Abstract
Critical for brain development, neurodevelopmental and network disorders, the GABRA1 gene encodes for the α1 subunit, an abundantly and developmentally expressed subunit of heteropentameric gamma-aminobutyric acid A receptors (GABAARs) mediating primary inhibition in the brain. Mutations of the GABAAR subunit genes including GABRA1 gene are associated with epilepsy, a group of syndromes, characterized by unprovoked seizures and diagnosed by integrative approach, that involves genetic testing. Despite the diagnostic use of genetic testing, a large fraction of the GABAAR subunit gene variants including the variants of GABRA1 gene is not known in terms of their molecular consequence, a challenge for precision and personalized medicine. Addressing this, one hundred thirty-seven GABRA1 gene variants of unknown clinical significance have been extracted from the ClinVar database and computationally analyzed for pathogenicity. Eight variants (L49H, P59L, W97R, D99G, G152S, V270G, T294R, P305L) are predicted as pathogenic and mapped to the α1 subunit's extracellular domain (ECD), transmembrane domains (TMDs) and extracellular linker. This is followed by the integration with relevant data for cellular pathology and severity of the epilepsy syndromes retrieved from the literature. Our results suggest that the pathogenic variants in the ECD of GABRA1 (L49H, P59L, W97R, D99G, G152S) will probably manifest decreased surface expression and reduced current with mild epilepsy phenotypes while V270G, T294R in the TMDs and P305L in the linker between the second and the third TMDs will likely cause reduced cell current with severe epilepsy phenotypes. The results presented in this study provides insights for clinical genetics and wet lab experimentation.
Keywords: GABA (A) receptors, Epilepsy, GABRA1, Alpha 1 subunit, ClinVar, Single nucleotide polymorphism, VUS, Variants of unknown significance, Personalized medicine, Genetic testing, In silico, Computational prediction tools, Non-synonymous, SNP, Variation
1. Introduction
The Homo sapiens gamma-aminobutyric acid A receptor alpha 1(GABRA1) gene encodes for the human alpha1 (α1) subunit of gamma-aminobutyric acid A receptors (GABAARs), the ligand gated ion channels acting as primary sites for inhibition in the mammalian central nervous system [[1], [2], [3], [4]]. Upon GABA binding, this heteropentameric channel opens temporarily leading to the chloride influx in adult neurons. Assembled from a large subunit pool (α1-α6, β1-β3, γ1-γ3, δ, ∈, θ, π and ρ1-ρ3) the GABAARs show enormous cellular, subcellular and molecular diversity depending on the subunit composition [3]. The α1 subunit and its isoforms (α1-α6) are differentially expressed in the brain. In the hippocampus α1βδ receptors are expressed mostly in hippocampal interneurons, whereas α4βδ receptors are expressed mostly in the dentate gyrus in granule cells (DGGCs) with presumably distinct physiological roles associated with distinct neural ensembles [5]. For example, receptors containing α1βδ subunits were shown to have a critical role in the maintenance of the in vitro γ oscillations in the CA3 region of the Hippocampus [6]. Hippocampus is also the region where subtle alterations related to GABAARs may predispose various pathophysiology such as status epilepticus [5,7,8] that α1 subunit's developmental changes have also been implicated [9].
In addition to its divergent role in the hippocampus, in the rest of the brain the α1 subunit co-assembles to multiple GABAAR subtypes expressed and regulated differentially in the molecular, cellular and sub cellular levels with distinct physiological functions, including α1βγ2 receptors that mediate phasic inhibition and α1β, α1βδ or α1α6βδ receptors, that mediate tonic inhibition [5,10,11]. These differential inhibitory actions of α1 containing GABAARs are further diversified by differential ligand binding affinities critical for clinical pharmacology [[12], [13], [14], [15], [16]], as well as regulation by posttranslational modifications such as phosphorylation [17]. Moreover, the α1 subunit knock out mice had increased seizure susceptibility and exhibited the loss of more than half of GABAARs underlying the importance of this subunit in the excitability of the brain [18]. Thus, even a minor alteration in GABRA1 gene that encodes α1 subunit may have profound effects on the multiple systems. This in turn, via specific mechanisms, such as distortion of excitation and inhibition balance and beyond, contribute to CNS diseases including epilepsy, a chronic neurological disorder characterized by recurrent unprovoked epileptic seizures, ranging from mild generalized forms to epileptic encephalopathies [[19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39]].
For GABRA1 gene alone more than hundreds of genetic variations, including nonsynonymous single nucleotide polymorphisms (nsSNPs), have been identified underlying the importance of relevant genetic variations in brain development, neurodevelopmental and network disorders [40]. Indeed, genetic testing has already become an integral part of the diagnosis for epilepsy, especially for unexplained epilepsy and pediatric epilepsy [41,42] and the phenotypic spectrum of GABRA1 gene such as infantile febrile seizures, bilateral‐tonic‐clonic seizures and atonic seizures has already been identified [32]. Moreover, in the short run, it appears that precision medicine will be widely used for diagnosis, personalized treatment, prognosis of epilepsy syndromes and comorbid states [43].
On the other hand, the number of variants with unknown clinical significance continuously increases with the availability of next‐generation sequencing (NGS), a trend that is also critical for personalized medicine and pharmacogenomics [[44], [45], [46], [47], [48]]. This challenge caused the necessity for variant interpretation that demands the integration of molecular biology, bioinformatic and clinical expertise combining basic and clinical data to address the significance of a variant and its potential to contribute the management of a given clinical condition. The American College of Medical Genetics and Genomics (ACMG) and Association for Molecular Pathology (AMP) provide a framework for interpreting the clinical significance of genetic variants [49]. This framework involves categorizing the variants as either Pathogenic or Likely Pathogenic, or as Benign or Likely Benign. Pathogenic variants are further classified based on their level of evidence, such as very strong (PVS1), strong (PS1-4), moderate (PM1-6), or supporting (PP1-5). Among these, the use of in silico methods to predict the consequence of the variants is considered as supporting evidence for pathogenicity which corresponds to criterion PP3 [49]. This criterion along with some other criteria can lead to the interpretation of a variant's pathogenicity, which has implications in precision and personalized treatments for epilepsy [41,50,51]. For instance, in cohort of 91 patients aged 19 years or less undertaking genetic testing with the NGS epilepsy gene panel, 16 patients had variants "pathogenic" or "likely pathogenic" according to ACMG-AMP criteria [52]. 63% of these patients had diagnosis benefited from the NGS epilepsy panel impacting on the clinical management such as avoiding unnecessary diagnostic procedures and potentially detrimental treatments [52]. As a result, predicting the pathogenicity of variants have become important to contribute to ACMG-AMP criteria for variant interpretation that could potentially guide the clinical management of epilepsy. Consequently, the accurate prediction of variant pathogenicity has emerged as a crucial factor contributing to the ACMG-AMP criteria for variant interpretation, potentially guiding the clinical management of epilepsy. In light of the paramount importance of GABAAR α1 subunit and its variants described so far, the present study focused on one hundred thirty-seven non-synonymous single-nucleotide polymorphisms (nsSNPs) within the coding region of the α1 subunit, which were categorized as variants of uncertain significance (VUS) in the ClinVar database [53,54]. These VUS were subjected to comprehensive in silico analysis, employing a range of well validated algorithms [55] (REVEL, ClinPred and FATHMM-XF) that encompassed diverse features such as sequence homology, evolutionary conservation, and folding stability to computationally assess the pathogenicity of the GABRA1 gene VUS. Subsequently, computational protein modeling was used for validation, followed by mapping the identified VUS to specific α1 subunit domains associated with molecular consequences, cellular pathology, and disease severity.
2. Methods
2.1. SNP data mining and data retrieval
The ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) was used as a primary data source for the identification nsSNPs in the coding region of GABRA1, as it provides the information for the specific relationships among human gene variations and phenotypes, with supporting evidence. [53,54] In order to retrieve all kind of variant information, the gene name (GABRA1) was entered in the search bar of the ClinVar server, which results in classification of variations according to different categories such as variation type and clinical significance. In order to filter the nsSNPs with unknown clinical significance (VUS), ‘single nucleotide variations’ and ‘missense’ and ‘uncertain significance’ were selected in the vertical menu bar. The retrieved data were downloaded as txt file and copied to an excel file by ‘use text import wizard’ that allows tab delimited excel file of variant data.
2.2. Evaluation of pathogenicity risks of GABRA1 VUS (nsSNPs)
The evaluation of the pathogenicity risks has been computed by two ensemble methods [55], namely ClinPred, [56] and REVEL [57], both of which are validated for ClinVar. In addition, another ClinVar validated, multiple feature predictor, FATHMM-XF [58], which incorporates 31 feature groups is used. ClinPred incorporates Allele Frequencies and 13 individual prediction scores from SIFT, PolyPhen-2 HDIV, PolyPhen-2 HVAR, LRT, MutationAssessor, PROVEAN, CADD, GERP, DANN, PhastCons, fitCons, PhyloP, and SiPhy while REVEL incorporates 18 individual pathogenicity scores from 13 in silico tools, namely MutPred, FATHMM, VEST, PolyPhen, SIFT, PROVEAN, MutationAssessor, MutationTaster, LRT, GERP, SiPhy, phyloP, and phastCons. All scores were obtained from dbNSFP database [59,60] accessed by the following web site: http://database.liulab.science/dbNSFP#database.
2.3. Conservation analysis by consurf
The structural information (PDB ID 6HUJD)-chain D for alpha 1 subunit-) was entered into Consurf server [61,62] (https://consurf.tau.ac.il/consurf_index.php). By default, the server runs a search for homologues collected from UNIREF90 database, a clustered version of the UniProt database.
2.4. Molecular modeling
The modeling and superimposition of amino acid mutations are generated by the use of HOPE server [60], which can be accessed from the following web address: (https://www3.cmbi.umcn.nl/hope/about/). The UniProt ID (P14867) of the GABRA1 protein was entered in the query tab followed by the entry of original and alternative amino acid positions to retrieve the superimpositions.
The 3D structures of GABAAR and α1 subunit was based on the electron microscopy.
(Resolution: 3.04 Å) of 6HUJ data [63] from RCSB (RCSB.org) [64]. The chains of A and D of 6HUJ data reflect the human α1 chimeric protein made up of residues of gene orthologue from Bos taurus (residues 2–27, UniProt ID P08219) in addition to the residues of Homo sapiens (28–456, UniProt ID P14867), the latter sequence being aligned with the mutational landscape identified in the present study. The 3D structures were reconstructed and visualized by UCSF Chimera 1.16 software [65]. By entering 6HUJ (PDB ID) [63], the structural data were imported by “Fetch by ID” command under the File Menu of the software. In order to visualize GABAARs subunits clearly, other structures such as picrotoxin, GABA and megabody 38 (Mb38), in complex with the receptor's structural data, were removed by using Select Menu and Actions Menu. Images were saved as png file with a size of 1280x720 pixels.
2.5. Determination of hot spots and functional domains
Hot spots and functional domains were determined by the use of different sources such as literature review, National Center for Biotechnology Information (NCBI) Conserved Domain Database and by UniProt database (https://www.uniprot.org/uniprotkb/P14867/entry) for the protein encoded by human GABRA1 (P14867). Also the ScanProsite motif search tool [66] hosted at the Expasy server (https://prosite.expasy.org/scanprosite/) was used to predict the functional motifs of the α1 subunit.
3. Results
3.1. Workflow
The data mining process in the ClinVar have led to the extraction of GABRA1 variants which were subjected to the in silico analysis. The in silico analysis have been performed by two meta predictors, namely ClinPred and REVEL, and a multiple feature predictor, FATHAMM-XF, which incorporates 31 feature groups. In addition, evolutionary conservation scores obtained by ConSurf to assess residue importance. This was followed by molecular modelling by HOPE server, and the motif scan by Expasy server. Also, the cellular pathology and disease phenotype prediction by using the information of patient mutations and experimental data mapped to the hot-spots and functional domains was performed in the final step. The overall workflow is shown in the Fig. 1.
