Abstract
Background
Skin cutaneous melanoma is characterized by high malignancy and prognostic heterogeneity. Immune cell networks are critical to the biological progression of melanoma through the tumor microenvironment. Thus, identifying effective biomarkers for skin cutaneous melanoma from the perspective of the tumor microenvironment may offer strategies for precise prognosis prediction and treatment selection.
Methods
A total of 470 cases from The Cancer Genome Atlas and 214 from the Gene Expression Omnibus were systematically evaluated to construct an optimal independent immune cell risk model with predictive value using weighted gene co-expression network analysis, Cox regression, and least absolute shrinkage and selection operator assay. The predictive power of the developed model was estimated through receiver operating characteristic curves and Kaplan-Meier analysis. The association of the model with tumor microenvironment status, immune checkpoints, and mutation burden was assessed using multiple algorithms. Additionally, the sensitivity of immune and chemotherapeutics was evaluated using the ImmunophenScore and pRRophetic algorithm. Furthermore, the expression profiles of risk genes were validated using gene expression profiling interactive analysis and Human Protein Atlas resources.
Results
The risk model integrated seven immune-related genes: ARNTL, N4BP2L1, PARP11, NUB1, GSDMD, HAPLN3, and IRX3. The model demonstrated considerable predictive ability and was positively associated with clinical and molecular characteristics. It can be utilized as a prognostic factor for skin cutaneous melanoma, where a high-risk score was linked to a poor prognosis and indicated an immunosuppressive microenvironment. Furthermore, the model revealed several potential target checkpoints and predicted the therapeutic benefits of multiple clinically used drugs.
Conclusion
Our findings provide a comprehensive landscape of the tumor immune microenvironment in skin cutaneous melanoma and identify prognostic markers that may serve as efficient clinical diagnosis and treatment selection tools.
Keywords: Skin cutaneous melanoma, Weighted gene co-expression network analysis, Tumor immune microenvironment, Prognosis, Immune checkpoint
1. Introduction
Skin cutaneous melanoma (SKCM) is a highly aggressive and life-threatening form of skin cancer, accounting for roughly 80% of skin cancer mortality [1]. Global epidemiology studies have revealed a rising trend in the incidence of melanoma, which increases by 3%–7% each year, causing a severe threat to human health [2]. Early-stage SKCM has a favorable 5-year survival rate of 80–90% after radical resection, but if local progression or metastasis occurs, the survival rate markedly deteriorates to below 20% [3,4]. Late-stage SKCM is notoriously characterized by a highly heterogeneous genetic background, inefficient therapeutic response, and prevalent drug resistance. As a result, its clinical efficacy and prognosis have not met expectations; thus, effective biomarkers for prognosis prediction and treatment selection are urgently needed.
Recently, the influence of the tumor microenvironment (TME) on the biological behavior of melanoma has been a topic of intense discussion. The tumor microenvironment is a complex network of tumor cells and the surrounding stromal cells. In particular, the crosstalk of various immune cells therein, with other components, sustains the plasticity of melanoma cells and extensively modulates melanomagenesis and progression [5]. Moreover, immunotherapies that stand as the mainstay of advanced melanoma treatment, especially immune checkpoint inhibitors, are greatly dictated by immune cell infiltration in TME [6,7]. Specifically, recent research has highlighted the importance of antigen-specific CD8+ T cells in the tumor-draining lymph nodes (TdLN-TTSM cells) as the primary cell population for positive PD1 immunotherapy outcomes. Adoptive transfer of TdLN-TTSM cells has demonstrated superior anti-tumor therapeutic efficacy [8]. Moreover, the chemokine CXCL12 has been identified as a regulator of antigen-specific CD8+ T cells' exit from tumors to lymphatic vessels, and its inhibition promotes T cell retention, resulting in enhanced melanoma control [9]. Therefore, a comprehensive evaluation of the TME is crucial for precisely managing SKCM.
Advances in sequencing technology and bioinformatics algorithms have enabled researchers to quantify the immuno-phenotype within the TME using marker genes efficiently. These cutting-edge approaches have led to identifying gene signatures that can serve as prognostic and drug response prediction tools for various types of tumors [[10], [11], [12], [13]]. Motivated by these discoveries, our study aimed to investigate the expression profile of immune-related genes and their prognostic significance in SKCM, to identify potential predictive biomarkers and therapeutic targets for the disease.
Here, a set of seven immune-related genes in combination with clinical data from several independent databases were employed to establish a prognostic signature using TME immune cell infiltration features as its core. Our risk model demonstrates the robust predictive ability for SKCM prognosis, and it represents a pivotal biomarker paradigm for immunotherapy and prognostic evaluation of SKCM patients.
2. Methods
2.1. Data source and immune infiltration analysis
The TCGA RNA-seq data (Counts) and clinical follow-up data of SKCM patients were obtained from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Additionally, microarray gene expression data of GSE65904 and its clinical follow-up data were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The TCGA dataset was used to investigate prognosis-related genes and potential risk score systems. The clinical data of TCGA and GEO samples were compiled in Table 1. The GEO datasets were used for external validation. The immune infiltration analysis of the TCGA datasets was performed using the limma package. The significantly different immune cells (SDICs) were selected using the Pheatmap and corrplot packages, and clustered heatmaps were generated.
Table 1.
Clinical feature of the included samples from both TCGA and GEO databases.
| TCGA | ||
|---|---|---|
| Feature | Data | total |
| Gender(Male/Female) | 290/180 | 470 |
| Age(years) | 58.22 ± 15.73 | |
| Tumor Stage(0/Ⅰ/Ⅱ/Ⅲ/Ⅳ/unknown) | 7/77/140/171/23/52 | |
| T(0/1/2/3/4/unknown) | 23/42/78/90/153/84 | |
| M(0/1/unknown) | 418/24/28 | |
| N(0/1/2/3/unknown) | 235/74/49/55/57 | |
| Patient status(life/death) | 248/222 | |
| Survival time(days) | 1837.48 ± 1937.10 | |
| GEO | ||
| Feature | Data | total |
| Gender(Male/Female/unknown) | 124/89/1 | 214 |
| Age(years) | 62.35 ± 14.40 | |
| Patient status(life/death/unknown) | 108/102/4 | |
| Survival time(days) | 981.40 ± 1222.82 | |
2.2. Weighted gene co-expression network analysis (WGCNA) and prognosis-related genes (PRGs)
WGCNA is a systems biology approach used to describe gene co-expression patterns between different samples. In contrast to focusing only on individual differentially expressed genes, WGCNA can identify co-expression modules at a genome-wide level and identify candidate biomarker genes or therapeutic targets based on the within-module connectivity and associations with phenotypes. Additionally, WGCNA is flexible and adaptive, as its analysis is not restricted to a specific type of gene expression data and does not require prior assumptions or the setting of thresholds or parameters [14].
