Abstract
Different regions of the gastrointestinal tract have specific functions and thus distinct motility patterns. Motility is primarily regulated by the enteric nervous system (ENS), an intrinsic network of neurons located within the gut wall. Under physiological conditions, the ENS is influenced by the central nervous system (CNS). However, by using ex vivo organ bath experiments, ENS regulation of gut motility can also be studied in the absence of CNS influences. The current technique enables the characterisation of small intestinal, caecal, and colonic motility patterns using an ex vivo organ bath and video imaging protocol. This approach is combined with the novel edge detection script GutMap, available in MATLAB, that functions across Windows and Mac platforms. Dissected intestinal segments are cannulated in an organ bath containing physiological saline with a camera mounted overhead. Video recordings of gut contractions are then converted to spatiotemporal heatmaps and analysed using the GutMap software interface. Using data analysed from the heatmaps, parameters of contractile patterns (including contraction propagation frequency and velocity as well as gut diameter) at baseline and in the presence of drugs/treatments/genetic mutations can be compared. Here, we studied motility patterns of female mice at baseline and in the presence of a nitric oxide synthase inhibitor (Nω-Nitro-L-arginine; NOLA) (nitric oxide being the main inhibitory neurotransmitter of gut motility) to showcase the application of GutMap. This technique is suitable for application to a broad range of animal models of clinical disorders to understand underlying biological pathways contributing to gastrointestinal dysfunction.
Key features
• Enhanced video imaging analysis of gut contractility in rodents using a novel software interface.
• New edge detection algorithm to accurately contour curvatures of the gastrointestinal tract.
• Allows for output of high-resolution spatiotemporal heatmaps across Windows and Mac platforms.
• Edge detection and analysis method makes motility measurements accessible in different gut regions including the caecum and stomach.
Keywords: Gastrointestinal motility, Enteric nervous system, Ex vivo gut motility, Edge detection, Software, Spatiotemporal heatmap
Graphical overview
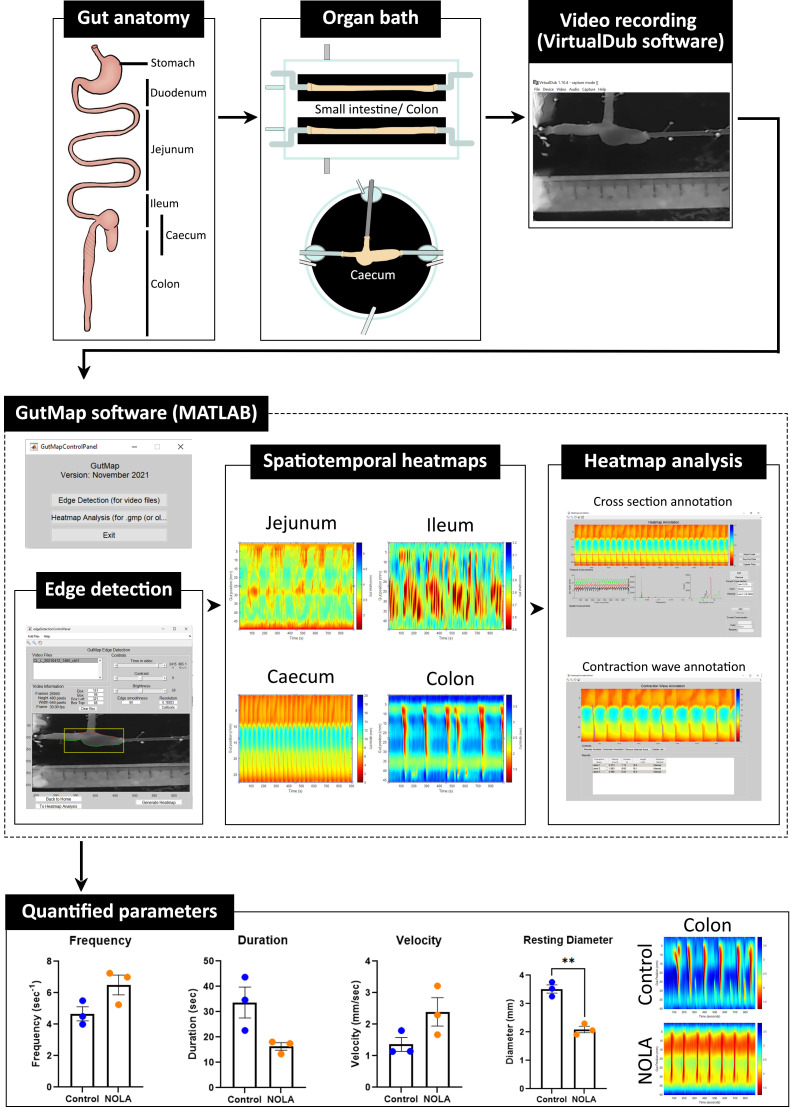
Graphical overview pipeline for the analysis of gastrointestinal motility using GutMap software. The mouse gastrointestinal tract can be divided into multiple regions. Tube-shaped segments, such as jejunum, ileum, and colon, can be placed in the small intestine/colon organ bath, which holds two preparations. The caecum preparation is mounted in the caecum organ bath. Fifteen-minute videos are recorded on the VirtualDub software. Using the GutMap user interface within MATLAB, the edge detection feature enables detection of the upper and lower boundaries of the gut preparation and analysis of the spatiotemporal heatmaps produced. GutMap allows for the visualisation and comparison of motility patterns of various regions along the gut. Data from these heatmaps can then be quantified using the cross-section annotation and contraction wave annotation function.
Background
