Abstract
Inflammatory bowel disease is a relapsing inflammatory disease seriously endanger human health. Wedelolactone (WED) is a major active ingredient from Eclipta prostrata (L.) L. and has shown anti-inflammatory effects. However, the mechanism of WED in treating inflammatory colitis remains unknown. We aimed to investigate the mechanisms of WED in treating ulcerative colitis through lipidomic study. Sixty male C57BL/6 mice were exposed to DSS to induce acute colitis. Disease progression was judged by the disease activity index (DAI) and pathological changes of colon tissue. An ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF-MS) method was performed for colon and plasma lipidomics analyses. Differential metabolites in the three groups were distinguished by univariate and multivariate analysis. WED exerted anti-inflammatory effects representing by body weight and DAI score. Three metabolites were identified in plasma and 20 in colon. According to pathway analysis, the effects of WED on colitis were associated with seven pathways. The glycerophospholipid metabolism and ether lipid metabolism were the primary pathways. The findings provide important insight of the mechanism of WED in treating DSS induced colitis through lipidomic perspective.
Keywords: Wedelolactone, Ulcerative colitis, UPLC-Q-TOF-MS, Lipidomics
Graphical abstract
Highlights
-
•
UPLC-Q-TOF-MS-based lipidomics of wedelolactone-treated DSS mice was analyzed.
-
•
Twenty-three metabolites were differentially expressed in the three study groups.
-
•
The effects of wedelolactone on colitis were associated with seven pathways.
-
•
Wedelolactone can potentially prevent IBD.
-
•
Untargeted lipidomics can help identify traditional pharmacological agents.
1. Introduction
Ulcerative colitis (UC), an inflammatory bowel disease (IBD), presents as a relapsing non-transmural inflammatory condition confined to the colon [1]. UC patients show marked growth of colorectal cancer, which can seriously endanger human health. 5-Aminosalicylic acid, corticosteroids, immunosuppressives, biologics, and Janus kinase inhibitors are the main therapeutic agents for UC [2]. However, the cur-rent medical therapies show different limitations and adverse effects, including hepatotoxicity and bone marrow suppression [3], highlighting the need for an effective drug for IBD.
In Asian traditional medicine, Eclipta prostrata (L.) L. is widely used to treat hemorrhage [4] and hyperlipidemia [5], and it has shown antitumor [6] and hepatoprotective [7] activities. Wedelolactone (WED), a polyphenolic compound (structure shown in Fig. 1A), is a major active ingredient extracted from the plant Eclipta prostrata (L.) L. [8] with excellent anti-cancer [9,10] and anti-inflammatory effects [[11], [12], [13]]. Moreover, one report suggested that WED had a definite therapeutic effect in experimental UC [14]. However, existing studies on the mechanism of action of WED have mainly focused on the inflammation-related IκK/IκB/NF-κB and AMPK/NLRP3/IL-1β pathways [[13], [14], [15], [16]], and they concluded that WED exerted its action only partially through these pathways [11,14]. Therefore, the mechanism of WED remains largely unknown, especially in treating inflammatory colitis.
Fig. 1.
Wedelolactone's protective effects on mouse colitis induced by DSS. (A) Wedelolactone molecular structure. (B) Flow chart of the experiment. (C) Changes in body weight. (D) DAI scores. (E) Colon length measurement. (F) Colon length visual diagram. (G) Mouse colon hematoxylin and eosin (HE) staining image (upper panel: 50X, lower panel: 200X). (H) Mucosal injury scores of the colon tissue. The data are expressed as the mean ± standard deviation (n = 10). *p < 0.05 compared to the control group; #p < 0.05 compared to the DSS group.
Lipids are important organic biomolecules that play vital roles in biological pathways and also serve as biomarkers or therapeutic targets for diseases [17]. Lipidomics is a branch of metabolomics involving the analysis of different lipid molecules in biological samples. Within the last decade, lipidomics has become a promising field of study for exploring the intervention mechanisms of drugs, determining pathogenic mechanisms, and identifying biomarkers to predict and monitor disease conditions [18]. Lipidomics has also been utilized to investigate the pathomechanisms of IBD, including inflammation, epithelial barrier function, and the gut microbiome [19,20].
In this study, we showed that WED treatment reversed intestinal injury in dextran sulfate sodium (DSS)-treated mice. We first used ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF-MS) lipidomics to obtain plasma and colon metabolic signatures of experimental mice. Then, we performed pattern recognition analysis, including principal component analysis (PCA) and orthogonal partial least-squares discriminant analysis (OPLS-DA), to identify the differential metabolites and provided important insights of the mechanism of WED in treating UC through lipidomic perspective.
2. Material and methods
2.1. Chemicals and reagents
Methanol and 2-propanol were obtained from Merck (Darmstadt, Germany); acetonitrile (ACN) was obtained from ANPEL Laboratory Technologies (Shanghai) Inc.; ammonium formate and formic acid were obtained from Sigma-Aldrich (St. Louis, MO, U.S.A.); and methyl tertbutyl ether (MTBE) was supplied by Aladdin (Shang Hai, China). DSS (36,000–50,000 Da) was obtained from Yeasen Biotechnology (Shanghai) Co., Ltd. A urine–fecal occult blood test kit was purchased from Yeasen Biotechnology (Shanghai) Co., Ltd. WED (Fig. 1A) (purity: HPLC ≥98%; Batch No. PQJZ20210511-1) was obtained from Nanjing & Autumn Biological Co, Ltd (Chengdu, China), and mesalazine was supplied by Aladdin (Shang Hai, China).
2.2. Mice
Protocols for all animal experiments were performed after obtaining approval from the Animal Care and Use Ethics Committee of the South China University of Technology with the approval number 2021017. Sixty C57BL/6 male mice aged 6 weeks were purchased from Hunan Slake Experimental Animal Co., Ltd. (Hunan, China). Mice were housed in a controlled room with unrestricted access to food and distilled water.
