Abstract
Inherited retinal dystrophies (IRDs) are a heterogeneous group of visual disorders caused by different pathogenic mutations in genes and regulatory sequences. The endoplasmic reticulum (ER) membrane protein complex (EMC) subunit 3 (EMC3) is the core unit of the EMC insertase that integrates the transmembrane peptides into lipid bilayers, and the function of its cytoplasmic carboxyl terminus remains to be elucidated. In this study, an insertional mutation c.768insT in the C-terminal coding region of EMC3 was identified and associated with dominant IRDs in a five-generation family. This mutation caused a frameshift in the coding sequence and a gain of an additional 16 amino acid residues (p.L256F-fs-ext21) to form a helix structure in the C-terminus of the EMC3 protein. The mutation is heterozygous with an incomplete penetrance, and cosegregates in all patients examined. This finding indicates that the C-terminus of EMC3 is essential for EMC functions and that EMC3 may be a novel candidate gene for retinal degenerative diseases.
Keywords: ER membrane protein complex (EMC), EMC3, Inherited retinal dystrophy (IRD), Dominant, Retinal pigment epithelia (RPE), Cone rod dystrophy (CRD), Retinitis pigmentosa (RP)
1. Introduction
Approximately a quarter of eukaryotic genes encode transmembrane peptides that need to be correctly inserted into the lipid bilayer membrane to function properly. The endoplasmic reticulum (ER) membrane protein complex (EMC) is the major insertase (also called protein transporter) for this purpose either co-translationally or post-translationally [[1], [2], [3], [4]]. The eukaryotic EMC is assembled by up to 10 subunits, including EMC1, EMC2, EMC3, EMC4, MMGT1, EMC6, EMC7, EMC8, EMC9 and EMC10, among which EMC8 and EMC9 are incompatible but can be substituted for each other, and only one of them is present in the complex. EMC3 is the core component and is assisted by other EMC subunits to exert the insertase function [1,2,5]. Although the structures of the EMC complexes have been elucidated using Cryo-EM by a few groups of scientists [[6], [7], [8], [9]], surprisingly very little new insight has been gained by simple deduction from the structures, while the function and mechanism of the EMC and the domains and functions of each subunit remain largely unknown.
Inherited retinal dystrophies (IRDs) represent a heterogeneous group of visual impairment caused by various pathogenic mutations in genes and regulatory sequences [10,11]. The majority of IRDs are a monogenic and progressive disease. Typical IRDs include retinitis pigmentosa (RP), cone-rod dystrophy (CRD), macular degeneration (MD), Leber congenital amaurosis (LCA) and others [[11], [12], [13], [14], [15]]. For example, RP is a group of IRDs characterized by primary degeneration of rod and cone photoreceptors caused mainly by defects in the neural retina and retinal pigment epithelium (RPE) [[15], [16], [17]]. It is characterized by night blindness in adolescents due to rod cell dysfunction, and central vision loss in adults due to loss of cone function. The hallmark of RP is the gradual accumulation of intraretinal deposits of melanin and/or bone spicule-like pigments. RP is mainly considered to be an inherited disease with genetic mutations in 70 identified genes. These genes contribute to retinal metabolism (RPE65, LRAT, etc.), phototransduction and structural integrity (RHO, ROM1, SAG, etc.), ciliary structure and function (BBSs, RPGR, etc.), transcription and translation (CRX, NR2E3, etc.), and others (https://sph.uth.edu/retnet) [16,18,19]. However, the current mutation spectrum can only explain about half of the cases in IRDs, and more genetic mutations are yet to be discovered with the use of next-generation sequencing technologies.
The association of gene mutations with diseases helps us identify the protein domains that are essential for protein function. Mutations in a few EMC genes have been found to be associated with IRDs and other neuropathies in humans and animals. Abu-Safieh et al. found that mutations in EMC1 along with 5 other genes were observed in IRD patients [20]. A homozygous splice acceptor site variant (c.679-1G > A) in EMC10 was found to be associated with developmental delay, intellectual disability and speech delay in a Saudi family [21]. Later, Shao et al. identified another recurrent homozygous EMC10 c.287delG variant to be associated with similar developmental defects in patient families [22]. In animal models, the first report by Harris et al. showed that a mutation in the Pob gene (the corresponding human EMC3, also known as TMEM111) causes retinal degeneration in zebrafish [23]. The same phenotype is also observed in EMC3 loss-of-function Drosophila and mouse models [[24], [25], [26], [27]]. Furthermore, loss-of-function of EMC5 and EMC6 also resulted in retinal degeneration in Drosophila and mouse [27].
As the core component of the EMC complex, EMC3 has not been associated with any human diseases. In this study, we report the first observation of an EMC3 mutation in a 5-generation Chinese pedigree with IRDs. All the examined patients have typical IRD symptoms, and a heterozygous nonsense point mutation, c.768insT (p.L256F-fs-ext21) in EMC3, was cosegregated with patients in an autosomal dominant manner. Our findings suggest that the C-terminus of EMC3 is indispensable for the integral function of the insertase, and that EMC is essential for the physiological function and survival of retinal cells.
2. Methods and materials
2.1. Subjects and clinical evaluations
This study was approved by the Ethics Committee of Wenzhou Medical University (KYK2015-18) in accordance with the Declaration of Helsinki. Informed consent was obtained from all participating subjects. Seven patients and two healthy family members from a five-generation Chinese family with autosomal dominant IRD were recruited. Medical histories were obtained and ophthalmic examinations were performed by professional ophthalmologists. Peripheral blood samples were collected and genomic DNAs were extracted with the Blood DNA Mini Kit (Simgen, China).