Gut motility is critical for digestive function, including fermentation and mixing, stool formation, and microbial composition. This contractile activity is predominantly regulated by the intrinsic network of neurons within the gut, the enteric nervous system (ENS) (Furness, 2012). Motility patterns vary in the different regions of the gastrointestinal (GI) tract. In the small intestine, contractile activity serves to maximise exposure to digestive enzymes and absorption of nutrients, whereas the colon is responsible for the formation and expulsion of faeces and absorption of water and electrolytes. Several studies have reported characteristics of contractile patterning in the small and large intestine of small animal species, e.g., rat, mouse, and guinea pig (Lyster et al., 1995; Tonini et al., 1996; Costa et al., 2013 and 2015; Swaminathan et al., 2016; Spencer et al., 2018; Li et al., 2019). However, contractile patterning of the caecum (the equivalent of the human appendix) is not well characterised. Caecal motility has been described in studies of chicken (Janssen et al., 2009; van Staaveren et al., 2020), guinea pig (Schulze‐Delrieu et al., 1996), and rabbit (Hulls et al., 2012 and 2016), but is yet to be modelled in mice or studied in humans.
GI dysmotility is a common comorbidity in many disorders including autism spectrum disorder (ASD; autism) (Gorrindo et al., 2012; McElhanon et al., 2014; Vuong and Hsiao, 2017; Lee et al., 2020) and Parkinson’s disease (Kupsky et al., 1987; Singaram et al., 1995; Cersosimo et al., 2013; Giancola et al., 2017). Given that changes in gut motility patterns can contribute to gastrointestinal symptoms that impact quality of life, such as constipation, diarrhoea, visceral pain, and/or bacterial overgrowth, the accurate measurement of motility patterns could assist in identifying potential therapeutic targets to treat these issues. It is therefore necessary to design and utilise unbiased methods to study gut motility patterns both to gain fundamental knowledge about the neural regulation of motility and to determine changes in these patterns in pre-clinical animal models of disease. Ex vivo motility assays enable the investigation of gut motility patterns in animal models (Roberts et al., 2007; Swaminathan et al., 2016). These techniques were first developed to investigate small intestinal peristalsis in guinea pigs (Hennig et al., 1999; Gwynne and Bornstein, 2007) and then expanded for use in the mouse colon (Swaminathan et al., 2016). This method has been primarily applied to segments of mouse and guinea pig colon (Roberts et al., 2007; Hosie et al., 2019; Leembruggen et al., 2020) and, in some studies, to segments of small bowel (Hennig et al., 1999; Gwynne et al., 2004; Neal et al., 2009). As mentioned previously, studies of caecal motility are restricted to rabbit and chicken (Schulze‐Delrieu et al., 1996; Janssen et al., 2009; Hulls et al., 2012 and 2016). Even though external innervation from the central nervous system (CNS) plays a role in modulating gut motility (Browning and Travagli, 2014), the ENS is capable of independently regulating GI function. Hence, a major advantage of the ex vivo video imaging technique reported here is the ability to measure gut motility in the absence of CNS inputs.