To perform WGCNA analysis, we utilized the WGCNA package, building upon the results of the SDIC analysis. Gene modules of |correlation coefficient|>0.5 and P value < 0.05 were considered strong immune cell-correlated modules. Module-trait genes (MTGs) with the lowest P value were defined to be used in the subsequent steps of the study, based on which univariate Cox analysis was used to select the PRGs. Herein, the filter was P value<0.0001.
2.3. Development of prognosis-related immune cells risk score (ICrisk)
The PRGs were inputted into the least absolute shrinkage and selection operator (LASSO) -Cox regression to identify the final risk genes (RGs) and establish the ICrisk. The formula calculated the risk score: ICrisk = ∑ βi*Expi, where βi is the risk coefficient of RGs, and Expi represents the gene expression value. According to the median of TCGA ICrisk, TCGA, and GEO samples were divided into high-risk and low-risk groups, respectively.
2.4. The receiver operating characteristic (ROC) curve and nomogram
The TimeROC package was used to draw the ROC curves and evaluate the predictive power of ICrisk along with the clinical features. The predictive power is considered with higher accuracy when area under the time-dependent receiver operating characteristic curve (AUC) > 0.5. Then, a nomogram was established to predict the survival time of SKCM patients. In addition, DCA curves were drawn using the ggDCA package to validate the accuracy of the nomogram.
2.5. Survival and independent prognostic analysis of the risk model
Kaplan-Meier (K-M) analysis was used to perform survival analysis between the high- and low- ICrisk groups. At the same time, the survival package was used to draw survival curves and perform univariate and multivariate independent prognostic analysis of ICrisk and other clinical features. Independent prognostic factors were defined as P value<0.05 in univariate and multivariate independent predictive analyses. In addition, the survival state diagram and risk heatmap were generated using the pheatmap package.
2.6. Tumor microenvironment and survival analysis of the risk model
Immune and stromal scores were calculated using the estimate package. The differences in immune and stromal scores between high- and low-risk groups were compared to explore the relation between ICrisk score and TME; the results were visualized using the ggpubr package. For the survival analysis of the RGs, The TCGA samples were divided into high- and low-expression groups based on the best cut-off value of RGs to explore the influences of RGs on the patients and perform the survival analysis between the two groups.
2.7. Tumor mutation burden (TMB) profiles of the risk model
The mutation data for SKCM patients were downloaded from the TCGA data portal and integrated using Perl (version 5.30.0) software. Waterfall plots were drawn to visualize mutation frequencies using the R package “maftools”, and the mutation status of SKCM patients was compared between high- and low-risk groups. Meanwhile, SKCM patients were divided into groups with different TMB statuses and risk scores according to the optimal cut-off value of TMB for survival analysis, respectively.
2.8. The relevance analysis of immune cells and RGs
Besides the relationship between immune cells and risk scores, the correlation between immune cells and the expression of RGs was analyzed. The results were finalized using the ggplot2 package with a filter of |correlation coefficient| >0.4 and P value < 0.001.
2.9. The relevance analysis of immune checkpoints and gene-set enrichment analysis (GSEA)
The relevance between immune checkpoint genes and RGs was analyzed to study the relationship between immune checkpoints and the ICrisk model. Then, the ggplot2 package was used to visualize this relevance. Furthermore, TCGA samples were divided into high- and low-expression groups according to the RGs’ median expression value. Gene ontology (GO) and kyoto encyclopedia of genes genomes (KEGG) were utilized using the clusterProfiler package to study the functional enrichment.
2.10. Clinical relevance heatmap
To allow the study of the correlation between the clinical features and patients’ prognosis, the ComplexHeatmap package was used to analyze and visualize the differences between high- and low- ICrisk groups.
2.11. The therapeutic sensitivity prediction of patients with different risk scores
At this step, the sensitivity of immunotherapy and other clinical used drugs was predicted. The potential response to immune treatment was represented by the immunophenscore (IPS), which was downloaded from The Cancer Immunome Database (TCIA) [15] (https://www.tcia.at/). TCIA provides results of comprehensive immunogenomic analyses of next-generation sequencing data (NGS) data for 20 solid cancers from the TCGA database and other data sources. A higher IPS predicts a better response to immunotherapy. In addition, the pRRophetic [16] package was used to calculate the 50% inhibiting concentration (IC50) value of 138 drugs.
Validation in the gene expression profiling interactive analysis (GEPIA) and human protein atlas (HPA) databases.
To validate the mRNA expression and prognosis of the identified IRGs in SKCM patients, we utilized the GEPIA database (http://gepia.cancer-pku.cn/). This web-based resource enables loading all identified IRGs onto the server and offers data from TCGA and Genotype-Tissue Expression (GTEx) for validation analysis. We employed the standard processing pipeline [17] for the screening modules and main analysis conditions. To further validate the protein expression levels of the identified IRGs in SKCM tissues and normal skin tissues, we employed the HPA portal (https://www.proteinatlas.org/), which currently stands as the most comprehensive protein database. Using transcriptome, proteomics, and other omics technologies, the HPA has mapped an extensive collection of human protein expression images of tissues, cells, and organs. All the data in this knowledge resource is open access to allow scientists to freely access the data for exploration of the human proteome.
2.12. Statistical analysis
All the before-mentioned packages and statistical analyses were performed in R.4.1.0 software. The K-M and Cox regression analyses were used to analyze the overall survival (OS) differences in categorical variables. For continuous variables, Cox regression was used to calculate the hazard ratio (HR) and the differences of the OS. For the relevance analyses, the Spearman method was applied to calculate the correlation coefficient and P value. The Wilcoxon test estimated the statistical difference between the two groups. Unless otherwise stated, the P values were two-sided, and P < 0.05 was considered statistically significant.
3. Results
3.1. Immune cells infiltration analysis
In the current study, we utilized the TCGA database to investigate immune cell infiltration in a cohort of 472 SKCM patients. Our results revealed that the central immune cells in SKCM were macrophages and T cells. In contrast, other immune cells, such as B cells and neutrophils, constituted only a small proportion of the infiltrated immune cells (Fig. 1A). We also examined the positive and negative correlations between different immune cells in the included samples. Our analysis showed that activated dendritic cells and neutrophils had the most positive correlation among immune cells, while CD8+ T cells and CD4+ T memory resting cells had the most negative correlation (Fig. 1B).