2.2. Whole exome sequencing and verification with sanger sequencing
Whole exome sequencing and subsequent procedures, including quality control and variation annotation, were performed as previously described [28,29]. Eight family members (6 patients, 2 non-patients) participated in the exome sequencing and Sanger sequencing while one patient only participated in the Sanger sequencing. Mutant alleles in known IRD genes were first filtered, and known IRD genes were gathered based on RetNet (https://sph.uth.edu/RetNet/). All candidate variants were confirmed by Sanger sequencing, and co-segregation analysis was performed in available subjects. The following PCR primers were used to validate the insertional mutation in EMC3: 5ʹ- AACATAAAGGGGAACCCAGCC-3ʹ (forward), 5ʹ- AACAATTCCCTCTCTTCCTGCC -3ʹ (reverse). A PCR product of 331bp was purified and subjected to the Sanger sequencing analysis.
2.3. Assessment of gene variants and their pathogenicity
Variants with a minor allele frequency in any ethnic population of greater than or equal to 1% for recessive genes and greater than 0.01% for dominant genes were excluded. The pathogenicity of candidate variants was evaluated by multiple online analysis programs including the Exome Variant Server (http://evs.gs.washington.edu/EVS); gnomAD (V3.1 & V2.1, gnomad-sg.org); 1000 Genomes, ExAC Release 1, UN10K ALSPAC & TWINSUK, dbSNP b155 V2 (https://www.ncbi.nlm.nih.gov/projects/sviewer/). The assessment result was categorized as pathogenic, likely pathogenic, uncertain significance, benign or likely benign according to ACMG/AMP classification guidelines [30]. The 3-dimensional structure of the mutant EMC3 (p.L256F-fs-ext21) was predicted by AlphaFold2 [31].
2.4. Bioinformatics analyses and data processing
The EMC3 HomoloGene protein sequences were downloaded from the National Center for Biotechnology Information (NCBI) databases, and the protein sequence alignment was performed using Muscle 3.8 at EMBL with default settings. The alignment file was saved as an SVG file and annotated with Adobe Illustrator (V2020). The polygenetic tree file was downloaded and processed with Figtree (V1.4.4).
After obtaining the RNA-seq datasets (mouse, accession number: PRJNA589677 [32]; human, tables from website [33]), the gene expression values were processed with the Graphpad Prism (V8.0). The statistical significance was evaluated with the nonparametric method Kruskai-Walis test, and a p-value less than 0.05 was considered statistically significant. The human EMC3 protein-protein-interaction (PPI) network was obtained from STRING (V11.5, https://string-db.org/) with a high confidence node score >0.70. The average local clustering coefficient was 0.982, the PPI enrichment p-value was 1.11e-16, and the false discovery rate of the local network cluster (STRING) was 7.48-e18.
3. Results
3.1. Patient family and clinical features
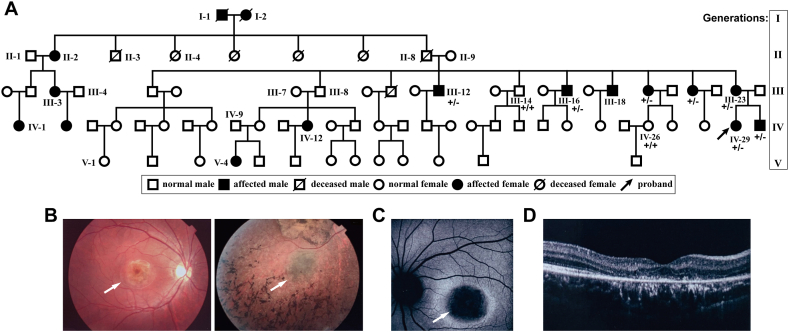
A five-generation pedigree with dominant IRDs was recruited in the rural area of northern China (Fig. 1A). The proband (IV-29) was a 19-year-old female at her first visit to our department who complained of photophobia and mild central vision loss. The funduscopy images and optical coherence tomography (OCT) images showed macular dystrophy in both eyes (Fig. S1). Twelve affected pedigree members are suffering a progressive loss of central vision and/or night blindness since their thirties. Male patients mainly complained of progressive loss of night vision. In contrast, female patients had a gradual loss of central vision, but they also developed night blindness at advanced stages. Seven of the affected individuals were available for the clinical examinations. Severe and diffuse retinal atrophy was observed in all the patients (Fig. 1B–D). Macular lesions were observed in all female patients but not in males. Peripheral pigment aggregation was observed only in patients with advanced disease. The better-corrected visual acuity (BCVA) of all the affected family members in their 40s and 50s had decreased to figures counting (FC). The proband was found to have macular dystrophy only and retained a relatively better BCVA of 0.12 at her first visit. Her younger brother (IV-30), aged 21 years, also had mild retinal degeneration by funduscopy examination, but no further examination was available. Their clinical features are summarized in the Table 1.
Fig. 1.
The pedigree tree and inherited eye diseases. (A) A 5-generation pedigree with inherited retinal dystrophy. The proband (IV-29) is indicated by the arrow. The genotype of the EMC3 mutation was indicated. +/+, wildtype; +/−, heterozygous. (B) The funduscopy images of the proband (left) and another patient (right). White arrows point to the macular regions. (C) The autofluorescence funduscopy image of a patient. The arrow points to the macular region. (D) The OCT image of the proband shows the retinal degeneration.
Table 1.
