Abstract
In recent years, many research groups have begun to utilize bioengineered in vitro models of cancer to study mechanisms of disease progression, test drug candidates, and develop platforms to advance personalized drug treatment options. Due to advances in cell and tissue engineering over the last few decades, there are now a myriad of tools that can be used to create such in vitro systems. In this review, we describe the considerations one must take when developing model systems that accurately mimic the in vivo tumor microenvironment (TME) and can be used to answer specific scientific questions. We will summarize the importance of cell sourcing in models with one or multiple cell types and outline the importance of choosing biomaterials that accurately mimic the native extracellular matrix (ECM) of the tumor or tissue that is being modeled. We then provide examples of how these two components can be used in concert in a variety of model form factors and conclude by discussing how biofabrication techniques such as bioprinting and organ-on-a-chip fabrication can be used to create highly reproducible complex in vitro models. Since this topic has a broad range of applications, we use the final section of the review to dive deeper into one type of cancer, glioblastoma, to illustrate how these components come together to further our knowledge of cancer biology and move us closer to developing novel drugs and systems that improve patient outcomes.
Graphical Abstract
1. Introduction
Improvements in cell culture techniques and tissue engineering technologies have led to increased acceptance of in vitro models of cancer for use in basic research on disease initiation and progression and the development and testing of novel therapeutics. Compared to animal models, these in vitro systems have traditionally been considered relatively simplistic. For many years, researchers have used simple cell culture models to study the mechanisms of cancer and to evaluate drug compounds. However, the lack of crucial components of the tumor microenvironment (TME), including the extracellular matrix (ECM) and a myriad of tumor-adjacent and tumor-infiltrating cell types, limits the clinical and translational utility of these systems.1,2 The majority of 3D tumor models are an improvement over these 2D cultures as they reproduce many of the complex cell-cell, and in some cases cell-matrix, interactions that occur in vivo. Advances in biomaterials development, biofabrication, and cellular engineering have promoted the development of more complex in vitro systems that more accurately mimic the native TME.3,4 Furthermore, realization of organ-on-a-chip technology not only allows us to study mechanisms and phenomenon such as invasion and migration within a single tumor, but can also be used to create “multi-organ” systems that can be used to study phenomena such as metastasis and off target drug metabolism.5,6 Techniques such as bioprinting now allow scientists to manufacture complex tissue structures outside the body with high levels of reproducibility.4 With recent advances in stem cell culture, we are now able to reliably generate many human cell types found in the TME and are moving the field closer to developing personalized cancer models.7,8 The development and implementation of these new techniques have led to new and exciting discoveries in the field of cancer biology. Yet, it can often be a daunting task to navigate these systems and choose the one that is appropriate for a particular research question.
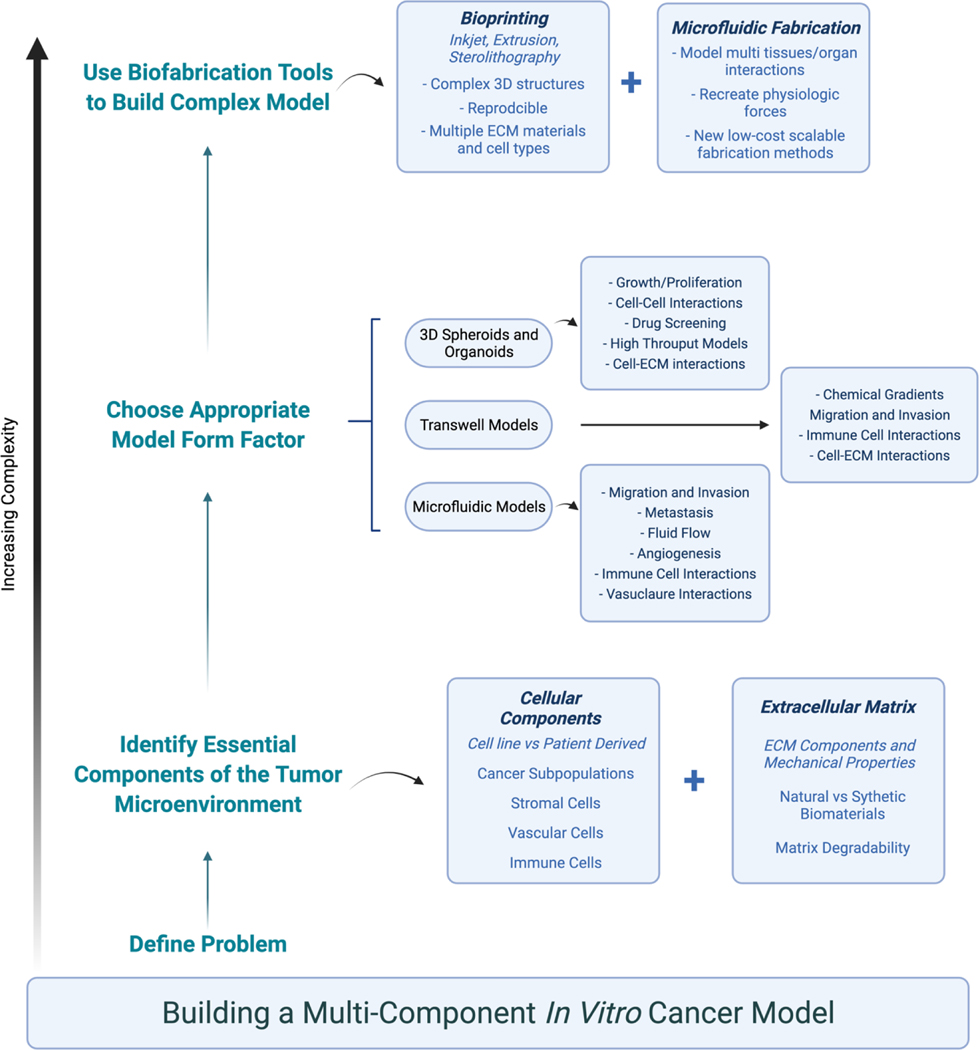
Our group is one of many groups utilizing these techniques to generate in vitro cancer models.9–12 In general, when we look to investigate a novel research question or develop a new model for a given cancer environment, we use the following workflow, which is represented in further detail in Figure 1. We first determine what cellular and extracellular matrix components are present in the tumor microenvironment (TME) of the tumor we would like to model. These include elements such as essential stromal cells, various tumor subpopulations, ECM proteins, and mechanical properties such as fluid flow or matrix stiffness. We then decide which components are most essential to answer our specific question and choose ECM-based materials needed to replicate those conditions. Finally, these components are combined in a specified model form factor. In some cases, biofabrication techniques are used to automate the process or create more complex systems. It is important to emphasize that when deciding what variables to incorporate and which form factor to use, complexity is not always better. In fact, there are many instances where simple systems have been used successfully. However, it has become increasingly clear that 2D cell culture of both cell lines and patient-derived tumor cells does not accurately mimic the patient tumor,1,2 so most of the models, examples, and form factors discussed herein pertain to 3D in vitro models.
Figure 1.
A visual representation of the workflow and considerations in building in vitro cancer model systems.
In this review, we will outline this approach and provide examples illustrating how a variety of tissue engineering and biofabrication technologies have been used to create multi-component in vitro models of cancer. We will conclude by describing how these tools have been used to study glioblastoma (GBM) in order to demonstrate the utility of these technologies in a specific cancer.
2. Choosing Essential Model Components: Cells and ECM
2.1. Cell source and cellular composition
When creating an in vitro cancer model, one must first consider the cell source or sources that will be used. For many decades, cancer cell lines were the preferred option for many researchers because they are easy to maintain in culture and can be propagated almost indefinitely. Since HeLa cells, cervical carcinoma cells removed from the tumor of Henrietta Lacks, were first cultured in 1951,13 hundreds of cell lines derived from a variety of cancers have been produced and characterized.14 These cells have proved invaluable in the study of cancer mechanisms and the development of new treatments.15 However, studies using cancer cell lines have been the subject of extensive criticism because many common cell lines have been contaminated with other cell sources.16,17 These problems can largely be addressed with routine genomic testing of cell lines from distributers and research labs that use the cells. More recently, many critics of cell lines have also argued that they are not accurate phenotypic and genetic representations of the in vivo tumor.18 These concerns are valid as studies have specifically shown long term culture on plastic can lead to genetic changes in cells and that patient derived cells are better indicators of a patient specific drug response.19 However, advances in genomics and proteomics and recent efforts such as the Cancer Cell Line Encyclopedia (CCLE) have made characterizing cancer cell lines much easier and more accessible.14,20 With these data, scientists are better able to evaluate possible clinical implications of a study using cell lines. For example, we recently used a set of four GBM cell lines paired with a hyaluronic acid-based ECM hydrogel to evaluate how environmental elastic modulus dynamically influenced cell proliferation.21 In the context of colorectal cancer, there have been numerous studies conducted in recent years developing a “multi-omics” profile of various commonly colorectal cancer cell lines.22–24 In breast cancer research, several cell lines have been used. There have been multiple efforts in recent years to gain a comprehensive understanding of their genomic and phenotypic signatures to insure that these lines are used appropriately.25–27 One particular study evaluated the utility of certain breast cancer cell lines for use in metastasis studies finding that some more closely matched in vivo signature than others.28 These studies have indicated that cell lines can be used to predict drug response when proteomic and genetic profiles of the cell lines are compared to that of patient tumors. This is certainly not always the case for all cancers, such as GBM, which we will address in a future section, for which predictive in vitro studies and omics methodologies have failed to translate into significant improvements in patients.29–31
Especially in the context of developing personalized treatments for cancer patients, the use of cells obtained directly from patient tumor biopsies in basic and translational research applications has continued to grow.32,33 However, it can be difficult to culture cancer cells from solid tumors in traditional 2D cultures, with successful culture rates reported in around 25% of samples.32,33 More recently, 3D culture techniques have expanded greatly. Higher success rates (60–90%) have been achieved for a variety of tissues using 3D cancer organoids which in this paper we will define as models in which patient derived cancer cells are grown in a 3D culture environment containing a supporting ECM hydrogel12,34,35. These 3D models will be addressed in greater detail later. Many studies have shown that these organoid cultures of primary cells replicate in vivo response to treatment.36 For example, our group has demonstrated this using mesothelioma and melanoma patient cells grown in organoids, while other groups have demonstrated similar outcomes in gastrointestinal tumor organoids.37–39 These platforms, while slightly more time consuming to generate, have proven extremely effective at replicating in vivo tumor response when treated with radiation, standard chemotherapeutics, and most recently, immunotherapies.20,40,41 the use of patient derived in vitro cancer models will continue to grow, and they will serve as essential tools to aid in the development and optimization of more targeted personalized therapies.
The cellular make-up of the TME is quite heterogenous, and it has become increasingly clear that tumor cells are not alone responsible for cancer initiation, progression, metastasis, and drug resistance. The stromal cells present in and around the tumor interact with cancerous cells through a variety of mechanisms including the secretion of growth factors and other bioactive molecules and ECM remodeling.42,43 These cells include cancer associated fibroblasts (CAFs), immune cells, endothelial cells, and pericytes. Inclusion of these cells in in vitro models, especially in 3D models which we will discuss in greater detail in the next section, has shown great promise in improving the accuracy of cancer models.8 For example, support cells, such as hepatic stellate cells, added to 3D in vitro models induce fibrotic changes in the ECM impacting tumor cell phenotype and reaction to chemotherapeutics.44 Including CAFs in 3D culture models has allowed researchers to investigate their impact on tumor cell migration and invasion.45 There are several research groups that have also used a similar approach and found that by adding CAFs to their model, the model more accurately mimics the native TME and serves a better predictor of in vivo drug response.46 The Tavana group has published a series of studies In which they used a high throughput co-culture model to investigate how the interactions between triple negative breast cancer cells and CXCL-12 secreting fibroblasts promoted cancer invasion and drug resistance.47–50 Most of the studies in this area have been conducted using cell lines, but there has been a recent push to start using patient-derived CAFs and other cells within these cancer models. In vitro models using patient-derived CAFs have been used to show that CAFs support the survival of cancer cells via the production of essential signaling factors and metabolites in a variety of cancers.51,52 Chung et al. created patient-derived 3D constructs containing tumor cells and CAFs isolated from a primary and a metastatic breast cancer tumor and showed that brain metastatic CAFs are a potential therapeutic target to prevent metastisis.53 A more thorough analysis of heterogeneity in engineered cancer models can be found in a previous review from our lab.54
There is also increased interest in adding other cells found in the TME to in vitro systems including immune cells. In vitro human models serve as great tools to test efficacy of novel immune therapies and study the impact of immune cell infiltration versus tumor growth.41,55 While maintaining and expanding many of these cells, such as T-cells, can be difficult, in vitro models provide a distinct advantage over more complicated animal models because all components are human derived, and they allow for the isolation of specific variables. In one model, researchers used 3D spheroid cultures to show pancreatic tumor cells and fibroblasts induce M2 macrophage polarization which suppresses T-cell proliferation.56 Importantly, this same study also indicated that the level of activation was cell line dependent, again emphasizing the need for patient specific assays.56 Some types of immune cells are particularly difficult to grow in vitro, but recent efforts have developed systems in which patient derived tumor organoids that contain native immune cells.38,57 Another recent study showed that tumor-reactive T cells can be enriched in vitro by patient tumor cells cultured in 3D with peripheral blood lymphocytes.58 This demonstrates utility of in vitro models, simple and complex, for mechanistic studies investigating tumor-immune interactions and clinical applications such as personalized medicine. Of course, to completely model tumor-immune interactions, one must include an active endothelial barrier including pericytes and endothelial cells. Advances in microfluidics, which we will discuss later, have allowed for the inclusion of these cell types in physiologically relevant systems to study processes such as angiogenesis, immune infiltration, and metastasis.59–61
Determining the ideal cell populations for a given in vitro tumor model is an important decision. With recent advances through large scale omics studies, cell lines can provide useful information on disease mechanisms and are easy to produce. While the inclusion of multiple cell types can help make these models more physiologically relevant, extended culture on 2D plastic prevents cancer cell lines from mimicking the native tumor cells.2 Patient cells are more difficult to grow, but they are a better option for personalized medicine applications, and when used in large scale studies, can provide invaluable mechanistic data about cancer biology that is impossible to achieve completely using cell lines. While not discussed in depth here, there has also been a recent push to start using induced pluripotent stem cell derived cell populations to improve model efficacy and create even better patient specific models.62 These cells can be combined in a variety of 3D systems to model different cancer mechanisms.
2.2. Materials to model the tumor microenvironment
The ECM provides both a structure for cells to grow and interact but is also intimately involved in cell signaling and tumor behavior and progression. Compared to healthy tissue microenvironments, the TME often manifests with significantly altered expression level ratios of ECM components, including collagens, hyaluronic acid, fibronectin, and laminins.63 This often leads to increased tissue stiffness and ECM density, as well as differences in composition, all of which result in altered matrix-to-cell signaling.64,65 The resident stromal cells within the tumor, notably CAFs, play a major role in remodeling the ECM, often spurred on by inflammation caused by both immune and tumor cells.8,66 Perhaps most importantly, changes in the ECM composition of tumors have been shown to impact treatment efficacy by physically shielding cells from therapies and initiating processes such as epithelial to mesenchymal transition (EMT) and metastasis.67,68 To create more accurate in vitro cancer models and study the ways in which the ECM affects cancer mechanisms and drug sensitivity, researchers use a variety of biomaterials in 3D models mimic physical alterations to the TME.