Fig. 1.
Construction of the immune cells risk score model (ICrisk). (A) The analysis of the immune cell infiltration from the TCGA database. (B) Immune cells correlation heatmap indicating the positive and the negative correlations between the different immune cells in the included sample. (C) The optimal soft threshold in WGCNA analysis was selected before the model establishment. (D) Gene modules identified by WGCNA. (E) Correlation between gene modules and immune cells. (F, G) The LASSO coefficient profiles about 7 IRGs, the lower X-axis indicates log (λ), the upper X-axis indicates the average number of OS-related genes, and the Y-axis shows the partial likelihood deviance error. Red dots indicate the average partial likelihood deviance about the model with a given λ, the vertical bars represent the range of the partial likelihood deviance errors, and the vertical black dotted lines meant the best fit with the optimal λ values.
3.2. Construction of the immune cells risk score model (ICrisk)
The first step of establishing the model was selecting the optimal soft threshold in WGCNA analysis, which was set to 10 (Fig. 1C). Based on WGCNA results (Fig. 1D), 19 co-expressed gene modules were identified (Fig. 1E). The brown module was selected from the module-trait genes relationships as it led to the least total P values. Within this module, a total of 1484 genes were included, then a univariate Cox analysis was applied, resulting in the preservation of 452 prognosis-related genes (Table S1). Finally, after the LASSO-COX regression (Fig. 1F and G), seven genes were identified as the risk genes to be used in constructing the risk score model. These risk genes are ARNTL (Aryl hydrocarbon receptor nuclear translocator like), N4BP2L1 (NEDD4 binding protein 2 like 1), PARP11 (Poly (ADP-ribose) polymerase family member 11), NUB1 (Negative regulator of ubiquitin like proteins 1), GSDMD (Gasdermin D), HAPLN3 (Hyaluronan and proteoglycan link protein 3), and IRX3 (Iroquois homeobox 3) (Fig. S1, Table S2). Therefore, the previously mentioned ICrisk formula was used to calculate the risk scores of each sample in TCGA and GEO databases, classifying them into high- and low-risk groups.
3.3. The predictive power of the risk to SKCM patients
After performing univariate and multivariate independent prognostic analyses, the tumor (T), node (N), and risk score were identified as independent prognostic factors through hazard ratio analysis, where the hazard ratios were around 1.4 with p-values <0.001 (Fig. 2A and B). The AUC values at 1, 3, or 5 years in the ROC curves were greater than 0.7 (Fig. 2C), illustrating the considerable value of the model in predicting the overall survival (OS) of SKCM patients. Moreover, comparing the AUC of the established risk model and other clinical features, such as stage, demonstrated that the established risk model had better prediction power (Fig. 2D). Subsequently, a nomogram was constructed to predict patients’ survival time after suffering SKCM, displaying the weight of each prognostic factor (Fig. 2E and F), and the corresponding DCA curves confirmed the effectiveness of the nomogram (Fig. 2G).
Fig. 2.
The univariate (A) and multivariate (B) analysis for the ICrisk (95% confidence interval (95% CI)). (C) The receiver operating characteristic (ROC) curves of ICrisk in predicting 1,3, and 5 year survival. (D) The ROC curves of ICrisk and other clinical features in predicting 1-year survival. Nomogram predicting the overall survival (E) and calibration plots for predicting survival at 1,3, and 5 years (F). The points for each of the six variables are obtained by drawing a line upwards from the value of each variable to the points line. The sum of points for the six variables is marked in the total points line, and the line drawn perpendicularly downward indicates the probability of survival at 1, 3, and 5 years. (G) Decision curve analysis (DCA) of the nomogram predicting 1, 3, and 5 years OS.
3.4. Survival analysis of the ICrisk in TCGA and GEO datasets
Based on the ICrisk, patients were stratified into low- and high-risk groups. K-M curves demonstrated that patients with high-risk scores had significantly lower OS than low-risk scores with a p-value below 0.0001 (Fig. 3A). Furthermore, the risk curve and the scatter diagram of TCGA samples indicated that the mortality of the high-risk group was higher than the low-risk group (Fig. 3C). Although surprisingly, six out of the risk model genes were upregulated in a low-risk group than the high-risk group, IRX3 was the only gene upregulated in the high-risk group (Fig. 3B). These correlation results were also speculated in GEO samples (Fig. 3D–F).
Fig. 3.
Survival analysis of the ICrisk in TCGA and GEO datasets. K-M curves of the high-risk and low-risk groups of TCGA (A) and GEO (D), respectively. Curves were compared using the log-rank test. The expression heatmap of identified genes in TCGA (B) and GEO samples (E). Risk survival status plot of TCGA (C) and GEO samples (F).
3.5. Different TME and TMB patterns in the ICrisk
To this end, we estimated both stromal and immune scores in TCGA samples. The stromal scores ranged from −1781.3 to 1886.2, while the immune scores ranged from −1473.7 to 3707.7. Notably, these estimated scores negatively correlated with tumor purity (Fig. 4A). Of note, the immune and stromal scores displayed distinct distributions in the high- and low-risk groups (Fig. 4B).
Fig. 4.
The association of ICrisk with TME and TMB. (A) The risk score's correlations between stromal score (left panel) and immune score (middle panel), and the correlation between estimate score and tumor purity (right panel). (B) Differences of TME scores in the ICrisk. The waterfall plots of most frequent somatic mutations in high-risk (C) and low-risk (D) groups. (E) TMB status in the ICrisk. (F, G) Survival analysis of risk groups with different TMB statuses.
The distribution of mutations in the low- and high-risk subgroups was compared, and the top five mutant genes in both groups were TTN, MUC16, BRAF, DNAH5, and PCLO (Fig. 4C and D). Notably, the low-risk group exhibited a higher mutational burden (Fig. 4E). The TMB is known to be positively correlated with immunotherapy efficacy, and consistent with this, the high-TMB group had a better probability of survival (Fig. 4F). In accordance with previous results, patients with high TMB and low-risk scores had the most favorable survival outcomes. Conversely, patients with low TMB and high-risk scores experienced the poorest survival outcomes (Fig. 4G).