Summarization of the clinical information of available individuals in the pedigree.
| ID | Gender | Diagnosis |
EMC3 variant |
Initial symptoms | Photophobia | Age (years) at |
BCVA OD/OS |
Macular atrophy | Peripheral pigment | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | Genotype | onset | visit | ||||||||
| III-12 | M | IRD | c.768insT (p.L256F-fs-ext21) | Het | Night blindness | No | In his 30s | 50s | OU FC | No | Yes |
| III-14 | M | Healthy | / | WT | / | No | / | / | NA | No | No |
| III-16 | M | IRD | c.768insT (p.L256F-fs-ext21) | Het | Night blindness | No | In his 30s | 50s | OU FC | No | Yes |
| III-19 | F | IRD | NA | NA | Central vision Loss, Night blindness |
Yes | In her 30s | 50 | OU FC | Yes | Yes |
| III-21 | F | IRD | c.768insT (p.L256F-fs-ext21) | Het | Central vision Loss, Night blindness |
Yes | In her 30s | 48 | OU FC | Yes | Yes |
| III-23 | F | IRD | c.768insT (p.L256F-fs-ext21) | Het | Central vision Loss, Night blindness |
Yes | In her 30s | 46 | OU FC | Yes | Yes |
| IV-26 | F | Healthy | / | WT | / | No | / | / | NA | No | No |
| IV-29 | F | IRD | c.768insT (p.L256F-fs-ext21) | Het | Central vision Loss |
Yes | 19 | 28 | OU 0.12 | Yes | No |
| IV-30 | M | IRD | c.768insT (p.L256F-fs-ext21) | Het | / | No | / | 21 | NA | NA | NA |
Abbreviations: BCVA, best-corrected visual acuity; FC, fingers counting; Het, heterozygous; IRD, inherited retinal disease; NA, not available; OU, oculus uterque (both eyes); WT, wildtype.
3.2. Identification of a cosegregated gene mutation in EMC3
Bioinformatic analysis was performed on exome sequencing data from 8 participants and variants/mutations in 5 genes, EMC3, NCAPH, PEX13, RNF213 and SLC43A2, were found to cosegregate in all the patients but not in the non-patients (see Table 2). Synonymous variants in PEX13 and SLC43A2 were excluded. The NCAPH and RNF213 mutations were associated with other characteristic symptoms and diseases that were not found in the pedigree. Therefore, EMC3 emerged as the most likely candidate gene responsible for the pathogenesis in the pedigree. The same causative disease in EMC3 loss-of-function animal models strengthened this proposition.
Table 2.
Cosegregated gene mutations in available patients.
| Chrom-osome | Position | Gene symbol | cDNA change in splicing isoforms | Amino acid change | Associated diseases | Inherit -ance |
Refer -ence |
|---|---|---|---|---|---|---|---|
| Chr3 | 100005771 | EMC3 | NM_018447:exon9:c.767insT | p.L256F-fs-ext21 | IRD in mouse, zebrafish and Drosophila; Not reported in human. |
AR | [[1], [2], [3]] |
| Chr2 | 97019101 | NCAPH |
NM_001281712:exon7:c.A560G NM_001281710:exon8:c.A935G NM_001281711:exon8:c.A896G NM_015341:exon8:c.A968G |
p.Q187R p.Q312R p.Q299R p.Q323R |
Primary Autosomal Recessive Microcephaly in human. | AR | [4] |
| Chr2 | 61275677 | PEX13 | NM_002618:exon4:c.G984A | p.A328A | Peroxisome Biogenesis Disorder, Infantile Refsum Disease, Zellweger Spectrum Disorder, and Neonatal Adrenoleukodystrophy in human | AR | [[5], [6], [7]] etc. |
| Chr17 | 78305973 | RNF213 | NM_001256071:exon21:c.G3685C | p.D1229H | Moyamoya disease in human | AR, AD | [8,9] etc. |
| Chr17 | 1520017 | SLC43A2 |
NM_001284498:exon3:c.C207T NM_001321364:exon3:c.C207T NM_001321365:exon3:c.C207T NM_152346:exon3:c.C207T |
p.H69H p.H69H p.H69H p.H69H |
Growth retardation and postnatal lethality in null mice. Not reported in human. |
NA | [10] |
Abbreviations: AD, autosomal dominant; AR, autosomal recessive; NA, not applicable.
[1] Cao, X., J. An, Y. Cao, J. Lv, J. Wang, Y. Ding, X. Lin, and X. Zhou, EMC3 Is Essential for Retinal Organization and Neurogenesis During Mouse Retinal Development. Invest Ophthalmol Vis Sci, 2021. 62(2): 31.
[2] Satoh, T., A. Ohba, Z. Liu, T. Inagaki, and A.K. Satoh, dPob/EMC is essential for biosynthesis of rhodopsin and other multi-pass membrane proteins in Drosophila photoreceptors. Elife, 2015. 4: e06306.
[3] Taylor, M.R., S. Kikkawa, A. Diez-Juan, V. Ramamurthy, K. Kawakami, P. Carmeliet, and S.E. Brockerhoff, The zebrafish pob gene encodes a novel protein required for survival of red cone photoreceptor cells. Genetics, 2005. 170(1): 263-73.
[4] Martin, C.A., J.E. Murray, P. Carroll, A. Leitch, K.J. Mackenzie, M. Halachev, A.E. Fetit, C. Keith, L.S. Bicknell, A. Fluteau et al., Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev, 2016. 30(19): 2158–2172.
[5] Al-Dirbashi, O.Y., R. Shaheen, M. Al-Sayed, M. Al-Dosari, N. Makhseed, L. Abu Safieh, T. Santa, B.F. Meyer, N. Shimozawa, and F.S. Alkuraya, Zellweger syndrome caused by PEX13 deficiency: report of two novel mutations. Am J Med Genet A, 2009. 149a(6): 1219–1223.
[6] Liu, Y., J. Björkman, A. Urquhart, R.J. Wanders, D.I. Crane, and S.J. Gould, PEX13 is mutated in complementation group 13 of the peroxisome-biogenesis disorders. Am J Hum Genet, 1999. 65(3): 621–634.