The same porous scaffolds that are used in many other areas of tissue engineering and regenerative medicine are applicable to in vitro cancer modelling.69–71 Generally, these materials are broken into 2 different categories: natural and synthetic. Natural materials are derived from polysaccharides or proteins purified from human tissue or produced by other biological processes. Synthetic hydrogels are made from polymers that are synthesized in the lab. Both natural and synthetic materials are often chemically modified to form a crosslinked network.72 Each system has their own advantages and disadvantages. For one, synthetic materials allow for greater control over physical and mechanical properties since the molecular weight of the components and crosslinking density can be tightly controlled. However, cells cannot always recognize and remodel the network. Integrin binding sequences and degradable crosslinkers have been added to these synthetic networks to increase cell binding and matrix remodeling.73,74 However, many cell-ECM interactions rely on specific protein conformations can be difficult to completely mimic with synthetic materials. Naturally-derived ECM biomaterials are comprised of one or more polysaccharides and proteins that occur natively in the extracellular environment which provide structure or can promote cell adhesion and cell signaling. However, these materials are not always completely defined making it difficult to control the chemical and mechanical properties of the network.75 For example, Matrigel, a commonly used matrix composed mainly of collagen IV, laminin, and entactin, contains an undefined array of growth factors and cytokines that enable spontaneous formation of acini or ductile epithelial structures. This unknown component combined with batch-to-batch variabilities act as confounding variables that cannot always be accounted for.76
Natural materials such as collagen, Matrigel, or ECM from decellularized tissues were used to create the first in vitro 3D cancer models.77,78 These were effective for early in vitro drug studies because most cell types are able to actively remodel theses environments and the stiffness can be modulated to a certain degree by simply altering the protein concentration. However, the ECM composition cannot be modulated. Therefore, many groups have used modified versions of these natural materials which has allowed them to design systems with specific mechanical properties and ECM binding and signaling molecules.72 Our group has used a modular hydrogel system composed of hyaluronic acid, gelatin, and collagen to create models of a variety of different cancers.9,12,79,80 The composition of these hydrogels can be tuned based on the tissue type. For example, our models of brain tumors do not contain collagen I as it is not present in large qualities in the brain.21 Recently, we have experimented with tuning the relative levels of synthetically modified fibronectin and laminin to further customize the TME, while keeping the system well-defined – an important characteristic for translation to clinically oriented applications. The stiffness of HA hydrogels can also be manipulated to match the desired tissue or study how changes affect cell behavior.81,82 Chemical modifications to HA and other materials also allows for the distribution and patterning of chemical factors to form gradients across the engineered tissue which can be used to study a variety of signaling cascades in cancer.83 Synthetic materials have been used in a similar manner, with researchers using them to investigate the impact of various properties of the ECM on cancer cell behavior and treatment response. In one major study, the stiffness, degradability, and ECM components in PEG hydrogels were manipulated to investigate how they affected the formation of intestinal stem cell and colorectal cancer organoids.84 It has also been shown that creating an engineered PEG hydrogel system that matches the properties of ovarian cancer accurately recapitulates in vivo drug response showing the importance of creating models with mechanical properties that mimic native tissue.85 PEG hydrogels have also been used to study the relationship between matrix degradation on cancer cell growth. For example, PEG hydrogel cultures were used to demonstrate that treatment melanoma cells with BRAF protein kinase inhibitor leads to increased MMP mediated degradation and migration.86 The modular nature of these hydrogel systems provides control over the environment allowing scientists to design systems for their specific tissue and experiment.
When designing a model, it is important to consider the composition of both the healthy and cancerous ECM. One must also consider the mechanical properties of the materials. It is likely that there are multiple options that could be used to produce a model to answer a specific set of questions. In that case, the scientists should take factors such as familiarity with the material, ease of production, cost, and ease of use into consideration. Once a subset of materials has been chosen, the scientific question that is being asked, the model system that is being used, and the ways in which that model will be fabricated will all impact the materials and the respective chemical manipulations that are needed.
3. Tumor model form factors
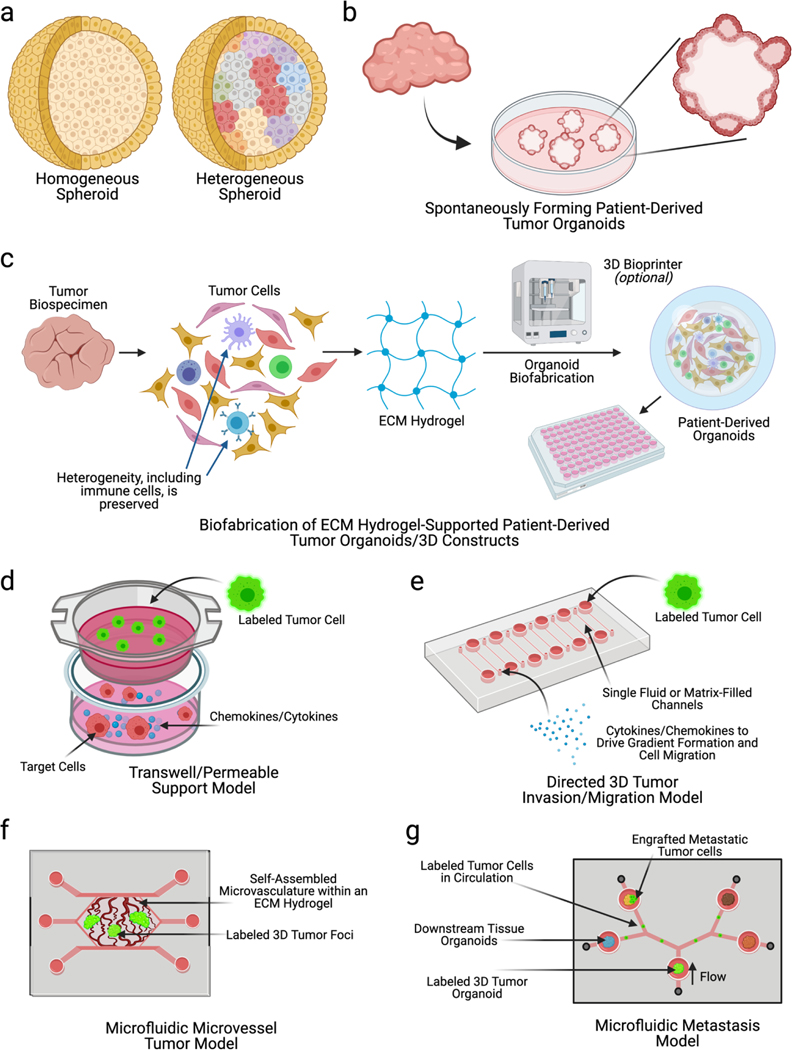
As previously stated, traditional 2D cell culture leads to genetic and phenotypic changes in cells and fails to mimic the complex 3-dimensoinal nature of the tumor-stroma-ECM interactions that occur within every solid tumor. As 3D culture has become more common, researchers now have many different model platforms at their disposal when moving from 2D culture to 3D systems. Each platform has its own advantages and disadvantages and are optimized to address specific scientific questions. Many of these systems were previously reviewed by Katt et al. in 2016.87 Here we describe these form factors, along with others, and provide some more recent examples from the literature. The form factors are also highlighted in Figure 2.
Figure 2: Overview of various form factors used in in vitro cancer modeling.
In vitro cancer models occupy a wide variety of forms. These range from relatively simple a) spheroids that can be homogeneous or heterogeneous to more complex b) organoids that spontaneously form in materials such as Matrigel. c) Other forms of organoids, or 3D tumor constructs, can be formed by encapsulating tumor cells, including those derived from a patient’s tumor, in an ECM hydrogel, which can then be subsequently deposited or bioprinted in small volumes to form organoids. d) Transwell inserts or permeable support wells have been widely used to assess tumor cell migration towards cytokines, chemokines, or other cell types. e-g) Microfluidic device technologies have rapidly advanced in cancer research, resulting in a wide variety of tumor-on-a-chip systems. These include e) directed 3D tumor cell invasion models, f) microvascular tissue chips containing self-organizing blood vessels and tumor foci, and g) metastasis-on-achip systems allowing tracking of tumor cells through microfluidic circulation to other tissue sites.
The most basic 3D model system is the spheroid, or an aggregate of cells. Traditionally, spheroids are produced using the hanging drop method where cells form aggregates in a suspended drop of fluid. The development of ultra-low attachment round bottom plates has made the process much simpler and replicable.88 The 3D structure of the spheroid replicates the cell-cell interactions seen in vivo. Early studies used spheroids to study cancer growth, and it was quickly revealed that these models were better representations of tumor biology than 2D cultures.2,87,89,90 For example, spheroid culture of colorectal and ovarian cancer cells produces higher levels of E-cadherin indicating that there are more physiologically relevant cell-cell interactions in these 3D cultures.91,92 However due to a lack of extracellular space to allow for sufficient diffusion kinetics, spheroids often develop a hypoxic core highlighting that these structures are limited in size.87,93 However, in some cases, this scenario can be leveraged to model the necrotic core often observed in solid tumors, with one study showing that the induction of a necrotic core in colorectal cancer spheroids better replicated in vivo drug response.94 Homogenous tumor spheroids have now been widely implemented for use in larger scale drug studies.93,95,96 Since the cells grown in spheroids secrete their own ECM, they can also be used to study ECM production and remodeling.97 However, spheroids do have some limitations. They are limited in size, there is little control over newly synthesized ECM, and there are often important cell types missing. It has also been observed that some cells are not capable of forming spheroids limiting the utility of the system. For example, in our team’s work with defined ECM hydrogel-based tumor organoids, we found that while we could easily encapsulate many patient-derived tumor cells in ECM hydrogels, thus forming this variety of organoid construct, Side experiments showed that many tumor types or grades did not readily form spheroids making this hydrogel based system advantageous for modeling certain tissue types.12,37,38,98,99
More complex 3D cultures have been created to address some of the problems with cancer spheroids. In these improved models, cells are suspended in a 3D hydrogel matrix as a single cell suspension, aggregates, or resected tissue chunks. Traditionally, these models were produced using Matrigel. At this point, we should note that over the last decade the term organoid has evolved, and has been used in a variety of different contexts to describe 3D culture models.12,37,38,98,100–106 As we previously mentioned, we primarily use the term “cancer organoid” or “tumor organoid” to describe models in which patient derived cancer cells are grown in a 3D culture environment containing a supporting ECM hydrogel. However, we and many other laboratories have published studies using the “organoid” nomenclature to also describe cell-matrix constructs in microfluidic devices – both primary cell and cell line-derived – as well as spheroids or hydrogel-based constructs containing multiple combinations of cell lines, cell lines and primary cells, and primary cells and induced pluripotent stem cell-derived differentiated cells. The driving feature that qualifies the term “organoid” being used is not the cellular makeup, but if advanced biological composition, physiology, and function can be accurately modeled using these platforms. Cells grown in organoids are able to remodel the matrix and produce a self-organized structure that can resemble the in vivo tumor, especially if other cell types such as CAFs are included.44,107 However as previously mentioned, the composition of Matrigel is somewhat undefined and when it is used, the researcher has limited control of the ECM composition of the tumor organoid.76 Therefore, many other groups have moved towards using more defined ECM hydrogels to create their cancer organoids.9,12,79,80 As previously discussed, the modular nature of these materials allows for precise manipulation of the ECM composition. By surrounding the a spheroid, tissue chunk, or cellularized hydrogel construct with an acellular hydrogel, cell migration can be studied in 3 dimensions.108 These ECM embedded structures serve as useful tools to continue to investigate mechanisms of cancer growth and produce more consistent platforms that can be used for testing drugs.
Invasion and migration can also be studied using a transwell based system (Figure 2d). These model systems have two distinct compartments, separated by a porous membrane. Therefore, it is relatively straightforward to create chemical or physical gradients across the membrane. The simplest of the transwell models consist simply of cells grown on one side of the semi-permeable membrane. When exposed to the given gradient, the cells may be stimulated to migrate through the pores to the other side.87,109–111 Movement of immune cells grown in suspension, such as T-cells or natural killer cells, can also be tested using a similar set-up.87,112–114 In many cases, the addition of an endothelial barrier to the transwell membrane is used to mimic the vascular endothelium. These models can be used to more accurately test factors that stimulate immune cell infiltration and cancer metastasis.115,116 The addition of a hydrogel on top of the transwell membrane can be used to more accurately simulate invasion of cancer cells, immune cells, or stromal cells through the ECM.117 These systems can be engineered to mimic the native tissue ECM or designed to investigate the effect of certain ECM molecules on cellular motility. As mentioned before, advances in hydrogel biomaterials engineering now allow for researchers to pattern various growth factors and proteins to form gradients in hydrogels which again could be used to investigate the mechanisms of invasion within these systems.83 Transwells have many valuable uses, but they are limited by their geometry and size and, because of the presence of a relatively stiff plastic membrane, do not always accurately mimic the physiological environment.
Over the past decade or so, the use of microfluidics, or organ-on-a-chip, platforms has increased dramatically. In general, organ-on-a-chip platforms have grown because they are able to produce features of the in vivo tissue environment which can be reproduced in the simpler models previously discussed.118–120 These conditions include fluid induced shear stresses, interstitial fluid flow, hydrostatic pressure, and controlled tissue deformation.5 Many microfluidic devices have been used to investigate similar problems discussed before including cell migration. These devices also consist of narrow, defined channels through which cell migration is driven by a chemo attractive, mechanical, or electrical gradients.121–124 By using a microfluidic device instead of an organoid or transwells based model, researchers have also been able to introduce other variables such as oxygen gradients and interstitial fluid flow.125,126 Analysis on microfluidics in real-time is also quite easy as most devices are optically transparent and can be imaged live. A variety of sensors have also been incorporated into these devices to take real time measurements of metrics such as electrical activity, protein concentrations, and mechanical forces.127,128
Microfluidics and organ-on-chip have also been used to model processes that require flow such as metastasis. For example, our group has generated microfluidic devices with flow in which circulating tumor cells from a central cancer organoid exit, enter fluid flow, and invade distal tissue hydrogel constructs (Figure 2g).11,129 This model and ones like it can be used to evaluate metastatic properties of a tumor and study tumor cell preferential homing to target tissue sites. Similar systems have also been designed to study premetastatic niche formation by evaluating the effect of cancer cells on matrix remodeling in a distal tissue hydrogel (with or without cells) in a separate chamber of the device.61,130,131 Increasing complexity further, many endothelialized vessel structures have been created within microfluidic models. These vascular models are formed in a few different ways. Some systems, namely those pioneered by the Kamm and George labs, consist of endothelial cells seeded with in an ECM matrix, often collagen I or Matrigel, which then spontaneously form an interconnected network of perusable vessels.132–134 Other systems have used engineered vessels formed on 2D membranes135 or in ECM hydrogels via molding with a rod or needle136, viscous finger patterning137, or the use of sacrificial hydrogels.138 The addition of the endothelial layer has been used to study immune cell infiltration and, of course, metastasis. For example, a self-assembling vascular model was used to investigate differences in breast cancer metastasis in different tissue types including bone and smooth muscle.139 A similar self-assembling vascular model was also used to investigate the relationship between M1 and M2 macrophages and cancer cell proliferation and tumor induced angiogeneisis.60 Nguyen et al. used a microfluidic vessel model consisting of 2 hollow lumen to study the mechanisms involved in endothelial ablation in pancreatic ductal adenocarcinoma.140 The differing devices used in these studies highlight how different methods for creating mimetic vasculature are used and implemented according to the specific problem at hand. Other more specialized models of tissue interfaces have been created such as lung-on-a-chip models with an air liquid interface to study lung cancer.141 There have also been several microfluidic models of the blood brain barrier (BBB) that are currently being adapted to study GBM which we will expand on later.142–144
When choosing which model format to use to investigate a given problem, it is important to consider whether the most complicated microfluidic device is necessary. As our group and many others have shown, spheroids or cancer organoids that can be produced in extremely large numbers with great reproducibility might be the best choice for a multi-drug screen or rapid personalized medicine applications. Microfluidic models may be necessary to investigate more complex questions such as factors that lead to ECM remodeling in the premetastatic niche or mechanisms of intravasation and extravasation in metastatic tumor models. A comprehensive understanding of the pros, cons, and previous uses of various biomaterials and model systems will inform the construction of a system that is problem specific. This will save time and money and produce elegant, streamlined results that can further add to the growing library of evidence supporting the effectiveness and utility of using in vitro models to study cancer.
4. Biofabrication Techniques to create in vitro models
The ways in which an in vitro model is fabricated can be just as important as the cells, the biomaterial, and the form factor chosen. Recent advances in biofabrication, or the processes involved in engineering with purpose 3D biological constructs through the use of cells, biomaterials, and technologies such as bioprinting, now give researchers even more flexibility and make producing 3D models more efficient.145 Informed use of these novel biofabrication tools can improve the model by increasing batch to batch consistency, facilitate high throughput screening of novel drug compounds, and allow for the production of models that better reproduce complex tissue structures. In the space of in vitro cancer modeling, bioprinting and organ-on-a-chip fabrication techniques are of particular interest. Streamlining both technologies making them accessible and affordable has the potential to increase implementation of complex in vitro models for research, development and production of novel therapeutics, and rapid personalized medicine.