2.3. Animal design and samples collection
Sixty mice were domesticated for 7 days, and then randomly divided into six groups (n = 10 per group): control group, DSS group, mesalazine (200 mg/kg) group, and WED (20, 40, 80 mg/kg) groups (WED-L, WED-M, WED-H, respectively). Each mouse received 3% DSS in the drinking water for five consecutive days followed by four days of treatment with 4% DSS. The same volume of distilled water was administered to control mice. Drinking water and DSS were freshly prepared, and the preparation of mesalazine solution, 10 mg mesalazine was dissolved in 5 ml water. For the preparation of WED-H solution, 2 mg WED was dissolved in 10 ml water, WED-M and WED-L solutions were prepared by equal proportion dilution of WED-H solution. All drugs made to dissolve as completely as possible by ultrasound and administered intragastric. The dosages of WED were chosen according to previous studies [14,21]. The body weight, feces consistency, and presence of gross blood were recorded daily.
On day 10, after 10 h of overnight fasting, the mice were sacrificed. Plasma and colon tissues were collected after sacrificed. Colon tissues were frozen at −80 °C after length measurement. Plasma was obtained by centrifugation at 800×g for 10 min, and then stored at −80 °C until lipidomics analysis.
2.4. Evaluation of disease severity
The disease activity index (DAI) score was computed as described previously [22]. Status of bleeding was tested using a urine–fecal occult blood test kit. The detailed DAI scoring rules are provided in Supporting Information Table S1. The middle part of the colon was fixed in 10% formalin, embedded in paraffin, cut into 4-μm-thick sections, and finally stained. Pathological changes in colon tissue were evaluated by Chiu's score. Five microscope fields (200X) of each section were randomly selected for the mucosal injury score, as described previously [23]. The detailed rules are provided in Supporting Information Table S2.
2.5. Extraction and preparation of lipid samples
Sample processing was performed as described previously [24]. Briefly, for plasma lipid extraction, 50 μL of plasma and 150 μL of methanol were exhaustively mixed by vortexing for 1 min. Next, 500 μL of MTBE was mixed into each sample and shaken at room temperature for 60 min, and 125 μL of H2O was then added to the solutions. After vortexing for another 1 min, the samples were spun for 10 min at 14,800×g, 4 °C. The upper-layer liquid was transferred to a low-retention EP tube. Next, the residue was extracted again using an MTBE/methanol/H2O (20:6:5v/v/v) mixture with the same procedure. The two parts of the organic phase were combined, dried under nitrogen flow, and redissolved with 40 μL of isopropanol and 40 μL of methanol. Next, 50 μL of the supernatant was obtained for UPLC-MS analysis, with 10 μL of each sample used as the QC sample.
For tissue lipid extraction, 20-mg colon samples were precisely weighed and homogenized with 300 μL of methanol and 100 μL of water. Next, 1 mL of MTBE was mixed into each sample, and the samples were deposited at-20 °C for 1 h. A total of 150 μL of H2O was added for another 15 min at −20 °C, followed by centrifugation at 14,800×g for 15 min at 4 °C. Then, the upper organic phase was collected in a low-retention EP tube. A pre-cooled methanol-MTBE-H2O mixture (400 μL, 20:6:5, v/v) was added to the remaining solution and incubated at −20 °C for 1 h. After centrifugation at 14,800×g for 15 min at 4 °C, the upper organic phase was combined together. The aliquots were dried under a nitrogen steam, and 40 μL of isopropanol and 40 μL of methanol were added for resolution. Next, 50 μL of the upper liquid was transferred to the injection bottle for UPLC-MS analysis, and 10 μL of each sample solution was added to the injection bottle as the QC sample.
2.6. UPLC-Q-TOF-MS conditions
The UPLC-Q-TOF-MS conditions were similar to those used in our previous study [25]. Sample analysis was performed on an Agilent Technologies 1290 II Infinity LC system coupled with a 6545 Q-TOF mass spectrometer. Five microliters of each sample solution were separated using an ACQUITY UPLC HSS T3 column (2.1 × 100 mm, 1.8 μm; Waters, Manchester). One QC sample was run after each of the five plasma or colon samples. Supporting Information Table S3 shows the detailed condition.
The MS was run under electrospray ionization positive (ESI+) and negative (ESI-) modes. The capillary voltage and source temperature were 3.0 kV and 350 °C, and the MS acquisition rate was 4 spectra/s. MS data in an m/z range of 50–1700 were collect-ed. The mass spectrometry data have been deposited to the MetaboLights (https://www.ebi.ac.uk/metabolights/login) through the MetaboLights partner repository. The data set identifier is MTBLS5086.
2.7. Statistical analysis
The raw UPLC-MS data were introduced to Agilent Masshunter Profinder (B.08.00) for molecular feature extraction (MFE), which was set at the mass of 10 ppm and retention time alignment of 0.25 min. The absolute height parameter of abundance was set at 300 to remove noise. Then a manual review was carried out to exclude abnormal molecular features. Like many studies using Agilent's compound annotation process [[26], [27], [28]], the qualified data extracted from non-target MFE were imported into Agilent MassHunter Mass Profiler Professional (MPP) (version B.14.9). The data were subjected to normalization, filtration, and recursion analysis.
All the data were normalized at 75% percentile to make sure the data were com-parable. Frequency analysis was set to 80% to ensure that 80% of the compounds are present in at least one study group. Then, the compounds with significant differences between DSS vs control group and WED vs DSS group were selected by t-test independent filtering (p values < 0.05) and fold change (FC) analysis (FC ≥ 1.5 or ≤0.67). The results were presented in the form of volcano plots.
Subsequently, principal component analysis (PCA) and orthogonal partial least-squares discriminant analysis (OPLS-DA) were performed on these normalized data by SIMCA-P 14.1 after Pareto-scaling (Par transformation). The quality of the OPLS-DA models is controlled by the R2 or Q2 values. To avoid model over-fitting, 200 cross-validations were performed. At the same time, the variable projected importance (VIP) was determined according to the OPLS-DA model to screen metabolic markers out. VIP (Variable influence on projection) values can be used to measure the metabolite accumulation difference of each sample classification discriminant ability of impact strength and explain. VIP values > 1.0 indicates a large differential contribution of this compound [[29], [30], [31]].
Differences in metabolites were determined by a combination of VIP values > 1 in OPLS-DA models, p values < 0.05 in Student's t-test for the comparison of peak area between groups, and fold change (FC) values of the corresponding peak area between groups ≥1.5 or ≤0.67. These criteria were employed in many studies [24,32,33].