[7] Shimozawa, N., Y. Suzuki, Z. Zhang, A. Imamura, R. Toyama, S. Mukai, Y. Fujiki, T. Tsukamoto, T. Osumi, T. Orii et al., Nonsense and temperature-sensitive mutations in PEX13 are the cause of complementation group H of peroxisome biogenesis disorders. Hum Mol Genet, 1999. 8(6): 1077–1083.
[8] Kamada, F., Y. Aoki, A. Narisawa, Y. Abe, S. Komatsuzaki, A. Kikuchi, J. Kanno, T. Niihori, M. Ono, N. Ishii et al., A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J Hum Genet, 2011. 56(1): 34–40.
[9] Miyatake, S., N. Miyake, H. Touho, A. Nishimura-Tadaki, Y. Kondo, I. Okada, Y. Tsurusaki, H. Doi, H. Sakai, H. Saitsu et al., Homozygous c.14576G > A variant of RNF213 predicts early-onset and severe form of moyamoya disease. Neurology, 2012. 78(11): 803–810.
[10] Guetg, A., L. Mariotta, L. Bock, B. Herzog, R. Fingerhut, S.M. Camargo, and F. Verrey, Essential amino acid transporter Lat4 (Slc43a2) is required for mouse development. J Physiol, 2015. 593(5): 1273–1289.
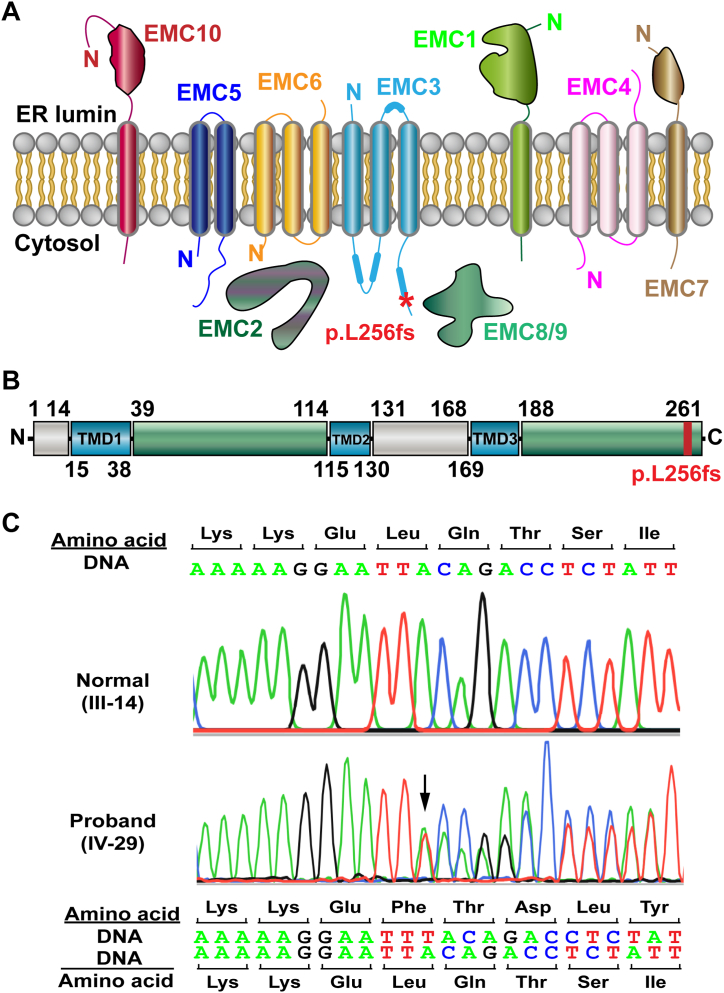
EMC3 is the core component of EMC (Fig. 2A). The human EMC3 protein consists of 261 amino acid residues (Fig. 2B), and the insertional mutation c.768insT causes a frameshift, resulting in a change in the amino acid sequence from the mutation site (Fig. 2C). The heterozygous mutation c.768insT in EMC3 was verified by Sanger sequencing in the proband. We further examined the mutation in all the other available patients and it was cosegregated in all the examined cases (III-12, 16, 19, 21, IV-23, 29, 30) but not in healthy (normal) family members (III-14, IV-26). As per ACGM standards and guidelines 2015 [30], this mutation was evaluated as PS3, PM2, PM4, PP1, PP3 and PP4, and it was classified as “pathogenic” by combining the criteria.
Fig. 2.
Identification of the EMC3 mutation. (A) An illustration of the EMC members in the complex. The mutation in EMC3 is indicated by a star symbol in red. (B) A diagram of the domains and their positions in the human EMC3 protein. The mutation site was close to the carboxyl end. TMD1-3, the transmembrane domain 1-3. (C) Sanger sequencing of the proband's genomic DNA of the peripheral blood cells. A thymine base was inserted at the position c.768 causing a frameshift in one of the EMC3 alleles.
The importance and function of a protein might be reflected by its conservation during animal evolution. We compared the amino acid sequences of EMC3 in vertebrates ranging from zebrafish (Danio rerio), frog (Xenopus tropicalis), chicken (Gallus gallus), rodents (Mus musculus and Rattus norvegicus), livestock (Bos taurus and Canis lupus familiaris), non-human primates (Macaca mulatta) to humans (Homo sapiens) (Fig. S2). These homologues share 89.27%–100% similarity (Fig. S3A). As shown in the phylogenetic tree (Fig. S3B), the EMC3 proteins are evolutionarily highly conserved in the vertebrates. Further examination of the region around the mutation showed that this region is also highly conserved, especially in higher animals (Fig. S2), suggesting that the EMC3 C-terminus is critical for EMC insertase structure and/or function.