4.1. Bioprinting
Bioprinting is the use of additive manufacturing techniques to produce biocompatible, and sometimes cellularized, 3D structures.146–149 Bioprinting technologies have improved significantly over the past 2 decades leading to greater ease of use and printed structures with higher resolution and complexity.150 Bioprinters printers were initially extremely expensive limiting the number of laboratories that could afford to develop and utilize the technology. In part because of the “maker revolution”, additive manufacturing tools have become much more affordable.75,151 Likewise, advances in biomaterial bioink technologies has enabled the use of biomimetic hydrogels as bioinks in bioprinting while better simulating native ECM.75,152 While not ubiquitous in the field of cancer engineering, more laboratories have embraced bioprinting because it allows for increased throughput of tumor model fabrication.70,104,153
3D bioprinting hardware can be broken down into 3 main categories: inkjet, extrusion, and stereolithography.154 Many of the first studies using bioprinting in the early 2000’s made use of commercial inkjet printers that were modified to dispense simple ECM materials and cell-laden bioinks with mediocre resolution.155,156 Conversely, inkjet printers operate by ejecting material through the printhead via thermal, mechanical, or piezo electrical processes producing small droplets.157 These processes are rapid and produce relatively high resolution features, but the ejection process can place high levels of stress on the cells.158 The hardware is also relatively inexpensive making the process accessible. Selection of a bioink for inkjet printing can be difficult because the material needs to have a low viscosity so that it can pass through the small printheads. This makes the formation of complex 3D structures difficult because most low viscosity materials fail to hold their shape post extrusion. Utilizing materials that can be crosslinked using UV light, changes in temperature, or changes in pH can address some of these problems.158,159 In the context of cancer modeling, inkjet printing has been used a variety of different systems. In one study, HeLa cells suspended in collagen matrix were printed on top of a layer fibroblasts to study how cell-cell interactions affect MMP production and drug response.160 While this study likely could have been conducted without the use of a bioprinter, it demonstrates that these machines can produce highly consistent samples and have utility in high throughput studies. The addition of multiple printheads to these printers also allows for the precise patterning of multiple cell types, such as cancer cells and CAFs, further improving the accuracy of these high throughput models.161
Extrusion printing is also widely used for bioprinting as it has become relatively affordable. Early extrusion printers were built by modifying standard non-biological 3D printers with new printheads, but now there are many bioprinting specific machines available commercially.75,162,163 These systems robotically control the extrusion of material which is deposited as a small bead on the print surface. The material can be pressed out of the printhead using either pneumatics or a screw piston.158 Similar to inkjet printing, the properties of the material used are extremely important. The bioink must be able to flow consistently through the printhead without clogging, but also hold its shape once deposited.159 Many standard biomaterials used to mimic the ECM such as Matrigel and Collagen I are not ideal for printing. Both materials have short “biofabrication windows”, or length of time in which they can flow through the printhead while still holding shape once deposited. To address this short window, materials like gelatin and alginate have been used because the crosslinking is controlled by change in temperature and exposure to calcium, respectively.164,165 However, these materials do not always support cell growth and/or mimic the native ECM. Therefore, there has been an increased effort to design modular bioinks that allow for successful printing and control over the chemical and mechanical properties. For example, our group has modified our hyaluronic acid-based hydrogels with alternative step-wise or reversible crosslinking methods to create shear thinning or thixotropic bioinks with extended biofabrication windows.75,104,162,166,167 Secondary crosslinking such as exposure to UV light can also be used to stabilize the extruded material.167 The Feinberg lab has also pioneered a technique called Freeform Reversible Embedding of Suspended Hydrogels (FRESH) in which material is extruded into a bath of shear thinning material, which can later be removed.168 These systems have still proven useful in the space of cancer modeling. As more research groups and companies begin to rely on spheroids and cancer organoids to test novel therapeutics, there has been a significant effort to develop methods of producing these constructs in a consistent high throughput manner. Our group and others have used extrusion bioprinters to create 3D constructs using patient derived tumor cells for high throughput drug screened.10 These techniques have even been used with patient derived tumor samples indicating that they could possibly be used in large scale personalized medicine applications in the future.153,169 However, extrusion printing is still limited by its resolution and a lack of extrudable bioinks with an appropriate level of complexity in terms of ECM components.
Light based bioprinting methods may address some of these challenges. These methods include, laser-assisted, stereolithography (SLA), digital light processing (DLP), and two-photon polymerization.158,170 All of these platforms require the use of a photo-crosslinkable bioink, but they do not require the same manipulation of fluid properties as inkjet and extrusion printing because they do not need to travel through small printhead apertures. With that being said, there are still several to these techniques. In general, light-based bioprinters can create complex geometries with high resolution. For this reason, many groups have proposed using SLA or DLP printing to create small perfusable vascularized tissue constructs.158,171 However, it can be difficult to achieve high resolution using softer hydrogels that are required to mimic many of the body’s tissues. There have not been many examples of cancer modeling as of yet, but there appear to be myriad possibilities. In one study, SLA printing was used to produce a bone scaffold to study interactions between osteocytes and breast cancer cells in bone metastasis.172 Another study used laser-assisted bioprinting to produce arrays of pancreatic spheroid organoids173, so these printing modalities can also be used to create high throughput assays for drug screening. As the cost of these printers continues to decrease, it is expected that there will be increased interest in developing compatible bioinks and use in cancer modeling.
Bioprinting for use in cancer modeling is still in its early stages. The studies highlighted above show the potential of these tools and techniques in the context of cancer engineering. As the field continues to grow, these systems will be applied to more cancers hopefully leading to the production of increasingly complex multi-component models that can be used to investigate novel questions.
4.2. Microfluidic models
Microfluidic devices have traditionally been produced using photolithography techniques originally developed for developing electrical and computer chips,174,175 hence the name organ-on-a-chip.5 Generally, a master mold is creating using a negative photoresist such as SU-8, which is then used to cast a soft polymer, usually polydimethylsiloxane (PDMS). Channel inlets and outlets are punched, and the PDMS mold is then bonded to a glass substrate. In these devices, cells are typically grown as a 2D monolayer after coating the channels with an ECM protein, or in 3D by injecting a hydrogel precursor solution into a specific hydrogel channel. To create more complex structures, photocrosslinkable hydrogels can be patterned in the device using a photomask.176 The uncrosslinked material is then washed out before adding media. We have used this method to create metastasis-on-a-chip models,100 a liver-on-a-chip for toxicity screens,177 and bone marrow-on-a-chip to evaluate healthy and malignant hematopoietic stem/progenitor cell homing to the bone marrow niche.178 Since the device materials – generally PDMS and glass – are transparent, images can be taken directly on the device via standard light and fluorescent microscopy, and even confocal microscopy. Endothelial barriers have also been created using variety of methods. In one commonly used device now commercially produced by Emulate, Inc., two channels are separated by a semi-permeable membrane on which cells can be seeded.179 Other groups have developed microfluidic platforms in which endothelial cells self-organize into a perfusable vascular network.132–134 These models have proven to be reliable, but the vessel network in each device is unique. To create more uniform vessels, many groups have taken to molding simple lumen into ECM hydrogels using needles or rods.136 Lumen have also been cast using PDMS rods to create bifurcated structures.180 Bioprinting can be used to create complex lumen structures either via extrusion bioprinting of a sacrificial hydrogel138 or by using light based printing.171 These advanced fabrication techniques have been combined to create some elegant models that have been used to study cancer. In one lung cancer-on-a-chip, a semipermeable membrane separating 2 channels was used to create an air-liquid interface. Two air channels separate from the cellular space were used create cyclical mechanical load on the membrane as it would experience in the human lung.141
Even with these incredible innovations, the advanced fabrication techniques required produce an insurmountable barrier to entry for many groups. Recent advances in low-cost high throughput fabrication techniques have allowed more researchers to apply microfluidics to their work. As 3D printing technologies continue to improve, some groups have begun using high-resolution SLA printing to produce their master molds. While these molds cannot yet produce the same high-resolution structures as traditional photolithography, they can be used repeatedly and are relatively low cost. Others have turned to more novel approaches using low-cost adhesives to form channels. One approach gaining popularity is based on patterning of adhesive films.181 A computer-controlled device such as a laser cutter or razor plotter is used to create a pattern in an adhesive film. These films are then attached to a glass substrate forming small channels. Multiple patterns can be layered on top of one another to create complex fluidic devices. These low-cost devices have been used extensively to create organ-on-a-chip models of many systems including cancer. These techniques, combined with photopatterning of hydrogels, has been to study the effects of drug treatment on colorectal cancer cell migration, to generate patient-derived mesothelioma-on-a-chip systems, and to support a multi-organoid “body-on-a-chip” platform to assess the side effects of liver organoid-facilitated metabolism of chemotherapy agents.37,105,182 Similar systems have also been used to create vessel lumen and could be used as an alternative to the more complex fabrication methods discussed earlier.183 Bioprinting is now also being used to pattern cells and ECM on these microfluidic devices,184 enabling researchers to create complex structures such as vasculature and pattern multiple cell types in physiologically relevant orientations. These techniques can be applied in concert with microfluidics fabrication to easily create models to study metastasis, tumor angiogenesis, drug delivery, and much more.
As we have repeatedly emphasized, advances in these biofabrication techniques will continue to drive the production of more physiologically relevant tissue models that can be used for both basic and translational research. Many have envisioned a future where in vitro models become so advanced that animal models are not necessary. To achieve this lofty goal, we must continue to investigate ways to reproduce in vivo microtissue architectures and vascularization. It is also essential that we create systems containing multiple tissue types and continue progressing towards a “body-on-a-chip” platform. It has already been demonstrated that systems containing 3D tissue constructs from multiple organs can be maintained together for many weeks and that the replicate simple in vivo processes.185,186 In the context of cancer research, developing these integrated models is key to studying areas such as pre-metastatic niche development, metastasis, and the effects drug metabolism on distal tissues. These systems should help propel us closer to the development of novel treatments and commercialization of in vitro models for drug testing and personalized medicine.
5.0. Using Multi-Component In Vitro Models to Study Glioblastoma
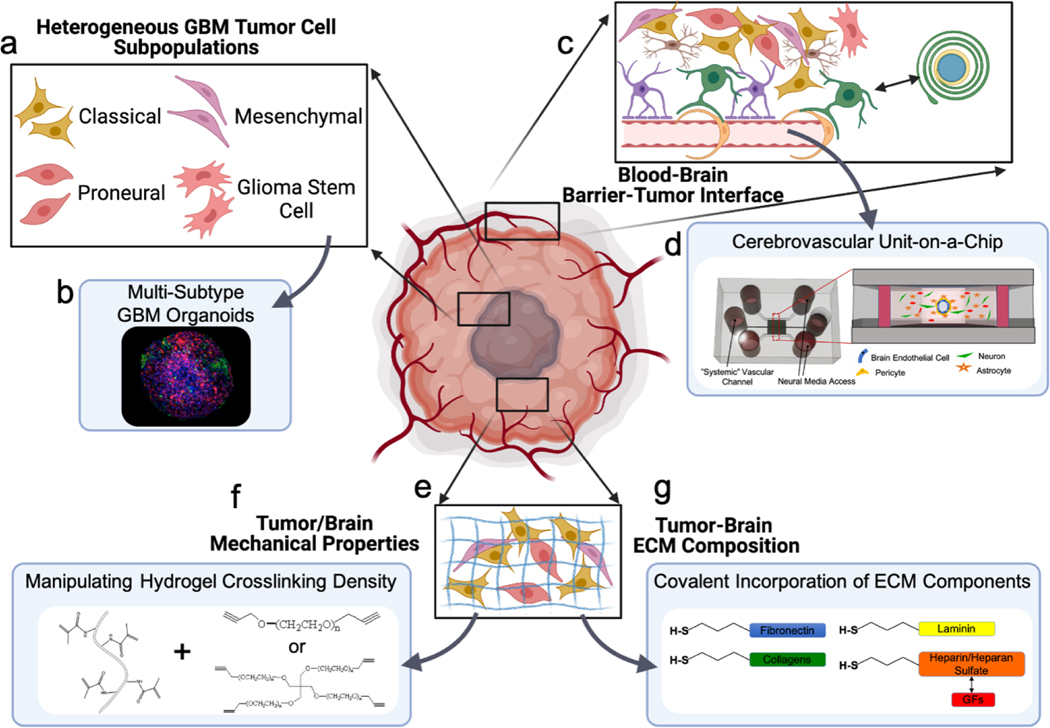
As with many cancers, the survival rate of glioblastoma patients continues to be abysmally low due in part to a lack of effective treatment. In the following section, we will outline the challenges that have faced researchers and clinicians studying GBM and highlight various examples of ways the tools we previously discussed have been used to create effective in vitro models. GBM specific considerations and model components are highlighted in Figure 3.
Figure 3. Overview of the components necessary to design complex multi-component models of glioblastoma.
a) Like many other cancers, the GBM TME consists of many unique cell types including tumor cells, glial cells, immune cells, and vascular cells. GBM tumors cells are also composed of multiple distinct subpopulations. b) These subpopulations can be labeled and combined in a spheroid or organoid culture and their growth and proliferation studied in response to treatment. c) The blood brain barrier plays a key role in regulating tumor growth and in the delivery of therapeutics to the tumor site. d) Microfluidic models have been created to model the BBB in vitro. e) The extracellular matrix in and around the tumor in GBM is different from that of the healthy brain. There is an increase in stiffness and deposition of certain ECM molecules such as hyaluronan and vitronectin. The stiffness (f) and the relative concentration of various ECM components (g) can be controlled via chemical modification of the hydrogel biomaterials used.
Glioblastoma (GBM, previously referred to as glioblastoma multiforme) is the most aggressive of the glial tumors and has a proclivity for necrosis, uncontrolled cellular proliferation, diffuse infiltration, increased angiogenesis, and widespread genomic heterogeneity.187 Current treatment strategies include chemotherapy, surgical resection, and radiotherapy which in most cases does not completely eradicate the disseminated infiltrated cancer cells resulting in a poor prognosis. The first choice of chemotherapy agent in these patients is temozolomide (TMZ). However, TMZ treatment invariably leads to development of resistance and increased likelihood of recurrence reducing the overall survival of diagnosed patients to 1.5 to 2 years.188
The blood brain barrier (BBB) is a physical and biochemical barrier that inhibits effective drug delivery in to both diseased and normal brain.189,190 Even though the BBB is disrupted in GBM, there is no evidence that this disruption increases drug bioavailability within the tumor with various studies reporting that a faction of the tumor cells resides in areas with intact BBB which limits access to drugs completely.191 The cancer cells induce neoangiogenesis with fenestrated endothelial cells lacking tight junction expression and co-opt the brain vasculature resulting in an anatomically and physiologically different barrier than the normal blood brain barrier.192 It has been established that the tumor compromises the integrity of the blood brain barrier surrounding it and promotes the formation of new poorly formed blood vasculature. This new barrier is referred to as the blood brain tumor barrier (BBTB) and it is quite different from the healthy BBB. Astrocytic endfeet displacement, aberrant pericyte distribution, and loss of astrocyte endfeet and neuronal connection along with leakier vasculature are the main characteristics of the blood tumor barrier. The BBTB/BBB structural and functional heterogeneity within the same tumor and among different patient tumors provides indicates that it is largely affected by the cellular, molecular, and the tumor subtype specific features of the glioblastoma. Various studies have demonstrated the effect of glioma on the blood brain barrier, so the inclusion of a functional BBTB/BBB in the model could help in developing more translational chemotherapeutics.193,194 In vitro models can be used to the characteristics of BBTB/BBB for the development of anticancer agents that can effectively cross them and improve treatment outcomes.195
The high degree of genetic, epigenetic, and cellular diversity observed in GBM tumors further complicates treatment. GBM tumors are classified into subtypes based on the common grouped set of neoplastic genetic alterations observed in them. The “original” four subtypes, proneural, neural, classical, and mesenchymal, were first established by Verhaak et al.196 Later, studies from the same group showed that the neural subtype is actually comprised of non-tumor cells which are thought to be from normal brain tissue contamination. 196,197 The classical subtype is characterized by amplification of the epidermal growth factor receptor (EGFR), inactivation of the retinoblastoma pathway, and high expression of neural stem markers. The mesenchymal subtype has a higher predilection for necrosis and inflammation with increased expression of families of genes involved in angiogenesis and tumor necrosis.198 More recently, it has been observed that this classification is not relevant to all the cells found in glioblastoma. Snuderl et al. showed that there is simultaneous amplification of multiple receptor tyrosine kinases such as EGFR and PDGFRA (platelet derived growth factor receptor A) in the same patient.197 Various subtypes are found in the same tumor with detrimental effects on treatment response.199 However, statistical analysis done by Coleman et al. demonstrated that the tumors dominated by the classical subtype had the best outcome in response to aggressive chemotherapy and radiotherapy whereas the mesenchymal subtype dominated tumors had the worst outcome.200 Heterogeneity is the main source of resistance observed in these tumors because certain parts of the tumor are more sensitive to treatments than others. This effect is amplified by the monotherapies usually used in the treatment of glioblastoma.201 The cellular heterogeneity observed in advanced cancers such as glioblastoma is caused in part by the glioma cancer stem cells. These cells are self-renewing and contribute to disease progression, treatment, and resistance.202 Interactions with the various components of the TME play an important role in glioma stem cell behavior and differentiation leading to variations in disease progression.203
Experimental in vitro models which are designed from a reductionistic standpoint to decipher specific aspects of tumor biology already risk having limited disease complexity relevance but simply become irrelevant in GBM because of the intense complexity involved.204 Successfully targeting glioblastoma heterogeneity requires an in depth understanding of the factors driving the sub- clonal variation such as vasculature, hypoxia, and inflammation which would require building these components into the model.205 The factors stated above impress upon the need for an in vitro model that can faithfully recapitulate the in vivo components that contribute to the complexity of the glioblastoma.