3.6. ICrisk predicts therapeutic benefits
IC50 values were calculated for 138 drugs in TCGA-SKCM patients to investigate the potency of ICrisk in predicting SKCM response to standard drugs, including chemotherapy, targeted therapy, and immunotherapy. The results indicated that patients in the low-risk group were more sensitive to imatinib, parthenolide, and sorafenib with p-values of 2.8e-09, 5.5e-05, and 9e-16, respectively (Fig. 5A–C). In contrast, patients in the high-risk group exhibited increased sensitivity to lenalidomide, metformin, and methotrexate, where the p-values were 2.22e-16, 4.4e-6, and 1.4e-14 (Fig. 5D–F). Regarding immunotherapy sensitivity, the low-risk group had a higher immunophenoscore (IPS), indicating that low-risk patients were more responsive to immunotherapies such as PD-1 and CTLA4 inhibitors (Fig. 5G–J).
Fig. 5.
The ICrisk's capacity in predicting therapeutic benefits. The predicted sensitivity of high- and low-risk patients to several clinically used drugs, including imatinib (A), parthenolide (B), sorafenib (C), lenalidomide (D), metformin (E), and methotrexate (F). (G–J) The association between IPS and ICrisk. IPS, IPS-PD1, IPS-CTLA4, and IPS-PD1/CTLA4 scores significantly increase in the low-risk group.
3.7. ICrisk is associated with the SKCM immune signature
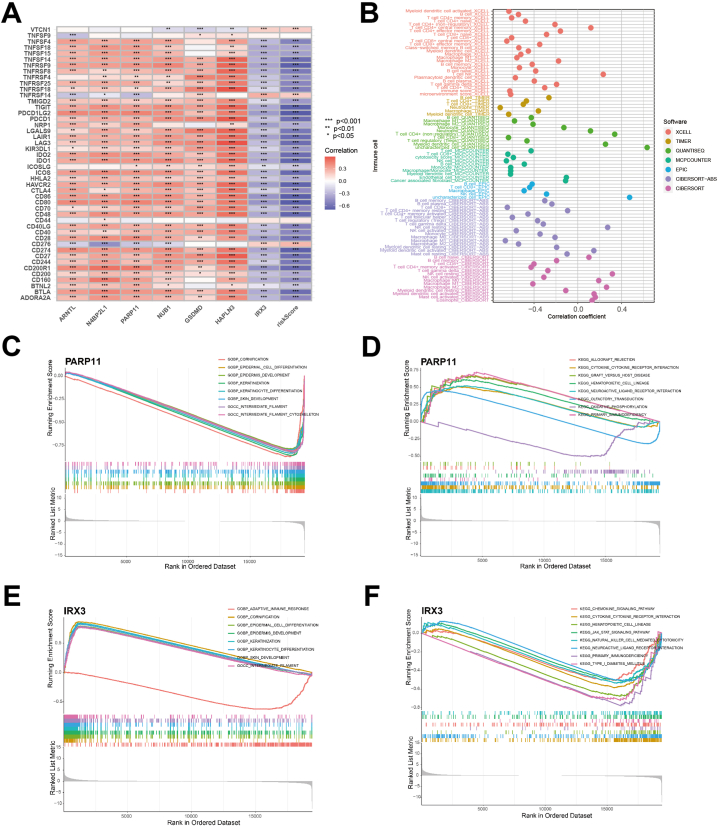
The interconnection between immune checkpoints and identified genes was further explored in this study. The analysis of immune checkpoints and risk genes revealed that most risk genes were positively correlated with immune checkpoints, except for IRX3, which showed a negative correlation with most immune checkpoints. Moreover, the risk score was also negatively correlated with most immune checkpoints, although a few immune checkpoints (VTCN1, TNFRSF14, CD276) did not follow this pattern (Fig. 6A). The next step was investigating the crosstalk between the risk genes and immune cells. The results indicated a strong association between risk genes and immune cells. For example, GSDMD was positively correlated with only one immune cell, while HAPLN3 was interconnected with 28 immune cell phenotypes (Table S3). In total, 88 immune cell phenotypes were found to be related to ICrisk, of which 16 exhibited positive relationships, while 72 were negatively related (Fig. 6B).
Fig. 6.
The ICrisk is associated with SKCM immune signature. (A) Immune checkpoint correlation analysis. (B) Crosstalk between immune cells and risk score, the color of the dots indicate the software used in the estimation. (C–F) GESA enrichment analysis explores correlation between risk genes and possible biological pathways, including GO (C for PARP11 and E for IRX3) and KEGG (D for PARP11 and F for IRX3).
To explore the potential mechanisms involved in the different clinical outcomes related to ICrisk, we performed GSEA enrichment analysis with GO and KEGG annotations for different expressions of identified IRGs. Surprisingly, we found that six risk genes (ARNTL, GSDMD, HAPLN3, N4BP2L1, NUB1, and PARP11) exhibited nearly uniform functional enrichment in low- and high-expression groups. The genes involved in leukocyte activation and regulation, immune function, adaptive immune response, cytokine and cytokine receptor interaction, primary immune efficiency, and hematopoietic cell lineage were enriched in the high-expression group. Conversely, in the low-expression group, genes related to skin development, such as cornified envelope and retinol metabolism, keratinocyte differentiation, and epidermis development, were enriched. However, consistent with the previous results of the immune checkpoint analysis, IRX3 exhibited an opposite trend (Fig. 6C–F, S2A-D).
3.8. Expression profiling of risk genes and prognosis analysis
Our ICrisk signature was validated in both GEPIA and HPA datasets. Analysis of the GEPIA transcriptional data revealed that all risk genes exhibited differential expression in mRNA between tumor and normal tissues, with GSDMD, HAPLN3, NUB1, and PARP11 being more highly expressed in tumor samples. At the same time, ARNTL, IRX3, and N4BP2L1 were more highly expressed in normal tissues (Fig. 7A–G). Immunohistochemistry analysis of protein expression patterns in the HPA demonstrated consistency with mRNA expression for ARNTL, GSDMD, and NUB1 but not for N4BP2L1, which showed the opposite trend. HAPLN3 and PARP11 displayed fewer cancer-specific expression patterns (Figs. S3A–F).
Fig. 7.
Risk genes expression and survival analysis. The scatter difference diagram of ARTNL (A), GSDMD (B), HAPLN3 (C), IRX3 (D), N4BP2L1 (E), NUB1 (F), and PARP11 (G) mRNA expression in tumor (T) and normal (N) skin tissue. (A–G) Corresponding K-M survival analysis of risk genes. (H) Clinical characteristics heatmap integrating ICrisk.