3.3. Expression of EMCs in the retina and RPE during tissue development and maintenance
To explore possible mechanisms of pathogenesis, we examined the expression of EMC genes in retina and RPE during tissue development and maintenance. First, we retrieved the protein-protein interaction network of EMC3 from the STRING database with high confidence node score (>0.70) (Fig. 3A). It showed that EMC3 has the strongest connection with other EMC components, and interacts with tryptophan-rich basic protein (WRB), also known as congenital heart disease protein 5 (CHD5), which is required for membrane insertion of tail-anchored proteins [34].
Fig. 3.
The protein interaction network and expression profiling of EMCs. (A) The STRING protein-protein interaction map of human EMC3. MMGT1 is also called EMC5. (B) Dynamic expression of Emc mRNAs in the mouse retina during different developmental stages. (C) Expression of EMC mRNAs in the macular, nasal, and temporal regions of the human retina. (D) Expression of EMC mRNAs in the corresponding macular, nasal, and temporal regions of human RPE-choroid.
Through the analyses of RNA-seq data [32] during mouse retinal development, we found that EMC3 was expressed at higher levels during early retinal development but was downregulated after postnatal day 13 (P13), whereas EMC2 showed the opposite trend (Fig. 3B). Other EMC subunits fluctuated without a clear pattern. Interestingly, EMC8 was consistently expressed, whereas the expression of EMC9, the paralogue and competitor of EMC8, was minimal or absent (Fig. 3B). To examine how EMC3 would be expressed in the human retina, we analyzed previous RNA sequencing (scRNA-seq) data from human retinas [33], and found that EMC3, along with other EMC subunits, was indiscriminately expressed in the macular, nasal, and temporal retinal regions (Fig. 3C). Contrary to the mouse retina, the expression of EMC9 was stronger than that of EMC8 in the human retina (Fig. 3C), reflecting an interspecies difference of unknown biological significance.
IRDs could also be caused by defects and dysfunction of the RPE. We examined the expression of EMCs in human RPE using RNA-seq data from human RPE and choroid [33]. Unexpectedly, the expression of EMC4 and EMC10 was significantly lower in the corresponding macular RPE-choroid region than that in the nasal and temporal RPE-choroid regions (Fig. 3D). Differences in the expression of EMC5, EMC7 and EMC9 were observed although they were not statistically significant due to the small sample size (n = 4) (Fig. 3D). The reason for regional differences in RPE-choroid but not in retina is unknown.
3.4. Structural and conformational changes in the mutant EMC3 protein
We investigated the structural and conformational changes of the mutant EMC3 protein at the primary, secondary, and tertiary structure levels. The c.768insT mutation caused a substitution of 6 amino acid residues in a peptide fragment (256LQTSIF261 to 256FTDLYF261), in addition to a gain of 16 additional residues (LKTEQGLAVSGTWSCT) in the carboxyl terminus (Fig. 4A). As predicted by AlphaFold2, these changes in amino acid residues introduced a secondary structure, a new α-helix, at the C-terminus (Fig. 4B). Rather than the hydrophilic nature of the wildtype C-terminus, the hydrophobicity of the α-helix would tend to integrate the terminus into the lipid membrane, which would likely interfere with normal insertase function.
Fig. 4.
Alterations of amino acid sequence and protein structure in mutated human EMC3. (A) The frameshift mutation caused a substitution in the last 6 amino acid residues and an addition of 16 additional residues. (B) The superimposed structures comparing the 3-dimensional changes between WT and mutant human EMC3. The alteration in C-terminus is highlighted by a red dashed circle. The structure of the mutant protein was predicted using AlphaFold2.
4. Discussion
4.1. Role of EMC in membrane protein homeostasis and diseases
The EMC is universally present in all cells and functions as an insertase for the translocation, insertion, folding, and assembly of membrane proteins in the ER lipid bilayer. In this sense, it can be classified as a cell housekeeper that maintains membrane protein homeostasis to sustain cell viability [1,2,35]. Membrane proteins can span the membrane via the α-helix (also known as a transmembrane domain (TMD)) or β-barrel structure, whose membrane insertion depends on different mechanisms [36].
Two major pathways, the GET and EMC pathways, overlap to some extent and are responsible for the membrane insertion of tail-anchored proteins. In addition to the signal recognition particle (SRP)-dependent and -independent pathways, which are considered backups of the GET pathway [37], an unknown number of TMD proteins can also insert spontaneously into a membrane without assistance. Therefore, only a fraction of membrane proteins depend on EMC for membrane insertion. However, the number of EMC-dependent proteins containing TMDs with polar and/or charged residues has never been systematically determined [38]. This may explain why mutation-induced EMC dysfunction can be reasonably tolerated in many cells and tissues.
Aberrant EMC function causes a constitutive stress response in the cells [37] and has been reported to cause various diseases. A few groups have shown through proteomic and other studies that many of the EMC-dependent proteins have been implicated in neurodevelopment disorders [4,[38], [39], [40]].
5. EMCs in retina and RPE
EMCs are abundantly expressed in the retina and RPE. There is no direct evidence that they participate in cell fate specification or determination during retinal and RPE development. Rather, skewed manifestations in these aspects are side effects of disturbed membrane protein homeostasis. Similarly, pleiotropic phenotypes in cell migration, synapse formation and regulation, axon guidance, angiogenesis, cell survival/death and other aspects could occur in various cell types.
Prior to this study, none EMC3 mutation has been found to cause retinal degenerative diseases in humans. In this study, we identified EMC3 as a novel candidate gene associated with dominant IRDs. The onset and progression of the disease varied significantly between individuals within the same family. Several obligate carriers were non-penetrant and asymptomatic.