5.1. Cells used in GBM models
For the last 30 years, several glioblastoma cell lines have been commonly used to study glioblastoma. Some have been quite valuable providing preliminary knowledge about the tumor.206 The most common among these cell lines is U87 which was established in 1968 at Uppsala University in Sweden and has more than 1900 citations in PubMed.206,207 Despite contributing abundant knowledge to glioblastoma research, these cell lines models are imperfect for several reasons. Namely, 2D culture of these cells has been shown to alter cell phenotype and genotype substantially so that they deviate appreciably from the in vivo disease conditions. The extracellular matrix that surrounds the cells in vivo provides cues that dictate cell differentiation, proliferation, mechano-responses, drug responses, and cell survival all of which are absent in the 2D cell culture.208 The culturing of these cells in 2D monolayer has been shown to cause considerable genetic drift, chromosomal aberration, and phenotypic alterations leading to poor reproducibility and low dependability of the data derived from these cultures. The best example of this phenomenon was observed in the U87 cell line. A recent genomic analysis has revealed that it has acquired many indels and translocations from being cultured long term on plastic. The cell line is immensely different from the original cells isolated in 1960.209 The in vitro plastic culture also applies selective pressure on these cells that does not reflect the original tumors that they have been isolated from. If these important variables are not acknowledged, the clinical utility of results obtained using cell lines in different laboratories may be incorrectly reported.210
As in other cancers, patient derived cells have the potneital to serve as a more accurate representations of the in vivo tumor if implemented correctly. The use of patient derived tumor cells in glioblastoma models facilitates precision medicine and can be used to observe individualized tumor progression and invasion.211 Patient derived tumor organoids were cultured in cerebral organoids to screen drugs, analyze tumor progression and invasion in a primitive human brain microenvironment.212 Patient derived organoids are used to study individual variability in the physical properties of the tumor and the different mechanical phenotypes of glioblastoma.213 Patient glioblastoma cells have been used to study the functional and genomic heterogeneity and also observe the intra-patient variability in response to various drug screens.214–216 Studies have demonstrated that 3D culturing methods such as spheroids of patient GBM cells effectively maintain EGFR upregulation and mutated form of EGFR-vIII. In contrast, these were gradually lost in adherent culture.217 3D culture encourages cell to cell interactions and cell to matrix interaction and allow for a more appropriate tissue physiology, anatomy and structure compared to 2D culture.218 The cells in our bodies respond to various spatial cues and stimulation from their neighboring cells and their surrounding three-dimensional environment. A model with an ECM mimetic microenvironment that provides the necessary biochemical and biomechanical cues allows researchers to study the mechanisms of cell proliferation, migration, matrix production and stem cell differentiation in the GBM microenvironment.
As with other cancer types, the inclusion of non-tumor cells in GBM models is also extremely important. The GBM microenvironment is highly complex, and it has been shown that many other cells in the GBM TME play important roles in tumor progression and growth.219–221 Engineered models are now being used to study these important cell-cell interactions. For example, multiple studies have used 3D culture systems to investigate the influence of microglia on GBM cell migration and treatment response.222,223 Similar studies have been conducted using co-cultures of GBM cells and astrocytes.224–226 The immune environment of GBM is also heterogeneous and plays an important role in progression and treatment resistance. One study used 3D spheroid cultures to evaluate the effect of tumor infiltrating macrophages on the abundance of mesenchymal cells in different populations of GBM cells.227 Another study used a bioprinted model to investigate glioma stem cell growth in the presence of various cell types including macrophages, astrocytes, and neural stem cells.170 Neural stem cells (NSC) are also of particular interest in GBM initiation, progression, and treatment.228 Therefore, one group created 3D in vitro models to investigate therapeutic efficacy of NSCs.229 GBM cells interact closely with the brain vasculature so simple co-culture models have been used to study how endothelial cells impact GBM cell behavior.230,231 More complex models of the blood brain tumor barrier (BBTB) will be discussed later. Finally, GBM cells also interact with the neural cells. Venkatesh et al. have shown that the transmission of nerve signals plays an important role in tumor progression and invasion, also evidenced by the fact that these tumors do not metastasize outside the central nervous system. Glioma cells can even form synapses with neurons, and the integrated neuron circuit provided the bidirectional signaling wherein neuron increased glioma growth and glioma cells increase neuronal activity.232 This discussion on the various cell interactions that can be studied using GBM in vitro models is certainly not exhaustive. However, it demonstrates the utility of these multi-component systems to discrete portions of the complex GBM microenvironment.
5.2. Engineering the GBM ECM
The extracellular composition of the human adult brain is unique in that collagen I, which is abundant in other tissues, is practically nonexistent in the brain. The brain has low levels of fibrous proteins such as fibronectin and vitronectin but high levels of glycosaminoglycans and aggregating proteoglycans called lecticans which includes neurocan, brevican, versican and aggrecan.233 An ECM structure composed of chondroitin sulfate proteoglycans, hyaluronic acid, tenascin-C and tenascin-R known as the perineuronal net (PNN), is exclusively found in the brain.234 S.L. et al. showed that the disruption of PNN is one of the main causes of seizures in primary brain tumor patients.235
The ECM in tumoral brain contains upregulated levels of hyaluronan and other ECM components such as vitronectin, osteopontin, and tenascin-C.236 Hyaluronan is increased fourfold in primary tumoral brain facilitating invasion and migration of tumor cells through its two receptors CD44 and CD168 or RHAMM (Receptor for Hyaluronan Mediated Motility). In high grade gliomas, CD44 is over expressed by some GBM subpopulations which helps CD44 positive cells to invade the brain parenchyma more efficiently.237 The elastic modulus of the brain tumor tissue has a twofold increase in modulus compared to the surrounding healthy tissues due to the formation of a more tightly packed cellular network and increased matrix deposition.238 Several studies have demonstrated that the mechanical properties of the substrate that the glioma cells are cultured on affect proliferation, migration, morphology, and invasion.239–244 An in vitro model system which was used to study the effect of hyaluronic acid and glioblastoma co-culture on endothelial cell network formation showed that this network formation in the hydrogels depended on the is dependent on the mechanical properties of the ECM and HA concentration which are critical parameters of a GBM microenvironment.230
The hallmark characteristics of glioblastoma are its infiltrative and invasive properties that enable the tumor cells to evade surgical resection and focal therapies. This invasion varies according to the subtype of GBM, and the mesenchymal subtype has been shown to harbor an increased potential to invade the tissue in comparison to proneural and classical subtypes.245 It has also been observed that the ECM of glioblastoma patients with a worse prognosis and poor response to the bevacizumab varies significantly in its composition from GBM patients with a better prognosis and who respond well to bevacizumab. There has been considerable interest to use this ECM compositional difference as a prognostic factor and also be used to develop anti-invasive therapies in GBM.246 Sood et al. have used bioengineered constructs with brain ECM and glioma cells and demonstrated that there is reciprocal signaling between tumor cells and the ECM microenvironment and an in depth understanding of this phenomenon is pertinent to understand tumor growth and invasion in the brain.247
These studies underscore the importance of accounting for the complex interplay between the tumor cells and the ECM along with the microenvironmental cues in glioblastoma models to understand tumor cell invasion and therapeutic resistance.
5.3. Form Factors for GBM models
As previously discussed in section 5.1, 2D culture of both cell lines and patient derived cells can lead to phenotypic and genetic changes. Therefore, there has been an increased interest in creating 3D models of GBM. By implementing many of the form factors discussed in section 3 to create complex multi-component models, many new questions have been investigated related to disease progression and heterogeneity.
Spheroids have emerged as a promising in vitro 3D model that can faithfully mimic many functional and structural properties of glioblastoma. Their ease of assembly and ability to provide high-throughput drug screening at considerably low cost, has made them one of the most popular scaffold free 3D models employed in cancer research.248 Cancer stem cells (CSC) are the most difficult to culture in vitro but play an important role in relapse of glioblastoma. Spheroid cultures often help maintain the stemness of the patient derived tumor cells and study the effect of stemness on drug resistance and relapse of disease.205,218,249 The tumor heterogeneity observed in glioblastoma has been replicated in spheroid cultures by creating multicellular tumor spheroids and employing them in drug studies to ascertain the effect of the heterogeneity on drug response in multiple studies.89,250-252Spheroid cell cultures provide advantages over 2D culture by better mimicking in vivo cell-cell interactions but still pose considerable challenges in terms of gradient access of nutrients, waste buildup and a necrotic core which prevented the spheroids size reaching more than 300 μm.208,253 Nevertheless, using spheroids comprised of multiple GBM cell lines, each labeled with a unique fluorescent tag, allowed us to create heterogeneous GBM spheroids in which we could track evolution of individual cell populations over time with and without drug interventions.254 While still a cell line-based system, this provided feasibility for current studies in which we are applying the same approach to track multiple individual GBM subtypes (classical, proneural, mesenchymal, and glioma stem cell subpopulations) within integrated organoid cultures.
To address the aforementioned hurdles using spheroids, more complex organoid models of GBM have been produced. 253 These systems allow for longer culturing period and better mimic the TME. Glioblastoma organoids are usually developed by embedding either intact tumor fragments or single-cell suspensions of tumor cells lines or cells obtained from patient tumors within an ECM hydrogel. Numerous studies have demonstrated that these materials provide a better in vitro environment for tumor cells mimicking the in vivo conditions. Commonly used naturally occurring biomaterials in glioblastoma models are collagen, hyaluronic acid, chitosan, alginate and Matrigel.241,255–257 GBM organoids in the recent years has also been developed by genetic modulation via transposon- and CRISPR-Cas9-mediated mutagenesis of cerebral organoids.258 The GLICO model, which is a GBM organoid model developed by co-culturing patient derived glioblastoma stem cells in human embryonic cells derived brain organoids, showed the deep invasion of tumor cells in the brain organoid with the aid of interconnected network of tumor microtubules.212
Transwells are used to model the neurovascular unit or the blood brain barriers in glioblastoma and also have been employed to study the effect of migration of tumor cells and microglia.259–263 The transwell system is useful in measuring the effect of tumor cells on the integrity of the BBB and testing the permeability of various chemotherapeutics across the BBB.264–268 Transwells have the advantage of ease of reproducibility, simple setup with minimal resources, access to apical and basal compartment for therapeutic testing, and easy visualization of cells. However, it is still only a monolayer culture setup without BBB microenvironmental features such as a mimetic ECM and physiologically accurate fluid flow and shear stress needed for proper endothelial polarization and tight junction formation.264,269 To address this limitations, microfluidic technology has been employed to model BBB/BTB and it can provide a highly controlled environment with incorporation of shear stresses that faithfully recapitulates the in vivo brain endothelium conditions. These models allow for high-resolution live imaging and high-throughput screening of new therapeutics which has revolutionized CNS disease modeling.143,144,270–273 To study the effect of invasive migratory tumor cells in glioblastoma microfluidic devices have been employed to determining which genetic mutations in GBM cells correlate to migration capacity in a confined space as well as assess the efficacy of drug treatment.274 One study by Han et al. makes use of an elegantly designed microfluidic chip to identify drug resistance mechanisms much more efficiently than in previous studies.275 The limitations of microfluidic devices are concerned with resolution and surface properties of many commercial materials used in devices such as acrylate, acrylonitrile butadiene styrene-based polymers, and PDMS can absorb lipids and proteins. These devices work with miniscule amount of molecules and lack of sustained nutrient and gas supply pose a challenge to long term assessment of biological processes.276 As previously stated, the cost and skills required to produce many microfluidic devices can be prohibitive to many groups, but the new low cost methods can be applied to studies of GBM, further advancing the field.
5.4. Advanced biofabrication techniques to produce GBM models
Bioprinting techniques can be used to develop more refined models of glioblastoma that contain anatomically distinct regions using patient specific components such as cells and ECM mimetic materials.277 In addition, bioprinting can be leveraged to ensure consistency between organoids and to enable high throughput organoid biofabrication.104,153 A variety of different models have been created to study GBM. These systems each use a unique combination of cells and bioinks to answer mechanistic questions related to GBM and illustrate the utility of bioprinting as an important tool moving forward.278
The majority of existing bioprinted GBM models utilize extrusion bioprinting. In the most basic cases, a single cell type is printed within a hydrogel bioink as demonstrated by the bioprinting of human glial cells with a hyaluronic acid based hydrogel bioink.279. In one series of studies, a 3D bioprinted systems was used to grow glioma stem cells in a gelatin, alginate, and fibrinogen based bioink. These studies show that these 3D constructs maintain stem cell populations, which are difficult to grow in vitro, and can be used to study cell differentiation and vasculogenic properties.280–282 Bioprinting has also been used to create models with multiple cell types to study cell-cell interactions in GBM. For example, an extrusion printer was used to print a GBM tumor within a hydrogel system containing macrophages to facilitate the study of infiltrating immune cells on tumor cell behavior and drug response.283 In another study, inkjet printing was used to pattern HepG2 and U251 GBM cells on a microfluidic device to show that prodrug metabolism into an active compound by the liver-derived HepG2 cells could induce effective U251 cell killing.284 Similarly, a bioprinted co-culture model of glioblastoma cells and astrocytes was used to identify an effective novel anticancer compound, a small molecule antagonist of the N-cadherin (NCAD) which demonstrated effective killing of glioma cells.285 Bioprinted glioblastoma tissue constructs have also been used to study the complex interactions between the immune cells and the tumor cells in a brain tumor microenvironment.170 Heinrich et al. demonstrated that a bioprinted glioblastoma model can be used to study interaction between macrophages and tumor cells and to test drugs targeting this interaction.286 This model was employed to show that the glioblastoma cells in this construct can recruit macrophages and polarize them in to glioblastoma associated macrophages which plays a role in tumor invasion and progression.286 Even more recently, a coaxial extrusion based bioprinting system was used to model glioma angiogeneisis.287 All of the highlighted bioprinted models exhibit moderately high cell viability and use mainly gelatin/collagen based systems. Extrusion and inkjet printing are limited by the bioinks that can be used and often induce high shear stresses on cells. A recent study printed hyaluronic acid-based hydrogels using digital light processing (DLP) bioprinting to study the interactions of glioma stem cells with astrocytes, macrophages and neural precursor cells.170 DLP printing uses a stereolithography-based approach to biofabrication, removing the need for printheads and associated shear stresses. These examples illustrate the utility of bioprinting in in vitro modeling of GBM. However, it is evident that the potential of this exciting technology has not yet been fully realized. As the availability of bioprinting systems and viable bioinks that better mimic the brain/GBM ECM increases, researchers will have the ability to create precise reproducible systems that can be used in further mechanistic studies or to test drugs. 3D bioprinting can also be used to scale production of patient derived models within a shorter time frame. It can be employed with ease in a clinical setting, and given the urgency of effective treatment in glioblastoma, this could be of remarkable advantage in guiding clinical decisions.104,153,288
Various microfluidic models have also been used to study glioblastoma.289 As previously discussed, microfluidic models have a wide variety of applications in the field of in vitro cancer modeling. GBM is no exception. Multiple studies have used microfluidic devices to create high throughput drug screening system for use with 3D spheroids or organoids composed of GBM cell lines or patient derived cells.290–292 As with other tumors, migration and invasion of GBM cell populations has also been investigated using microfluidic devices patterned with small channels.293 In one particular model, the microfluidic device was elegantly designed such that the effect of chemical gradients and ECM stiffness on cell migration could be studied.294 Recently, this style of microfluidic device has also been used to create a diagnostic tool for GBM. Wong et al. developed a microfluidic device, which they refer to as a microfluidic assay for quantification of cell invasion (MAqCI), that can be used to quantify the migratory ability of GBM cells. Using these migratory outputs combined with the proliferative potential of the resulting cell populations, the researchers showed that they were able to accurately predict patient tumor recurrence.295 Microfluidic devices with ECM hydrogels have also been used to study mechanisms of invasion by incorporating other important cell types including endothelial cells and other important cells found within the perivascular niche296,297 and simulating interstitial fluid flow through the ECM.298 Now, more groups are beginning to create 3D microfluidic models of the BBB, so logically it makes sense to continue to address these mechanistic questions on slightly more complex systems. Many microfluidic models of the BBB have been developed with the goal of using them to study BBB dysfunction and modeling effective systemic drug delivery in a variety of brain diseases.143,144,299–302 Some have been implemented in studying GBM, including one model using small micro-vessel structures created using 2-photon lithography.303 In another recent study, bioprinting was used to create a GBM-on-a-chip model to evaluate the effect of chemoradiotherapy on patient-derived GBM cells. Tumor and vascular bioinks were printed in a concentric ring structure using extrusion printing. This printed circular GBM model was able to sustain a radial oxygen gradient helping to mimic the central hypoxia, biochemical, biophysical characteristics observed in GBM patients.304
These models highlight the promise of these advanced biofabrication techniques for studying glioblastoma. Existing bioprinting techniques and microfluidic models can be used to answer important mechanistic questions. However, these techniques are still limited by the availability of bioinks that mimic the GBM microenvironment. Microfluidic models have already proven to be extremely useful in studying GBM. Low-cost fabrication methods combined with high-throughput fabrication techniques such as bioprinting could make these devices more applicable for large scale drug screening and personalized medicine applications.