Additionally, we individually conducted the survival analysis of the risk genes, revealing a significant difference in OS between high- and low-expression groups for all genes, underscoring their prognostic relevance. Specifically, K-M curves showed that patients in the high-expression group for IRX3 had a poorer OS, while patients in the high-expression group for the other risk genes had a better OS (Fig. 7A–G). Finally, comparing clinical features, We observed significant differences in age, stage, and tumor (T) between the high- and low-risk groups (Fig. 7H).
4. Discussion
The high malignancy and prognostic discrepancy of cutaneous melanoma have underscored the pressing need for effective biomarkers to accurately predict survival and therapeutic response. Recent advances in sequencing technologies have made it possible to systematically identify critical genetic and epigenetic alterations in various types of melanoma. Additionally, the availability of bulk transcriptome information from cancer genome databases has enabled the mining and translating data from multiple genomics aspects [18,19]. In this study, we leverage several landmark datasets to construct an immune-based prognostic signature through an extensive network analysis of immune-related gene profiles in SKCM.
Herein, our study demonstrates consistent results that underscore the effectiveness of our prognostic model. Specifically, we developed the ICrisk signature, which uses a risk-score system to classify SKCM patients into high-risk and low-risk groups with significantly different survival probabilities. Both univariate and multivariate Cox regression analyses identified the risk score as an independent prognostic factor. Furthermore, we validated the robustness of our risk model in two independent datasets. Importantly, our clinical relevance Heatmap revealed that the risk score was independent of conventional clinical characteristics. We also found that the risk score demonstrated higher sensitivity and better clinical applicability than conventional criteria for predicting melanoma survival, as evidenced by our ROC curves and nomogram analysis. These results highlight the potential of our model for improving clinical decision-making in managing cutaneous melanoma patients.
Our ICrisk signature has integrated 7 IRGs (ARNTL, GSDMD, HAPLN3, IRX3, N4BP2L1, NUB1, PARP11). A subset of these identified genes are not solely defined to be associated with carcinogenesis but rather to play a regulatory role in non-malignant physiological or pathological processes, such as ARNTL controlling circadian rhythms [20], GSDMD regulating inflammatory responses in cardiac and lung injury pyroptosis [21,22], and IRX3 regulating obesity-related metabolism [23]. In recent years, studies have started supporting their roles in cancer pathology (Table 2). However, evidence relating to melanoma is still lacking. Consistent with K-M survival results, ARNTL and NUB1 exhibited tumor-suppressing phenotypes in several in vitro studies. Conversely, GSDMD, HAPLN3, and N4BP2L1 are indicated to correlate with poor tumor prognosis, suggesting they might function distinctively in melanoma. Regarding PARP11, a recent critical study uncovers its link to TME immunosuppression. Specifically, PARP11 has been shown to downregulate IFNAR1 (type I interferon receptor) on CD8+ cytotoxic T lymphocytes, which results in inhibition of the tumor-eliminating ability of these cells, as well as decreased efficacy of CAR-T (chimeric antigen receptor T-cell) immunotherapy [33]. Although PARP11 was defined as less cancer-specific in the HPA, our risk model, coupled with existing literature, still supports its close correlation with TME and the prognosis of SKCM.
Table 2.
The role of identified IRGs in cancer pathology.
| Identified genes | Roles in cancer pathology |
|---|---|
| ARNTL | Promoting apoptosis and inhibiting cell viability via autophagy in oral cancer [24]; Suppressing cell proliferation and enhancing sensitivity to cisplatin in ovarian cancer cells [25] and nasopharyngeal carcinoma by activating CDK5 [26] |
| GSDMD | Highly expressed in bladder cancer tissue and linked with a higher risk of local tumor recurrence [27] |
| IRX3 | Regulating the differentiation of Wilms tumor by activating WNT/β-catenin-signalling [28]; Enhancing the morphologic and phenotypic differentiation block of acute myeloid leukemia cells [29] |
| N4BP2L1 | Promoting oral squamous cell carcinoma invasion by activating miR-448 [30] |
| NUB1 | Inhibiting proliferation and invasion of gastric cancer cells through upregulation of p27Kip1 and inhibition of epithelial-mesenchymal transition [31]; Exerting IFN-a induced antimitogenic function in renal cell carcinoma [32] |
| PARP11 | Downregulating type I interferon receptor and promoting immunosuppression in the TME [33] |
| HAPLN3 | Highly expressed in breast cancer tissue and positively correlated to human epidermal receptor 2 level [34] |
Out of our gene set, HAPLN3 has already been integrated into other prediction models, two of which were concerning melanoma [35,36]. These studies have concluded that HAPLN3 is a potential signature that could be used in prognosis, risk assessment, and prediction of immunotherapy response in melanoma patients, especially in combination with specific clinical features. In addition, a pan-cancer analysis has examined several pathological roles of the GSDM family in kidney renal clear cell carcinoma (KIRC). In particular, GSDMD is considered the most promising biomarker for evaluating KIRC prognosis and drug sensitivity [37]. Overall, the aforementioned genes in melanoma have received comparatively less in-depth research attention, particularly regarding their specific roles in melanoma TME and immunity, which still need to be well-established. To date, most evidence of their involvement in melanoma pathogenesis is based on bioinformatics studies. Therefore, additional research efforts are essential to comprehensively understand the contribution of these identified genes to melanoma immunity and prognosis.
SKCM treatment is constantly evolving, with researchers consistently updating current therapies and exploring novel treatment strategies. Currently, immunotherapy remains the primary focus of research, with efforts being made to maximize its potential. The combination of CTLA4 and PD1 blockage has demonstrated a significant breakthrough in treating advanced melanoma [38]. Furthermore, postoperative adjuvant therapy utilizing PD1 blockage has shown significant survival benefits in early-stage melanoma patients [39]. Moreover, ongoing research investigating novel immune targets, such as LAG-3 and TIM-3, exhibits promising potential for therapeutic improvement [40,41]. In our study, the risk model's immunological characteristics and drug sensitivity can offer alternative insights into the treatment strategy for SKCM.