The fact that multipass (multiple TMDs) proteins, such as GPCRs, are more dependent on EMC for membrane insertion makes photoreceptors more susceptible to EMC dysfunction, since photoreceptors depend on rhodopsin and other GPCRs for their normal visual functions [24,27]. Another reason is that the proteins in the outer segments and the visual pathway are constantly renewed, which places a heavy burden on the EMC and other pathways to maintain protein homeostasis.
5.1. Compromised EMC function underlying the pathogenesis of EMC3 mutation
In this article, we identified the first EMC3 mutation associated with human disease. The gnomAD database reports 6 heterozygous LoF variants for EMC3 in normal controls, with an associated pLI of 0.29, indicating that this gene is very much tolerant to haploinsufficiency. However, the pitfalls of interpreting pLI scores must be carefully avoided, and there are too many exceptions [41]. In the case of p.L256F-fs-ext21, the insertional mutation is close to the stop codon in the last exon, causing the frameshift with the gain of extra amino acids and an α-helix, making the pLI very unreliable in this scenario [41]. Moreover, the addition of the extra α-helix in the C-terminus made EMC3 a fusion-like protein that dominantly acquired abnormal function. In fact, a mutant (Q192ter) EMC3 lacking the C-terminus in Drosophila clearly demonstrated that the C-terminus of EMC3 is required for photoreceptor viability [27].
We propose that three mechanisms may underlie the pathogenesis of the EMC3 mutation. First, the loss of critical residues in the C-terminus could abolish important interactions with other partners to confer EMC function. Second, introduction of a predominantly hydrophobic side chain of a 15–30 amino acid long α-helix would be likely to establish a new TMD that could insert into the membrane to disrupt the EMC complex structure [42]. Third, the C-terminus of EMC3 is known to interact with EMC2/4/5/8/9 and stabilize the EMC conformation, so the mutation could dismantle the conformation to cause EMC dysfunction [9]. As a result, the disturbance of protein homeostasis caused ER stress and consequent cascade reactions leading to apoptosis in retinal cells. Nonsense mediated mRNA decay (NMD) has been found in the pathogenesis of many eye and retinal diseases [43]. In patients with dominant mutations, NMD-mediated degradation of mRNA strand with nonsense mutations would cause haploinsufficiency of gene function, leading to retinal degeneration [44,45]. The nonsense mutation c.768InsT is close to the 3’ proximity of the EMC3 mRNA and is unlikely to induce the NMD response. Indeed, the NMD Classifier rules out the NMD events of the mutation (Fig. S4) [46]. The unfolded protein response and ER stress are likely to be the major forces triggering photoreceptor cell death.
However, this mutation seems to cause some phenotypic heterogeneity in patients. First, although all patients suffer from IRDs, there is a sex difference in symptoms. Male patients present with RP-like symptoms such as night blindness and peripheral pigment deposits in the retinas. In contrast, female patients initially exhibit macular dystrophy symptoms such as central vision loss, however, they display both macular dystrophy and RP-like symptoms with disease progression (Table 1). We hypothesize that the transport of sex hormone receptors and related proteins is compromised in retinal cells, leading to the sex difference in symptoms. Indeed, sex difference has been found in many common ocular diseases, including cataract, glaucoma, macular hole and diabetic retinopathy [47]. Estrogen signaling has been suggested to protect the macula from damage. Tamoxifen, an estrogen antagonist, is correlated with macular degeneration [48]. Finally, the phenotype of the mutation is not completely penetrant, as shown in both male and female obligate carriers (II-3, III-2, 8, and IV-10). Incomplete penetrance and variable expressivity are common in plants and animals, especially in autosomal dominant disorders [[49], [50], [51]]. Many factors, such as variants in cis- or trans-regulatory elements, epigenetic modifications, age-, sex-, and environment-dependent modifiers, splicing isoforms, and genetic compensation [49,[51], [52], [53]], contribute to these phenomena.
6. Limitations of the study and future perspectives
There are several limitations in this study. First, we demonstrate that EMC3 mutation is associated with retinal degeneration, but the association needs to be further verified in future studies, i.e., applying animal models with gain-of-function and loss-of-function approaches. Second, based on the gene expression profiles, we speculate that the photoreceptor loss may be caused by direct defects of EMC3 in the photoreceptors in a cell-autonomous manner, or alternatively by dysfunctions in the supporting RPE cells in a cell non-autonomous manner, or both. Further studies are needed to distinguish between these possibilities by tissue-specific misexpression of the wildtype and mutant genes. Third, the pathogenesis underlying the dominant phenotypes could be a result of EMC3 gain-of-function or the dominant-negative effect of truncated EMC3. In Drosophila, the dPob/EMC3 mutation causes mislocalization of rhodopsin and Na + -K+ ATPase and consequent retinal degeneration [24]. In Emc3 knockout mice, loss of Emc3 led to mislocalization of cell junction and cell polarity proteins [25]. It remains to be investigated whether such changes are also present in human EMC3 mutation. Finally, exome sequencing can only reveal those mutations in the exons, and mutations in introns and regulatory regions (i.e., enhancers) could be identified by genome sequencing. Solving these puzzles will not only provide insights into the function and mechanism of EMC, but will also be very helpful in the development of novel therapeutics and drugs for related human diseases.
Author contribution statement
Yan-Ping Li: Performed the experiments.
Ren-Juan Shen: Analyzed and interpreted the data.
You-Min Cheng: Performed the experiments.
Qingqing Zhao: Performed the experiments.
Kangxin Jin: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Zi-Bing Jin: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Shaodan Zhang: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data.