6. Future Directions
In this review, we have attempted to summarize the components necessary for creating an effective multi-component in vitro tumor model. This engineering approach is extremely useful as it encourages researchers to utilize cells, biomaterials, and biofabrication techniques together to best mimic the native TME. Using this approach and the tools discussed has and will continue to facilitate the creation of even more complex in vitro models, it is also extremely useful for creating simple reductionist models that can be used to effectively screen for drugs or answer simple mechanistic questions. As in vitro models become more common in research and clinical applications, is important that the field agree on a set of benchmarks for model design and data analysis. In many cases, in vitro have been shown to be much better representative models of human disease than small animal models. However, rigorous standards will be required for in vitro systems to take the place of animal models in the clinical pipeline.
Acknowledgements:
The authors acknowledge funding support through National Cancer Institute grant R21CA229027, the Ohio State University Comprehensive Cancer Center, and the Pelotonia foundation.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kapałczyńska M. et al. 2D and 3D cell cultures – a comparison of different types of cancer cell cultures. Archives of Medical Science, doi: 10.5114/aoms.2016.63743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duval K. et al. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology (Bethesda) 32, 266–277, doi: 10.1152/physiol.00036.2016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 3.Thakuri PS, Liu C, Luker GD & Tavana H. Biomaterials-Based Approaches to Tumor Spheroid and Organoid Modeling. Advanced Healthcare Materials 7, 1700980, doi: 10.1002/adhm.201700980 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datta P, Dey M, Ataie Z, Unutmaz D. & Ozbolat IT 3D bioprinting for reconstituting the cancer microenvironment. npj Precision Oncology 4, 18, doi: 10.1038/s41698-020-0121-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sontheimer-Phelps A, Hassell BA & Ingber DE Modelling cancer in microfluidic human organs-on-chips. Nature Reviews Cancer 19, 65–81, doi: 10.1038/s41568-018-0104-6 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Tsai H-F, Trubelja A, Shen AQ & Bao G. Tumour-on-a-chip: microfluidic models of tumour morphology, growth and microenvironment. Journal of The Royal Society Interface 14, 20170137, doi: 10.1098/rsif.2017.0137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papapetrou EP Patient-derived induced pluripotent stem cells in cancer research and precision oncology. Nature Medicine 22, 1392–1401, doi: 10.1038/nm.4238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodrigues J, Heinrich MA, Teixeira LM & Prakash J. 3D In Vitro Model (R)evolution: Unveiling Tumor–Stroma Interactions. Trends in Cancer 7, 249–264, doi: 10.1016/j.trecan.2020.10.009 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Mazzocchi AR, Rajan SAP, Votanopoulos KI, Hall AR & Skardal A. In vitro patient-derived 3D mesothelioma tumor organoids facilitate patient-centric therapeutic screening. Scientific reports 8, 2886–2886, doi: 10.1038/s41598-018-21200-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maloney E. et al. Immersion Bioprinting of Tumor Organoids in Multi-Well Plates for Increasing Chemotherapy Screening Throughput. Micromachines 11, doi: 10.3390/mi11020208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skardal A, Devarasetty M, Forsythe S, Atala A. & Soker S. A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnology and Bioengineering 113, 2020–2032, doi: 10.1002/bit.25950 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Votanopoulos KI et al. Appendiceal Cancer Patient-Specific Tumor Organoid Model for Predicting Chemotherapy Efficacy Prior to Initiation of Treatment: A Feasibility Study. Ann Surg Oncol 26, 139–147, doi: 10.1245/s10434-018-7008-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scherer WF, Syverton JT & Gey GO Studies on the propagation in vitro of poliomyelitis viruses. IV. Viral multiplication in a stable strain of human malignant epithelial cells (strain HeLa) derived from an epidermoid carcinoma of the cervix. J Exp Med 97, 695–710, doi: 10.1084/jem.97.5.695 (1953). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barretina J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607, doi: 10.1038/nature11003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mirabelli P, Coppola L. & Salvatore M. Cancer Cell Lines Are Useful Model Systems for Medical Research. Cancers (Basel) 11, doi: 10.3390/cancers11081098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucey BP, Nelson-Rees WA & Hutchins GM Henrietta Lacks, HeLa cells, and cell culture contamination. Arch Pathol Lab Med 133, 1463–1467, doi: 10.1043/1543-2165-133.9.1463 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Wilding JL & Bodmer WF Cancer Cell Lines for Drug Discovery and Development. Cancer Research 74, 2377–2384, doi: 10.1158/0008-5472.Can-13-2971 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Gillet JP, Varma S. & Gottesman MM The clinical relevance of cancer cell lines. J Natl Cancer Inst 105, 452–458, doi: 10.1093/jnci/djt007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-David U. et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 560, 325–330, doi: 10.1038/s41586-018-0409-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pauli C. et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discovery 7, 462, doi: 10.1158/2159-8290.CD-16-1154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sivakumar H, Strowd R. & Skardal A. Exploration of Dynamic Elastic Modulus Changes on Glioblastoma Cell Populations with Aberrant EGFR Expression as a Potential Therapeutic Intervention Using a Tunable Hyaluronic Acid Hydrogel Platform. Gels 3, doi: 10.3390/gels3030028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berg KCG et al. Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Molecular Cancer 16, 116, doi: 10.1186/s12943-017-0691-y (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S-C et al. Establishment and characterization of 18 human colorectal cancer cell lines. Scientific Reports 10, 6801, doi: 10.1038/s41598-020-63812-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J. et al. Colorectal Cancer Cell Line Proteomes Are Representative of Primary Tumors and Predict Drug Sensitivity. Gastroenterology 153, 1082–1095, doi: 10.1053/j.gastro.2017.06.008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai X, Cheng H, Bai Z. & Li J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. Journal of Cancer 8, 3131–3141, doi: 10.7150/jca.18457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holliday DL & Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Research 13, 215, doi: 10.1186/bcr2889 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smart CE et al. In Vitro Analysis of Breast Cancer Cell Line Tumourspheres and Primary Human Breast Epithelia Mammospheres Demonstrates Inter- and Intrasphere Heterogeneity. PLOS ONE 8, e64388, doi: 10.1371/journal.pone.0064388 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu K. et al. Evaluating cell lines as models for metastatic breast cancer through integrative analysis of genomic data. Nature Communications 10, 2138, doi: 10.1038/s41467-019-10148-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stupp R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352, 987–996, doi: 10.1056/NEJMoa043330 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Rybinski B. & Yun K. Addressing intra-tumoral heterogeneity and therapy resistance. Oncotarget 7, 72322–72342, doi: 10.18632/oncotarget.11875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malkki H. Trial Watch: Glioblastoma vaccine therapy disappointment in Phase III trial. Nat Rev Neurol 12, 190, doi: 10.1038/nrneurol.2016.38 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Kodack DP et al. Primary Patient-Derived Cancer Cells and Their Potential for Personalized Cancer Patient Care. Cell Reports 21, 3298–3309, doi: 10.1016/j.celrep.2017.11.051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aboulkheyr Es H, Montazeri L, Aref AR, Vosough M. & Baharvand H. Personalized Cancer Medicine: An Organoid Approach. Trends in Biotechnology 36, 358–371, doi: 10.1016/j.tibtech.2017.12.005 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Sachs N. et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 172, 373–386.e310, doi: 10.1016/j.cell.2017.11.010 (2018). [DOI] [PubMed] [Google Scholar]
- 35.van de Wetering M. et al. Prospective Derivation of a Living Organoid Biobank of Colorectal Cancer Patients. Cell 161, 933–945, doi: 10.1016/j.cell.2015.03.053 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilazieva Z, Ponomarev A, Rutland C, Rizvanov A. & Solovyeva V. Promising Applications of Tumor Spheroids and Organoids for Personalized Medicine. Cancers (Basel) 12, doi: 10.3390/cancers12102727 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazzocchi AR, Rajan SAP, Votanopoulos KI, Hall AR & Skardal A. In vitro patient-derived 3D mesothelioma tumor organoids facilitate patient-centric therapeutic screening. Sci Rep 8, 2886, doi: 10.1038/s41598-018-21200-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Votanopoulos KI et al. Model of Patient-Specific Immune-Enhanced Organoids for Immunotherapy Screening: Feasibility Study. Ann Surg Oncol, doi: 10.1245/s10434-019-08143-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan HHN et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell 23, 882–897 e811, doi: 10.1016/j.stem.2018.09.016 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Driehuis E. et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proceedings of the National Academy of Sciences 116, 26580–26590, doi: 10.1073/pnas.1911273116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Votanopoulos KI et al. Model of Patient-Specific Immune-Enhanced Organoids for Immunotherapy Screening: Feasibility Study. Ann Surg Oncol 27, 1956–1967, doi: 10.1245/s10434-019-08143-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whiteside TL The tumor microenvironment and its role in promoting tumor growth. Oncogene 27, 5904–5912, doi: 10.1038/onc.2008.271 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson NM & Simon MC The tumor microenvironment. Current Biology 30, R921–R925, doi: 10.1016/j.cub.2020.06.081 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dominijanni A, Devarasetty M. & Soker S. Manipulating the Tumor Microenvironment in Tumor Organoids Induces Phenotypic Changes and Chemoresistance. iScience 23, 101851, doi: 10.1016/j.isci.2020.101851 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Truong DD et al. A Human Organotypic Microfluidic Tumor Model Permits Investigation of the Interplay between Patient-Derived Fibroblasts and Breast Cancer Cells. Cancer Research 79, 3139–3151, doi: 10.1158/0008-5472.Can-18-2293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S-A, Lee EK & Kuh H-J Co-culture of 3D tumor spheroids with fibroblasts as a model for epithelial–mesenchymal transition in vitro. Experimental Cell Research 335, 187–196, doi: 10.1016/j.yexcr.2015.05.016 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Singh S, Tran S, Putman J. & Tavana H. Three-dimensional models of breast cancer–fibroblasts interactions. Experimental Biology and Medicine 245, 879–888, doi: 10.1177/1535370220917366 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ham SL et al. Three-dimensional tumor model mimics stromal - breast cancer cells signaling. Oncotarget 9, 249–267, doi: 10.18632/oncotarget.22922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh S. et al. Organotypic breast tumor model elucidates dynamic remodeling of tumor microenvironment. Biomaterials 238, 119853, doi: 10.1016/j.biomaterials.2020.119853 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plaster M, Singh S. & Tavana H. Fibroblasts Promote Proliferation and Matrix Invasion of Breast Cancer Cells in Co-Culture Models. Advanced Therapeutics 2, 1900121, doi: 10.1002/adtp.201900121 (2019). [DOI] [Google Scholar]
- 51.Sakamoto A. et al. Pyruvate secreted from patient-derived cancer-associated fibroblasts supports survival of primary lymphoma cells. Cancer Sci 110, 269–278, doi: 10.1111/cas.13873 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao H. et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 5, e10250, doi: 10.7554/eLife.10250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung B. et al. Human brain metastatic stroma attracts breast cancer cells via chemokines CXCL16 and CXCL12. NPJ Breast Cancer 3, 6, doi: 10.1038/s41523-017-0008-8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chakraborty S, DePalma TJ & Skardal A. Increasing Accuracy of In Vitro Cancer Models: Engineering Stromal Complexity into Tumor Organoid Platforms. Advanced NanoBiomed Research n/a, 2100061, doi: 10.1002/anbr.202100061 (2021). [DOI] [Google Scholar]
- 55.Cattaneo CM et al. Tumor organoid–T-cell coculture systems. Nature Protocols 15, 15–39, doi: 10.1038/s41596-019-0232-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuen J, Darowski D, Kluge T. & Majety M. Pancreatic cancer cell/fibroblast co-culture induces M2 like macrophages that influence therapeutic response in a 3D model. PLoS One 12, e0182039, doi: 10.1371/journal.pone.0182039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neal JT et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 175, 1972–1988.e1916, doi: 10.1016/j.cell.2018.11.021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dijkstra KK et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 174, 1586–1598.e1512, doi: 10.1016/j.cell.2018.07.009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujimoto T. et al. Pericytes Suppress Brain Metastasis from Lung Cancer In Vitro. Cellular and Molecular Neurobiology 40, 113–121, doi: 10.1007/s10571-019-00725-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bi Y. et al. Tumor-on-a-chip platform to interrogate the role of macrophages in tumor progression. Integr Biol (Camb) 12, 221–232, doi: 10.1093/intbio/zyaa017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bersini S. et al. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials 35, 2454–2461, doi: 10.1016/j.biomaterials.2013.11.050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Papapetrou EP Patient-derived induced pluripotent stem cells in cancer research and precision oncology. Nat Med 22, 1392–1401, doi: 10.1038/nm.4238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu P, Weaver VM & Werb Z. The extracellular matrix: A dynamic niche in cancer progression. Journal of Cell Biology 196, 395–406, doi: 10.1083/jcb.201102147 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu T, Zhou L, Li D, Andl T. & Zhang Y. Cancer-Associated Fibroblasts Build and Secure the Tumor Microenvironment. Frontiers in Cell and Developmental Biology 7, doi: 10.3389/fcell.2019.00060 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seewaldt V. ECM stiffness paves the way for tumor cells. Nature Medicine 20, 332–333, doi: 10.1038/nm.3523 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Erdogan B. & Webb DJ Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochem Soc Trans 45, 229–236, doi: 10.1042/bst20160387 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henke E, Nandigama R. & Ergün S. Extracellular Matrix in the Tumor Microenvironment and Its Impact on Cancer Therapy. Frontiers in Molecular Biosciences 6, doi: 10.3389/fmolb.2019.00160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winkler J, Abisoye-Ogunniyan A, Metcalf KJ & Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nature Communications 11, doi: 10.1038/s41467-020-18794-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Devarasetty M, Mazzocchi AR & Skardal A. Applications of Bioengineered 3D Tissue and Tumor Organoids in Drug Development and Precision Medicine: Current and Future. BioDrugs 32, 53–68, doi: 10.1007/s40259-017-0258-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mazzocchi AR, Soker S. & Skardal A. in Tumor Organoids Vol. In Press (eds Soker S. & Skardal A) 51–70 (Springer Nature, 2017). [Google Scholar]
- 71.Skardal A. in Biopolymers for Medical Applications (eds Ruso JM & Messina PV) (CRC Press, 2016). [Google Scholar]
- 72.Muir VG & Burdick JA Chemically Modified Biopolymers for the Formation of Biomedical Hydrogels. Chemical Reviews, doi: 10.1021/acs.chemrev.0c00923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sridhar BV et al. Development of a cellularly degradable PEG hydrogel to promote articular cartilage extracellular matrix deposition. Advanced Healthcare Materials 4, 702–713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Galarza S, Crosby AJ, Pak C. & Peyton SR Control of Astrocyte Quiescence and Activation in a Synthetic Brain Hydrogel. Advanced Healthcare Materials 9, 1901419, doi: 10.1002/adhm.201901419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skardal A. Perspective: “Universal” bioink technology for advancing extrusion bioprinting-based biomanufacturing. Bioprinting 10, doi: 10.1016/j.bprint.2018.e00026 (2018). [DOI] [Google Scholar]