Fig. 1.
The workflow of identifying deleterious/pathogenic nsSNPs of the GABRA1 gene in ClinVar (GABRA1: GABA A Receptor alpha 1 gene; nsSNPs: nonsynonymous (missense) Single Nucleotide Polymorphisms; VUS: Variants of unknown significance).
3.2. Data mining in ClinVar
Located in the chromosome 5, the human GABRA1 gene has 13 exons encoding the protein transcribed from variant templates (transcript variant 1–7) The NCBI gene records five of these variants (https://www.ncbi.nlm.nih.gov/gene/2554). For our analysis we take NCBI Reference Sequence: NP_001121116.1 (MANE Select) of the transcript variant 3 (NM_001127644.2) UniProt ID: P14867.
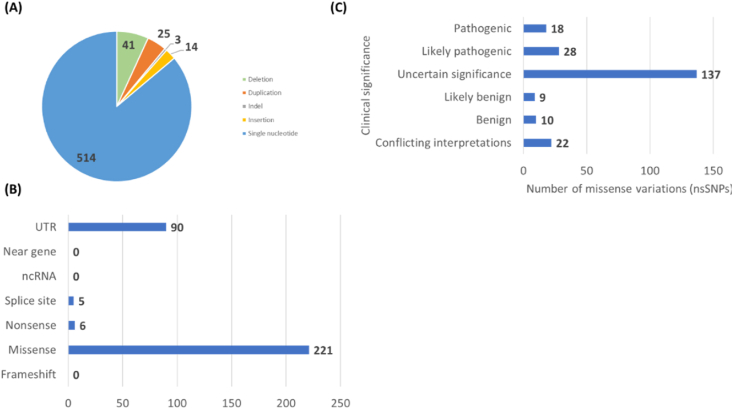
(https://www.ncbi.nlm.nih.gov/protein/NP_001121116.1) since this is the standard transcript across all databases (MANE select). The data mining process for the transcript variant 3 (NM_001127644.2) in the ClinVar have led to the extraction of variants of GABRA1. As of February 18, 2023, there are 514 single nucleotide variations, corresponding to the majority of the variants while the other variants are 41 deletions, 25 duplications, 3 indels, and 14 insertions (Fig. 2 A).
Fig. 2.
Summary of the data retrieved from ClinVar. (A) Variation types of GABRA1 gene (B) Molecular consequence of GABRA1 variations. (C) Classification of missense variations of GABRA1 gene according to clinical significance.
These variations show different molecular consequences as summarized in the Fig. 2 B. Among the different categories, the highest number is observed in the missense category, which corresponds to 221 variations. Among the missense variations there are 18 “pathogenic variants”, 28 “likely pathogenic variants”, 137 variants with “uncertain clinical significance”, 9 variants as “likely benign”, 10 variants as “benign” and 22 variants as “conflicting interpretations” (Fig. 2C). In order to determine the missense (nonsynonymous) variations (nsSNPs) with uncertain clinical significance (VUS), we selected “Single nucleotide” for Variation Type, “Missense” for Molecular Consequence and “Uncertain Significance” for Clinical Significance in the vertical menu of the ClinVar server. As of February 18, 2023, this identified 137 nsSNPs as VUS (Fig. 2C) (Supplementary File S1) which were subjected to in silico analytical instruments to predict their risk of pathogenicity.
3.3. Evaluation of pathogenicity risks of 137 GABRA1 nsSNPs of unknown significance
The evaluation of the pathogenicity risks has been computed by FATHMM-XF and two ensemble methods [[55], [56], [57]] [[55], [56], [57]] [[55], [56], [57]] namely ClinPred and REVEL both of which are validated for ClinVar. ClinPred incorporates Allele Frequencies and 13 individual prediction scores from SIFT, PolyPhen-2 HDIV, PolyPhen-2 HVAR, LRT, MutationAssessor, PROVEAN, CADD, GERP, DANN, PhastCons, fitCons, PhyloP, and SiPhy while REVEL incorporates 18 individual pathogenicity scores from 13 in silico tools, namely MutPred, FATHMM, VEST, PolyPhen, SIFT, PROVEAN, MutationTaster, MutationAssessor, LRT, GERP, SiPhy, phyloP, and phastCons. 62% of the VUS was predicted as pathogenic according to REVEL while this ratio was 82% and 83% for ClinPred and Fathmm-XF [58] respectively (Fig. 3). The full data are given in the S2 Supplementary File 2 and S3 Supplementary File 3. Among the consensus, variants (nsSNPs) with REVEL score >0.95 were selected as the variants that have the highest pathogenicity risks. These nsSNPs are listed in the Table 1.
Fig. 3.
Percentage of the pathogenicity risks predicted by FATHAMM-XF and two Ensemble + predictors, REVEL & ClinPred.
Table 1.
The list of variants of GABRA1 nsSNPs classified as “Uncertain (Clinical) Significance” in ClinVar and selected as having the highest probability of pathogenicity. The list shows the consensus evaluation by three methods (REVEL, ClinPred and Fathmm-XF) and variants with REVEL score >0.95. (nsSNP: nonsynonymous Single Nucleotide Polymorphism, T: Tolerated, D: Deleterious, N: Neutral).
| Accession | Aminoacid change | Condition (ClinVar) | REVEL score | ClinPred Prediction |
FATHAMM-XF Prediction |
|---|---|---|---|---|---|
| VCV002293869 | L49H | Inborn genetic diseases | 0.961 | D | D |
| VCV000423472 | P 59 L | Idiopathic generalized epilepsy|Epilepsy, idiopathic generalized, susceptibility to, 13|Epilepsy, childhood absence 4|not provided | 0.951 | D | D |
| VCV001810645 | W 97 R | Not provided | 0.958 | D | D |
| VCV000938214 | D 99 G | Idiopathic generalized epilepsy|Epilepsy, idiopathic generalized, susceptibility to, 13|Epilepsy, childhood absence 4 | 0.958 | D | D |
| VCV001338962 | G 152 S | Not provided | 0.962 | D | D |
| VCV001514122 | V 270 G | Idiopathic generalized epilepsy|Epilepsy, idiopathic generalized, susceptibility to, 13|Epilepsy, childhood absence 4 | 0.954 | D | D |
| VCV001026791 | T 294 R | Epilepsy, idiopathic generalized, susceptibility to, 13|Epilepsy, childhood absence 4|Idiopathic generalized epilepsy | 0.964 | D | D |
| VCV000938690 | P 305 L | Idiopathic generalized epilepsy|Epilepsy, idiopathic generalized, susceptibility to, 13|Epilepsy, childhood absence 4 | 0.984 | D | D |
3.3.1. Conservation profile of variants in the GABRA1 protein
Comparison of amino acid sequences of homolog proteins might signal particularly crucial residues, as they would have survived the purifying natural selection, if the functional consequence also to be conserved. In order to calculate and rank the conservation profile of the GABRA1 subunit, the Consurf server was used. Consurf, by default runs a search for homologues collected from UNIREF90 database, a clustered version of the UniProt database. The process ends up with homologues for multiple sequence comparison by which the conservation of each amino acid of the GABRA1 protein is calculated in a range of 1–9, 9 being the most conserved. Table 2 shows the conservation scores of identified amino acids as deleterious in the previous section. While V270 and T 294 are less conserved, the remaining of all amino acids predicted as deleterious/pathogenic in the previous section have the highest conservation degree. The summary of Consurf data is given in the (S4 Supplementary File 4).
Table 2.
Conservation scores of GABRA1 amino acid residues predicted as deleterious.
| Amino acid residue | L49 | P 59 | W 97 | D 99 | G 152 | V 270 | T 294 | P 305 |
|---|---|---|---|---|---|---|---|---|
| Conservation Score | 9 | 9 | 9 | 9 | 9 | 7 | 7 | 9 |
3.4. Comparative modeling of native GABRA1 protein and its mutants
The predicted effect of the identified nsSNPs should be considered in the context of their corresponding amino acid positions in the secondary and tertiary structure. Thus in the 3D structure of the human alpha 1 subunit (UniProt ID: P14867, PDB ID: 6HUJ, chain: A) [63] (Fig. 4), where the native amino acid residues are shown as green, the amino acid variants (mutations) are observed as enlarged superimpositions The mutation of Leucine into Histidine at position 49 is shown in the Fig. 4A. The wild type Leucine is smaller, a more hydrophobic residue buried in the core of the protein, at the alpha helix. This mutation will cause loss of hydrophobic interactions in the core of the protein. Thus, the bigger mutant Histidine residue will most likely not fit at this position in the alpha helix and will not favor it as a secondary structure.
Fig. 4.
Comparative modeling of native GABRA1 protein and its variants, which shows the position of native amino acids (in the center) and enlarged view of their superimpositions with the variants. The superimpositions are shown as green for the wild type and red for the mutant residues: leu and histidine at the position 49 (A), proline and leucine at the position 59 (B), tryptophan and arginine at the position (97) (C), aspartic acid and arginine at the position 99 (D), glycine and serine at the position 152 E, valine and glycine at the position 270 (F), threonine and arginine at the position 294 (G), proline and leucine at the position 305 (H).
The mutation of Proline into Leucine at position of 59 is shown in the Fig. 4B. The mutant residue is bigger than the wild-type residue Proline buried in the core of the protein. Prolines are rigid residues and proline in this position may serve as a basis for a critical conformation thus might be essential in the position 59. This effect might be disrupted by a mutation to Leucine, since its bigger size will probably not compensate the conformational stability created by Proline residue.
The mutation of Tryptophan into a smaller amino acid Arginine at position 97 is shown in the Fig. 4C. Tryptophan at this position is located in its favored secondary structure, a β-strand, as annotated in UniProt, while Arginine deviates from this secondary structure orientation as seen in the Figure. Also, the difference between the hydrophobicity of these two residues will cause the loss of hydrophobic interactions in the core of the protein. The smaller size of the mutant Arginine residue will likely cause an empty space in the core of the protein. The mutant Arginine at position 97 will introduce a charge in a buried residue which can interfere with protein folding.
The mutation of Aspartic acid into Arginine at position 99 is shown in the Fig. 4D. The wild-type Aspartic acid, the negatively charged residue forms a hydrogen bond with Tyrosine at position 53. It also makes salt bridges with Arginine residue at position 64 and with another Arginine residue at position 101. These interactions will likely be disrupted in the case of positively charged, bigger mutant Arginine residue. The size difference between the two residues makes that the new residue is not in the correct position to make the same hydrogen bond as the original wild-type residue did. The charge difference between the two residues makes that the new residue will not maintain the same non-covalent interactions in the salt bridges as the wild-type residue did. Besides, in the Figure showing the 3D-structure, it can be seen that the wild-type Aspartic acid is located in its preferred secondary structure favoring a turn, which is unlikely supported by the mutant arginine residue favoring a different orientation in the secondary structure. Thus, the original conformation will not be favored fully in the case of mutant Arginine residue.
The mutation of the small buried Glycine at the core of the protein into Serine residue at position of 152 is shown in the Fig. 4E. The wild-type Glycine residue, being the most flexible of all amino acids, might be essential for the protein function, which can be disrupted by mutations. The torsion angles at this specific position are unusual, a feature that can only be maintained by a flexible Glycine. A mutation at this point will force the local backbone into an unfavorable conformation and thus will alter the local structure. The V270 is annotated as transmembrane domain (TMD) (the first transmembrane domain, TM1) in the UniProt database (with the inclusion of signal peptide sequence).