Data availability statement
Data associated with this study has been deposited at https://www.cncb.ac.cn/with access number: HRA002930.
Supplementary content related to this article has been published online at [URL].
Ethics statement
This study was approved by the Ethics Committee of Wenzhou Medical University (KYK2015-18) in accordance with the Declaration of Helsinki.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We would like to thank Dr. Jinyu Wu for his assistance with bioinformatic analyses, and Ms. Yihan Wang for polishing the paper.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e20146.
Contributor Information
Kangxin Jin, Email: jinkx@mail.ccmu.edu.cn.
Zi-Bing Jin, Email: jinzb502@ccmu.edu.cn.
Shaodan Zhang, Email: shaodan_zhang@eye.ac.cn.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
figs1.
figs2.
figs3.
figs4.
References
- 1.Chitwood P.J., Hegde R.S. The role of EMC during membrane protein biogenesis. Trends Cell Biol. 2019;29(5):371–384. doi: 10.1016/j.tcb.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Hegde R.S. The function, structure, and origins of the ER membrane protein complex. Annu. Rev. Biochem. 2022;91:651–678. doi: 10.1146/annurev-biochem-032620-104553. [DOI] [PubMed] [Google Scholar]
- 3.Guna A., et al. The ER membrane protein complex is a transmembrane domain insertase. Science. 2018;359(6374):470–473. doi: 10.1126/science.aao3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shurtleff M.J., et al. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. Elife. 2018;7 doi: 10.7554/eLife.37018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dederer V., Lemberg M.K. Transmembrane dislocases: a second chance for protein targeting. Trends Cell Biol. 2021;31(11):898–911. doi: 10.1016/j.tcb.2021.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Bai L., et al. Structure of the ER membrane complex, a transmembrane-domain insertase. Nature. 2020;584(7821):475–478. doi: 10.1038/s41586-020-2389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller-Vedam L.E., et al. Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients. Elife. 2020;9 doi: 10.7554/eLife.62611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Donnell J.P., et al. The architecture of EMC reveals a path for membrane protein insertion. Elife. 2020;9 doi: 10.7554/eLife.57887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pleiner T., et al. Structural basis for membrane insertion by the human ER membrane protein complex. Science. 2020;369(6502):433–436. doi: 10.1126/science.abb5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider N., et al. Inherited retinal diseases: linking genes, disease-causing variants, and relevant therapeutic modalities. Prog. Retin. Eye Res. 2022;89 doi: 10.1016/j.preteyeres.2021.101029. [DOI] [PubMed] [Google Scholar]
- 11.Manley A., et al. Cellular and molecular mechanisms of pathogenesis underlying inherited retinal dystrophies. Biomolecules. 2023;13(2):271. doi: 10.3390/biom13020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider N., et al. Inherited retinal diseases: linking genes, disease-causing variants, and relevant therapeutic modalities. Prog. Retin. Eye Res. 2021 doi: 10.1016/j.preteyeres.2021.101029. [DOI] [PubMed] [Google Scholar]
- 13.Rozing M.P., et al. Age-related macular degeneration: a two-level model hypothesis. Prog. Retin. Eye Res. 2020;76 doi: 10.1016/j.preteyeres.2019.100825. [DOI] [PubMed] [Google Scholar]
- 14.Kumaran N., et al. Leber congenital amaurosis/early-onset severe retinal dystrophy: clinical features, molecular genetics and therapeutic interventions. Br. J. Ophthalmol. 2017;101(9):1147–1154. doi: 10.1136/bjophthalmol-2016-309975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin Z.B., et al. Stemming retinal regeneration with pluripotent stem cells. Prog. Retin. Eye Res. 2019;69:38–56. doi: 10.1016/j.preteyeres.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Dias M.F., et al. Molecular genetics and emerging therapies for retinitis pigmentosa: basic research and clinical perspectives. Prog. Retin. Eye Res. 2018;63:107–131. doi: 10.1016/j.preteyeres.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Verbakel S.K., et al. Non-syndromic retinitis pigmentosa. Prog. Retin. Eye Res. 2018;66:157–186. doi: 10.1016/j.preteyeres.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Newton F., Megaw R. Mechanisms of photoreceptor death in retinitis pigmentosa. Genes. 2020;11(10):1120. doi: 10.3390/genes11101120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daiger S.P., Sullivan L.S., Bowne S.J. Genes and mutations causing retinitis pigmentosa. Clin. Genet. 2013;84(2):132–141. doi: 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abu-Safieh L., et al. Autozygome-guided exome sequencing in retinal dystrophy patients reveals pathogenetic mutations and novel candidate disease genes. Genome Res. 2013;23(2):236–247. doi: 10.1101/gr.144105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umair M., et al. EMC10 homozygous variant identified in a family with global developmental delay, mild intellectual disability, and speech delay. Clin. Genet. 2020;98(6):555–561. doi: 10.1111/cge.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao D.D., et al. A recurrent, homozygous EMC10 frameshift variant is associated with a syndrome of developmental delay with variable seizures and dysmorphic features. Genet. Med. 2021;23(6):1158–1162. doi: 10.1038/s41436-021-01097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor M.R., et al. The zebrafish pob gene encodes a novel protein required for survival of red cone photoreceptor cells. Genetics. 2005;170(1):263–273. doi: 10.1534/genetics.104.036434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satoh T., et al. dPob/EMC is essential for biosynthesis of rhodopsin and other multi-pass membrane proteins in Drosophila photoreceptors. Elife. 2015;4 doi: 10.7554/eLife.06306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao X., et al. EMC3 is essential for retinal organization and Neurogenesis during mouse retinal development. Invest. Ophthalmol. Vis. Sci. 2021;62(2):31. doi: 10.1167/iovs.62.2.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu X., et al. Loss of the ER membrane protein complex subunit Emc3 leads to retinal bipolar cell degeneration in aged mice. PLoS One. 2020;15(9) doi: 10.1371/journal.pone.0238435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong L., et al. ER complex proteins are required for rhodopsin biosynthesis and photoreceptor survival in Drosophila and mice. Cell Death Differ. 2020;27(2):646–661. doi: 10.1038/s41418-019-0378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin Z.B., et al. Identification of de novo germline mutations and causal genes for sporadic diseases using trio-based whole-exome/genome sequencing. Biol. Rev. Camb. Phil. Soc. 2018;93(2):1014–1031. doi: 10.1111/brv.12383. [DOI] [PubMed] [Google Scholar]