- 76.Aisenbrey EA & Murphy WL Synthetic alternatives to Matrigel. Nature Reviews Materials 5, 539–551, doi: 10.1038/s41578-020-0199-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoshiba T. Decellularized Extracellular Matrix for Cancer Research. Materials (Basel) 12, doi: 10.3390/ma12081311 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benton G, Kleinman HK, George J. & Arnaoutova I. Multiple uses of basement membrane-like matrix (BME/Matrigel) in vitro and in vivo with cancer cells. International Journal of Cancer 128, 1751–1757, doi: 10.1002/ijc.25781 (2011). [DOI] [PubMed] [Google Scholar]
- 79.Mazzocchi A, Devarasetty M, Huntwork R, Soker S. & Skardal A. Optimization of collagen type I-hyaluronan hybrid bioink for 3D bioprinted liver microenvironments. Biofabrication 11, 015003, doi: 10.1088/1758-5090/aae543 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Forsythe S. et al. Development of a Colorectal Cancer 3D Micro-tumor Construct Platform From Cell Lines and Patient Tumor Biospecimens for Standard-of-Care and Experimental Drug Screening. Ann Biomed Eng 48, 940–952, doi: 10.1007/s10439-019-02269-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skardal A. et al. A hydrogel bioink toolkit for mimicking native tissue biochemical and mechanical properties in bioprinted tissue constructs. Acta Biomaterialia 25, 24–34, doi: 10.1016/j.actbio.2015.07.030 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Shen Y-I et al. Hyaluronic acid hydrogel stiffness and oxygen tension affect cancer cell fate and endothelial sprouting. Biomaterials Science 2, 655–665, doi: 10.1039/C3BM60274E (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vega SL et al. Combinatorial hydrogels with biochemical gradients for screening 3D cellular microenvironments. Nature Communications 9, 614, doi: 10.1038/s41467-018-03021-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gjorevski N. et al. Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564, doi: 10.1038/nature20168 (2016). [DOI] [PubMed] [Google Scholar]
- 85.Brooks EA, Gencoglu MF, Corbett DC, Stevens KR & Peyton SR An omentum-inspired 3D PEG hydrogel for identifying ECM-drivers of drug resistant ovarian cancer. APL Bioengineering 3, 026106, doi: 10.1063/1.5091713 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leight JL, Tokuda EY, Jones CE, Lin AJ & Anseth KS Multifunctional bioscaffolds for 3D culture of melanoma cells reveal increased MMP activity and migration with BRAF kinase inhibition. Proceedings of the National Academy of Sciences 112, 5366–5371, doi: 10.1073/pnas.1505662112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Katt ME, Placone AL, Wong AD, Xu ZS & Searson PC In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Frontiers in bioengineering and biotechnology 4, 12–12, doi: 10.3389/fbioe.2016.00012 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bresciani G. et al. Evaluation of Spheroid 3D Culture Methods to Study a Pancreatic Neuroendocrine Neoplasm Cell Line. Frontiers in Endocrinology 10, doi: 10.3389/fendo.2019.00682 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Durand RE Multicell spheroids as a model for cell kinetic studies. Cell Tissue Kinet 23, 141–159, doi: 10.1111/j.1365-2184.1990.tb01111.x (1990). [DOI] [PubMed] [Google Scholar]
- 90.Sutherland RM, McCredie JA & Inch WR Growth of multicell spheroids in tissue culture as a model of nodular carcinomas. J Natl Cancer Inst 46, 113–120 (1971). [PubMed] [Google Scholar]
- 91.Xu S. et al. Construction and characteristics of an E-cadherin-related three-dimensional suspension growth model of ovarian cancer. Scientific Reports 4, 5646, doi: 10.1038/srep05646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liang X, Xu X, Wang F, Li N. & He J. E-cadherin increasing multidrug resistance protein 1 via hypoxia-inducible factor-1α contributes to multicellular resistance in colorectal cancer. Tumor Biology 37, 425–435, doi: 10.1007/s13277-015-3811-6 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Friedrich J, Seidel C, Ebner R. & Kunz-Schughart LA Spheroid-based drug screen: considerations and practical approach. Nat Protoc 4, 309–324, doi: 10.1038/nprot.2008.226 (2009). [DOI] [PubMed] [Google Scholar]
- 94.Däster S. et al. Induction of hypoxia and necrosis in multicellular tumor spheroids is associated with resistance to chemotherapy treatment. Oncotarget 8, 1725–1736, doi: 10.18632/oncotarget.13857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sant S. & Johnston PA The production of 3D tumor spheroids for cancer drug discovery. Drug Discov Today Technol 23, 27–36, doi: 10.1016/j.ddtec.2017.03.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mehta G, Hsiao AY, Ingram M, Luker GD & Takayama S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. Journal of Controlled Release 164, 192–204, doi: 10.1016/j.jconrel.2012.04.045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nederman T, Norling B, Glimelius B, Carlsson J. & Brunk U. Demonstration of an extracellular matrix in multicellular tumor spheroids. Cancer Res 44, 3090–3097 (1984). [PubMed] [Google Scholar]
- 98.Forsythe S. et al. Development of a Colorectal Cancer 3D Micro-tumor Construct Platform From Cell Lines and Patient Tumor Biospecimens for Standard-of-Care and Experimental Drug Screening. Ann Biomed Eng, doi: 10.1007/s10439-019-02269-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mazzocchi A. et al. Pleural Effusion Aspirate for use in 3D Lung Cancer Modeling and Chemotherapy Screening. ACS Biomater Sci Eng 5, 1937–1943, doi: 10.1021/acsbiomaterials.8b01356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aleman J. & Skardal A. A multi-site metastasis-on-a-chip microphysiological system for assessing metastatic preference of cancer cells. Biotechnol Bioeng, doi: 10.1002/bit.26871 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Devarasetty M. et al. Simulating the human colorectal cancer microenvironment in 3D tumor-stroma co-cultures in vitro and in vivo. Sci Rep 10, 9832, doi: 10.1038/s41598-020-66785-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Devarasetty M, Skardal A, Cowdrick K, Marini F. & Soker S. Bioengineered Submucosal Organoids for In Vitro Modeling of Colorectal Cancer. Tissue Eng Part A 23, 1026–1041, doi: 10.1089/ten.tea.2017.0397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Forsythe SD et al. Environmental Toxin Screening Using Human-Derived 3D Bioengineered Liver and Cardiac Organoids. Front Public Health 6, 103, doi: 10.3389/fpubh.2018.00103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maloney E. et al. Immersion Bioprinting of Tumor Organoids in Multi-Well Plates for Increasing Chemotherapy Screening Throughput. Micromachines (Basel) 11, doi: 10.3390/mi11020208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rajan SAP et al. Probing prodrug metabolism and reciprocal toxicity with an integrated and humanized multi-tissue organ-on-a-chip platform. Acta Biomater 106, 124–135, doi: 10.1016/j.actbio.2020.02.015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Skardal A, Devarasetty M, Rodman C, Atala A. & Soker S. Liver-Tumor Hybrid Organoids for Modeling Tumor Growth and Drug Response In Vitro. Ann Biomed Eng 43, 2361–2373, doi: 10.1007/s10439-015-1298-3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tsai S. et al. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer 18, 335, doi: 10.1186/s12885-018-4238-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Puls TJ et al. Development of a Novel 3D Tumor-tissue Invasion Model for High-throughput, High-content Phenotypic Drug Screening. Scientific Reports 8, 13039, doi: 10.1038/s41598-018-31138-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Omar Zaki SS, Kanesan L, Leong MYD & Vidyadaran S. The influence of serum-supplemented culture media in a transwell migration assay. Cell Biology International 43, 1201–1204, doi: 10.1002/cbin.11122 (2019). [DOI] [PubMed] [Google Scholar]
- 110.van de Merbel AF, van der Horst G, Buijs JT & van der Pluijm G. Protocols for Migration and Invasion Studies in Prostate Cancer. Methods Mol Biol 1786, 67–79, doi: 10.1007/978-1-4939-7845-8_4 (2018). [DOI] [PubMed] [Google Scholar]
- 111.Skardal A. et al. Bioprinted amniotic fluid-derived stem cells accelerate healing of large skin wounds. Stem Cells Transl Med 1, 792–802, doi: 10.5966/sctm.2012-0088 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.SenGupta S. et al. Triple-Negative Breast Cancer Cells Recruit Neutrophils by Secreting TGF-β and CXCR2 Ligands. Front Immunol 12, 659996, doi: 10.3389/fimmu.2021.659996 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kremer V. et al. Genetic engineering of human NK cells to express CXCR2 improves migration to renal cell carcinoma. Journal for ImmunoTherapy of Cancer 5, 73, doi: 10.1186/s40425-017-0275-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Böttcher JP et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 172, 1022–1037.e1014, doi: 10.1016/j.cell.2018.01.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pignatelli J. et al. Invasive breast carcinoma cells from patients exhibit MenaINV- and macrophage-dependent transendothelial migration. Sci Signal 7, ra112, doi: 10.1126/scisignal.2005329 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zen K. et al. CD44v4 Is a Major E-Selectin Ligand that Mediates Breast Cancer Cell Transendothelial Migration. PLOS ONE 3, e1826, doi: 10.1371/journal.pone.0001826 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rahn JJ et al. MUC1 mediates transendothelial migration in vitro by ligating endothelial cell ICAM-1. Clin Exp Metastasis 22, 475–483, doi: 10.1007/s10585-005-3098x (2005). [DOI] [PubMed] [Google Scholar]
- 118.Bhise NS et al. Organ-on-a-chip platforms for studying drug delivery systems. J Control Release 190, 82–93, doi: 10.1016/j.jconrel.2014.05.004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Polini A. et al. Organs-on-a-chip: a new tool for drug discovery. Expert opinion on drug discovery 9, 335–352, doi: 10.1517/17460441.2014.886562 (2014). [DOI] [PubMed] [Google Scholar]
- 120.Skardal A, Shupe T. & Atala A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov Today 21, 1399–1411, doi: 10.1016/j.drudis.2016.07.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Garg AA et al. Electromagnetic fields alter the motility of metastatic breast cancer cells. Communications Biology 2, 303, doi: 10.1038/s42003-019-0550-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Irimia D. & Toner M. Spontaneous migration of cancer cells under conditions of mechanical confinement. Integrative Biology 1, 506–512, doi: 10.1039/b908595e (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zou H. et al. Microfluidic Platform for Studying Chemotaxis of Adhesive Cells Revealed a Gradient-Dependent Migration and Acceleration of Cancer Stem Cells. Anal Chem 87, 7098–7108, doi: 10.1021/acs.analchem.5b00873 (2015). [DOI] [PubMed] [Google Scholar]
- 124.Gallego-Perez D. et al. Microfabricated mimics of in vivo structural cues for the study of guided tumor cell migration. Lab on a Chip 12, 4424–4432, doi: 10.1039/C2LC40726D (2012). [DOI] [PubMed] [Google Scholar]
- 125.Shang M, Soon RH, Lim CT, Khoo BL & Han J. Microfluidic modelling of the tumor microenvironment for anti-cancer drug development. Lab on a Chip 19, 369–386, doi: 10.1039/C8LC00970H (2019). [DOI] [PubMed] [Google Scholar]
- 126.Huang YL, Tung C. k., Zheng A, Kim BJ & Wu M. Interstitial flows promote amoeboid over mesenchymal motility of breast cancer cells revealed by a three dimensional microfluidic model. Integrative Biology 7, 1402–1411, doi: 10.1039/c5ib00115c (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang B, Korolj A, Lai BFL & Radisic M. Advances in organ-on-a-chip engineering. Nature Reviews Materials 3, 257–278, doi: 10.1038/s41578-018-0034-7 (2018). [DOI] [Google Scholar]
- 128.Zhang YS et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci U S A 114, E2293–E2302, doi: 10.1073/pnas.1612906114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aleman J. & Skardal A. A multi-site metastasis-on-a-chip microphysiological system for assessing metastatic preference of cancer cells. Biotechnol Bioeng 116, 936–944, doi: 10.1002/bit.26871 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Aguado BA, Bushnell GG, Rao SS, Jeruss JS & Shea LD Engineering the pre-metastatic niche. Nature Biomedical Engineering 1, 0077, doi: 10.1038/s41551-017-0077 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim H. et al. Macrophages-Triggered Sequential Remodeling of Endothelium-Interstitial Matrix to Form Pre-Metastatic Niche in Microfluidic Tumor Microenvironment. Advanced Science 6, 1900195, doi: 10.1002/advs.201900195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Campisi M. et al. 3D self-organized microvascular model of the human blood-brain barrier with endothelial cells, pericytes and astrocytes. Biomaterials 180, 117–129, doi: 10.1016/j.biomaterials.2018.07.014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen MB, Whisler JA, Jeon JS & Kamm RD Mechanisms of tumor cell extravasation in an in vitro microvascular network platform. Integrative Biology 5, 1262, doi: 10.1039/c3ib40149a (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Moya M, Tran D. & George SC An integrated in vitro model of perfused tumor and cardiac tissue. Stem Cell Res Ther 4 Suppl 1, S15, doi: 10.1186/scrt376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Choi Y. et al. A microengineered pathophysiological model of early-stage breast cancer. Lab on a Chip 15, 3350–3357, doi: 10.1039/c5lc00514k (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Polacheck WJ, Kutys ML, Tefft JB & Chen CS Microfabricated blood vessels for modeling the vascular transport barrier. Nature Protocols 14, 1425–1454, doi: 10.1038/s41596-019-0144-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bischel LL, Lee SH & Beebe DJ A practical method for patterning lumens through ECM hydrogels via viscous finger patterning. Journal of laboratory automation 17, 96–103, doi: 10.1177/2211068211426694 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Song KH, Highley CB, Rouff A. & Burdick JA Complex 3D-Printed Microchannels within Cell-Degradable Hydrogels. Advanced Functional Materials 28, 1801331, doi: 10.1002/adfm.201801331 (2018). [DOI] [Google Scholar]
- 139.Jeon JS et al. Human 3D vascularized organotypic microfluidic assays to study breast cancer cell extravasation. Proc Natl Acad Sci U S A 112, 214–219, doi: 10.1073/pnas.1417115112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nguyen D-HT et al. A biomimetic pancreatic cancer on-chip reveals endothelial ablation via ALK7 signaling. Science Advances 5, eaav6789, doi: 10.1126/sciadv.aav6789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hassell BA et al. Human Organ Chip Models Recapitulate Orthotopic Lung Cancer Growth, Therapeutic Responses, and Tumor Dormancy In Vitro. Cell Reports 21, 508–516, doi: 10.1016/j.celrep.2017.09.043 (2017). [DOI] [PubMed] [Google Scholar]
- 142.Nzou G. et al. Human Cortex Spheroid with a Functional Blood Brain Barrier for High-Throughput Neurotoxicity Screening and Disease Modeling. Scientific Reports, 1–10, doi: 10.1038/s41598-018-25603-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Herland A. et al. Distinct Contributions of Astrocytes and Pericytes to Neuroinflammation Identified in a 3D Human Blood-Brain Barrier on a Chip. PLOS ONE 11, e0150360, doi: 10.1371/journal.pone.0150360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Faley SL et al. iPSC-Derived Brain Endothelium Exhibits Stable, Long-Term Barrier Function in Perfused Hydrogel Scaffolds. Stem Cell Reports 12, 474–487, doi: 10.1016/j.stemcr.2019.01.009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mazzocchi A, Soker S. & Skardal A. in Tumor Organoids (eds Shay Soker & Aleksander Skardal) 51–70 (Springer International Publishing, 2018). [Google Scholar]
- 146.Skardal A. in Essentials of 3D Biofabrication and Translation (eds Atala A. & Yoo JJ) (Elsevier, 2015). [Google Scholar]
- 147.Mironov V, Boland T, Trusk T, Forgacs G. & Markwald RR Organ printing: computer-aided jet-based 3D tissue engineering. Trends Biotechnol 21, 157–161, doi: 10.1016/S0167-7799(03)00033-7 (2003). [DOI] [PubMed] [Google Scholar]
- 148.Mironov V, Kasyanov V, Drake C. & Markwald RR Organ printing: promises and challenges. Regen Med 3, 93–103, doi: 10.2217/17460751.3.1.93 (2008). [DOI] [PubMed] [Google Scholar]
- 149.Murphy SV & Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol 32, 773–785, doi: 10.1038/nbt.2958 (2014). [DOI] [PubMed] [Google Scholar]
- 150.Sun W. et al. The bioprinting roadmap. Biofabrication 12, 022002, doi: 10.1088/1758-5090/ab5158 (2020). [DOI] [PubMed] [Google Scholar]
- 151.Thayer P, Martinez H. & Gatenholm E. in 3D Bioprinting: Principles and Protocols (ed Crook Jeremy M.) 3–18 (Springer US, 2020). [Google Scholar]