Our findings suggest that high ICrisk indicates an immunosuppressed status, characterized by low frequency of TMB, subdued immune cell activities, and overall downregulation of immune checkpoints. Consequently, the high-risk group may have fewer immunotherapy options and benefits compared to the low-risk group. Notably, CD276 was the immune checkpoint that demonstrated the most significant positive association with the risk score. CD276, also known as B7–H3, belongs to the B7 family of immunoregulatory proteins that play a crucial role in regulating T-cell activation and autoimmunity. Previous pre-clinical studies have shown that targeting CD276 can improve the therapeutic control of melanoma by increasing patient-derived cells' sensitivity to chemotherapy, MAP kinase (MAPK), and AKT/mTOR inhibitors [42]. Combining the blockade of PD1 with CD276 can lead to further enhanced efficacy [43,44]. Therefore, enoblituzumab, a humanized anti-CD276 mAb (monoclonal antibody), has been designed and is currently undergoing intensive clinical trials for various advanced cancers [[45], [46], [47]].
Furthermore, the drug sensitivity analysis indicates that the high-risk group may benefit from lenalidomide, methotrexate, and metformin. In particular, recent research suggests that metformin can enhance the efficacy of anti-PD1 Ab in melanoma models by stimulating the production of mitochondrial reactive oxygen species, which robustly activates CD8+ infiltrating T lymphocytes' proliferation [48]. These findings suggest synergistic immune checkpoints and immune activation may offer potential therapeutic options for SKCM patients in the immunosuppressed high-risk group. Ultimately, we aim to optimize combination regimens of chemotherapy, targeted therapy, and immunotherapy based on our ICrisk model, thereby providing new insights for SKCM treatment strategies.
Before our research, various prognostic immune-related gene signatures for skin melanoma had been identified. These signatures have different numbers of marker genes, ranging from 2 to 33 [35,36,[49], [50], [51], [52]]. While a larger number of marker genes is usually considered to enhance the specificity and predictive power of prognostic models, an excessive number of marker genes can also complicate the data, introduce noise, and increase the cost of gene testing, thus impeding clinical translation. In this context, our study developed the ICrisk model with an appropriate number of marker genes, achieving AUC values of 0.7 for predicting 1-, 3-, and 5-year survival. Significantly, we expanded the pool of SKCM biomarkers by identifying novel prognostic genes that have not been previously evaluated as a complete set in published prognostic models for SKCM. Moreover, we comprehensively assessed ICrisk in multiple aspects, including the tumor microenvironment, mutational burden, drug sensitivity, immune checkpoints, and biological pathways. These analyses provided a more thorough understanding of the model's performance and potential clinical relevance.
However, our study has several limitations that need to be acknowledged. Firstly, our study is based primarily on existing database resources, where the sample cohorts are retrospective. Therefore, the robustness of the predictive value of the risk model needs to be validated with additional external clinical samples and real-world data. Secondly, the datasets' clinical information was limited in terms of surgery and radiation therapy, which prevented us from incorporating these clinical characteristics into the assessment of the predictive value of the ICrisk. Thirdly, as previously mentioned, the biological functions of the identified genes in SKCM, particularly in TME regulation, are not yet clearly defined and require further examination using experimental methods.
5. Conclusion
Our study identifies prognostic immune genes associated with SKCM and establishes a network related to prognosis. Furthermore, we highlight the crucial role of seven identified IRGs in regulating the immune microenvironment, disease progression, and drug sensitivity in SKCM patients. These findings provide potential strategies for guiding personalized and precise therapies for SKCM.
Author contribution statement
Xixi Lin, Razan Hessenow: Analyzed and interpreted the data; Performed the experiments; Wrote the paper.
Siling Yang, Dongjie Ma: Analyzed and interpreted the data; Wrote the paper.
Sijie Yang: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
1 - Conceived and designed the experiments.
2 - Performed the experiments.
3 - Analyzed and interpreted the data.
4 - Contributed reagents, materials, analysis tools or data.
5 - Wrote the paper.
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e20234.
Contributor Information
Xixi Lin, Email: xixi.lin1994@gmail.com.
Razan Hessenow, Email: razan.hessenow@uk-essen.de.
Siling Yang, Email: yang_siling1994@yeah.net.
Dongjie Ma, Email: madongjiesj@sina.com.
Sijie Yang, Email: yangsijiemdj@sina.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
figs2.
figs3.
References
- 1.Schadendorf D., van Akkooi A.C.J., Berking C., Griewank K.G., Gutzmer R., Hauschild A., Stang A., Roesch A., Ugurel S. Melanoma. Lancet (London, England) 2018;392(10151):971–984. doi: 10.1016/S0140-6736(18)31559-9. [DOI] [PubMed] [Google Scholar]
- 2.Felton S., Taylor R.S., Srivastava D. Excision margins for melanoma in situ on the head and neck. Dermatol. Surg. 2016;42(3):327–334. doi: 10.1097/DSS.0000000000000648. 10.1097/DSS.0000000000000648. [DOI] [PubMed] [Google Scholar]
- 3.Haydu L.E., Lo S.N., McQuade J.L., Amaria R.N., Wargo J., Ross M.I.…Davies M.A. Cumulative incidence and predictors of CNS metastasis for patients with American Joint Committee on Cancer 8th Edition stage III melanoma. J. Clin. Oncol. 2020;38(13):1429. doi: 10.1200/JCO.19.01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gershenwald J.E., Scolyer R.A., Hess K.R., Sondak V.K., Long G.V., Ross M.I.…Sommariva A. Melanoma staging: evidence‐based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA A Cancer J. Clin. 2017;67(6):472–492. doi: 10.3322/caac.21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marzagalli M., Ebelt N.D., Manuel E.R. vol. 59. Academic Press; 2019, December. Unraveling the crosstalk between melanoma and immune cells in the tumor microenvironment; pp. 236–250. (Seminars in Cancer Biology). [DOI] [PubMed] [Google Scholar]