Entities obtained were identified using an ID browser from MPP. This software used a database from Metlin. The mass error was set at 15 ppm and the limitation of the charge state was set to a maximum of two. Metabolites were identified with the top 10 best matched depending on the score. Then the annotation was performed by searching databases, including the Human Metabolome Database (HMDB), Kyoto Encyclopedia of Genes and Genomes (KEGG), MassBank, and LIPID MAPS (http://www.lipidmaps.org). The identity of metabolites was confirmed by matching the experimental MS/MS spectra to MS/MS spectra from databases. The validation followed the criteria of the Metabolomics Standards Initiative level 2 [34].
The MetaboAnalyst 5.0 online system (http://www.%20Metaboanalyst.%20ca) was used to perform pathway analysis. Correlation analysis between DAI and lipid biomarkers was performed using Pearson correlation coefficients for data that followed a normal distribution or Spearman correlation for data that are non-normally distributed. The results are presented as mean ± standard deviation (SD). SPSS 26.0 was used for statistical analysis of the experimental data and GraphPad Prism 8.0 for statistical drawing. Significance was defined by a P-value of 0.05 or less.
3. Results
3.1. Effects of WED on DSS in C57BL/6 mice
Fig. 1A shows the chemical structure of WED. To assess the effects of WED on DSS-induced UC, we generated the mouse model by feeding the mice DSS solution for 9 days (Fig. 1B). As shown in Fig. 1C-F, mice in the DSS group showed significant body weight loss, colon length reduction, and DAI score increments in comparison with those in the control group. In contrast, the findings for mice in the mesalazine and WED (20, 40, 80 mg/kg) groups showed various degrees of improvement in comparison with the DSS group. The histological analysis indicated the protective effects of WED (Fig. 1G). The DSS group showed significant histological injuries such as loss of epithelial crypts, disruption of the mucosal barrier, and infiltration of inflammatory cells, while the WED (20, 40, 80 mg/kg) groups showed obvious improvements in these findings. Similarly, in comparison with the Chiu's score for control mice, the scores for DSS mice indicated obvious mucosal lesions, but these scores were reduced after mesalazine and WED treatment (Fig. 1H), indicating that the treatment ameliorated histological injuries. Together, these data indicate that oral administration of WED (20, 40 mg/kg) had better effects on DSS-induced colitis than mesalazine. However, the effects of WED were not dose-dependent, since the low dose (WED-L) group showed the best results in the current study. Therefore, the control group, DSS group, and WED-L groups were chosen for the following lipidomics experiments.
3.2. Plasma lipidomic profiling
The representative total ion chromatograms (TICs) of plasma lipidomics are illustrated in Fig. S1 (A, B). The overlapping TICs of the plasma QC samples, which provide robust evidence to assess instrument stability and data quality, are shown in Fig. S2A.
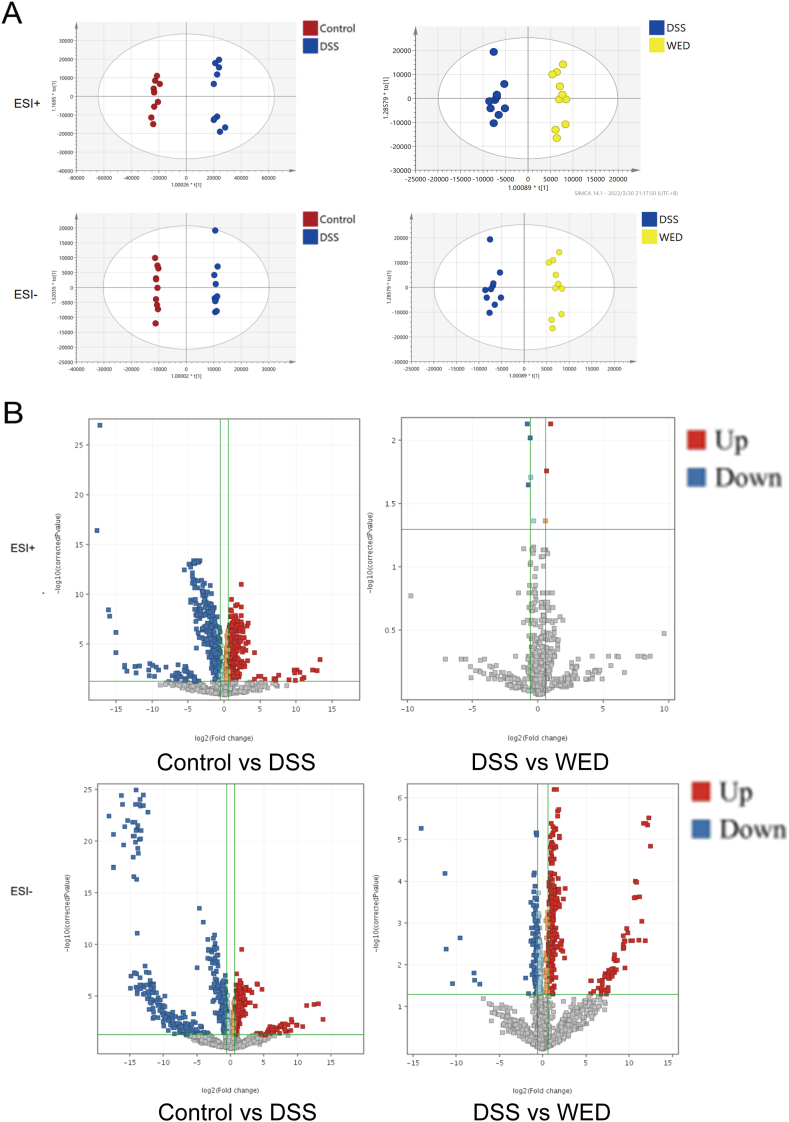
To observe the separation trend among groups, we established PCA and OPLS-DA model. The PCA scatter plots in the ESI+ and ESI- modes are presented in Fig. 2A. The metabolic features of each group (represented as differently colored dots) were well clustered, indicating that the DSS mice were obviously different from the control mice, and the WED group tended to be distinct from the DSS group, showing that WED has definite medical effects of UC. The OPLS-DA model was conducted in Fig. 3A. In both ion modes, the DSS group was significantly different from the controls, while the WED group was significantly different from the DSS group. The parameters of the OPLS-DA model are listed in Table S4. Fig. S3A shows the permutation tests for the OPLS-DA model of plasma samples. The result suggested that the OPLS-DA model was credible.