- 152.Osidak EO, Kozhukhov VI, Osidak MS & Domogatsky SP Collagen as Bioink for Bioprinting: A Comprehensive Review. Int J Bioprint 6, 270, doi: 10.18063/ijb.v6i3.270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mazzocchi A, Soker S. & Skardal A. 3D bioprinting for high-throughput screening: Drug screening, disease modeling, and precision medicine applications. Applied Physics Reviews 6, 011302 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Oztan YC et al. Recent advances on utilization of bioprinting for tumor modeling. Bioprinting 18, e00079, doi: 10.1016/j.bprint.2020.e00079 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Roth EA et al. Inkjet printing for high-throughput cell patterning. Biomaterials 25, 3707–3715, doi: 10.1016/j.biomaterials.2003.10.052 (2004). [DOI] [PubMed] [Google Scholar]
- 156.Nakamura M. et al. Biocompatible Inkjet Printing Technique for Designed Seeding of Individual Living Cells. Tissue Engineering 11, 1658–1666, doi: 10.1089/ten.2005.11.1658 (2005). [DOI] [PubMed] [Google Scholar]
- 157.Mazzocchi A, Soker S. & Skardal A. 3D bioprinting for high-throughput screening: Drug screening, disease modeling, and precision medicine applications. Applied Physics Reviews 6, 011302, doi: 10.1063/1.5056188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Murphy SV & Atala A. 3D bioprinting of tissues and organs. Nature Biotechnology 32, 773–785, doi: 10.1038/nbt.2958 (2014). [DOI] [PubMed] [Google Scholar]
- 159.Murphy SV, Skardal A. & Atala A. Evaluation of hydrogels for bio-printing applications. Journal of Biomedical Materials Research Part A 101A, 272–284, doi: 10.1002/jbm.a.34326 (2013). [DOI] [PubMed] [Google Scholar]
- 160.Park TM et al. Fabrication of In Vitro Cancer Microtissue Array on Fibroblast-Layered Nanofibrous Membrane by Inkjet Printing. Int J Mol Sci 18, doi: 10.3390/ijms18112348 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xu F. et al. A three-dimensional in vitro ovarian cancer coculture model using a high-throughput cell patterning platform. Biotechnol J 6, 204–212, doi: 10.1002/biot.201000340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Skardal A, Zhang J, McCoard L, Oottamasathien S. & Prestwich GD Dynamically crosslinked gold nanoparticle - hyaluronan hydrogels. Adv Mater 22, 4736–4740, doi: 10.1002/adma.201001436 (2010). [DOI] [PubMed] [Google Scholar]
- 163.Skardal A, Zhang J. & Prestwich GD Bioprinting vessel-like constructs using hyaluronan hydrogels crosslinked with tetrahedral polyethylene glycol tetracrylates. Biomaterials 31, 6173–6181, doi: 10.1016/j.biomaterials.2010.04.045 (2010). [DOI] [PubMed] [Google Scholar]
- 164.Bertassoni LE et al. Direct-write bioprinting of cell-laden methacrylated gelatin hydrogels. Biofabrication 6, 024105, doi: 10.1088/1758-5082/6/2/024105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bertassoni LE et al. Hydrogel bioprinted microchannel networks for vascularization of tissue engineering constructs. Lab Chip 14, 2202–2211, doi: 10.1039/c4lc00030g (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Skardal A. et al. A hydrogel bioink toolkit for mimicking native tissue biochemical and mechanical properties in bioprinted tissue constructs. Acta Biomater 25, 24–34, doi: 10.1016/j.actbio.2015.07.030 (2015). [DOI] [PubMed] [Google Scholar]
- 167.Skardal A. et al. Photocrosslinkable hyaluronan-gelatin hydrogels for two-step bioprinting. Tissue Eng Part A 16, 2675–2685, doi: 10.1089/ten.TEA.2009.0798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hinton TJ et al. Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Science Advances 1, e1500758, doi: 10.1126/sciadv.1500758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Langer EM et al. Modeling Tumor Phenotypes In Vitro with Three-Dimensional Bioprinting. Cell Reports 26, 608–623.e606, doi: 10.1016/j.celrep.2018.12.090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tang M. et al. Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res 30, 833–853, doi: 10.1038/s41422-020-0338-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhu W. et al. Direct 3D bioprinting of prevascularized tissue constructs with complex microarchitecture. Biomaterials 124, 106–115, doi: 10.1016/j.biomaterials.2017.01.042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhu W. et al. A 3D printed nano bone matrix for characterization of breast cancer cell and osteoblast interactions. Nanotechnology 27, 315103, doi: 10.1088/0957-4484/27/31/315103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hakobyan D. et al. Laser-assisted 3D bioprinting of exocrine pancreas spheroid models for cancer initiation study. Biofabrication 12, 035001, doi: 10.1088/1758-5090/ab7cb8 (2020). [DOI] [PubMed] [Google Scholar]
- 174.McDonald JC & Whitesides GM Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc Chem Res 35, 491–499 (2002). [DOI] [PubMed] [Google Scholar]
- 175.Qin D, Xia Y. & Whitesides GM Soft lithography for micro- and nanoscale patterning. Nat Protoc 5, 491–502, doi: 10.1038/nprot.2009.234 (2010). [DOI] [PubMed] [Google Scholar]
- 176.Tentori AM & Herr AE Photopatterned materials in bioanalytical microfluidic technology. J Micromech Microeng 21, 54001, doi: 10.1088/0960-1317/21/5/054001 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Skardal A, Devarasetty M, Soker S. & Hall AR In situ patterned micro 3D liver constructs for parallel toxicology testing in a fluidic device. Biofabrication 7, 031001, doi: 10.1088/1758-5090/7/3/031001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Aleman J. et al. Deconstructed Microfluidic Bone Marrow On-A-Chip to Study Normal and Malignant Hemopoietic Cell-Niche Interactions. Small 15, e1902971, doi: 10.1002/smll.201902971 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Novak R. et al. Scalable Fabrication of Stretchable, Dual Channel, Microfluidic Organ Chips. J Vis Exp, doi: 10.3791/58151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jimenez-Torres JA, Peery SL, Sung KE & Beebe DJ LumeNEXT: A Practical Method to Pattern Luminal Structures in ECM Gels. Adv Healthc Mater 5, 198–204, doi: 10.1002/adhm.201500608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cooksey GA & Atencia J. Pneumatic valves in folded 2D and 3D fluidic devices made from plastic films and tapes. Lab Chip 14, 1665–1668, doi: 10.1039/c4lc00173g (2014). [DOI] [PubMed] [Google Scholar]
- 182.Rajan SAP, Skardal A. & Hall AR Multi-Domain Photopatterned 3D Tumor Constructs in a Micro-Physiological System for Analysis, Quantification, and Isolation of Infiltrating Cells. Advanced Biosystems 4, 1900273, doi: 10.1002/adbi.201900273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cortes-Medina M. et al. Microfluidic Prototyping by Xurography to Engineer Fully-lumenized Microvessels In Vitro. The FASEB Journal 34, 1-1, doi: 10.1096/fasebj.2020.34.s1.06369 (2020). [DOI] [Google Scholar]
- 184.Miri AK et al. Bioprinters for organs-on-chips. Biofabrication 11, 042002, doi: 10.1088/1758-5090/ab2798 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rajan SAP et al. Probing prodrug metabolism and reciprocal toxicity with an integrated and humanized multi-tissue organ-on-a-chip platform. Acta Biomaterialia 106, 124–135, doi: 10.1016/j.actbio.2020.02.015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Skardal A. et al. Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication 12, 025017, doi: 10.1088/1758-5090/ab6d36 (2020). [DOI] [PubMed] [Google Scholar]
- 187.Kesari S. Understanding Glioblastoma Tumor Biology: The Potential to Improve Current Diagnosis and Treatments. Seminars in Oncology 38, S2–S10, doi: 10.1053/j.seminoncol.2011.09.005 (2011). [DOI] [PubMed] [Google Scholar]
- 188.Bahadur S, Sahu AK, Baghel P. & Saha S. Current promising treatment strategy for glioblastoma multiform: A review. Oncol Rev 13, 417, doi: 10.4081/oncol.2019.417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fokas E, Steinbach JP & Rödel C. Biology of brain metastases and novel targeted therapies: time to translate the research. Biochim Biophys Acta 1835, 61–75, doi: 10.1016/j.bbcan.2012.10.005 (2013). [DOI] [PubMed] [Google Scholar]
- 190.Theodorakis PE, Müller EA, Craster RV & Matar OK Physical insights into the blood-brain barrier translocation mechanisms. Phys Biol 14, 041001, doi: 10.1088/1478-3975/aa708a (2017). [DOI] [PubMed] [Google Scholar]
- 191.Sarkaria JN et al. Is the blood–brain barrier really disrupted in all glioblastomas? A critical assessment of existing clinical data. Neuro-Oncology 20, 184–191, doi: 10.1093/neuonc/nox175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sprowls SA et al. Improving CNS Delivery to Brain Metastases by Blood-Tumor Barrier Disruption. Trends Cancer 5, 495–505, doi: 10.1016/j.trecan.2019.06.003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Deligne C. et al. Development of a human in vitro blood–brain tumor barrier model of diffuse intrinsic pontine glioma to better understand the chemoresistance. Fluids and Barriers of the CNS 17, 37, doi: 10.1186/s12987-020-00198-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen L. et al. Blood–Brain Barrier- and Blood–Brain Tumor Barrier-Penetrating Peptide-Derived Targeted Therapeutics for Glioma and Malignant Tumor Brain Metastases. ACS Applied Materials & Interfaces 11, 41889–41897, doi: 10.1021/acsami.9b14046 (2019). [DOI] [PubMed] [Google Scholar]
- 195.Arvanitis CD, Ferraro GB & Jain RK The blood–brain barrier and blood–tumour barrier in brain tumours and metastases. Nature Reviews Cancer 20, 26–41, doi: 10.1038/s41568-019-0205-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Verhaak RGW et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110, doi: 10.1016/j.ccr.2009.12.020 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Snuderl M. et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 20, 810–817, doi: 10.1016/j.ccr.2011.11.005 (2011). [DOI] [PubMed] [Google Scholar]
- 198.Zhang P, Xia Q, Liu L, Li S. & Dong L. Current Opinion on Molecular Characterization for GBM Classification in Guiding Clinical Diagnosis, Prognosis, and Therapy. Frontiers in Molecular Biosciences 7, doi: 10.3389/fmolb.2020.562798 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sottoriva A. et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proceedings of the National Academy of Sciences 110, 4009–4014, doi: 10.1073/pnas.1219747110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Colman H. et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol 12, 49–57, doi: 10.1093/neuonc/nop007 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bonavia R, Inda M.-d.-M., Cavenee WK & Furnari FB. Heterogeneity Maintenance in Glioblastoma: A Social Network. Cancer Research 71, 4055–4060, doi: 10.1158/0008-5472.Can-11-0153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mitchell K, Troike K, Silver DJ & Lathia JD The evolution of the cancer stem cell state in glioblastoma: emerging insights into the next generation of functional interactions. Neuro-Oncology 23, 199–213, doi: 10.1093/neuonc/noaa259 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Reinhard J, Brösicke N, Theocharidis U. & Faissner A. The extracellular matrix niche microenvironment of neural and cancer stem cells in the brain. The International Journal of Biochemistry & Cell Biology 81, 174–183, doi: 10.1016/j.biocel.2016.05.002 (2016). [DOI] [PubMed] [Google Scholar]
- 204.Robertson FL, Marqués-Torrejón M-A, Morrison GM & Pollard SM Experimental models and tools to tackle glioblastoma. Disease Models & Mechanisms 12, doi: 10.1242/dmm.040386 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Prager BC, Bhargava S, Mahadev V, Hubert CG & Rich JN Glioblastoma Stem Cells: Driving Resilience through Chaos. Trends Cancer 6, 223–235, doi: 10.1016/j.trecan.2020.01.009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Xie Y. et al. The Human Glioblastoma Cell Culture Resource: Validated Cell Models Representing All Molecular Subtypes. EBioMedicine 2, 1351–1363, doi: 10.1016/j.ebiom.2015.08.026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Pontén J. & Macintyre EH Long term culture of normal and neoplastic human glia. Acta Pathol Microbiol Scand 74, 465–486, doi: 10.1111/j.1699-0463.1968.tb03502.x (1968). [DOI] [PubMed] [Google Scholar]
- 208.Duval K. et al. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology (Bethesda) 32, 266–277, doi: 10.1152/physiol.00036.2016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 209.Allen M, Bjerke M, Edlund H, Nelander S. & Westermark B. Origin of the U87MG glioma cell line: Good news and bad news. Sci Transl Med 8, 354re353, doi: 10.1126/scitranslmed.aaf6853 (2016). [DOI] [PubMed] [Google Scholar]
- 210.Torsvik A. et al. U-251 revisited: genetic drift and phenotypic consequences of long-term cultures of glioblastoma cells. Cancer Med 3, 812–824, doi: 10.1002/cam4.219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gómez-Oliva R. et al. Evolution of Experimental Models in the Study of Glioblastoma: Toward Finding Efficient Treatments. Frontiers in Oncology 10, doi: 10.3389/fonc.2020.614295 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Linkous A. et al. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep 26, 3203–3211.e3205, doi: 10.1016/j.celrep.2019.02.063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Foss A. et al. Patient-derived glioblastoma cells (GBM) exhibit distinct biomechanical profiles associated with altered activity in the cytoskeleton regulatory pathway. bioRxiv, 2020.2007.2016.207233, doi: 10.1101/2020.07.16.207233 (2020). [DOI] [Google Scholar]
- 214.Liu Y, Li C. & Lin J. STAT3 as a Therapeutic Target for Glioblastoma. Anticancer Agents Med Chem 10, 512–519, doi: 10.2174/187152010793498636 (2010). [DOI] [PubMed] [Google Scholar]
- 215.D’Alessandris QG et al. The clinical value of patient-derived glioblastoma tumorspheres in predicting treatment response. Neuro Oncol 19, 1097–1108, doi: 10.1093/neuonc/now304 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Meyer M. et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci U S A 112, 851–856, doi: 10.1073/pnas.1320611111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Witusik-Perkowska M. et al. Glioblastoma-derived spheroid cultures as an experimental model for analysis of EGFR anomalies. J Neurooncol 102, 395–407, doi: 10.1007/s11060-010-0352-0 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zhang C. et al. 3D culture technologies of cancer stem cells: promising ex vivo tumor models. Journal of Tissue Engineering 11, 2041731420933407, doi: 10.1177/2041731420933407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Schiffer D, Annovazzi L, Casalone C, Corona C. & Mellai M. Glioblastoma: Microenvironment and Niche Concept. Cancers 11, 5, doi: 10.3390/cancers11010005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cha J. & Kim P. Biomimetic Strategies for the Glioblastoma Microenvironment. Frontiers in Materials 4, doi: 10.3389/fmats.2017.00045 (2017). [DOI] [Google Scholar]
- 221.Wolf KJ, Chen J, Coombes JD, Aghi MK & Kumar S. Dissecting and rebuilding the glioblastoma microenvironment with engineered materials. Nature Reviews Materials 4, 651–668, doi: 10.1038/s41578-019-0135-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Chen J-WE et al. Crosstalk between microglia and patient-derived glioblastoma cells inhibit invasion in a three-dimensional gelatin hydrogel model. Journal of Neuroinflammation 17, doi: 10.1186/s12974-020-02026-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Leite DM et al. A human co-culture cell model incorporating microglia supports glioblastoma growth and migration, and confers resistance to cytotoxics. Faseb j 34, 1710–1727, doi: 10.1096/fj.201901858RR (2020). [DOI] [PubMed] [Google Scholar]
- 224.Yang N. et al. A co-culture model with brain tumor-specific bioluminescence demonstrates astrocyte-induced drug resistance in glioblastoma. Journal of Translational Medicine 12, 278, doi: 10.1186/s12967-014-0278-y (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Herrera-Perez RM et al. Presence of stromal cells in a bioengineered tumor microenvironment alters glioblastoma migration and response to STAT3 inhibition. PLOS ONE 13, e0194183, doi: 10.1371/journal.pone.0194183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Neufeld L. et al. Microengineered perfusable 3D-bioprinted glioblastoma model for in vivo mimicry of tumor microenvironment. Science Advances 7, eabi9119, doi:doi: 10.1126/sciadv.abi9119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hara T. et al. Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell 39, 779–792.e711, doi: 10.1016/j.ccell.2021.05.002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Matarredona ER & Pastor AM Neural Stem Cells of the Subventricular Zone as the Origin of Human Glioblastoma Stem Cells. Therapeutic Implications. Frontiers in Oncology 9, doi: 10.3389/fonc.2019.00779 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Carey-Ewend AG et al. Developing Bioinspired Three-Dimensional Models of Brain Cancer to Evaluate Tumor-Homing Neural Stem Cell Therapy. Tissue Engineering Part A 27, 857–866, doi: 10.1089/ten.tea.2020.0113 (2020). [DOI] [PubMed] [Google Scholar]