Video imaging combined with spatiotemporal mapping enables the quantitative assessment of gastrointestinal motility in animal models. Using these techniques, multiple contractility parameters such as propagation speed, magnitude, length, duration, diameter, and frequency of gastrointestinal contractions can be measured. However, the software platform we previously utilised for this purpose (Swaminathan et al., 2016) had limited functionality. A lack of compatibility with current software platforms and closed source code that prevented flexibility were major limitations. We found that although suitable for assessing intestinal motility patterns, the edge detection function within the previous version of the MATLAB-based software (Analyse2) could not accurately detect edges of tissue segments from irregular shaped gut regions, such as the caecum, due to the curved/nonlinear anatomy of this organ. In addition, the quality of the heatmaps generated using the previous edge detection function (Swaminathan et al., 2016) was lower, as 16-bit unsigned integer arrays led to a quantisation and pixelation of the resultant spatiotemporal heatmaps. Importantly, this edge detection code was not compatible across different operating systems (i.e., Windows and Macintosh), which reduced user accessibility. Here, we highlight the utility of the new MATLAB-based software interface, entitled GutMap, that enables i) accurate edge detection in multiple gut regions, including in combination with a novel mouse caecal motility protocol (which can additionally be applied to stomach motility), ii) output of high-resolution spatiotemporal heatmaps, and iii) usage across different operating systems.
GutMap is a user-friendly enhanced software interface (available on request) that enables a sensitive and robust approach for visualising and analysing regional gut motility patterns in rodents (with potential applications in other preclinical models following some modification). Data from video recordings of gut contractility patterns acquired from different regions of the GI tract (i.e., the small intestine, caecum, and colon) are converted to high-resolution spatiotemporal heatmaps. The GutMap resolution, measured in μm/pixel or mm/pixel as appropriate to the length scale of the experimental preparation, is entirely dependent on the resolution of the input video file. The spatiotemporal heatmaps generated are high quality as the edge data is stored in double precision arrays, providing a near-continuum of edge locations and thus greater precision of gut width measurements.
Spatiotemporal heatmaps display gut diameter and gut position plotted as a function of time and assign warm colours denoting gut contractions and cool colours indicating gut relaxation. GutMap can be used to measure multiple contractile parameters to characterise subtle changes in motility patterns and ENS activity. Novel features of GutMap include a video calibration function, real-time video tagging information that shows the video properties and edge detection dimensions, as well as a file queueing function for edge detection processing. These features ensure increased accuracy and consistency of measures (i.e., based on the initial tissue size calibration) for each video recording file, as well as streamlined heatmap generation. Heatmap analysis using GutMap enables full functionality of the previous Analyse2 module (Swaminathan et al., 2016), plus an additional novel analysis function that enables measurement of overall contractility of the gut tissue segment.
Here, we provide a comprehensive guide for investigating ex vivo gastrointestinal motility in rodent models and highlight the novelty of our caecal motility measurement protocol in female mouse GI preparations, using the novel video analysis software, GutMap. A detailed explanation is provided for the experimental setup and execution, which are adapted from studies performed in mouse colon (Swaminathan et al., 2016) and rabbit caecum (Hulls et al., 2012 and 2016). Here, we also outline detailed steps for generating and analysing spatiotemporal heatmaps using GutMap. We demonstrate that these processes enable the comparison of motility patterns in different gut regions (i.e., the small intestine, caecum, and colon) both in physiological conditions and in response to drug administration [i.e., the nitric oxide synthase (NOS) inhibitor drug, Nω-Nitro-L-arginine (NOLA)].
The method can be further extended for use in various conditions including examining gut motility in response to different treatments and in other preclinical models of disease.