The mutation of Valine into a smaller, less hydrophobic and flexible residue, Glycine at position 270 is shown in the Fig. 4F. Being the most flexible of all amino acids, Glycine's flexibility can alter the essential firmness of the protein at this position. Since the wild-type residue is in TMD (TM1), the differences in size and hydrophobicity between the two wild type and mutant residues will make that the mutant residue can not interact with the membrane lipids as the wild-type residue did. The T294R is located at the end of the second transmembrane domain (TM2) as annotated in the UniProt database.
The mutation of Threonine into bigger, positively charged Arginine residue at position of 294 is shown in the Fig. 4G. The mutant amino acid is bigger than the wild-type. This size difference can alter the contacts with the lipid-membrane. The wild-type amino acid is more hydrophobic than the mutant amino acid. This differences in hydrophobicity can change the hydrophobic interactions with the membrane lipids. The wild-type Threonine residue forms a hydrogen bond with Valine at position 290. The size difference between wild-type and mutant residue makes that the new residue is not in the correct position to make the same hydrogen bond as the original wild-type residue did. The wild-type Threonine residue is more hydrophobic than the mutant Arginine residue. The difference in hydrophobicity will affect hydrogen bond formation.
The mutation of Proline into Leucine at position 305 is shown in the Fig. 4H. The mutation P305L corresponds to the region between the second and the third transmembrane domain (TM2-TM3 region), the region between the amino acids 301–312 (with the inclusion of signal peptide sequence). The wild-type proline is smaller and buried in the center of the protein. Being bigger in size, the mutant Leucine at position 305 probably will not fit at this core. Due to their cyclic side chains Prolines are very rigid and thus they introduce a special conformational backbone which might be required at this position. A mutation might disrupt this conformational state.
3.5. Mapping the variants to the α1 subunit domains and hot spots
Typically, all GABAAR subunits have a large N terminus extracellular domain (ECD), four transmembrane spanning domains (TMD: TM1, TM2, TM3, TM4), a large intracellular domain/loop between TM3 and TM4 (TM3-TM4 loop or ICD) and a short extracellular C terminus (Fig. 5A and D). In addition, there are relatively short linkers between the TM1 and TM2 as well as TM2 and TM3. Besides, TM1, TM3 and TM4 separate the channel lining TM2 from the lipid membrane [3]. Many of the GABAAR subunits have the amino acid sequence (TTVLTMTT) in the TM2 domain appears to be critical for the ion channel [3]. In the UniProt database, the ECD of the alpha1 subunit is located in the region between the amino acids 56 and 249 (with the inclusion of signal peptide sequence), thus L49H, P59L, W97R, D99G, G152S mutations fall into the region of large extracellular N terminus of the α1 subunit, while V270G is located in the TM1, T294R is located in the TM2 and P305L is located in between the TM2-TM3 loop (Fig. 5A). N-terminal sequences of GABAAR subunits (Fig. 5B and C) are important for expression, assembly and intracellular trafficking ](Luscher et al., 2011). This can happen via different mechanisms. For example, mutations at this region may prevent the subunits trafficked to the plasma membrane and cause them degraded intracellularly, or that the mutation inhibited proper folding and assembly to form functional pentameric GABAARs. On the other hand, studies of functional analysis show that mutations in the N terminal extracellular sequence distort other features of the receptor function as well. This is usually characterized by altered channel gating properties due to the reduction of the receptor's activation, deactivation and desensitization causing a decreased channel opening probability [3,67].
Fig. 5.
Structural features of GABAAR showing the assembly with α1 subunit and its domains. (A) The protein diagram of α1 subunit showing the domain specific location of 8 VUS identified as having the highest risk of pathogenicity. The amino acid positions of each domain are shown in parentheses in the diagram. (B) 3D reconstruction of GABA (A) receptor colored by subunit (top view) showing the position of α1 subunit in the receptor co-assembly. (C) 3D reconstruction of GABA (A) receptor, showing the position of α1 subunit in the receptor co-assembly (side view). (D) 3D reconstruction of α1 subunit of GABA (A) receptor (side view, extended version) showing the positions of the wild type residues (green), where the pathogenic variants are detected (ECD: Extracellular domain; ICD: Intracellular domain; TMD: Transmembrane domain; TM1: Transmembrane domain 1; TM2: Transmembrane domain 2; TM3: Transmembrane domain 3; TM4: Transmembrane domain 4). Image not to scale.
Inherited or de novo mutations of GABAARs in GABRA1-6, GABRB1-3, GABRG1-3, and GABRD subunit genes have been identified as one of the primary genetic causes of epilepsy [38,68]. However, the association between epilepsy syndromes (specific epilepsy types characterized by different conditions) and GABAA receptor variants is not understood fully, a challenge for the development of effective treatment. For example, specific genotype phenotype relation has not been reported in between the GABRA1 gene variants and epilepsy phenotypes and the genetic mutants of GABAARs are widespread among different subunits [32,37,38,69]. Nevertheless, the mutations located in the GABAAR subunit domains, characterized by structural and functional significance, are critical and often associated with the severity of the epileptic phenotypes [29,37,38]. Moreover, among the epilepsy associated variants, typically clustered at the ECD, TM2, and TM3 regions of GABAAR subunits, the inherited variants in the GABRG2 and GABRA1 subunit genes are associated with relatively mild forms of monogenic epilepsies while de novo variants in several GABAAR subunit genes cause severe developmental and epileptic encephalopathies [29,37,38].
Consequently, we wanted to examine our results in the light of the current evidence for the genetic variants to predict the possible impact on cellular pathology and disease phenotype. Selected examples for the domain specific locations of GABRA1 mutations show that they are associated with the cellular pathology and disease phenotype (Supplementary File 5) and thus the mutations identified in this study, which are mapped to the ECD, TM1, TM2 and TM2-TM3 (Fig. 5A and C), can be matched with the variants in the same regions for further reflections. For example, in vitro studies have shown that R214C, a de novo mutation in a child with epileptic encephalopathy (EE), which is located in the ECD of the GABRA1, have reduced surface and total GABAA receptor expression and reduced whole-cell GABA-evoked currents (Supplementary File 5). Thus, for the mutations L49H, P59L, W97R, D99G, G152S in the N-terminal ECD of the α1 subunit, the molecular consequence of pathogenicity in the cell may be reflected at least one of the followings: reduced trafficking, ER retention, reduced cell surface expression, reduced GABAergic currents, reduced GABA sensitivity. Formed by ten beta strands and two alpha helices, the ECD, which is about 200 amino acids long and shaped by a cysteine disulfide bridge (the “Cys-loop”), is important for receptor assembly and trafficking [3]. Thus, one mechanism of reduced current appears to be the consequence of decreased cell surface expression due to the mutations in the ECD. Moreover, two GABA binding sites are located in between β−α interfaces and one Benzodiazepine binding site located at the α-γ interface both located in the ECD [[63], [70], [71], [72], [73], [74]]. [[,70] [[63], [70], [71], [72], [73], [74]] [[63], [70], [71], [72], [73], [74]] Based on homology to other members of type-A gamma-aminobutyric acid receptors and mutational analysis, the residues R64 and F92 is critical for GABA binding at the extracellular interface of the α and β subunits [3,70,73,75].
Also, residues Asp 57 and Asp 149 in the ECD region couple the ligand binding with channel gating [76]. Thus, ECD mutations P59L and G152S in the proximity of these residues may also alter the ligand binding leading to the altered channel conductance and kinetics since it is proposed that the receptor operates through a complex “lock and pull” mechanism [63] that GABA binding locks the β and α1 subunits, which pull on the ECDs, rotating them counterclockwise. This bends the TMDs, spreading the ion channel [63]. As a result, reduced GABAergic currents may not only derive from the reduced surface expression but also altered receptor kinetics caused by the mutations in the ECD. Another structural hot spot of the ECD is the group of residues that are found in the pentamer interface. These residues are retrieved from the NCBI Conserved Domain Database and made up of a combination of highly conserved residues. Among these, residues F92, R94, and Q95 are in close proximity to the variants W97R and D99G. Also, the variant G152S are in the close proximity of residues R147 and T157 of the subunit interface. These altogether can thus interfere with the receptor oligomerization as well as cell current transmitted by the mutant receptors.
As GABAAR subunit gene variants located in the transmembrane regions are likely to be associated with a more severe phenotype, compared to those located in the ECD [37], we expect that L49H, P59L, W97R, D99G, G152S in the ECD will likely cause milder symptoms of the epilepsy syndromes compared to V270G, T294R which are located in the TMDs namely TM1 and TM2 respectively (Fig. 5). Typically, the four TMDs, each formed by four alpha helices, control the opening and closing of the channel aperture through the movement of the helical structure of the TM2 that aligns with the central pore to form the selective ion channel with a partial contribution from the TM1. In addition, the TM1 domain harbors a neurosteroid binding site [77] in that mutations in this domain of GABRA1 could alter interaction with neurosteroids and reduce activity of GABAARs. Patient (patient with infantile epileptic spasms and West syndrome) mutations in the TM1 are characterized by reduced GABA-evoked current without decreasing GABA potency (S5 Supplementary File 5). Thus, the variant V270G, which is found in this region will probably impact on the GABAergic currents. In the cases for early onset epileptic encephalopathies, Childhood absence epilepsy and Dravet syndrome, patient mutations in the TM2 is characterized by reduced current & desensitization with normal total and surface subunit expression level (S5 Supplementary File 5). Thus, the variant T294R, which is found in this region will probably have similar cellular pathology. The TM2-TM3 extracellular linker may be important in pore opening. [78] In a case of Myoclonic astatic epilepsy, patient mutation K306T in the TM2-TM3 loop is characterized by reduced current and decreased GABA sensitivity (S5 Supplementary File 5), which may be seen in the cellular pathology for the variant P305L since it is adjacent to K306T residue.
In order to further identify if any mutation is possibly located in the functionally critical protein regions or hot spots the GABRA1 sequence was scanned against the ScanProsite collection of motifs provided by Expasy server (see Methods section). This collection of motifs includes profiles and patterns to identify protein domains, families and functional sites. N-myristoylation sites, protein kinase C phos [75]phorylation sites, Casein kinase II phosphorylation site, N-glycosylation site, Amidation site, ATP/GTP-binding site motif A (P-loop), Cell attachment sequence, Leucine zipper pattern were predicted in the specific positions (S5 Supplementary File 6). Among the six predicted protein kinase C phosphorylation sites, four of them were in the N terminus ECD (positions 22–24, 96–98, 157–159, 161–163), one near or in the TM1 (299–301) and one in the intracellular domain between the TM3 and TM4 (338–340). Among these, the motif 96–98 in the ECD corresponds to the mutation W97R and D99G. None of the remaining predicted sites overlap with the identified mutations. The overall summary of the profile of pathogenic variants identified in this study is given in the Table 3.
Table 3.
Summary of GABRA1 variants identified as pathogenic and characterized according to its molecular domain in the three dimensional structure of the α1 subunit, and according to predicted molecular consequence, cellular pathology and Epilepsy phenotype severity. Molecular Consequence is deduced from the data from modeling (HOPEserver) and other in silico tools (Prosite, Expasy). Cellular pathology and epilepsy phenotype severity are identified according to literature results for the phenotypic spectrum of GABRA1 gene for epilepsy (See also S5 Supplementary File 5). (ECD: Extracellular domain (N terminus), TM1: First transmembrane domain, TM2: Second transmembrane domain, TM2-TM3 Loop: segment between the first and the second transmembrane domains).
| Mutation | Domain | Molecular Consequence |
Cellular pathology<listaend> | Disease severity |
|---|---|---|---|---|
| L49H | ECD |
|
|
Mild |
| P59L | ECD |
|
|
Mild |
| W97R | ECD |
|
|
Mild |
| D99G | ECD |
|
|
Mild |
| G152S | ECD |
|
|
Mild |
| V270G | TM1 |
|
|
Severe |
| T294R | TM2 |
|
|
Severe |
| P305L | TM2-TM3 Loop |
|
|
Severe |
4. Discussion
In the present study, eight variants of unknown clinical significance (L49H, P59L, W97R, D99G, G152S, V270G, T294R, P305L) were predicted as pathogenic based on the integrative and computational analysis. Our data show that the identified variants are mapped to the distinct molecular domains of the epilepsy associated GABRA1 protein, potentially linked with specific cellular pathophysiology and epilepsy symptom severity.