Fig. 2.
PCA score plots (n = 10/group); control group is expressed as red dots, WED-L group is expressed as yellow dots, while DSS group is expressed as blue dots. (A) Plots of PCA score for plasma samples. (B) Plots of PCA score for colon samples.
Fig. 3.
(A) OPLS-DA analysis of plasma (n = 10/group); control group is expressed as red dots, WED group is expressed as yellow dots, while DSS group is expressed as blue dots. (B) The most significant ions for differentiation between DSS group and control group, WED-treated group and DSS group were acquired by volcano plots.
The result of univariate analysis was displayed in the form of volcano plots (Fig. 3B). They were established to display the FC (≥1.5 or ≤0.67) and P-value (<0.05) indicators. Meanwhile, to appraise the protective impact of WED on UC, the VIP value (>1.0) of OPLS-DA model, P-value (<0.05), and FC (≥1.5 or ≤0.67) were used to screen the potential biomarkers. Twelve molecules that presented opposite trends in the model and WED groups were screened out. Including ten glycerophospholipids (GPs), one sphingolipid (SP), and one fatty acyls (FA). Table S8 shows the detailed in-formation.
The compound was further confirmed on the MS/MS information of the experiment. The specific identification process was shown in Supplementary Material 2. Finally, 3 differential lipid metabolites in plasma (Table 1) were screened out.
Table 1.
Differential lipid metabolites in plasma.
| No. | Compound | Ionization mode | Retention time | Mass | DSS vs. Control |
WED vs. DSS |
||
|---|---|---|---|---|---|---|---|---|
| Trend | FC | Trend | FC | |||||
| 1 | PE (P-18:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | + | 12.47 | 775.5517 | ↓ | −1.79 | ↑ | 1.52 |
| 2 | SM (d20:1/19:0) | + | 14.45 | 772.6436 | ↓ | −2.47 | ↑ | 1.68 |
| 3 | PG (P-16:0/18:4(6Z,9Z,12Z,15Z)) | – | 12.65 | 726.4831 | ↑ | 4.40 | ↓ | −2.55 |
FC:Fold change.
3.3. Colon lipidomics profiling
Similar to plasma lipidomics analysis, the representative TIC of colon lipidomics is shown in Fig. S1 (C and D). The overlaid TIC of colon QC samples is shown in Fig. S2B, demonstrating the good stability and reproducibility of this method. Fig. 2, Fig. 4A show the PCA and OPLS-DA scatter graphs, respectively. Both graphs indicated that the control group vs. DSS group and DSS group vs. WED group had good separation. Volcano plots (Fig. 4B) were established, and the parameters (R2 and Q2 in Table S5) and the permutation test (Fig. S3B) indicated that the OPLS-DA model of colon samples was reliable. Fifty-three molecules that presented opposite trends were screened out. Including 18 fatty acyls (FAs), 16 glycerophospholipids (GPs), 13 sterol lipids (STs), 3 glycerolipids (GLs), one polyketide (PK), one sphingolipid (SP), and one LysoPC (LPC). Table S9 shows the detailed information. According to MS/MS information, 20 different variables were identified in colon lipidomics. Identification process was shown in Supplementary Material 2. Table 2 shows the detailed metabolites.
Fig. 4.
(A) OPLS-DA analysis of colon (n = 10/group); control group is expressed as red dots, WED group is expressed as yellow dots, while DSS group is expressed as blue dots. (B) The most significant ions for differentiation between DSS group and control group, WED-treated group and DSS group were acquired by volcano plots.
Table 2.
Differential lipid metabolites in colon.
| No. | Compound | Ionization mode | Retention time | Mass | DSS vs. Control |
WED vs. DSS |
||
|---|---|---|---|---|---|---|---|---|
| Trend | FC | Trend | FC | |||||
| 1 | Behenoyl-EA | + | 7.11 | 383.3762 | ↑ | 1.45 | ↓ | −1.89 |
| 2 | PC (18:3(9Z,12Z,15Z)/19:1(9Z)) | + | 14.09 | 795.584 | ↓ | −12580.9 | ↑ | 13665.72 |
| 3 | PE (22:5(4Z,7Z,10Z,13Z,16Z)/P-18:1(11Z)) | + | 11.40 | 775.552 | ↓ | −10.19 | ↑ | 10.15 |
| 4 | PC (18:4(6Z,9Z,12Z,15Z)/P-18:1(11Z)) | + | 9.21 | 763.5506 | ↓ | −1.72 | ↑ | 5.97 |
| 5 | PC (14:0/14:0) | + | 8.18 | 677.9325 | ↓ | −1.68 | ↑ | 5.71 |
| 6 | DG (16:1(9Z)/22:2(13Z,16Z)/0:0) | + | 17.28 | 646.5542 | ↓ | −1.83 | ↑ | 1.40 |
| 7 | PC (20:0/14:0) | + | 15.47 | 761.5934 | ↓ | −4454.09 | ↑ | 23525.96 |
| 8 | PE (P-18:0/18:0) | + | 16.67 | 731.5821 | ↓ | −2.64 | ↑ | 5.57 |
| 9 | PC (18:1(9Z) e/2:0) | + | 3.85 | 549.3798 | ↓ | −2.04 | ↑ | 1.63 |
| 10 | PC (17:1(9Z)/0:0) | + | 2.37 | 507.3329 | ↓ | −2.52 | ↑ | 1.61 |
| 11 | Minabeolide-2 | + | 3.94 | 466.2728 | ↓ | −2.25 | ↑ | 5.54 |
| 12 | 9-Octadecene | + | 2.81 | 252.2817 | ↑ | 1.88 | ↓ | −2.25 |
| 13 | PG (16:0/0:0) | + | 2.86 | 484.2826 | ↓ | −1.68 | ↑ | 3.33 |
| 14 | Phytosphingosine | + | 2.07 | 318.3003 | ↑ | 2.40 | ↓ | −2.05 |
| 15 | LysoPC (14:0/0:0) | + | 2.02 | 468.3018 | ↓ | −1.63 | ↑ | 1.39 |
| 16 | LysoPE (0:0/22:2(13Z,16Z)) | – | 3.57 | 533.3481 | ↓ | −2.42 | ↑ | 1.67 |
| 17 | 7-Tridecynoic acid | – | 1.08 | 210.1615 | ↓ | −3.88 | ↑ | 4.07 |
| 18 | Dimorphecolic acid | – | 2.96 | 296.2348 | ↑ | 2.49 | ↓ | −2.22 |
| 19 | DL-2-hydroxy stearic acid | – | 2.60 | 300.2661 | ↑ | 11.34 | ↓ | −2.01 |
| 20 | 4Z,7Z,10Z,13Z,16Z,19Z,22Z,25Z-octacosaoctaenoic acid | – | 6.60 | 408.3027 | ↓ | −2.56 | ↑ | 7.32 |
FC: Fold change.