- 230.Ngo MT & Harley BA The Influence of Hyaluronic Acid and Glioblastoma Cell Coculture on the Formation of Endothelial Cell Networks in Gelatin Hydrogels. Advanced Healthcare Materials 6, 1700687, doi: 10.1002/adhm.201700687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ngo MT & Harley BAC Perivascular signals alter global gene expression profile of glioblastoma and response to temozolomide in a gelatin hydrogel. Biomaterials 198, 122–134, doi: 10.1016/j.biomaterials.2018.06.013 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Venkatesh HS et al. Electrical and synaptic integration of glioma into neural circuits. Nature 573, 539–545, doi: 10.1038/s41586-019-1563-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ruoslahti E. Brain extracellular matrix. Glycobiology 6, 489–492, doi: 10.1093/glycob/6.5.489 (1996). [DOI] [PubMed] [Google Scholar]
- 234.Bandtlow CE & Zimmermann DR Proteoglycans in the developing brain: new conceptual insights for old proteins. Physiol Rev 80, 1267–1290, doi: 10.1152/physrev.2000.80.4.1267 (2000). [DOI] [PubMed] [Google Scholar]
- 235.Tewari BP et al. Perineuronal nets decrease membrane capacitance of peritumoral fast spiking interneurons in a model of epilepsy. Nature Communications 9, 4724, doi: 10.1038/s41467-018-07113-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bellail AC, Hunter SB, Brat DJ, Tan C. & Van Meir EG Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. The International Journal of Biochemistry & Cell Biology 36, 1046–1069, doi: 10.1016/j.biocel.2004.01.013 (2004). [DOI] [PubMed] [Google Scholar]
- 237.Delpech B. et al. Hyaluronan and hyaluronectin in the extracellular matrix of human brain tumour stroma. European Journal of Cancer 29, 1012–1017, doi: 10.1016/S0959-8049(05)80214-X (1993). [DOI] [PubMed] [Google Scholar]
- 238.Stewart DC, Rubiano A, Dyson K. & Simmons CS Mechanical characterization of human brain tumors from patients and comparison to potential surgical phantoms. PloS one 12, e0177561-e0177561, doi: 10.1371/journal.pone.0177561 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhu D, Trinh P, Li J, Grant GA & Yang F. Gradient hydrogels for screening stiffness effects on patient-derived glioblastoma xenograft cellfates in 3D. Journal of Biomedical Materials Research Part A 109, 1027–1035, doi: 10.1002/jbm.a.37093 (2021). [DOI] [PubMed] [Google Scholar]
- 240.Thomas TW & DiMilla PA Spreading and motility of human glioblastoma cells on sheets of silicone rubber depend on substratum compliance. Med Biol Eng Comput 38, 360–370, doi: 10.1007/bf02347059 (2000). [DOI] [PubMed] [Google Scholar]
- 241.Kaufman LJ et al. Glioma expansion in collagen I matrices: analyzing collagen concentration-dependent growth and motility patterns. Biophys J 89, 635–650, doi: 10.1529/biophysj.105.061994 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ulrich TA, de Juan Pardo EM & Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer research 69, 4167–4174, doi: 10.1158/0008-5472.CAN-08-4859 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kim SN et al. ECM stiffness regulates glial migration in Drosophila and mammalian glioma models. Development 141, 3233–3242, doi: 10.1242/dev.106039 (2014). [DOI] [PubMed] [Google Scholar]
- 244.Sivakumar H, Strowd R. & Skardal A. Exploration of Dynamic Elastic Modulus Changes on Glioblastoma Cell Populations with Aberrant EGFR Expression as a Potential Therapeutic Intervention Using a Tunable Hyaluronic Acid Hydrogel Platform. Gels 3, 28, doi: 10.3390/gels3030028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Vollmann-Zwerenz A, Leidgens V, Feliciello G, Klein CA & Hau P. Tumor Cell Invasion in Glioblastoma. Int J Mol Sci 21, 1932, doi: 10.3390/ijms21061932 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Virga J. et al. Extracellular matrix differences in glioblastoma patients with different prognoses. Oncol Lett 17, 797–806, doi: 10.3892/ol.2018.9649 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Sood D. et al. 3D extracellular matrix microenvironment in bioengineered tissue models of primary pediatric and adult brain tumors. Nature Communications 10, 4529, doi: 10.1038/s41467-019-12420-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Mirab F, Kang YJ & Majd S. Preparation and characterization of size-controlled glioma spheroids using agarose hydrogel microwells. PloS one 14, e0211078-e0211078, doi: 10.1371/journal.pone.0211078 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hubert CG et al. A Three-Dimensional Organoid Culture System Derived from Human Glioblastomas Recapitulates the Hypoxic Gradients and Cancer Stem Cell Heterogeneity of Tumors Found In Vivo. Cancer Research 76, 2465–2477, doi: 10.1158/0008-5472.Can-15-2402 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Au - Guyon J. et al. A 3D Spheroid Model for Glioblastoma. JoVE, e60998, doi: 10.3791/60998 (2020). [DOI] [PubMed] [Google Scholar]
- 251.Sivakumar H, Devarasetty M, Kram DE, Strowd RE & Skardal A. Multi-Cell Type Glioblastoma Tumor Spheroids for Evaluating Sub-Population-Specific Drug Response. Frontiers in Bioengineering and Biotechnology 8, doi: 10.3389/fbioe.2020.538663 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Günther W, Pawlak E, Damasceno R, Arnold H. & Terzis AJ Temozolomide induces apoptosis and senescence in glioma cells cultured as multicellular spheroids. Br J Cancer 88, 463–469, doi: 10.1038/sj.bjc.6600711 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Azzarelli R. Organoid Models of Glioblastoma to Study Brain Tumor Stem Cells. Frontiers in Cell and Developmental Biology 8, doi: 10.3389/fcell.2020.00220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sivakumar H, Devarasetty M, Kram DE, Strowd RE & Skardal A. Multi-Cell Type Glioblastoma Tumor Spheroids for Evaluating Sub-Population-Specific Drug Response. Front Bioeng Biotechnol 8, 538663, doi: 10.3389/fbioe.2020.538663 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yang Y. l., Motte S. & Kaufman LJ Pore size variable type I collagen gels and their interaction with glioma cells. Biomaterials 31, 5678–5688, doi: 10.1016/j.biomaterials.2010.03.039 (2010). [DOI] [PubMed] [Google Scholar]
- 256.Kievit FM et al. Chitosan–alginate 3D scaffolds as a mimic of the glioma tumor microenvironment. Biomaterials 31, 5903–5910, doi: 10.1016/j.biomaterials.2010.03.062 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Erickson AE, Lan Levengood SK, Sun J, Chang F-C & Zhang M. Fabrication and Characterization of Chitosan–Hyaluronic Acid Scaffolds with Varying Stiffness for Glioblastoma Cell Culture. Advanced Healthcare Materials 7, 1800295, doi: 10.1002/adhm.201800295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Bian S. et al. Genetically engineered cerebral organoids model brain tumor formation. Nat Methods 15, 631–639, doi: 10.1038/s41592-018-0070-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Leite DM et al. A human co-culture cell model incorporating microglia supports glioblastoma growth and migration, and confers resistance to cytotoxics. The FASEB Journal 34, 1710–1727, doi: 10.1096/fj.201901858RR (2020). [DOI] [PubMed] [Google Scholar]
- 260.Li L-Y, Xiao J, Liu Q. & Xia K. Parecoxib inhibits glioblastoma cell proliferation, migration and invasion by upregulating miRNA-29c. Biology Open 6, 311–316, doi: 10.1242/bio.021410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Hira VVV, Breznik B, Van Noorden CJF, Lah T. & Molenaar RJ 2D and 3D in vitro assays to quantify the invasive behavior of glioblastoma stem cells in response to SDF-1α. BioTechniques 69, 339–346, doi: 10.2144/btn-2020-0046 (2020). [DOI] [PubMed] [Google Scholar]
- 262.Louca M. et al. Ras suppressor-1 (RSU-1) promotes cell invasion in aggressive glioma cells and inhibits it in non-aggressive cells through STAT6 phospho-regulation. Scientific Reports 9, 7782, doi: 10.1038/s41598-019-44200-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wolff A, Antfolk M, Brodin B. & Tenje M. In Vitro Blood-Brain Barrier Models-An Overview of Established Models and New Microfluidic Approaches. J Pharm Sci 104, 2727–2746, doi: 10.1002/jps.24329 (2015). [DOI] [PubMed] [Google Scholar]
- 264.Prashanth A. et al. Are In Vitro Human Blood-Brain-Tumor-Barriers Suitable Replacements for In Vivo Models of Brain Permeability for Novel Therapeutics? Cancers (Basel) 13, doi: 10.3390/cancers13050955 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Stone NL, England TJ & O’Sullivan SE A Novel Transwell Blood Brain Barrier Model Using Primary Human Cells. Frontiers in Cellular Neuroscience 13, doi: 10.3389/fncel.2019.00230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Bagchi S. et al. In-vitro blood-brain barrier models for drug screening and permeation studies: an overview. Drug Des Devel Ther 13, 3591–3605, doi: 10.2147/dddt.S218708 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Aday S, Cecchelli R, Hallier-Vanuxeem D, Dehouck MP & Ferreira L. Stem Cell-Based Human Blood-Brain Barrier Models for Drug Discovery and Delivery. Trends Biotechnol 34, 382–393, doi: 10.1016/j.tibtech.2016.01.001 (2016). [DOI] [PubMed] [Google Scholar]
- 268.Parodi A. et al. Established and Emerging Strategies for Drug Delivery Across the Blood-Brain Barrier in Brain Cancer. Pharmaceutics 11, 245, doi: 10.3390/pharmaceutics11050245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Alcendor DJ et al. Neurovascular unit on a chip: implications for translational applications. Stem Cell Res Ther 4 Suppl 1, S18–S18, doi: 10.1186/scrt379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Oddo A. et al. Advances in Microfluidic Blood-Brain Barrier (BBB) Models. Trends Biotechnol 37, 1295–1314, doi: 10.1016/j.tibtech.2019.04.006 (2019). [DOI] [PubMed] [Google Scholar]
- 271.Prabhakarpandian B. et al. SyM-BBB: a microfluidic blood brain barrier model. Lab Chip 13, 1093–1101, doi: 10.1039/C2LC41208J (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Yeon JH et al. Reliable permeability assay system in a microfluidic device mimicking cerebral vasculatures. Biomedical Microdevices 14, 1141–1148, doi: 10.1007/s10544-012-9680-5 (2012). [DOI] [PubMed] [Google Scholar]
- 273.Xu Z. et al. Design and Construction of a Multi-Organ Microfluidic Chip Mimicking the in vivo Microenvironment of Lung Cancer Metastasis. ACS Applied Materials & Interfaces 8, 25840–25847, doi: 10.1021/acsami.6b08746 (2016). [DOI] [PubMed] [Google Scholar]
- 274.Bui L, Bhuiyan SH, Hendrick A, Chuong C-J & Kim Y -t. Role of key genetic mutations on increasing migration of brain cancer cells through confinement. Biomedical Microdevices 19, 56, doi: 10.1007/s10544-017-0197-9 (2017). [DOI] [PubMed] [Google Scholar]
- 275.Han J. et al. Rapid emergence and mechanisms of resistance by U87 glioblastoma cells to doxorubicin in an in vitro tumor microfluidic ecology. Proceedings of the National Academy of Sciences 113, 14283–14288, doi: 10.1073/pnas.1614898113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Cai X. et al. Application of microfluidic devices for glioblastoma study: current status and future directions. Biomedical Microdevices 22, 60, doi: 10.1007/s10544-020-00516-1 (2020). [DOI] [PubMed] [Google Scholar]
- 277.Choi Y-J, Yi H-G, Kim S-W & Cho D-W 3D Cell Printed Tissue Analogues: A New Platform for Theranostics. Theranostics 7, 3118–3137, doi: 10.7150/thno.19396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Tang M, Rich JN & Chen S. Biomaterials and 3D Bioprinting Strategies to Model Glioblastoma and the Blood–Brain Barrier. Advanced Materials 33, 2004776, doi: 10.1002/adma.202004776 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Ma L. et al. 3D bioprinted hyaluronic acid-based cell-laden scaffold for brain microenvironment simulation. Bio-Design and Manufacturing 3, 164–174, doi: 10.1007/s42242-020-00076-6 (2020). [DOI] [Google Scholar]
- 280.Wang X. et al. Bioprinting of glioma stem cells improves their endotheliogenic potential. Colloids and Surfaces B: Biointerfaces 171, 629–637, doi: 10.1016/j.colsurfb.2018.08.006 (2018). [DOI] [PubMed] [Google Scholar]
- 281.Wang X. et al. 3D bioprinted glioma cell-laden scaffolds enriching glioma stem cells via epithelial–mesenchymal transition. Journal of Biomedical Materials Research Part A 107, 383–391, doi: 10.1002/jbm.a.36549 (2019). [DOI] [PubMed] [Google Scholar]
- 282.Dai X, Ma C, Lan Q. & Xu T. 3D bioprinted glioma stem cells for brain tumor model and applications of drug susceptibility. Biofabrication 8, 045005, doi: 10.1088/17585090/8/4/045005 (2016). [DOI] [PubMed] [Google Scholar]
- 283.Tang M. et al. Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res 30, 833–853, doi: 10.1038/s41422-020-0338-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Zhang J. et al. A novel approach for precisely controlled multiple cell patterning in microfluidic chips by inkjet printing and the detection of drug metabolism and diffusion. Analyst 141, 2940–2947, doi: 10.1039/c6an00395h (2016). [DOI] [PubMed] [Google Scholar]
- 285.Smits IPM, Blaschuk OW & Willerth SM Novel N-cadherin antagonist causes glioblastoma cell death in a 3D bioprinted co-culture model. Biochemical and Biophysical Research Communications 529, 162–168, doi: 10.1016/j.bbrc.2020.06.001 (2020). [DOI] [PubMed] [Google Scholar]
- 286.Heinrich MA et al. 3D-Bioprinted Mini-Brain: A Glioblastoma Model to Study Cellular Interactions and Therapeutics. Advanced Materials 31, 1806590, doi: 10.1002/adma.201806590 (2019). [DOI] [PubMed] [Google Scholar]
- 287.Wang X. et al. Coaxially Bioprinted Cell-Laden Tubular-Like Structure for Studying Glioma Angiogenesis. Frontiers in bioengineering and biotechnology 9, 761861–761861, doi: 10.3389/fbioe.2021.761861 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Parra-Cantu C, Li W, Quiñones-Hinojosa A. & Zhang YS 3D bioprinting of glioblastoma models. Journal of 3D Printing in Medicine 4, 113–125, doi: 10.2217/3dp-2019-0027 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Logun M, Zhao W, Mao L. & Karumbaiah L. Microfluidics in Malignant Glioma Research and Precision Medicine. Adv Biosyst 2, doi: 10.1002/adbi.201700221 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Fan Y, Nguyen DT, Akay Y, Xu F. & Akay M. Engineering a Brain Cancer Chip for High-throughput Drug Screening. Sci Rep 6, 25062, doi: 10.1038/srep25062 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Chang TC et al. Parallel microfluidic chemosensitivity testing on individual slice cultures. Lab Chip 14, 4540–4551, doi: 10.1039/c4lc00642a (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Han J. et al. Rapid emergence and mechanisms of resistance by U87 glioblastoma cells to doxorubicin in an in vitro tumor microfluidic ecology. Proc Natl Acad Sci U S A 113, 14283–14288, doi: 10.1073/pnas.1614898113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Sheykhzadeh S. et al. Transferrin-targeted porous silicon nanoparticles reduce glioblastoma cell migration across tight extracellular space. Scientific Reports 10, 2320, doi: 10.1038/s41598-020-59146-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Dou J, Mao S, Li H. & Lin J-M Combination Stiffness Gradient with Chemical Stimulation Directs Glioma Cell Migration on a Microfluidic Chip. Analytical Chemistry 92, 892–898, doi: 10.1021/acs.analchem.9b03681 (2020). [DOI] [PubMed] [Google Scholar]
- 295.Wong BS et al. A microfluidic cell-migration assay for the prediction of progression-free survival and recurrence time of patients with glioblastoma. Nature Biomedical Engineering 5, 26–40, doi: 10.1038/s41551-020-00621-9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Truong D. et al. A three-dimensional (3D) organotypic microfluidic model for glioma stem cells – Vascular interactions. Biomaterials 198, 63–77, doi: 10.1016/j.biomaterials.2018.07.048 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Chonan Y, Taki S, Sampetrean O, Saya H. & Sudo R. Endothelium-induced three-dimensional invasion of heterogeneous glioma initiating cells in a microfluidic coculture platform. Integrative Biology 9, 762–773, doi: 10.1039/c7ib00091j (2017). [DOI] [PubMed] [Google Scholar]
- 298.Namba N. et al. Heterogeneous Glioma Cell Invasion Under Interstitial Flow Depending on Their Differentiation Status. Tissue Engineering Part A 27, 467–478, doi: 10.1089/ten.tea.2020.0280 (2021). [DOI] [PubMed] [Google Scholar]
- 299.Jamieson JJ, Linville RM, Ding YY, Gerecht S. & Searson PC Role of iPSC-derived pericytes on barrier function of iPSC-derived brain microvascular endothelial cells in 2D and 3D. Fluids Barriers CNS 16, 15, doi: 10.1186/s12987-019-0136-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Osaki T, Sivathanu V. & Kamm RD Engineered 3D vascular and neuronal networks in a microfluidic platform. Sci Rep 8, 5168, doi: 10.1038/s41598-018-23512-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Maoz BM et al. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nature Biotechnology 36, 865–874, doi: 10.1038/nbt.4226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Brown JA et al. Metabolic consequences of inflammatory disruption of the blood-brain barrier in an organ-on-chip model of the human neurovascular unit. Journal of Neuroinflammation, 1–17, doi: 10.1186/s12974-016-0760-y (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Marino A. et al. A 3D Real-Scale, Biomimetic, and Biohybrid Model of the Blood-Brain Barrier Fabricated through Two-Photon Lithography. Small 14, doi: 10.1002/smll.201702959 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Yi HG et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat Biomed Eng 3, 509–519, doi: 10.1038/s41551-019-0363-x (2019). [DOI] [PubMed] [Google Scholar]