The genetic spectrum of epilepsies is complex [79]. The complexity is reflected by multiple genes causing specific epilepsy syndromes that are clinically indistinguishable as well as genetic variants which may impact on the severity of the disease phenotypes [45]. Mutations of GABAARs have been identified as one of the primary genetic causes of epilepsy [20,22,25,25,28,[32], [33], [34], [35],38,42,45,48,80,80,81], but this phenomenon is not well understood, which is a challenge for the development of effective personalized treatment and management of epilepsy conditions. The advancements in neuroimaging and genetics have revealed that, while essential, the electro-clinical characterization of epilepsy syndromes alone is insufficient for personalized treatment strategies and a deeper comprehension of the fundamental mechanisms causing seizures [82]. Thus, according to International League Against Epilepsy (ILAE), classification of the Epilepsies is a multi-level system including seizure types (focal onset, generalized onset and unknown onset), epilepsy types (Generalized, Focal, Combined Generalized and Focal, Unknown) and in the third level the epilepsy syndromes that are integrated with comorbidity and etiology which also includes genetic etiology [83]. An epilepsy syndrome, such as Dravet syndrome or West syndrome, is a cluster of characteristics that comprise specific seizure types, electroencephalogram (EEG) patterns, and imaging features, which tend to co-occur. It frequently exhibits age-related aspects, such as the age at which it begins and ends (if applicable), seizure triggers, variations throughout the day, and occasionally, the expected course or outcome, in addition to distinctive co-morbidities such as intellectual and psychiatric dysfunction. Thus, an epilepsy syndrome may have associated etiological, prognostic and treatment implications. Additionally, it might present unique co-existing conditions such as cognitive and mental impairments, alongside specific observations on EEG and imaging examinations. It could also have implications for identifying the underlying cause, predicting the prognosis, and determining appropriate treatments. Importantly, an epilepsy syndrome does not necessarily align directly with a specific etiological diagnosis and serves a distinct purpose, such as guiding the management of a specific condition [83]. The effective utilization of genetic data has the potential to significantly enhance patient outcomes by providing valuable guidance for managing the condition. For instance, a case of five-year-old boy who had a history of unprovoked hemiclonic seizure with a progression to bilateral tonic-clonic activity with normal EEG and MRI results represents such a condition [50]. The first line treatment, levetiracetam, and other treatment options, topiramate, clobazam, valproic acid, as well as lifestyle adjustment (the ketogenic diet) did not help the patient. When an epilepsy gene panel was performed, it identified a new heterozygous “likely pathogenic variant” in the SCN1A gene, one of the most common epilepsy genes. This has aided the diagnosis and successful treatment decision (fenfluramine) that the diagnosis of Dravet Syndrome was confirmed by combining patient data with the variant interpretation (“likely pathogenic”) for SCN1A gene, encoding the sodium voltage-gated channel alpha subunit one [50]. This and other examples [84] show the significance of variant interpretation aiding the precision medicine for diagnosis and treatment of epilepsy syndromes. Consequently, variant interpretation for GABAAR will potentially generate such opportunities for personalized treatment and management of epilepsy conditions. Phenobarbital, benzodiazepines, clobazam are antiepileptic drugs enhancing the GABA A receptor activity [85]. Interpretation of genetic variants in GABAAR may indicate a potential responsiveness to these drugs guiding the treatment decision as a part of comprehensive assessment of the individual's clinical presentation, seizure types, and response to previous treatments, in addition to genetic testing results. Thus, our data have important implications since predicting pathogenicity of specific genetic variants associated with epilepsy syndromes will contribute to variant interpretation for clinically unknown genetic variants, which provides important insights into the underlying causes of the condition and guiding treatment decisions, such as selecting targeted therapies.
In this context, it is essential to reflect our results in terms of ACMG-AMP guidelines for variant interpretation which is a framework for established criteria for interpreting sequence variants (Richards et al., 2015). This framework involves categorizing the variants as either Pathogenic or Likely Pathogenic, or as Benign or Likely Benign. Pathogenic variants are further classified based on their level of evidence, such as very strong (PVS1), strong (PS1-4), moderate (PM1-6), or supporting (PP1-5). Similarly, each benign criterion is assigned a weight, either stand-alone (BA1), strong (BS1-4), or supporting (BP1-6). The numerical labels within each category do not indicate varying degrees of importance but are provided for easier reference to the different criteria. When assessing a specific variant, the user selects the relevant criteria based on the observed evidence for that variant. In silico analyses are considered as “Supporting evidence of pathogenicity”, the PP3 criterion, which refers to multiple lines of computational evidence supporting a deleterious effect on the gene or gene product [49]. According to this scheme of ACMG-AMP, our results computationally identifying the pathogenicity of GABRA1 VUS can be interpreted as “Supporting evidence of pathogenicity” (PP3). The pathogenicity assertion for each genetic variant further requires the combining rules for these criteria (Richards et al., 2015). Thus, our computational results meet the PP3 criterion, which helps interpretation of relevant GABRA1 variants as “pathogenic” or “likely pathogenic” depending on the availability of other criteria. For instance, Moderate pathogenicity requires at least four supporting criteria such as PP3 criterion in addition to one of the “Moderate evidence of pathogenicity criteria” (PM1–PM6) (Richards et al., 2015). Among the (PM1–PM6), the PM1 represents a variant located in “mutational hot spot and/or critical and well-established functional domain (e.g. active site of an enzyme) without benign variation”. Since in our integrative approach, findings from mutagenesis studies, mutational landscape of epilepsy patients and domain specific data were incorporated into the results of computational analysis, our data could meet PM1 criterion in addition to PP3 criterion. Especially, the functional significance of N terminus extracellular domain of GABAAR subunits is well established since this domain is critical for ligand binding, receptor assembly, interaction with other proteins and modulation of receptor activity. For instance, the N-terminal extracellular domain of the GABRA1 protein binds to GABA, primary inhibitory neurotransmitter, initiating receptor activation and downstream signaling in addition to interaction with various allosteric modulators, such as benzodiazepines and barbiturates modulating the GABAergic signaling [3] 5 out of 8 VUS of GABRA1 gene, which are predicted as pathogenic are located in the ECD: L49H, P59L, W97R, D99G, G152S. Among the remaining variants, V270G is located in the first transmembrane domain (TM1) and the T294R in the second transmembrane domain (TM2). The latter domain plays a crucial role in the formation and gating of the ion channel pore, determining ion selectivity and permeation, and influencing receptor pharmacology. These functions are essential for the proper functioning of the receptor in mediating inhibitory neurotransmission in the central nervous system. While the primary role of ion channel gating resides in the second transmembrane domain, the TM1 can also influence the gating process in directly. It contributes to the overall structural stability of the receptor complex and helps maintain the appropriate conformation required for efficient channel opening and closing. Especially, the TM1 domain is responsible for anchoring the α1 subunit within the cell membrane. It spans the lipid bilayer, providing stability and proper positioning of the receptor in the neuronal membrane. Besides, this domain also participates in the assembly of the GABAAR complex. It interacts with corresponding transmembrane domains of other subunits, such as beta and gamma subunits, enabling the formation of a functional receptor unit. Thus, our predicted pathogenic variants derived from multi-level computational and integrative analysis contribute variant classification criteria based on the ACMG-AMP guidelines.
The integration of in silico results with the data from patient mutations and experimental studies is crucial for identification of variants in molecular hotspots. Additionally, search for hot spots was also performed computationally, that the GABRA1 sequence was scanned against the ScanProsite collection of motifs provided by Expasy server: Six-protein kinase C phosphorylation sites were predicted. Four of them were in the N terminus ECD (positions 22–24, 96–98, 157–159, 161–163), one near or in the TM1 (299–301) and one in the intracellular domain between the TM3 and TM4 (338–340). Among these, the motif 96–98 in the N-terminus ECD corresponds to the pathogenic variants W97R and D99G. Among the GABAARs subunits, mostly β and γ2 subunits are phosphorylated in the intracellular domain between TM3 and TM4 (ICD) which contains consensus sites for serine/threonine and tyrosine protein kinases and α1 subunit may be phosphorylated by Calcium/calmodulin-dependent kinase II [86,87]. On the other hand, the existence of consensus sequences for a specific kinase does not necessarily refer that the protein will undergo phosphorylation in vivo [87] and bona fide phosphorylation sites may not match with the consensus sites [88]. Nevertheless, GABAARs subunits, especially β and γ2 subunits are phosphorylated in the intracellular domain between TM3 and TM4 (ICD) that phosphorylation and dephosphorylation are mechanisms for receptor surface expression and endocytic internalization, respectively [87]. However, in our study we have not predicted any pathogenic variant in this domain. In addition, none of the remaining ScanProsite predicted sites overlap with the identified mutations. Nevertheless, the integration of our multi-level analysis confirms that the identified variants have high risk of pathogenicity presumably by cellular mechanisms that cause distortion of the interactions required for the structural and functional efficacy in the respective protein domains of the α1 subunit that is supported by the variant mapping to the specific protein domains and hot spots based on the data from the epilepsy patient mutations and experimental studies describing cellular pathology.