3.4. Metabolic pathway
Through MS/MS identification, a total of 23 compounds from plasma and colon were eventually identified as differential compounds. Fig. 5A shows the hierarchical clustering of 23 WED-affected metabolites screened from plasma and colon lipidomic analyses. The clustering heatmap showed that WED group and control group had relatively similar metabolite profiles and expression levels, while the corresponding findings for the DSS model group were obviously different.
Fig. 5.
(A) The heatmap of potential biomarkers in control group, WED group and DSS group. Each row represents a metabolite and each column represents a sample. The firebrick color indicates high abundance and navy indicates low abundance. (B) Pathway analysis of 23 differential metabolites identified based on plasma or colon lipidomics. Different colors represent different p-values (y-axis), which augment as red deepens. The size of the circle represents the pathway influence (x-axis), and the magnitude of the influence climbs up as the area of the circle becomes larger.
To understand the functions of the 23 differential metabolites recognized by plasma or colon lipidomics, metabolic pathway analysis was conducted using MetaboAnalyst 5.0. Pathway impact was weight calculations based on topological analysis of relative-betweeness centrality and -log(p) was conducted by Hypergeometric Test. As shown in Fig. 5B, seven metabolic pathways—glycerophospholipid metabolism, ether lipid metabolism, linoleic acid metabolism, alpha-linoleic acid metabolism, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, sphingolipid metabolism, and arachidonic acid metabolism—were found. The details of the pathways analysis are shown in Table S6.
As shown in Table S7, six metabolites were involved in multiple pathways, and these metabolites could be considered as key molecules in the therapeutic effects of WED on DSS-induced colitis. Therefore, the correlation of the six metabolites with the DAI score was analyzed. As shown in Table 3, all metabolites showed good correlations with the DAI scores. Phytosphingosine showed positive associations with DAI (r = 0.545, p < 0.001), while the others showed negative associations with DAI (r between −0.798 and −0.432 and p values between 0.001 and 0.017). Additionally, the relative peak areas for the six identified key molecules in each group are represented in Fig. S4.
Table 3.
Correlation of the peak area and DAI score.
| Variables | r | p-value |
|---|---|---|
| PE (22:5(4Z,7Z,10Z,13Z,16Z)/P-18:1(11Z)) | −0.432* | 0.017 |
| PC (18:1(9Z) e/2:0) | −0.612*** | 0.000 |
| PC (18:4(6Z,9Z,12Z,15Z)/P-18:1(11Z)) | −0.798*** | 0.000 |
| Phytosphingosine | 0.545** | 0.002 |
| PC (14:0/14:0) | −0.626*** | 0.000 |
| LysoPC (14:0/0:0) | −0.535** | 0.002 |
*p < 0.05, **p < 0.01, ***p < 0.001.
To obtain further insights into the potential therapeutic effects of WED on DSS-induced acute colitis, the correlation networks of the key biomarkers that are abnormally expressed in DSS mice on KEGG are shown in Fig. 6.
Fig. 6.
Potential metabolic pathways regulated in DSS mice after treatment with WED. The metabolites marked in red were significantly reduced in the DSS group in comparison with the control group but were increased in the WED group in comparison with the DSS group.
4. Discussion
Lipids constitute most of the cellular membrane bilayer and can regulate diverse biological processes. Thus, the important role of lipid function in IBD is obvious. To our knowledge, the present study is the first attempt to evaluate the lipidomic profile of DSS-induced colitis after WED treatment.
The simple flowchart of this experiment is shown in Fig. S5. In this study, we used DSS to induce colitis, and compared the anti-inflammatory effect of WED to the effect of mesalazine, a commonly used drug for UC. We found that WED showed better protective effects against the symptoms of experimental colitis such as loss of epithelial crypts and disruption of the mucosal barrier. However, the protective effect of WED on colitis was dose-independent, similar to the findings obtained by Wei et al. (2017). UPLC-Q-TOF-MS-based plasma and colon lipidomics revealed disturbances in the intestinal metabolism, while metabolic pathway enrichment analysis revealed that WED regulated seven metabolic pathways involving six metabolites, including four glycerophospholipids (GPs), one sphingolipid (SP), and one LysoPC (LPC). Among them, PE (22:5(4Z,7Z,10Z,13Z,16Z)/P-18:1(11Z)), and PC (18:4(6Z,9Z,12Z,15Z)/P-18:1(11Z)) involved in multiple pathways. These six metabolites correlated well with DAI scores, indicating that they are key molecules affected by WED in DSS-induced colitis.
In our study, pathway analysis of lipidomics showed that the glycerophospholipid metabolism was the most crucial pathway. Glycerophospholipid has been reported to exert a critical effect during immune and inflammatory responses. Phosphoethanolamine (PE), also called kephalin, and phosphocholine (PC), also called lecithin, play crucial roles in biological membranes. In this study, we obtained three hits, PE (22:5(4Z,7Z,10Z,13Z,16Z)/P-18:1(11Z)), PC(14:0/0:0), and LysoPC(14:0/0:0), involved in glycerophospholipid metabolism. As the material components in glycerophospholipid metabolism, the contents of these three metabolites in DSS mice were lower than those in control mice, but increased after WED treatment. Similar to a previous report in which PCs and LPCs decreased in UC patients [35,36]. PCs increased hydrophobicity on the luminal side of the mucus layer and may motivate an anti-inflammatory response at the mucosal barrier [37]. Many studies reveal that therapeutic addition of PC using slow-release preparations is beneficial [38,39]. Thus, WED may exert its function through reversal of PC reduction.