- 6.Camisaschi C., Vallacchi V., Castelli C., Rivoltini L., Rodolfo M. Immune cells in the melanoma microenvironment hold information for prediction of the risk of recurrence and response to treatment. Expert Rev. Mol. Diagn. 2014;14(6):643–646. doi: 10.1586/14737159.2014.928206. [DOI] [PubMed] [Google Scholar]
- 7.Falcone I., Conciatori F., Bazzichetto C., Ferretti G., Cognetti F., Ciuffreda L., Milella M. Tumor microenvironment: implications in melanoma resistance to targeted therapy and immunotherapy. Cancers. 2020;12(10):2870. doi: 10.3390/cancers12102870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Q., Wu X., Wang Z., Chen X., Wang L., Lu Y.…Ye L. The primordial differentiation of tumor-specific memory CD8+ T cells as bona fide responders to PD-1/PD-L1 blockade in draining lymph nodes. Cell. 2022;185(22):4049–4066. doi: 10.1136/jitc-2021-004022. [DOI] [PubMed] [Google Scholar]
- 9.Steele M.M., Jaiswal A., Delclaux I., Dryg I.D., Murugan D., Femel J.…Lund A.W. T cell egress via lymphatic vessels is tuned by antigen encounter and limits tumor control. Nat. Immunol. 2023:1–12. doi: 10.1038/s41590-023-01443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S., Yu H., Li J., et al. Identification of prognostic and tumor microenvironment by shelterin complex-related signatures in oral squamous cell carcinoma[J] Oxid. Med. Cell. Longev. 2022:2022. doi: 10.1155/2022/6849304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong M., Cui X., Wang G., Zhang Q., Li X. Development of a prognostic signature based on immune-related genes and the correlation with immune microenvironment in breast cancer. Aging (Albany NY) 2022;14(13):5427. doi: 10.18632/aging.204158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng N., Guo C., Wang Y., et al. Characterization of aging-related genes to predict prognosis and evaluate the tumor immune microenvironment in malignant melanoma. Journal of oncology. 2022;2022 doi: 10.1155/2022/1271378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng N., Ma L., Cheng Y., Xia Q., et al. Construction of a ferroptosis-related gene signature for predicting survival and immune microenvironment in melanoma patients. Int. J. Gen. Med. 2021;14:6423–6438. doi: 10.2147/IJGM.S327348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9(1):1–13. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2016;18:248–262. doi: 10.1016/j.celrep.2016.12.019. 2017. [DOI] [PubMed] [Google Scholar]
- 16.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harbers L., Agostini F., Nicos M., Poddighe D., Bienko M., Crosetto N. Somatic copy number alterations in human cancers: an analysis of publicly available data from the cancer genome atlas. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.700568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pane A.A., Kordaß T., Hotz‐Wagenblatt A., Dickes E., Kopp‐Schneider A., Will R.…Eichmüller S.B. MicroRNAs affecting the susceptibility of melanoma cells to CD8+ T cell‐mediated cytolysis. Clin. Transl. Med. 2023;13(2):e1186. doi: 10.1002/ctm2.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martchenko S.E., Martchenko A., Biancolin A.D., et al. L-cell Arntl is required for rhythmic glucagon-like peptide-1 secretion and maintenance of intestinal homeostasis[J] Mol. Metabol. 2021;54 doi: 10.1016/j.molmet.2021.101340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie J., Zhu C.L., Wan X.J., Zhao Z.Z., Meng Y., Li P.…Wang J.F. GSDMD‐mediated NETosis promotes the development of acute respiratory distress syndrome. Eur. J. Immunol. 2022 doi: 10.1002/eji.202250011. [DOI] [PubMed] [Google Scholar]
- 22.Ye X., Zhang P., Zhang Y., Luan J., Xu C., Wu Z.…Hu W. GSDMD contributes to myocardial reperfusion injury by regulating pyroptosis. Front. Immunol. 2022:5418. doi: 10.3389/fimmu.2022.893914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Son J.E., Dou Z., Kim K.H., Hui C.C. Deficiency of Irx5 protects mice from obesity and associated metabolic abnormalities. Int. J. Obes. 2022;46(11):2029–2039. doi: 10.1038/s41366-022-01221-0. [DOI] [PubMed] [Google Scholar]
- 24.Li H., Li M., Chen K., Li Y., Yang Z., Zhou Z. The circadian clock gene ARNTL overexpression suppresses oral cancer progression by inducing apoptosis via activating autophagy. Med. Oncol. 2022;39(12):1–9. doi: 10.1007/s12032-022-01832-7. [DOI] [PubMed] [Google Scholar]
- 25.Yeh C.M., Shay J., Zeng T.C., Chou J.L., Huang T.H.M., Lai H.C., Chan M.W. Epigenetic silencing of ARNTL, a circadian gene and potential tumor suppressor in ovarian cancer. Int. J. Oncol. 2014;45(5):2101–2107. doi: 10.3892/ijo.2014.2627. [DOI] [PubMed] [Google Scholar]
- 26.Peng H., Zhang J., Zhang P.P., Chen L., Tang L.L., Yang X.J.…Ma J. ARNTL hypermethylation promotes tumorigenesis and inhibits cisplatin sensitivity by activating CDK5 transcription in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2019;38(1):1–14. doi: 10.1186/s13046-018-0997-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Gamal R., Abdelrahim M., El-Sherbiny M., Enan E.T., El-Nablaway M. Gasdermin D: a potential mediator and prognostic marker of bladder cancer. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.972087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmquist Mengelbier L., Lindell‐Munther S., Yasui H., Jansson C., Esfandyari J., Karlsson J.…Gisselsson D. The Iroquois homeobox proteins IRX3 and IRX5 have distinct roles in Wilms tumour development and human nephrogenesis. J. Pathol. 2019;247(1):86–98. doi: 10.1002/path.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somerville T.D., Simeoni F., Chadwick J.A., Williams E.L., Spencer G.J., Boros K.…Somervaille T.C. Derepression of the Iroquois homeodomain transcription factor gene IRX3 confers differentiation block in acute leukemia. Cell Rep. 2018;22(3):638–652. doi: 10.1016/j.celrep.2017.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasahira T., Kurihara M., Nishiguchi Y., Fujiwara R., Kirita T., Kuniyasu H. NEDD 4 binding protein 2-like 1 promotes cancer cell invasion in oral squamous cell carcinoma. Virchows Arch. 2016;469(2):163–172. doi: 10.1007/s00428-016-1955-4. [DOI] [PubMed] [Google Scholar]