Materials and reagents
Two-chamber organ bath [manufactured by The University of Melbourne, Department of Physics (Swaminathan et al., 2016)] or equivalent, such as glass Petri dish, 14 cm diameter (e.g., Sterilin 140 mm Petri dish, Thermo Scientific, catalog number: 501V)
-
Organ bath tubing
-
Krebs solution inflow tubes
i. Polyethylene tubing 3.00 mm × 2.00 mm (Microtube Extrusions, North Rocks NSW, catalog number: PE300200)
ii. 1 cm of Masterflex Platinum-cured silicone tubing (John Morris Group, catalog number: 96410-14; L/S 14, 25 ft)
iii. Silicone rubber tubing 0.078" ID × 0.125" OD × 50 ft Sil-Med Corporation
iv. Tube connector (200 μL pipette tip) (Axygen, catalog number: AX-T-200-Y)
-
Vacuum and carbogen tubes
Laboratory tubing 1.02 mm ID × 2.16 mm OD (Silastic, catalog number: 508-005)
-
Front and back pressure cannulation inflow tubes
i. Single lumen polyethylene tube OD 2.00 mm × ID 1.00 mm used as cannula (catalog number: 112074)
ii. 0.2 cm of laboratory tubing 1.02 mm ID × 2.16 mm OD to create grip at the end of cannula (Silastic, catalog number: 508-005)
iii. Masterflex L/S® precision pump tubing, platinum-cured silicone (John Morris Group, catalog number: L/S 16 96410-16) to connect to 3-way stopcock and silicone tube connector
iv. Masterflex L/S® precision pump tubing, platinum-cured silicone (John Morris Group, catalog number: L/S 14 96410-14) to connect cannula and tube connector
v. Tube connector (200 μL pipette tip) (Axygen, catalog number: AX-T-200-Y)
-
Circulating water bath tubes
i. Platinum-cured silicone Masterflex L/S® precision pump tubing (John Morris Group, catalog number: L/S 15 96410-15)
-
Luer Lock tip syringe, 60 mL (Livingstone, catalog number: DSL050MLLCL)
Luer Slip tip tuberculin syringe, 1 mL (Livingstone, catalog number: DSL001MLSC)
Luer Slip tip syringe, 10 mL (Livingstone, catalog number: DS100MTL)
Rubber stopper (Mad About Science, 10 pack, SKU: MAS-03121-1)
Capillary glass tubing, 8 mm diameter, 1.5 mm wall thickness
Multi-purpose sealant [732 Dow Corning (clear) or equivalent]
Suture thread (white cotton thread, Woolworths Supermarket, Woolworths Group, Australia)
Nω-Nitro-L-arginine (NOLA) (Sigma-Aldrich, catalog number: N5501-5G)
-
Sylgard
184 Silicone elastomer base (DOWSIL)
184 Silicone elastomer curing agent (DOWSIL)
3-way stopcocks (B Braun Discofix, catalog number: 16494C)
Glass media bottle, clear, 500 mL with hose connection at base
Activated charcoal (White Glo)
Filter flask 2 L (Kimax Kimble, catalog number: 27060)
Sodium chloride (NaCl) (Chem-Supply, catalog number: SA046-5KG)
Potassium chloride (KCl) (Ajax Finechem, catalog number: AJA383-500G)
Sodium dihydrogen orthophosphate dihydrate (NaH2PO4·2H2O) (Chem-Supply, catalog number: SA328-500G)
Magnesium sulphate heptahydrate (MgSO4·H2O) (Chem-Supply, catalog number: MA048-500G)
Calcium chloride dihydrate (CaCl2·2H2O) (Ajax Finechem, catalog number: 127-500G)
D-glucose anhydrous (Chem-Supply, catalog number: GA018-500G)
Sodium hydrogen carbonate (NaHCO3) (Chem-Supply, catalog number: SA001-5KG)
Hydrochloric acid (HCl) (Merck Millipore, SKU: 100313)
Distilled H2O (dH2O)
Krebs 10× stock solution in 2 L (4 °C) (see Recipes)
1× Krebs solution in 2 L (4 °C) (see Recipes)
NOLA solution 100 mM (4 °C) 5 mL (see Recipes)
100 μM NOLA-Krebs solution in 500 mL (see Recipes)
Recipes
-
Krebs 10× stock solution in 2 L (4 °C)
Reagent Final concentration Quantity Sodium chloride (NaCl) 1.18 M 138 g Potassium chloride (KCl) 47.6 mM 7.1 g Sodium dihydrogen orthophosphate dihydrate (NaH2PO4·2H2O) 10 mM 3.1 g Magnesium sulphate heptahydrate (MgSO4·H2O) 12 mM 5.9 g Calcium chloride dihydrate (CaCl2·2H2O) 32.99 mM 9.7 g dH2O n/a 2 L -
1× Krebs solution in 2 L (4 °C)
Reagent Final concentration Quantity Krebs 10× stock solution 10% 200 mL dH2O n/a 2 L D-glucose anhydrous 11.1 mM 4.0 g Sodium hydrogen carbonate (NaHCO3) 25 mM 4.2 g -