Ether lipids are key components of cell membranes, as well as biological signaling molecules. Recent studies have demonstrated that ether lipid associated with neurodegenerative diseases, cancer, and metabolic disorders [40,41]. In our study, the degree of ether lipid metabolism disorder was also serious, PC (18:1(9Z) e/2:0) was involved in it. On the basis of these results, we can speculate that WED is a potential novel therapeutic drug against colitis. However, an integration of the interaction of transcriptome-level, protein-level, and microbial-level findings in systems biology studies is recommended to validate the mechanism of action of WED on IBD. And the biological significance of these biomarkers should be further experimentally verified in the follow-up research as well.
5. Conclusions
A UPLC-Q-TOF-MS method based on lipidomics technology was developed to explore the therapeutic effects of WED in a DSS-induced colitis mouse model. Our findings showed that WED can improve body weight, DAI score, and Chiu's score and play an anti-inflammatory role, providing evidence for the therapeutic effects of WED in UC. We identified three potential biomarkers in plasma lipidomics, 20potential biomarkers in colon lipidomics, and seven pathways based on these biomarkers. Overall, this study provides a theoretical basis for the use of WED as a natural product to prevent and treat intestinal inflammation, and the novel strategy based on untargeted lipidomics analysis proposed in this paper can facilitate the development of traditional pharmacological agents.
Author contribution statement
Yuanyuan Gou: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper. Zichen Wang; Jiaxin Huang: Performed the experiments. Liping Zhou: Conceived and designed the experiments; Analyzed and interpreted the data. Jinpan Du: Conceived and designed the experiments; Wrote the paper. Jing Li:Contributed reagents, materials, analysis tools or data. Xuyu Zhang: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data. Su Guan: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Data availability statement
Data associated with this study has been deposited at The mass spectrometry data have been deposited to the MetaboLights (https://www.ebi.ac.uk/metabolights/login) through the MetaboLights partner repository. The data set identifier is MTBLS5086.
Funding
The study was supported by grants from National Natural Science Foundation of China (No. 82072204 and No.81701874), and by grants from Natural Science Foundation of Guangdong Province, China (No. 2021A1515010990 and No. 2019A1515012233).
Declarations
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Animal Care and Use Ethics Committee of the South China University of Technology with the approval number 2021017.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e20162.
Contributor Information
Xuyu Zhang, Email: zhangxuy@mail.sysu.edu.cn.
Su Guan, Email: guansu@scut.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Material 1: Supplementary Fig. S1: Total ion chromatography (TIC) of lipidomics (A) ESI + mode of plasma, (B) ESI- mode of plasma, (C) ESI + mode of colon, (D) ESI- mode of colon; Supplementary Fig. S2: Overlapped chromatograms of five replicates of quality controls in plasma analysis (A) and colon analysis (B); SupplementaryFig. S3: The permutation test of OPLS-DA scatter plots for plasma (A) and colon (B); Supplementary Fig. S4: Changes in the relative contents of target metabolites identified by KEGG pathway in different groups; Supplementary Table S1: DAI scoring rules; Supplementary Table S2: Chiu's scoring rules; Supplementary Table S3: UPLC gradient for lipidomics analysis; Supplementary Table S4: The parameters of OPLS-DA models for plasma lipidomics; Supplementary Table S5: The parameters of OPLS-DA models for colon lipidomics; Supplementary Table S6: Summary of pathway analysis; Supplementary Table S7: Pathway analysis of the identified metabolites; Supplementary Table S8: Total of 12 differential metabolites of plasma in both positive and negative ion mode; Supplementary Table S9: Total of 53 differential metabolites of colon in both positive and negative ion mode.
Supplementary Material 2: MS/MS identification of differential metabolites.
References
- 1.a Baumgart D.C., Sandborn W.J. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641–1657. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]; b Baumgart D.C., Sandborn W.J. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641–1657. doi: 10.1016/S0140-6736(07)60751-X. [DOI] [PubMed] [Google Scholar]
- 2.Burri E., Maillard M.H., Schoepfer A.M., Seibold F., Van Assche G., Rivière P., Laharie D., Manz M. Treatment algorithm for mild and moderate-to-severe ulcerative colitis: an update. Digestion. 2020;101:2–15. doi: 10.1159/000504092. [DOI] [PubMed] [Google Scholar]
- 3.Jing W., Dong S., Luo X., Liu J., Wei B., Du W., Yang L., Luo H., Wang Y., Wang S., et al. Berberine improves colitis by triggering AhR activation by microbial tryptophan catabolites. Pharmacol. Res. 2021:164. doi: 10.1016/j.phrs.2020.105358. [DOI] [PubMed] [Google Scholar]
- 4.Feng L., Zhai Y.-Y., Xu J., Yao W.-F., Cao Y.-D., Cheng F.-F., Bao B.-H., Zhang L. A review on traditional uses, phytochemistry and pharmacology of Eclipta prostrata (L.) L. J. Ethnopharmacol. 2019;245 doi: 10.1016/j.jep.2019.112109. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y., Peng L., Lu W., Wang Y., Huang X., Gong C., He L., Hong J., Wu S., Jin X. Effect of Eclipta prostrata on lipid metabolism in hyperlipidemic animals. Exp. Gerontol. 2015;62:37–44. doi: 10.1016/j.exger.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 6.Yadav N.K., Arya R.K., Dev K., Sharma C., Hossain Z., Meena S., Arya K.R., Gayen J.R., Datta D., Singh R.K. Alcoholic extract of Eclipta alba shows in vitro antioxidant & anticancer activity without exhibiting toxicological effects. Oxid. Med. Cell. Longev. 2017;2017 doi: 10.1155/2017/9094641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thirumalai T., David E., Therasa V., Elumalai E.K. Restorative effect of Eclipta alba in CCl 4 induced hepatotoxicity in male albino rats. Asian Pacific J. Trop. Dis. 2011;1 doi: 10.1016/S2222-1808(11)60072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melo P.A., Do Nascimento M.C., Mors W.B., Suarez-Kurtz G. Inhibition of the myotoxic and hemorrhagic activities of crotalid venoms by Eclipta prostrata (asteraceae) extracts and constituents. Toxicon. 1994;32:595–603. doi: 10.1016/0041-0101(94)90207-0. [DOI] [PubMed] [Google Scholar]
