.virol.2023.01.003, PMID: [DOI] [PubMed] [Google Scholar]
- Zhou T., Wang M., Ruan P., Fan D., Cheng A., Zhang W., et al. (2023). Research note: duck plague virus pUL48 is a late protein that plays an important role in viral replication. Poult. Sci. 102:102358. doi: 10.1016/j.psj.2022.102358, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Zheng C. (2020). The race between host antiviral innate immunity and the immune evasion strategies of herpes simplex virus 1. Microbiol. Mol. Biol. Rev. 84:e00099. doi: 10.1128/MMBR.00099-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]