The nature of the cellular pathology caused by the variations found in the α1 subunit, that is manifested as distorted cell surface expression and GABA evoked currents, may involve various mechanisms due to reduced mRNA transcription or stability, improper protein folding, altered subunit co-assembly, distorted receptor trafficking by different mechanisms such as dephosphorylation and/or by direct molecular alteration of GABA efficacy, sensitivity and desensitization characteristics of the receptors [19,33,89]. These in turn may converge on various epilepsy syndromes ranging from mild forms of monogenic epilepsies to severe developmental and epileptic encephalopathies in a way that may be associated with the symptom severity depending on the location of the genetic variant [37]. In the present study 8 VUS of GABRA1 gene are predicted as pathogenic variants that may be critical for epilepsy syndromes and disease severity, which have phenotypes and symptoms ranging from mild to severe, depending on various factors such as the underlying cause, type of seizure, and individual differences. For instance, Juvenile Myoclonic Epilepsy (JME) is a type of epilepsy that typically begins in adolescence and is characterized by mild indications such as myoclonic jerks, which are brief, involuntary muscle twitches. Febrile seizures are generally considered as relatively benign and self-limiting condition, making them classified as a relatively mild form of seizures. The phenotypic profile of these mild or more severe cases are determined by electroclinical and neuroimaging data classifying patients as having these and other mild syndromes such as photosensitive idiopathic generalized epilepsy, generalized epilepsy with febrile seizures plus, moderately severe phenotypes such as myoclonic-astatic epilepsy, Dravet syndrome and severe epileptic encephalopathies [29,30]. A comparison of scientific literature and various patient data, including clinical, electrophysiological, therapeutic, and molecular information, pertaining to GABAAR subunit variants (specifically GABRA1, GABRB2, GABRB3, and GABRG2), has revealed that these variants are linked to diverse and highly variable phenotypes, despite their close molecular and physiological proximity [37]. Remarkably, none of the mentioned genes were found to be specifically associated with a particular phenotype. However, it has been observed that the specific location of the variant on the protein [37] may serve as an indicator of the severity of the condition reflected by its symptom(s), specific manifestation or indication of the underlying condition experienced by the individual and epilepsy phenotype. A phenotype definition involves employing epidemiological, biological, molecular, or computational techniques to systematically identify characteristics of a disorder that could arise from different genetic influences [90]. However, this phenomenon is complex in epilepsy. In numerous genetic epilepsies, the range of observable characteristics can be wide, even among individuals with the same genetic mutations. Additionally, patients with distinct genetic causes may exhibit seemingly similar clinical features. For instance, while the majority of patients with Dravet syndrome have mutations in SCN1A, similar phenotypes may also be observed in patients with mutations in PCDH19, CHD2, SCN8A, or, in rare instances, GABRA1 and STXBP1 [91]. Nevertheless, it is possible to describe the severity of the condition which represents the burden of the symptom or phenotype such as seizure burden, and the epileptiform activity, the distinctive EEG waves found in patients with epilepsy [91]. Regarding this, Human Phenotype Ontology (HPO) [92] (https://hpo.jax.org/app/) provides a uniform set of terms and descriptions for abnormal physical and clinical characteristics observed in human diseases. According to HPO, EEG with generalized epileptiform discharges (UMLS:C4023476) is described as spikes, which are brief waveforms lasting less than 70 ms, and sharp waves, which typically last between 70 and 200 ms. Naturally, the duration of waveforms and sharp waves would be a factor determining severity of this condition, for example. Thus in our study, depending on the specific location of the variant on the GABRA1 protein [37], a manifestation of either mild or severe phenotypes that would be associated with the specific conditions such as febrile seizures [27,32,38], JME [32], or epileptic encephalopathies [37] could be predicted. However, what exactly does this mean? Let's clarify with an example: In the platform of HPO [92], epileptic encephalopathy (HP:0200134) is defined as a condition in which epileptiform abnormalities are thought to contribute to the gradual disruption of brain function. It is characterized by several key features: (1) the presence of intense and often aggressive electrographic EEG paroxysmal activity, (2) seizures that are typically diverse and resistant to treatment, (3) persistent cognitive, behavioral, and neurological impairments that may worsen over time, and (4) in some cases, early mortality. Therefore, our results suggest that the position of the identified GABRA1 variants may potentially predict the intensity of one or more of these clinical features, reflecting the severity of the condition. In this case, GABRA1 variants in the extracellular domain (L49H, P59L, W97R, D99G, G152S) will likely manifest with less severity, while variants in the transmembrane domain 1 (V270G), transmembrane domain 2 (T294R), and the linker between transmembrane domain 2 and 3 (P305L) may predict a more severe disease condition.
The associated cellular pathology is expected as decreased cell surface expression and reduced current based on the patient mutations and experimental studies in the literature (Supplementary file 5). As described in the above paragraph, we have also predicted that variants could be associated with severe epilepsy phenotypes of any type ranging from benign to severe forms characterized by several conditions such as Generalized Tonic-Clonic Seizures (formerly known as Grand Mal Seizures) [93]. These seizures involve the entire brain and are characterized by loss of consciousness, convulsions, muscle rigidity, and often followed by a period of confusion or fatigue. Other conditions that implicate disease severity include Status Epilepticus [94], characterized by continuous or recurrent seizures lasting for an extended period or seizures occurring in rapid succession without the person regaining consciousness in between. Status epilepticus requires immediate medical attention as it can lead to significant complications and neurological damage. Epileptic Encephalopathy [95] refers to another group of severe epilepsy disorders that are associated with cognitive and developmental impairments. These conditions often manifest early in life and are characterized by frequent seizures, developmental deterioration, intellectual disability, and other neurological abnormalities. Consequently, our comparative analysis integrating patient mutations show that V270G in the TM1, T294R in the TM2 and P305L in the linker between the TM2 and TM3 is presumably associated cellular pathology characterized as reduced current and likely connected with the severity of the epilepsy syndromes -although not necessarily associated with specific syndrome- [37].
Many in silico tools are used for the computational analysis of the pathogenicity of VUS but their criteria may differ and thus posing the problem of selecting the best approach for the prediction of variant effect. Among the available methods, the “ensemble” methods are meta predictors that combine the preexisting methods to evaluate pathogenic or benign effects of the variants [55]. The “ensemble methods” are among the top-ranked predictors with highest accuracy compared to popular widely used tools such as PolyPhen2 and SIFT [55]. Considering these, in the present study two ensemble methods namely ClinPred and REVEL, both of which are validated for ClinVar was used. ClinPred incorporates Allele Frequencies and 13 individual prediction scores from SIFT, PolyPhen-2 HDIV, PolyPhen-2 HVAR, LRT, MutationAssessor, CADD, PROVEAN, GERP, DANN, PhastCons, fitCons, PhyloP, and SiPhy while REVEL incorporates 18 individual pathogenicity scores from 13 in silico tools, namely MutPred, FATHMM, VEST, PolyPhen, SIFT, PROVEAN, MutationTaster, MutationAssessor, LRT, GERP, SiPhy, phyloP, and phastCons. In addition, another ClinVar validated, multiple feature predictor, FATHAMM-XF, which incorporates 31 feature groups is used. These include 27 features from ENCODE (The ENCODE Project Consortium, 2012) and NIH Roadmap Epigenomics as well as additional feature groups from conservation scores, the Variant Effect Predictor, annotated gene models, and the DNA sequence [58]. Indeed, the ACMG-AMP guidelines recommend the use of multiple computational tools to predict variant pathogenicity [49]. Since our approach employs meta predictors made up of different algorithms and databases leading to the prediction based on a wide variety of features such as sequence conservation, structural features, genomic position, physicochemical properties, our study provides a comprehensive assessment of the variant's impact. Furthermore, our data derived from the robust predictive tools are validated by molecular modeling. Moreover, we have integrated data from known patient mutations and associated experimental findings, which altogether help better evaluate the pathogenicity of a variant. In this context, consistent predictions from multiple databases, algorithms and sources reinforce the classification of a variant according to the ACMG-AMP guidelines. Consequently, our integrative approach and the use of ensemble predictors and meta predictors represent a methodological strength of the present study.
There are several limitations of this study. First, although, we have adopted well validated and high accuracy methods as described above, in silico tools and the reference sequence impact on the prediction accuracy [96]. Thus, they must be used cautiously, and generalizations should be avoided based on single studies. Second, in ensemble predictors such as REVEL, setting appropriate cutoffs is important for balancing the trade-off between sensitivity and specificity and ensuring accurate classification of variants. In the present study a relatively higher 0.95 REVEL cutoff was chosen. One reason to choose this cutoff was to prioritize high-confidence predictions of pathogenic variants. This approach helps reduce the chances of false positives and minimizes the inclusion of variants that may not have a significant impact on disease development or progression. By setting a higher cutoff, such as 0.95, one can focus on a smaller number of variants with a higher probability of being pathogenic according to the REVEL algorithm. Thus, a higher cutoff may enhance specificity. However, this may cost some sensitivity that some truly pathogenic variants may not be captured. On the other hand, working with small but more confidently predicted number of variants better align with the objectives of the present study that higher cutoff was chosen. Since this raises the possibility that some pathogenic variants of GABRA1 were not captured by the analysis of this study, some true positive pathogenic variants with REVEL scores slightly below the cutoff may be missed or classified as non-pathogenic. Third, we have specifically focused on the nonsynonymous point mutations found in the coding region of the α1 subunit. However, other variants such as variants in the splice sites, 5′ or 3’ UTRs (untranslated regions or noncoding regions) of GABRA1 might be critical for the pathogenesis of epilepsy syndromes or other channelopathies of GABAARs. For instance, according to computational studies, the 5′ UTR region of GABAAR subunit genes has some consensus sites for transcription factors [97], that variation in these elements may impact on subunit expression. It will be interesting to analyze the noncoding region rare variants of the GABRA1 gene by using new tools and frameworks, which allow gene-centric analysis of such variants [98,99]. Besides, GABRA1 gene variant analysis can be further elaborated by the use of new methodologies to facilitate the prioritization of causal variants influencing human phenotypes [100]. Moreover, other in silico studies identified three microRNAs, namely miR-181, miR-216, and miR-203 targeting α1-subunit [101]. Experimental studies show that miR-181a downregulate α1 subunit expression [102]. In another study, a down-regulation of mRNA levels of GABAAR subunits in glioblastomas, a major type of the most common form of primary brain tumor (gliomas), have been detected [103]. miR-139-5p has been reported to impact on glioma malignant biological behavior via targeting α1 subunit [104]. Thus, variations other than those in the coding sequence of GABRA1 gene are also important.
In conclusion, the findings of this study hold significant importance in the context of genetic testing and precision medicine for epilepsy syndromes [51,75,[105], [106], [107]]. As the field increasingly relies on next-generation sequencing [43,45,52,79,107], [[43], [45], [52], [79], [107], [108], [109]] [[43], [45], [52], [79], [107], [108], [109]] the number of variants with unknown molecular consequences continues to grow, posing a challenge for precision medicine. Classifying these genetic variants as benign or pathogenic necessitates the estimation of their impact, which is a complex task considering the vast number of variants with low frequencies in human exomes [110]. Conducting clinical associations and experimental studies for all variants simultaneously is not feasible. Therefore, the utilization of in silico tools to predict variant consequences has become crucial and is recommended by the American College of Medical Genetics and Genomics (ACMG-AMP). In alignment with these challenges, the present study offers valuable insights into the pathogenicity of clinically unknown variants in the GABRA1 gene, which are associated with epilepsy. Inherited or de novo mutations in the α1-6, β1-3, γ1-3, and δ subunit isoforms of GABAARs have been identified as significant genetic causes of epilepsy. However, the comprehensive understanding of the relationship between epilepsy syndromes and GABAAR variants remains limited, hindering the development of effective and personalized interventions. Addressing this gap, the present study computationally analyzed 137 variants of GABRA1 to assess their pathogenicity. This analysis led to the in silico identification of eight pathogenic variants (L49H, P59L, W97R, D99G, G152S, V270G, T294R, P305L), which are mapped to the extracellular domain (ECD) and transmembrane domains (TMDs) of the GABAARs. These variants have been linked to cellular pathology and disease severity, providing valuable insights for understanding the pathogenic mechanisms underlying epilepsy. By elucidating the pathogenicity of these GABRA1 variants through a combination of patient mutation data and experimental studies, this study contributes to the advancement of personalized interventions and improved treatment strategies for individuals with epilepsy.
Furthermore, the results of this study have implications for guiding wet lab experimentation in the field of epilepsy and GABAARs research. The identification of eight pathogenic variants in the GABRA1 gene (L49H, P59L, W97R, D99G, G152S, V270G, T294R, P305L) and their association with cellular pathology and disease severity provide crucial insights into the underlying mechanisms of epilepsy besides to variant effects on the functionality of GABAARs. Our findings offer a starting point for designing targeted wet lab experiments to further investigate the functional consequences of these specific variants with a focus on their efforts on studying the impact of these variants on GABAARs, elucidating their effects on receptor function, neuronal excitability, and synaptic transmission. Understanding the precise molecular mechanisms underlying these pathogenic variants could contribute to the development of novel therapeutic strategies aimed at modulating GABAAR activity and potentially mitigating the epileptic phenotype. Additionally, the computational analysis performed in this study demonstrates the utility of in silico tools in predicting variant pathogenicity. This highlights the potential of utilizing such tools in future wet lab experiments to streamline the screening and prioritization of variants for functional characterization. By leveraging computational predictions alongside wet lab investigations, researchers can optimize their resources and focus on variants with a higher likelihood of pathogenicity, accelerating the discovery of critical insights into variants of GABAAR subunits and epilepsy mechanisms.
Author contribution statement
Ayla Arslan: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. </p>
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The author would like to express gratitude for the time and effort invested by the Reviewers in evaluating the work and providing constructive feedback
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e20218.
List of Abbreviations
- ACMG
The American College of Medical Genetics and Genomics.