- 29.Huang X.F., et al. Unraveling the genetic cause of a consanguineous family with unilateral coloboma and retinoschisis: expanding the phenotypic variability of RAX mutations. Sci. Rep. 2017;7(1):9064. doi: 10.1038/s41598-017-09276-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jumper J., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen K., et al. Widespread translational control regulates retinal development in mouse. Nucleic Acids Res. 2021;49(17):9648–9664. doi: 10.1093/nar/gkab749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitmore S.S., et al. Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-Seq. Exp. Eye Res. 2014;129:93–106. doi: 10.1016/j.exer.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vilardi F., Lorenz H., Dobberstein B. WRB is the receptor for TRC40/Asna1-mediated insertion of tail-anchored proteins into the ER membrane. J. Cell Sci. 2011;124(Pt 8):1301–1307. doi: 10.1242/jcs.084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guna A., et al. A TAle of two pathways: tail-anchored protein insertion at the endoplasmic reticulum. Cold Spring Harbor Perspect. Biol. 2023;15(3):a041252. doi: 10.1101/cshperspect.a041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diederichs K.A., Buchanan S.K., Botos I. Building better barrels - β-barrel biogenesis and insertion in bacteria and mitochondria. J. Mol. Biol. 2021;433(16) doi: 10.1016/j.jmb.2021.166894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casson J., et al. Multiple pathways facilitate the biogenesis of mammalian tail-anchored proteins. J. Cell Sci. 2017;130(22):3851–3861. doi: 10.1242/jcs.207829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian S., et al. Proteomic analysis identifies membrane proteins dependent on the ER membrane protein complex. Cell Rep. 2019;28(10):2517–2526.e5. doi: 10.1016/j.celrep.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whittsette A.L., Wang Y.J., Mu T.W. The endoplasmic reticulum membrane complex promotes proteostasis of GABA(A) receptors. iScience. 2022;25(8) doi: 10.1016/j.isci.2022.104754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung H.L., et al. De novo variants in EMC1 lead to neurodevelopmental delay and cerebellar degeneration and affect glial function in Drosophila. Hum. Mol. Genet. 2022;31(19):3231–3244. doi: 10.1093/hmg/ddac053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziegler A., et al. A snapshot of some pLI score pitfalls. Hum. Mutat. 2019;40(7):839–841. doi: 10.1002/humu.23763. [DOI] [PubMed] [Google Scholar]
- 42.Zhao G., London E. An amino acid “transmembrane tendency” scale that approaches the theoretical limit to accuracy for prediction of transmembrane helices: relationship to biological hydrophobicity. Protein Sci. 2006;15(8):1987–2001. doi: 10.1110/ps.062286306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X., Gregory-Evans C.Y. Nonsense suppression therapies in ocular genetic diseases. Cell. Mol. Life Sci. 2015;72(10):1931–1938. doi: 10.1007/s00018-015-1843-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rio Frio T., et al. Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay. J. Clin. Invest. 2008;118(4):1519–1531. doi: 10.1172/JCI34211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roman-Sanchez R., Wensel T.G., Wilson J.H. Nonsense mutations in the rhodopsin gene that give rise to mild phenotypes trigger mRNA degradation in human cells by nonsense-mediated decay. Exp. Eye Res. 2016;145:444–449. doi: 10.1016/j.exer.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu M.-K., Lin H.-Y., Chen F.-C. NMD Classifier: a reliable and systematic classification tool for nonsense-mediated decay events. PLoS One. 2017;12(4) doi: 10.1371/journal.pone.0174798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nuzzi R., et al. Gonadal hormones and retinal disorders: a review. Front. Endocrinol. 2018;9:66. doi: 10.3389/fendo.2018.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung S.E., et al. Estrogen antagonist and development of macular hole. Kor. J. Ophthalmol. 2010;24(5):306–309. doi: 10.3341/kjo.2010.24.5.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kingdom R., Wright C.F. Incomplete penetrance and variable expressivity: from clinical studies to population cohorts. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.920390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marchant A., Evans K.O. In: Encyclopedia of Animal Cognition and Behavior. Vonk J., Shackelford T., editors. Springer International Publishing; Cham: 2019. Incomplete penetrance; pp. 1–2. [Google Scholar]
- 51.Cooper D.N., et al. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum. Genet. 2013;132(10):1077–1130. doi: 10.1007/s00439-013-1331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El-Brolosy M.A., Stainier D.Y.R. Genetic compensation: a phenomenon in search of mechanisms. PLoS Genet. 2017;13(7) doi: 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Einson J., et al. Genetic control of mRNA splicing as a potential mechanism for incomplete penetrance of rare coding variants. Genetics. 2023:iyad115. doi: 10.1093/genetics/iyad115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study has been deposited at https://www.cncb.ac.cn/with access number: HRA002930.
Supplementary content related to this article has been published online at [URL].