- 9.Nehybova T., Smarda J., Daniel L., Brezovsky J., Benes P. Wedelolactone induces growth of breast cancer cells by stimulation of estrogen receptor signalling. J. Steroid Biochem. Mol. Biol. 2015;152:9–18. doi: 10.1016/j.jsbmb.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 10.Kučírková T., Stiborek M., Dúcka M., Navrátilová J., Bogdanović Pristov J., Popović-Bijelić A., Vojvodić S., Preisler J., Kanický V., Šmarda J., et al. Anti-cancer effects of wedelolactone: interactions with copper and subcellular localization. Metallomics. 2018;10:1524–1531. doi: 10.1039/c8mt00191j. [DOI] [PubMed] [Google Scholar]
- 11.Pan H., Lin Y., Dou J., Fu Z., Yao Y., Ye S., Zhang S., Wang N., Liu A., Li X., et al. Wedelolactone facilitates ser/thr phosphorylation of NLRP3 dependent on PKA signalling to block inflammasome activation and pyroptosis. Cell Prolif. 2020;53 doi: 10.1111/cpr.12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng M., Lin J., Li C., Zhao W., Yang H., Lv L., Che C. Wedelolactone suppresses IL-1β maturation and neutrophil infiltration in Aspergillus fumigatus keratitis. Int. Immunopharm. 2019;73:17–22. doi: 10.1016/j.intimp.2019.04.050. [DOI] [PubMed] [Google Scholar]
- 13.Zhu M. mao, Wang L., Yang D., Li C., Pang S. ting, Li X. hua, Li R., Yang B., Lian Y. pei, Ma L., et al. Wedelolactone alleviates doxorubicin-induced inflammation and oxidative stress damage of podocytes by iκk/iκb/NF-κb pathway. Biomed. Pharmacother. 2019;117 doi: 10.1016/j.biopha.2019.109088. [DOI] [PubMed] [Google Scholar]
- 14.Wei W., Ding M., Zhou K., Xie H., Zhang M., Zhang C. Protective effects of wedelolactone on dextran sodium sulfate induced murine colitis partly through inhibiting the NLRP3 inflammasome activation via AMPK signaling. Biomed. Pharmacother. 2017;94:27–36. doi: 10.1016/j.biopha.2017.06.071. [DOI] [PubMed] [Google Scholar]
- 15.Yuan F., Chen J., Sun P.P., Guan S., Xu J. Wedelolactone inhibits LPS-induced pro-inflammation via NF-KappaB pathway in RAW 264.7 cells. J. Biomed. Sci. 2013;20 doi: 10.1186/1423-0127-20-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen P., Yang X., Jiang J., Wang X., Liang T., He L. Wedelolactone from Eclipta alba inhibits lipopolysaccharide-enhanced cell proliferation of human renal mesangial cells via NF-κb signaling pathway. Am. J. Transl. Res. 2017;9:2132–2142. [PMC free article] [PubMed] [Google Scholar]
- 17.Züllig T., Trötzmüller M., Köfeler H.C. Lipidomics from sample preparation to data analysis: a primer. Anal. Bioanal. Chem. 2020;412:2191–2209. doi: 10.1007/s00216-019-02241-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno L.O., Sánchez P.N., Abalo R. Lipidomics as tools for finding biomarkers of intestinal pathology: from irritable bowel syndrome to colorectal cancer. Curr. Drug Targets. 2021;23:636–655. doi: 10.2174/1389450122666210707122151. [DOI] [PubMed] [Google Scholar]
- 19.Titz B., Gadaleta R.M., Sasso G. Lo, Elamin A., Ekroos K., Ivanov N.V., Peitsch M.C., Hoeng J. Proteomics and lipidomics in inflammatory bowel disease research: from mechanistic insights to biomarker identification. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19092775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diab J., Hansen T., Goll R., Stenlund H., Ahnlund M., Jensen E., Moritz T., Florholmen J., Forsdahl G. Lipidomics in ulcerative colitis reveal alteration in mucosal lipid composition associated with the disease state. Inflamm. Bowel Dis. 2019;25:1780–1787. doi: 10.1093/ibd/izz098. [DOI] [PubMed] [Google Scholar]
- 21.Luo Q., Ding J., Zhu L., Chen F., Xu L. Hepatoprotective effect of wedelolactone against concanavalin A-induced liver injury in mice. Am. J. Chin. Med. 2018;46:819–833. doi: 10.1142/S0192415X1850043X. [DOI] [PubMed] [Google Scholar]
- 22.Cooper H.S., Murthy S.N.S., Shah R.S., Sedergran D.J. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab. Invest. 1993;69:238–250. doi: 10.1016/s0021-5198(19)41298-5. [DOI] [PubMed] [Google Scholar]
- 23.Chiu C.J., McArdle A.H., Brown R., Scott H.J., Gurd F.N. Intestinal mucosal lesion in low-flow states: I. A morphological, hemodynamic, and metabolic reappraisal. Arch. Surg. 1970;101:478–483. doi: 10.1001/archsurg.1970.01340280030009. [DOI] [PubMed] [Google Scholar]
- 24.Liu K., Jia B., Zhou L., Xing L., Wu L., Li Y., Lu J., Zhang L., Guan S. Ultraperformance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry-based metabolomics and lipidomics identify biomarkers for efficacy evaluation of mesalazine in a dextran sulfate sodium-induced ulcerative colitis mouse model. J. Proteome Res. 2021;20:1371–1381. doi: 10.1021/acs.jproteome.0c00757. [DOI] [PubMed] [Google Scholar]
- 25.Guan S., Jia B., Chao K., Zhu X., Tang J., Li M., Wu L., Xing L., Liu K., Zhang L., et al. UPLC-QTOF-MS-Based plasma lipidomic profiling reveals biomarkers for inflammatory bowel disease diagnosis. J. Proteome Res. 2020;19:600–609. doi: 10.1021/acs.jproteome.9b00440. [DOI] [PubMed] [Google Scholar]
- 26.Yu J.S., Nothias L.-F., Wang M., Kim D.H., Dorrestein P.C., Kang K. Bin, Yoo H.H. Tandem mass spectrometry molecular networking as a powerful and efficient tool for drug metabolism studies. Anal. Chem. 2022;94:1456–1464. doi: 10.1021/acs.analchem.1c04925. [DOI] [PubMed] [Google Scholar]