- AMP
Association for Molecular Pathology
- ECD
Extracellular domain (N terminus)
- GABRA1
Gamma-aminobutyric acid A receptor alpha 1 subunit gene
- GABAARs
Gamma-aminobutyric acid A receptors
- TM1
First transmembrane domain
- TM2
Second transmembrane domain
- TM2-TM3 Loop:
segment between the first and the second transmembrane domains
- VUS:
Variants of uncertain significance
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Schofield P.R., Darlison M.G., Fujita N., Burt D.R., Stephenson F.A., Rodriguez H., Rhee L.M., Ramachandran J., Reale V., Glencorse T.A. Sequence and functional expression of the GABA A receptor shows a ligand-gated receptor super-family. Nature. 1987;328(6127):221–227. doi: 10.1038/328221a0. [DOI] [PubMed] [Google Scholar]
- 2.Schofield P.R., Pritchett D.B., Sontheimer H., Kettenmann H., Seeburg P.H. Sequence and expression of human GABAA receptor alpha 1 and beta 1 subunits. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1989;244(2):361–364. doi: 10.1016/0014-5793(89)80563-0. [DOI] [PubMed] [Google Scholar]
- 3.Goetz T., Arslan A., Wisden W., Wulff P. GABA(A) receptors: structure and function in the basal ganglia. Prog. Brain Res. 2007;160:21–41. doi: 10.1016/S0079-6123(06)60003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeburg P.H., Wisden W., Verdoorn T.A., Pritchett D.B., Werner P., Herb A., Lüddens H., Sprengel R., Sakmann B. The GABAA receptor family: molecular and functional diversity. Cold Spring Harbor Symp. Quant. Biol. 1990;55:29–40. doi: 10.1101/sqb.1990.055.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Arslan A. Extrasynaptic δ-subunit containing GABAA receptors. J. Integr. Neurosci. 2021;20(1):173–184. doi: 10.31083/j.jin.2021.01.284. [DOI] [PubMed] [Google Scholar]
- 6.Ferando I., Mody I. Altered gamma oscillations during pregnancy through loss of δ subunit-containing GABA(A) receptors on parvalbumin interneurons. Front. Neural Circ. 2013;7:144. doi: 10.3389/fncir.2013.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y.Q., Yu F., Liu W.H., He X.H., Peng B.W. Dysfunction of hippocampal interneurons in epilepsy. Neurosci. Bull. 2014;30(6):985–998. doi: 10.1007/s12264-014-1478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dengler C.G., Coulter D.A. Normal and epilepsy-associated pathologic function of the dentate gyrus. Prog. Brain Res. 2016;226:155–178. doi: 10.1016/bs.pbr.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanaumi T., Takashima S., Iwasaki H., Mitsudome A., Hirose S. Developmental changes in the expression of GABAA receptor alpha 1 and gamma 2 subunits in human temporal lobe, hippocampus and basal ganglia: an implication for consideration on age-related epilepsy. Epilepsy Res. 2006;71(1):47–53. doi: 10.1016/j.eplepsyres.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Farrant M., Nusser Z. Variations on an inhibitory theme: phasic and tonic activation of GABA(A) receptors. Nat. Rev. Neurosci. 2005;6(3):215–229. doi: 10.1038/nrn1625. [DOI] [PubMed] [Google Scholar]
- 11.Arslan A., Engelhardt J., Wisden W. Cytoplasmic domain of δ subunit is important for the extra-synaptic targeting of GABAA receptor subtypes. J. Integr. Neurosci. 2014;13(4):617–631. doi: 10.1142/S0219635214500228. [DOI] [PubMed] [Google Scholar]
- 12.Pritchett D.B., Sontheimer H., Shivers B.D., Ymer S., Kettenmann H., Schofield P.R., Seeburg P.H. Importance of a novel GABAA receptor subunit for benzodiazepine pharmacology. Nature. 1989;338(6216):582–585. doi: 10.1038/338582a0. [DOI] [PubMed] [Google Scholar]
- 13.Korpi E.R., Mattila M.J., Wisden W., Lüddens H. GABA(A)-receptor subtypes: clinical efficacy and selectivity of benzodiazepine site ligands. Ann. Med. 1997;29(4):275–282. doi: 10.3109/07853899708999348. [DOI] [PubMed] [Google Scholar]
- 14.Rudolph U., Crestani F., Benke D., Brünig I., Benson J.A., Fritschy J.M., Martin J.R., Bluethmann H., Möhler H. Benzodiazepine actions mediated by specific gamma-aminobutyric acid(A) receptor subtypes. Nature. 1999;401(6755):796–800. doi: 10.1038/44579. [DOI] [PubMed] [Google Scholar]
- 15.Smart T.G., Stephenson F.A. A half century of γ-aminobutyric acid. Brain and neurosci. adv. 2019;3 doi: 10.1177/2398212819858249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engin E. GABAA receptor subtypes and benzodiazepine use, misuse, and abuse. Front. Psychiatr. 2023;13 doi: 10.3389/fpsyt.2022.1060949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moss S.J., Gorrie G.H., Amato A., Smart T.G. Modulation of GABAA receptors by tyrosine phosphorylation. Nature. 1995;377(6547):344–348. doi: 10.1038/377344a0. [DOI] [PubMed] [Google Scholar]
- 18.Kralic J.E., Korpi E.R., O'Buckley T.K., Homanics G.E., Morrow A.L. Molecular and pharmacological characterization of GABA(A) receptor alpha1 subunit knockout mice. J. Pharmacol. Exp. Therapeut. 2002;302(3):1037–1045. doi: 10.1124/jpet.102.036665. [DOI] [PubMed] [Google Scholar]
- 19.Terunuma M., Xu J., Vithlani M., Sieghart W., Kittler J., Pangalos M., Haydon P.G., Coulter D.A., Moss S.J. Deficits in phosphorylation of GABA(A) receptors by intimately associated protein kinase C activity underlie compromised synaptic inhibition during status epilepticus. J. Neurosci. : The Off. J. Soc. Neurosci. 2008;28(2):376–384. doi: 10.1523/JNEUROSCI.4346-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cossette P., Liu L., Brisebois K., Dong H., Lortie A., Vanasse M., Saint-Hilaire J.M., Carmant L., Verner A., Lu W.Y., Wang Y.T., Rouleau G.A. Mutation of GABRA1 in an autosomal dominant form of juvenile myoclonic epilepsy. Nat. Genet. 2002;31(2):184–189. doi: 10.1038/ng885. [DOI] [PubMed] [Google Scholar]
- 21.Gallagher M.J., Song L., Arain F., Macdonald R.L. The juvenile myoclonic epilepsy GABA(A) receptor alpha1 subunit mutation A322D produces asymmetrical, subunit position-dependent reduction of heterozygous receptor currents and alpha1 subunit protein expression. J. Neurosci. : The Off. J. Soc. Neurosci. 2004;24(24):5570–5578. doi: 10.1523/JNEUROSCI.1301-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krampfl K., Maljevic S., Cossette P., Ziegler E., Rouleau G.A., Lerche H., Bufler J. Molecular analysis of the A322D mutation in the GABA receptor alpha-subunit causing juvenile myoclonic epilepsy. Eur. J. Neurosci. 2005;22(1):10–20. doi: 10.1111/j.1460-9568.2005.04168.x. [DOI] [PubMed] [Google Scholar]
- 23.Kodera H., Ohba C., Kato M., Maeda T., Araki K., Tajima D., Matsuo M., Hino-Fukuyo N., Kohashi K., Ishiyama A., Takeshita S., Motoi H., Kitamura T., Kikuchi A., Tsurusaki Y., Nakashima M., Miyake N., Sasaki M., Kure S.…Matsumoto N. De novo GABRA1 mutations in Ohtahara and West syndromes. Epilepsia. 2016;57(4):566–573. doi: 10.1111/epi.13344. [DOI] [PubMed] [Google Scholar]
- 24.Bradley C.A., Taghibiglou C., Collingridge G.L., Wang Y.T. Mechanisms involved in the reduction of GABAA receptor alpha1-subunit expression caused by the epilepsy mutation A322D in the trafficking-competent receptor. J. Biol. Chem. 2008;283(32):22043–22050. doi: 10.1074/jbc.M801708200. [DOI] [PubMed] [Google Scholar]
- 25.Ding L., Feng H.J., Macdonald R.L., Botzolakis E.J., Hu N., Gallagher M.J. GABA(A) receptor alpha1 subunit mutation A322D associated with autosomal dominant juvenile myoclonic epilepsy reduces the expression and alters the composition of wild type GABA(A) receptors. J. Biol. Chem. 2010;285(34):26390–26405. doi: 10.1074/jbc.M110.142299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lachance-Touchette P., Brown P., Meloche C., Kinirons P., Lapointe L., Lacasse H., Lortie A., Carmant L., Bedford F., Bowie D., Cossette P. Novel α1 and γ2 GABAA receptor subunit mutations in families with idiopathic generalized epilepsy. Eur. J. Neurosci. 2011;34(2):237–249. doi: 10.1111/j.1460-9568.2011.07767.x. [DOI] [PubMed] [Google Scholar]
- 27.Carvill G.L., Weckhuysen S., McMahon J.M., Hartmann C., Møller R.S., Hjalgrim H., Cook J., Geraghty E., O'Roak B.J., Petrou S., Clarke A., Gill D., Sadleir L.G., Muhle H., Spiczak S., Nikanorova M., Hodgson B.L., Gazina E.V., Suls A.…Mefford H.C. GABRA1 and STXBP1: novel genetic causes of Dravet syndrome. Neurology. 2014;82(14):1245–1253. doi: 10.1212/WNL.0000000000000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez C.C., XiangWei W., Hu N., Shen D., Shen W., Lagrange A.H., Zhang Y., Dai L., Ding C., Sun Z., Hu J., Zhu H., Jiang Y., Macdonald R.L. Altered inhibitory synapses in de novo GABRA5 and GABRA1 mutations associated with early onset epileptic encephalopathies. Brain : J. Neurol. 2019;142(7):1938–1954. doi: 10.1093/brain/awz123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez C.C., Macdonald R.L. A structural look at GABAA receptor mutations linked to epilepsy syndromes. Brain Res. 2019;1714:234–247. doi: 10.1016/j.brainres.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Hernandez C.C., Tian X., Hu N., Shen W., Catron M.A., Yang Y., Chen J., Jiang Y., Zhang Y., Macdonald R.L. Dravet syndrome-associated mutations in GABRA1, GABRB2 and GABRG2 define the genetic landscape of defects of GABAA receptors. Brain Commun. 2021;3(2):33. doi: 10.1093/braincomms/fcab033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hernandez C.C., Klassen T.L., Jackson L.G., Gurba K., Hu N., Noebels J.L., Macdonald R.L. Deleterious rare variants reveal risk for loss of GABAA receptor function in patients with genetic epilepsy and in the general population. PLoS One. 2016;11(9) doi: 10.1371/journal.pone.0162883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johannesen K., Marini C., Pfeffer S., Møller R.S., Dorn T., Niturad C.E., Gardella E., Weber Y., Søndergård M., Hjalgrim H., Nikanorova M., Becker F., Larsen L.H., Dahl H.A., Maier O., Mei D., Biskup S., Klein K.M., Reif P.S.…Maljevic S. Phenotypic spectrum of GABRA1: from generalized epilepsies to severe epileptic encephalopathies. Neurology. 2016;87(11):1140–1151. doi: 10.1212/WNL.0000000000003087. [DOI] [PubMed] [Google Scholar]
- 33.Bai Y.F., Chiu M., Chan E.S., Axerio-Cilies P., Lu J., Huh L., Connolly M.B., Guella I., Farrer M.J., Xu Z.D., Liu L., Demos M., Wang Y.T. Pathophysiology of and therapeutic options for a GABRA1 variant linked to epileptic encephalopathy. Mol. Brain. 2019;12(1):92. doi: 10.1186/s13041-019-0513-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X., Durisic N., Lynch J.W., Keramidas A. Inhibitory synapse deficits caused by familial α1 GABAA receptor mutations in epilepsy. Neurobiol. Dis. 2017;108:213–224. doi: 10.1016/j.nbd.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 35.Hannan S., Affandi A.H.B., Minere M., Jones C., Goh P., Warnes G., Popp B., Trollmann R., Nizetic D., Smart T.G. Differential coassembly of α1-GABAARs associated with epileptic encephalopathy. J. Neurosci. : The Off. J. Soc. Neurosci. 2020;40(29):5518–5530. doi: 10.1523/JNEUROSCI.2748-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riaz M., Abbasi M.H., Sheikh N., Saleem T., Virk A.O. GABRA1 and GABRA6 gene mutations in idiopathic generalized epilepsy patients. Seizure. 2021;93:88–94. doi: 10.1016/j.seizure.2021.10.013. [DOI] [PubMed] [Google Scholar]