- 31.Zhang D., Wu P., Zhang Z., An W., Zhang C., Pan S.…Xu H. Overexpression of negative regulator of ubiquitin-like proteins 1 (NUB1) inhibits proliferation and invasion of gastric cancer cells through upregulation of p27Kip1 and inhibition of epithelial-mesenchymal transition. Pathol. Res. Pract. 2020;216(8) doi: 10.1016/j.prp.2020.153002. [DOI] [PubMed] [Google Scholar]
- 32.Hosono T., Tanaka T., Tanji K., Nakatani T., Kamitani T. NUB1, an interferon-inducible protein, mediates anti-proliferative actions and apoptosis in renal cell carcinoma cells through cell-cycle regulation. Br. J. Cancer. 2010;102(5):873–882. doi: 10.1038/sj.bjc.6605574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H., Yu P., Tomar V.S., Chen X., Atherton M.J., Lu Z.…Fuchs S.Y. Targeting PARP11 to avert immunosuppression and improve CAR T therapy in solid tumors. Nature Cancer. 2022:1–13. doi: 10.1038/s43018-022-00383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo S.J., Chien S.Y., Lin C., Chan S.E., Tsai H.T., Chen D.R. Significant elevation of CLDN16 and HAPLN3 gene expression in human breast cancer. Oncol. Rep. 2010;24(3):759–766. doi: 10.3892/or_00000918. [DOI] [PubMed] [Google Scholar]
- 35.Mei Y., Chen M.J.M., Liang H., Ma L. A four-gene signature predicts survival and anti-CTLA4 immunotherapeutic responses based on immune classification of melanoma. Commun. Biol. 2021;4(1):1–12. doi: 10.1038/s42003-021-01911-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C., Dang D., Wang Y., Cong X. A nomogram combining a four-gene biomarker and clinical factors for predicting survival of melanoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.593587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng Y., Yuan D., Zhang F., Tang R. A systematic pan-cancer analysis of the gasdermin (GSDM) family of genes and their correlation with prognosis, the tumor microenvironment, and drug sensitivity. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.926796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolchok J.D., Chiarion-Sileni V., Gonzalez R., Grob J.J., Rutkowski P., Lao C.D.…Hodi F.S. Long-term outcomes with nivolumab plus ipilimumab or nivolumab alone versus ipilimumab in patients with advanced melanoma. J. Clin. Oncol. 2022;40(2):127–137. doi: 10.1200/JCO.21.02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long G.V., Luke J.J., Khattak M.A., de la Cruz Merino L., Del Vecchio M., Rutkowski P.…Ascierto P.A. Pembrolizumab versus placebo as adjuvant therapy in resected stage IIB or IIC melanoma (KEYNOTE-716): distant metastasis-free survival results of a multicentre, double-blind, randomised, phase 3 trial. Lancet Oncol. 2022;23(11):1378–1388. doi: 10.1016/S1470-2045(22)00559-9. [DOI] [PubMed] [Google Scholar]
- 40.Yu X., Huang X., Chen X., Liu J., Wu C., Pu Q.…Zhou L. vol. 11. Taylor & Francis; 2019. Characterization of a novel anti-human lymphocyte activation gene 3 (LAG-3) antibody for cancer immunotherapy; pp. 1139–1148. (MAbs). 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dixon K.O., Tabaka M., Schramm M.A., Xiao S., Tang R., Dionne D.…Kuchroo V.K. TIM-3 restrains anti-tumour immunity by regulating inflammasome activation. Nature. 2021;595(7865):101–106. doi: 10.1038/s41586-021-03626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flem‐Karlsen K., Tekle C., Andersson Y., Flatmark K., Fodstad Ø., Nunes‐Xavier C.E. Immunoregulatory protein B7‐H3 promotes growth and decreases sensitivity to therapy in metastatic melanoma cells. Pigment cell & melanoma research. 2017;30(5):467–476. doi: 10.1111/pcmr.12599. [DOI] [PubMed] [Google Scholar]
- 43.Picarda E., Ohaegbulam K.C., Zang X. Molecular pathways: targeting B7-H3 (CD276) for human cancer ImmunotherapyCancer immunotherapies against B7-H3. Clin. Cancer Res. 2016;22(14):3425–3431. doi: 10.1158/1078-0432.CCR-15-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bao R., Wang Y., Lai J., Zhu H., Zhao Y., Li S.…Liu Z. Enhancing anti-PD-1/PD-L1 immune checkpoint inhibitory cancer therapy by CD276-targeted photodynamic ablation of tumor cells and tumor vasculature. Mol. Pharm. 2018;16(1):339–348. doi: 10.1021/acs.molpharmaceut.8b00997. [DOI] [PubMed] [Google Scholar]
- 45.Aggarwal C., Prawira A., Antonia S., Rahma O., Tolcher A., Cohen R.B.…Lakhani N.J. Dual checkpoint targeting of B7-H3 and PD-1 with enoblituzumab and pembrolizumab in advanced solid tumors: interim results from a multicenter phase I/II trial. Journal for immunotherapy of cancer. 2022;10(4) doi: 10.1136/jitc-2021-004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shenderov E., Demarzo A., Boudadi K., Allaf M., Wang H., Chapman C.…Harb R. Phase II neoadjuvant and immunologic study of B7-H3 targeting with enoblituzumab in localized intermediate-and high-risk prostate cancer. J. Clin. Oncol. 2018;36:15. doi: 10.1200/JCO.2018.36.15. [DOI] [Google Scholar]
- 47.Rizvi N.A., Loo D., Baughman J.E., Yun S., Chen F., Moore P.A.…Tolcher A.W. 2016. A Phase 1 Study of Enoblituzumab in Combination with Pembrolizumab in Patients with Advanced B7-H3-Expressing Cancers. [DOI] [Google Scholar]
- 48.Nishida M., Yamashita N., Ogawa T., Koseki K., Warabi E., Ohue T.…Udono H. Mitochondrial reactive oxygen species trigger metformin-dependent antitumor immunity via activation of Nrf2/mTORC1/p62 axis in tumor-infiltrating CD8T lymphocytes. Journal for immunotherapy of cancer. 2021;9(9) doi: 10.1136/jitc-2021-002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao M., Zeng F., Li Y., Gao Q., Yin M., Deng G., et al. A novel predictive model incorporating immune-related gene signatures for overall survival in melanoma patients. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-69330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen X., Shang L., Han J., Zhang Y., Niu W., Liu H., Shi H. Immune-related gene signature associates with immune landscape and predicts prognosis accurately in patients with skin cutaneous melanoma. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.1095867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang R., Mao M., Lu Y., Yu Q., Liao L. A novel immune-related genes prognosis biomarker for melanoma: associated with tumor microenvironment. Aging (Albany NY) 2020;12(8):6966. doi: 10.18632/aging.103054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meng L., He X., Zhang X., Zhang X., Wei Y., Wu B.…Xiao Y. Predicting the clinical outcome of melanoma using an immune-related gene pairs signature. PLoS One. 2020;15(10) doi: 10.1371/journal.pone.0240331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.