- 27.Avula B., Smillie T.J., Wang Y.H., Zweigenbaum J., Khan I.A. Authentication of true cinnamon (cinnamon verum) utilising direct analysis in real time (DART)-QToF-MS. FOOD Addit. Contam. Part A-Chemistry Anal. Control Expo. RISK Assess. 2015;32:1–8. doi: 10.1080/19440049.2014.981763. [DOI] [PubMed] [Google Scholar]
- 28.Lucini L., Rouphael Y., Cardarelli M., Canaguier R., Kumar P., Colla G. The effect of a plant-derived biostimulant on metabolic profiling and crop performance of lettuce grown under saline conditions. Sci. Hortic. 2015;182:124–133. doi: 10.1016/j.scienta.2014.11.022. [DOI] [Google Scholar]
- 29.Zhang H., Zhuo S., Song D., Wang L., Gu J., Ma J., Gu Y., Ji M., Chen M., Guo Y. Icariin inhibits intestinal inflammation of DSS-induced colitis mice through modulating intestinal flora abundance and modulating p-P65/P65 molecule. Turkish J. Gastroenterol. Off. J. Turkish Soc. Gastroenterol. 2021;32:382–392. doi: 10.5152/tjg.2021.20282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma L., Shen Q., Lyu W., Lv L., Wang W., Yu M., Yang H., Tao S., Xiao Y. Clostridium butyricum and its derived extracellular vesicles modulate gut homeostasis and ameliorate acute experimental colitis. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.01368-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z., Tan M., Deng H., Yang X., Yu Y., Zhou D., Dong H. Geographical origin differentiation of rice by LC–MS-based non-targeted metabolomics. Foods. 2022;11 doi: 10.3390/foods11213318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu L., Lai Q.H., Feng Q., Li Y.M., Feng J.F., Xu B. Serum metabolic profiling analysis of chronic gastritis and gastric cancer by untargeted metabolomics. Front. Oncol. 2021:11. doi: 10.3389/fonc.2021.636917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ban Y.N., Ran H.Y., Chen Y., Ma L. Lipidomics analysis of human follicular fluid form normal-weight patients with polycystic ovary syndrome: a pilot study. J. Ovarian Res. 2021:14. doi: 10.1186/s13048-021-00885-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sumner L.W., Amberg A., Barrett D., Beale M.H., Beger R., Daykin C.A., Fan T.W.M., Fiehn O., Goodacre R., Griffin J.L., et al. Proposed minimum reporting standards for chemical analysis. Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braun A., Treede I., Gotthardt D., Tietje A., Zahn A., Ruhwald R., Schoenfeld U., Welsch T., Kienle P., Erben G., et al. Alterations of phospholipid concentration and species composition of the intestinal mucus barrier in ulcerative colitis: a clue to pathogenesis. Inflamm. Bowel Dis. 2009;15:1705–1720. doi: 10.1002/ibd.20993. [DOI] [PubMed] [Google Scholar]
- 36.Santoru M.L., Piras C., Murgia A., Palmas V., Camboni T., Liggi S., Ibba I., Lai M.A., Orrù S., Blois S., et al. Cross sectional evaluation of the gut-microbiome Metabolome Axis in an Italian cohort of IBD patients. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-10034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider H., Braun A., Füllekrug J., Stremmel W., Ehehalt R. Lipid based therapy for ulcerative colitis-modulation of intestinal mucus membrane phospholipids as a tool to influence inflammation. Int. J. Mol. Sci. 2010;11:4149–4164. doi: 10.3390/ijms11104149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roediger W.E.W. The colonic epithelium in ulcerative colitis: an energy-deficiency disease? Lancet. 1980;316:712–715. doi: 10.1016/S0140-6736(80)91934-0. [DOI] [PubMed] [Google Scholar]
- 39.Li Q.Q., Chen G., Zhu D., Zhang W.W., Qi S.Z., Xue X.F., Wang K., Wu L.M. Effects of dietary phosphatidylcholine and sphingomyelin on DSS-induced colitis by regulating metabolism and gut microbiota in mice. J. Nutr. Biochem. 2022;105 doi: 10.1016/j.jnutbio.2022.109004. [DOI] [PubMed] [Google Scholar]
- 40.Dorninger F., König T., Scholze P., Berger M.L., Zeitler G., Wiesinger C., Gundacker A., Pollak D.D., Huck S., Just W.W., et al. Disturbed neurotransmitter homeostasis in ether lipid deficiency. Hum. Mol. Genet. 2019;28:2046–2061. doi: 10.1093/hmg/ddz040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dean J.M., Lodhi I.J. Structural and functional roles of ether lipids. Protein Cell. 2018;9:196–206. doi: 10.1007/s13238-017-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Supplementary Fig. S1: Total ion chromatography (TIC) of lipidomics (A) ESI + mode of plasma, (B) ESI- mode of plasma, (C) ESI + mode of colon, (D) ESI- mode of colon; Supplementary Fig. S2: Overlapped chromatograms of five replicates of quality controls in plasma analysis (A) and colon analysis (B); SupplementaryFig. S3: The permutation test of OPLS-DA scatter plots for plasma (A) and colon (B); Supplementary Fig. S4: Changes in the relative contents of target metabolites identified by KEGG pathway in different groups; Supplementary Table S1: DAI scoring rules; Supplementary Table S2: Chiu's scoring rules; Supplementary Table S3: UPLC gradient for lipidomics analysis; Supplementary Table S4: The parameters of OPLS-DA models for plasma lipidomics; Supplementary Table S5: The parameters of OPLS-DA models for colon lipidomics; Supplementary Table S6: Summary of pathway analysis; Supplementary Table S7: Pathway analysis of the identified metabolites; Supplementary Table S8: Total of 12 differential metabolites of plasma in both positive and negative ion mode; Supplementary Table S9: Total of 53 differential metabolites of colon in both positive and negative ion mode.
Supplementary Material 2: MS/MS identification of differential metabolites.
Data Availability Statement
Data associated with this study has been deposited at The mass spectrometry data have been deposited to the MetaboLights (https://www.ebi.ac.uk/metabolights/login) through the MetaboLights partner repository. The data set identifier is MTBLS5086.