- 37.Maillard P.Y., Baer S., Schaefer É., Desnous B., Villeneuve N., Lépine A., Fabre A., Lacoste C., El Chehadeh S., Piton A., Porter L.F., Perriard C., Wardé M.A., Spitz M.A., Laugel V., Lesca G., Putoux A., Ville D., Mignot C.…Milh M. Molecular and clinical descriptions of patients with GABAA receptor gene variants (GABRA1, GABRB2, GABRB3, GABRG2): a cohort study, review of literature, and genotype-phenotype correlation. Epilepsia. 2022;63(10):2519–2533. doi: 10.1111/epi.17336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L., Liu X. Clinical phenotype and genotype of children with GABAA receptor α1 subunit gene-related epilepsy. Front. Neurol. 2022;13 doi: 10.3389/fneur.2022.941054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bryson A., Reid C., Petrou S. Fundamental Neurochemistry Review: GABAA receptor neurotransmission and epilepsy: principles, disease mechanisms and pharmacotherapy. J. Neurochem. 2023 doi: 10.1111/jnc.15769. [DOI] [PubMed] [Google Scholar]
- 40.Ben-Ari Y., Khalilov I., Kahle K.T., Cherubini E. The GABA excitatory/inhibitory shift in brain maturation and neurological disorders. The Neuroscientist: a review journal bringing neurobiology. Neurol. Psychiatr. 2012;18(5):467–486. doi: 10.1177/1073858412438697. [DOI] [PubMed] [Google Scholar]
- 41.Striano P., Minassian B.A. From genetic testing to precision medicine in epilepsy. Neurotherapeutics : The J. Am. Soc. Exp. NeuroTher. 2020;17(2):609–615. doi: 10.1007/s13311-020-00835-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruggiero S.M., Xian J., Helbig I. The current landscape of epilepsy genetics: where are we, and where are we going? Curr. Opin. Neurol. 2023;36(2):86–94. doi: 10.1097/WCO.0000000000001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGinn R.J., Stein E.L., Summers Stromberg J.E., Li Y. Precision medicine in epilepsy. Progress in mol. biol. transl. sci. 2022;190(1):147–188. doi: 10.1016/bs.pmbts.2022.04.001. [DOI] [PubMed] [Google Scholar]
- 44.Xie X., Gu J., Zhuang D., Chen X., Zhou Y., Shen W., Li L., Liu Y., Xu W., Hong Q., Chen W., Zhou W., Liu H. Association study of genetic polymorphisms in GABRD with treatment response and dose in methadone maintenance treatment. Pers. Med. 2021;18(5):423–430. doi: 10.2217/pme-2021-0063. [DOI] [PubMed] [Google Scholar]
- 45.Thakran S., Guin D., Singh P., Singh P., Kukal S., Rawat C., Yadav S., Kushwaha S.S., Srivastava A.K., Hasija Y., Saso L., Ramachandran S., Kukreti R. Genetic landscape of common epilepsies: advancing towards precision in treatment. Int. J. Mol. Sci. 2020;21(20):7784. doi: 10.3390/ijms21207784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J., Yao Y., He H., Shen J. Clinical interpretation of sequence variants. Curr. Protoc. Hum. Genet. 2020;106(1):98. doi: 10.1002/cphg.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O'Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Tukiainen T., Birnbaum D.P., Kosmicki J.A., Duncan L.E., Estrada K., Zhao F., Zou J., Pierce-Hoffman E., Berghout J., Cooper D.N. Exome aggregation Consortium. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium E., Project E.P., Allen A.S., Berkovic S.F., Cossette P., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T., Goldstein D.B., Han Y., Heinzen E.L., Hitomi Y., Howell K.B., Johnson M.R., Kuzniecky R., Lowenstein D.H., Lu Y.F.…Winawer M.R. De novo mutations in epileptic encephalopathies. Nature. 2013;501(7466):217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H.L., Committee, A. C. M. G. L. Q. A Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet. Med. : Off. J. Am. Coll. Med. Genet. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trowbridge S., Poduri A., Olson H. Early diagnosis and experimental treatment with fenfluramine via the Investigational New Drug mechanism in a boy with Dravet syndrome and recurrent status epilepticus. Epileptic Disord. : Int. Epilepsy J. Videotape. 2021;23(6):954–956. doi: 10.1684/epd.2021.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johannesen K.M., Nikanorova N., Marjanovic D., Pavbro A., Larsen L.H.G., Rubboli G., Møller R.S. Utility of genetic testing for therapeutic decision-making in adults with epilepsy. Epilepsia. 2020;61(6):1234–1239. doi: 10.1111/epi.16533. [DOI] [PubMed] [Google Scholar]
- 52.Hoelz H., Herdl C., Gerstl L., Tacke M., Vill K., von Stuelpnagel C., Rost I., Hoertnagel K., Abicht A., Hollizeck S., Larsen L.H.G., Borggraefe I. Impact on clinical decision making of next-generation sequencing in pediatric epilepsy in a tertiary epilepsy referral center. Clin. EEG Neurosci. 2020;51(1):61–69. doi: 10.1177/1550059419876518. [DOI] [PubMed] [Google Scholar]
- 53.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., Karapetyan K., Katz K., Liu C., Maddipatla Z., Malheiro A., McDaniel K., Ovetsky M., Riley G., Zhou G.…Maglott D.R. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):1062–1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harrison S.M., Riggs E.R., Maglott D.R., Lee J.M., Azzariti D.R., Niehaus A., Ramos E.M., Martin C.L., Landrum M.J., Rehm H.L. Using ClinVar as a resource to support variant interpretation. Curr. Protoc. Hum. Genet. 2016;89(8 16 1) doi: 10.1002/0471142905.hg0816s89. 8 16 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katsonis P., Wilhelm K., Williams A., Lichtarge O. Genome interpretation using in silico predictors of variant impact. Hum. Genet. 2022;141(10):1549–1577. doi: 10.1007/s00439-022-02457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alirezaie N., Kernohan K.D., Hartley T., Majewski J., Hocking T.D. ClinPred: prediction tool to identify disease-relevant nonsynonymous single-nucleotide variants. Am. J. Hum. Genet. 2018;103(4):474–483. doi: 10.1016/j.ajhg.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., Cannon-Albright L.A., Teerlink C.C., Stanford J.L., Isaacs W.B., Xu J., Cooney K.A., Lange E.M., Schleutker J., Carpten J.D.…Sieh W. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 2016;99(4):877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rogers M.F., Shihab H.A., Mort M., Cooper D.N., Gaunt T.R., Campbell C. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinf. (Oxford. 2018;34(3):511–513. doi: 10.1093/bioinformatics/btx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X., Jian X., Boerwinkle E. DbNSFP: a lightweight database of human non-synonymous SNPs and their functional predictions. Hum. Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu X., Li C., Mou C., Dong Y., Tu Y. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020;12(103) doi: 10.1186/s13073-020-00803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venselaar H., Te Beek T.A., Kuipers R.K., Hekkelman M.L., Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinf. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glaser F., Pupko T., Paz I., Bell R.E., Bechor-Shental D., Martz E., Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19(1):163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 63.Masiulis S., Desai R., Uchański T., Serna Martin I., Laverty D., Karia D., Malinauskas T., Zivanov J., Pardon E., Kotecha A., Steyaert J., Miller K.W., Aricescu A.R. GABAA receptor signalling mechanisms revealed by structural pharmacology. Nature. 2019;565(7740):454–459. doi: 10.1038/s41586-018-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., & Bourne, P. E. (n.d.). The Protein Data Bank. Nucleic acids research. (1)(28), 235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed]
- 65.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004 doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 66.Castro E., Sigrist C.J., Gattiker A., Bulliard V., Langendijk-Genevaux P.S., Gasteiger E., Bairoch A., Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkl124. Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaczor P.T., Wolska A.D., Mozrzymas J.W. Α1 subunit histidine 55 at the interface between extracellular and transmembrane domains affects preactivation and desensitization of the GABAA receptor. ACS Chem. Neurosci. 2021;12(3):562–572. doi: 10.1021/acschemneuro.0c00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fisher R.S., Emde Boas W., Blume W., Elger C., Genton P., Lee P., Engel J., Jr. Epileptic seizures and epilepsy: definitions proposed by the international League against epilepsy (ILAE) and the international bureau for epilepsy (IBE. Epilepsia. 2005;46(4):470–472. doi: 10.1111/j.0013-9580.2005.66104.x. [DOI] [PubMed] [Google Scholar]
- 69.Maljevic S., Krampfl K., Cobilanschi J., Tilgen N., Beyer S., Weber Y.G., Schlesinger F., Ursu D., Melzer W., Cossette P., Bufler J., Lerche H., Heils A. A mutation in the GABA(A) receptor alpha(1)-subunit is associated with absence epilepsy. Ann. Neurol. 2006;59(6):983–987. doi: 10.1002/ana.20874. [DOI] [PubMed] [Google Scholar]
- 70.Sieghart W., Ramerstorfer J., Sarto-Jackson I., Varagic Z., Ernst M. A novel GABA(A) receptor pharmacology: drugs interacting with the α(+) β(-) interface. Br. J. Pharmacol. 2012;166(2):476–485. doi: 10.1111/j.1476-5381.2011.01779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sigel E., Baur R., Kellenberger S., Malherbe P. Point mutations affecting antagonist affinity and agonist dependent gating of GABAA receptor channels. EMBO J. 1992;11(6):2017–2023. doi: 10.1002/j.1460-2075.1992.tb05258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boileau A.J., Evers A.R., Davis A.F., Czajkowski C. Mapping the agonist binding site of the GABAA receptor: evidence for a beta-strand. J. Neurosci. : The Off. J. Soc. Neurosci. 1999;19(12):4847–4854. doi: 10.1523/JNEUROSCI.19-12-04847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller P.S., Aricescu A.R. Crystal structure of a human GABAA receptor. Nature. 2014;512(7514):270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim J.J., Gharpure A., Teng J., Zhuang Y., Howard R.J., Zhu S., Noviello C.M., Walsh R.M., Jr., Lindahl E., Hibbs R.E. Shared structural mechanisms of general anaesthetics and benzodiazepines. Nature. 2020:303–308. doi: 10.1038/s41586-020-2654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim S.Y., Jang S.S., Kim H., Hwang H., Choi J.E., Chae J.H., Kim K.J., Lim B.C. Genetic diagnosis of infantile-onset epilepsy in the clinic: application of whole-exome sequencing following epilepsy gene panel testing. Clin. Genet. 2021;99(3):418–424. doi: 10.1111/cge.13903. [DOI] [PubMed] [Google Scholar]
- 76.Kash T.L., Jenkins A., Kelley J.C., Trudell J.R., Harrison N.L. Coupling of agonist binding to channel gating in the GABA(A) receptor. Nature. 2003;421(6920):272–275. doi: 10.1038/nature01280. [DOI] [PubMed] [Google Scholar]
- 77.Bracamontes J.R., Li P., Akk G., Steinbach J.H. A neurosteroid potentiation site can be moved among GABAA receptor subunits. J. Physiol. 2012;590(22):5739–5747. doi: 10.1113/jphysiol.2012.237255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nors J.W., Gupta S., Goldschen-Ohm M.P. A critical residue in the α1M2-M3 linker regulating mammalian GABAA receptor pore gating by diazepam. Elife. 2021;10 doi: 10.7554/eLife.64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Møller R.S., Dahl H.A., Helbig I. The contribution of next generation sequencing to epilepsy genetics. Expert Rev. Mol. Diagn. 2015;15(12):1531–1538. doi: 10.1586/14737159.2015.1113132. [DOI] [PubMed] [Google Scholar]
- 80.Kocaaga A., Yimenicioglu S. Identification of novel gene variants in children with drug-resistant epilepsy: expanding the genetic spectrum. Pediatr. Neurol. 2023;139:7–12. doi: 10.1016/j.pediatrneurol.2022.11.005. [DOI] [PubMed] [Google Scholar]
- 81.Hancili S., Önal Z.E., Ata P., Karatoprak E.Y., Gürbüz T., Bostancı M., Paçal Y., Nuhoğlu Ç., Ceran Ö. The GABAA receptor γ2 subunit (R43Q) mutation in febrile seizures. Pediatr. Neurol. 2014;50(4):353–356. doi: 10.1016/j.pediatrneurol.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 82.Koutroumanidis, M., Arzimanoglou, A., Caraballo, R., et al. (n.d.). The role of EEG in the diagnosis and classification of the epilepsy syndromes: A tool for clinical practice by the ILAE Neurophysiology Task Force (Part 1). Epileptic Disorders : Int. Epilepsy J. Videotape,. 10.1684/epd.2017.0935. [DOI] [PubMed]
- 83.Scheffer, I. E., Berkovic, S. et al. (n.d.). ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. 10.1111/epi.13709. [DOI] [PMC free article] [PubMed]
- 84.Perucca P., Scheffer I.E., Harvey A.S., James P.A., Lunke S., Thorne N., Gaff C., Regan B.M., Damiano J.A., Hildebrand M.S., Berkovic S.F., O'Brien T.J., Kwan P. Real-world utility of whole exome sequencing with targeted gene analysis for focal epilepsy. Epilepsy Res. 2017;131:1–8. doi: 10.1016/j.eplepsyres.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 85.Stafstrom C.E., Carmant L. Seizures and epilepsy: an overview for neuroscientists. Cold Spring Harb. Perspect. Med. 2015;5(6):a022426. doi: 10.1101/cshperspect.a022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Churn S.B., Rana A., Lee K., Parsons J.T., Blas A., Delorenzo R.J. Calcium/calmodulin-dependent kinase II phosphorylation of the GABAA receptor alpha1 subunit modulates benzodiazepine binding. J. Neurochem. 2002;82(5):1065–1076. doi: 10.1046/j.1471-4159.2002.01032.x. [DOI] [PubMed] [Google Scholar]
- 87.Nakamura Y., Darnieder L.M., Deeb T.Z., Moss S.J. vol. 72. 2015. pp. 97–146. (Regulation of GABAARs by Phosphorylation). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ubersax J.A., Ferrell J.E., Jr. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007;8(7):530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 89.Macdonald R.L., Kang J.Q., Gallagher M.J. Mutations in GABAA receptor subunits associated with genetic epilepsies. J. Physiol. 2010;588(Pt 11):1861–1869. doi: 10.1113/jphysiol.2010.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Winawer M.R. Phenotype definition in epilepsy. Epilepsy Behav. : E&B. 2006 doi: 10.1016/j.yebeh.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 91.Helbig, I., & Tayoun, A. A. (n.d.). Understanding Genotypes and Phenotypes in Epileptic Encephalopathies. Mol. Syndromol., 7(4), 172–181. 10.1159/000448530. [DOI] [PMC free article] [PubMed]
- 92.Sebastian Köhler, Michael Gargano, Nicolas Matentzoglu, Leigh C Carmody, David Lewis-Smith, Nicole A Vasilevsky, Daniel Danis, Ganna Balagura, Gareth Baynam, Amy M Brower, Tiffany J Callahan, Christopher G Chute, Johanna L Est, Peter D Galer, Shiva Ganesan, Matthias Griese, Matthias Haimel, Julia Pazmandi, Marc Hanauer, Nomi L Harris, Michael J Hartnett, Maximilian Hastreiter, Fabian Hauck, Yongqun He, Tim Jeske, Hugh Kearney, Gerhard Kindle, Christoph Klein, Katrin Knoflach, Roland Krause, David Lagorce, Julie A McMurry, Jillian A Miller, Monica C Munoz-Torres, Rebecca L Peters, Christina K Rapp, Ana M Rath, Shahmir A Rind, Avi Z Rosenberg, Michael M Segal, Markus G Seidel, Damian Smedley, Tomer Talmy, Yarlalu Thomas, Samuel A Wiafe, Julie Xian, Zafer Yüksel, Ingo Helbig, Christopher J Mungall, Melissa A Haendel, Peter N Robinson/. (2021). The Human Phenotype Ontology. Sebast Nucleic Acids Research, D1207–D1217. 10.1093/nar/gkaa1043. [DOI]
- 93.Kodankandath T.V., Theodore D., Samanta D. StatPearls Publishing; 2022. Generalized Tonic-Clonic Seizure.https://pubmed.ncbi.nlm.nih.gov/32119383/ [PubMed] [Google Scholar]
- 94.Betjemann J.P., Lowenstein D.H. Status epilepticus in adults. Lancet Neurol. 2015;14(6):615–624. doi: 10.1016/S1474-4422(15)00042-3. [DOI] [PubMed] [Google Scholar]
- 95.Specchio N., Curatolo P. Developmental and epileptic encephalopathies: what we do and do not know. Brain : J. Neurol. 2021;144(1):32–43. doi: 10.1093/brain/awaa371. [DOI] [PubMed] [Google Scholar]
- 96.Vihinen M. Systematic errors in annotations of truncations, loss-of-function and synonymous variants. Front. Genet. 2023;14 doi: 10.3389/fgene.2023.1015017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.J J.C. In silico comparative genomic analysis of GABAA receptor transcriptional regulation. BMC Genom. 2007;8:203. doi: 10.1186/1471-2164-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li Z., Li X., Zhou H., Gaynor S.M., Selvaraj M.S., Arapoglou T., Quick C., Liu Y., Chen H., Sun R., Dey R., Arnett D.K., Auer P.L., Bielak L.F., Bis J.C., Blackwell T.W., Blangero J., Boerwinkle E., Bowden D.W., Brody J.A.…Lin X. A framework for detecting noncoding rare-variant associations of large-scale whole-genome sequencing studies. Nat. Methods. 2022;19(12):1599–1611. doi: 10.1038/s41592-022-01640-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li, X., Li, Z., Zhou, H., Gaynor, S. M., Liu, Y., Chen, H., Sun, R., Dey, R., Arnett, D. K., Aslibekyan, S., Ballantyne, C. M., Bielak, L. F., Blangero, J., Boerwinkle, E., Bowden, D. W., Broome, J. G., Conomos, M. P., Correa, A., Cupples, L. A., Curran, J. E., … Lin, X. (n.d.). Dynamic incorporation of multiple in silico functional annotations empowers rare variant association analysis of large whole-genome sequencing studies at scale. Nat. Genet., 52(9), 969–983.. [DOI] [PMC free article] [PubMed]
- 100.Zhou, H., Arapoglou, T., Li, X., Li, Z., Zheng, X., Moore, J., Asok, A., Kumar, S., Blue, E. E., Buyske, S., Cox, N., Felsenfeld, A., Gerstein, M., Kenny, E., Li, B., Matise, T., Philippakis, A., Rehm, H. L., Sofia, H. J., Snyder, G., … Lin, X. (n.d.). FAVOR: functional annotation of variants online resource and annotator for variation across the human genome. Nucleic Acids Res., 51(D1), D1300–D1311. 10.1093/nar/gkac966. [DOI] [PMC free article] [PubMed]
- 101.Zhao C., Huang C., Weng T., Xiao X., Ma H., Liu L. Computational prediction of MicroRNAs targeting GABA receptors and experimental verification of miR-181, miR-216 and miR-203 targets in GABA-A receptor. BMC Res. Notes. 2012;5:91. doi: 10.1186/1756-0500-5-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sengupta J.N., Pochiraju S., Kannampalli P., Bruckert M., Addya S., Yadav P., Miranda A., Shaker R., Banerjee B. MicroRNA-mediated GABA Aα-1 receptor subunit down-regulation in adult spinal cord following neonatal cystitis-induced chronic visceral pain in rats. Pain. 2013;154(1):59–70. doi: 10.1016/j.pain.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smits A., Jin Z., Elsir T., Pedder H., Nistér M., Alafuzoff I., Dimberg A., Edqvist P.H., Pontén F., Aronica E., Birnir B. GABA-A channel subunit expression in human glioma correlates with tumor histology and clinical outcome. PLoS One, 2012. 2012;7(5) doi: 10.1371/journal.pone.0037041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang L., Liu Y., Yu Z., Gong J., Deng Z., Ren N., Zhong Z., Cai H., Tang Z., Cheng H., Chen S., He Z. Mir-139-5p inhibits glioma cell proliferation and progression by targeting GABRA1. J. Transl. Med. 2021;19(1):213. doi: 10.1186/s12967-021-02880-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Balciuniene J., DeChene E.T., Akgumus G., Romasko E.J., Cao K., Dubbs H.A., Mulchandani S., Spinner N.B., Conlin L.K., Marsh E.D., Goldberg E., Helbig I., Sarmady M., Abou Tayoun A. Use of a dynamic genetic testing approach for childhood-onset epilepsy. JAMA Netw. Open. 2019;2(4) doi: 10.1001/jamanetworkopen.2019.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bayat A., Fenger C.D., Techlo T.R., Højte A.F., Nørgaard I., Hansen T.F., Rubboli G., Møller R.S., Group D.C.C.R.S. Impact of genetic testing on therapeutic decision-making in childhood-onset epilepsies-a study in a tertiary epilepsy center. Neurotherapeutics : The J. Am. Soc. Exp. NeuroTher. 2022;19(4):1353–1367. doi: 10.1007/s13311-022-01264-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Striano P., Minassian B.A. From genetic testing to precision medicine in epilepsy. Neurotherapeutics. 2020 doi: 10.1007/s13311-020-00835-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lerche H., Weckhuysen S., Whittemore V., Berkovic S.F., Lowenstein D.H. Precision medicine for genetic epilepsy on the horizon: recent advances, present challenges, and suggestions for continued progress. Epilepsia. 2022;63(10):2461–2475. doi: 10.1111/epi.17332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Salinas V., Martínez N., Maturo J.P., Rodriguez-Quiroga S.A., Zavala L., Medina N., Amartino H., Sfaello I., Agosta G., Serafín E.M., Morón D.G., Kauffman M.A., Vega P. Clinical next generation sequencing in developmental and epileptic encephalopathies: diagnostic relevance of data re-analysis and variants re-interpretation. Eur. J. Med. Genet. 2021;64(12) doi: 10.1016/j.ejmg.2021.104363. [DOI] [PubMed] [Google Scholar]
- 110.MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B., Albers C.A., Zhang Z.D., Conrad D.F., Lunter G., Zheng H., Ayub Q., DePristo M.A., Banks E., Hu M.…Tyler-Smith C. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335(6070):823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.