NOLA solution 100 mM (4 °C) 5 mL
Reagent Final concentration Quantity Nω-Nitro-L-Arginine (NOLA) 100 mM 109.6 mg Hydrochloric acid (HCl) 1 M 400 μL dH2O n/a 4.6 mL -
100 μM NOLA-Krebs solution in 500 mL
Reagent Final concentration Quantity NOLA solution 100 μM 500 μL 1×Krebs l solution n/a 500 mL
Equipment
Webcam (Logitech Carl Zeiss Tessar HD 1080p or equivalent)
Dry block heater (Ratek, model PB)
Circulating water bath 8.5 L (ELMI TW-2.03)
Carbogen, BOC Limited
Vacuum (e.g., laboratory vacuum system)
Retort-stand with clamp (e.g., Australian Scientific, catalog number: SKU: 91055006, 91055028, 120015)
Dumont tweezer, style 1 (Fine Science Tools, catalog number: T01-111)
Dumont forceps, style 5 (Fine Science Tools, catalog number: 11252-30)
Curved forceps (Fisher Scientific, catalog number: 17-467-201)
Fine dissecting scissors (Fine Science Tools, catalog number: 14060-09)
Vannas spring scissors (Fine Science Tools, catalog number: 15000-08)
Amber scintillation 250 μL glass vial (Agilent, catalog number: 5183-2085) for light-sensitive NOLA solution 100 mM
Insect pins (Australian Entomological Supplies, catalog number E-157)
Thermometer (Westlab, catalog number: 072002-0131)
Software
VirtualDub version 1.10.4 64-bit
GutMap (must be downloaded into the MATLAB program files folder)
-
MATLAB 2021b (or any version of MATLAB that is compatible with Mac or Windows devices):
Curve fitting toolbox
Image processing toolbox
Logitech HD Pro Webcam C920 Driver
Procedure
-
Dissection and tissue collection
Cull the mouse via cervical dislocation. Exposure to drugs, such as ketamine/xylazine, should be avoided to reduce the potential influence on gut motility.
Weigh and record mouse body weight.
Disinfect the site of incision by spraying the abdomen with 70% ethanol.
Using curved dissecting forceps and small scissors, hold the skin at the mid-lower abdominal area and make an incision. Starting from this initial incision, cut a straight line to the sternum.
Make lateral incisions to open the abdominal cavity and expose the intestinal tract. If required, pour 1× Krebs solution onto the tissue immediately after opening the abdomen to prevent the tissue from drying out during the dissection process.
Lift the liver to expose the stomach and snip the oesophagus to separate it from the stomach. Next, orientate the fine dissecting scissors parallel to the tissue and carefully cut the mesentery along the gastrointestinal tract, while continuing to grasp the gastrointestinal tract by the stomach (using curved forceps), taking care to avoid puncturing the tissue preparation.
Cut the pelvic bone using fine dissecting scissors and carefully remove the mesentery with spring scissors to expose the colon.
-
Submerge the excised gut in a Petri dish containing approximately 10 mL of 1× Krebs solution and carefully unravel the gastrointestinal tract and cut the required segments for motility. Handle carefully to avoid stretching gut as this can skew results.
Identify the duodenum segment by cutting a 5 cm length of the gastrointestinal tract distal to the stomach. From that point, cut 6 cm of proximal jejunum tissue.
Measure 2 cm proximal to the ileo-caecal junction and 2 cm distal to the caeco-colonic junction to excise the caecum.
From the distal end of the small intestine, measure and cut 5 cm of ileum tissue.
Immediately place the dissected segments in a 100 mL beaker filled with approximately 30 mL of 1× Krebs solution on ice and bubbled with carbogen gas (95% O2, 5% CO2) until the setup is ready for the cannulation step. For best motility results, it is recommended that gastrointestinal segments are cannulated, and video recording begins within 1 h of dissection.
-
Setting up the organ bath
-
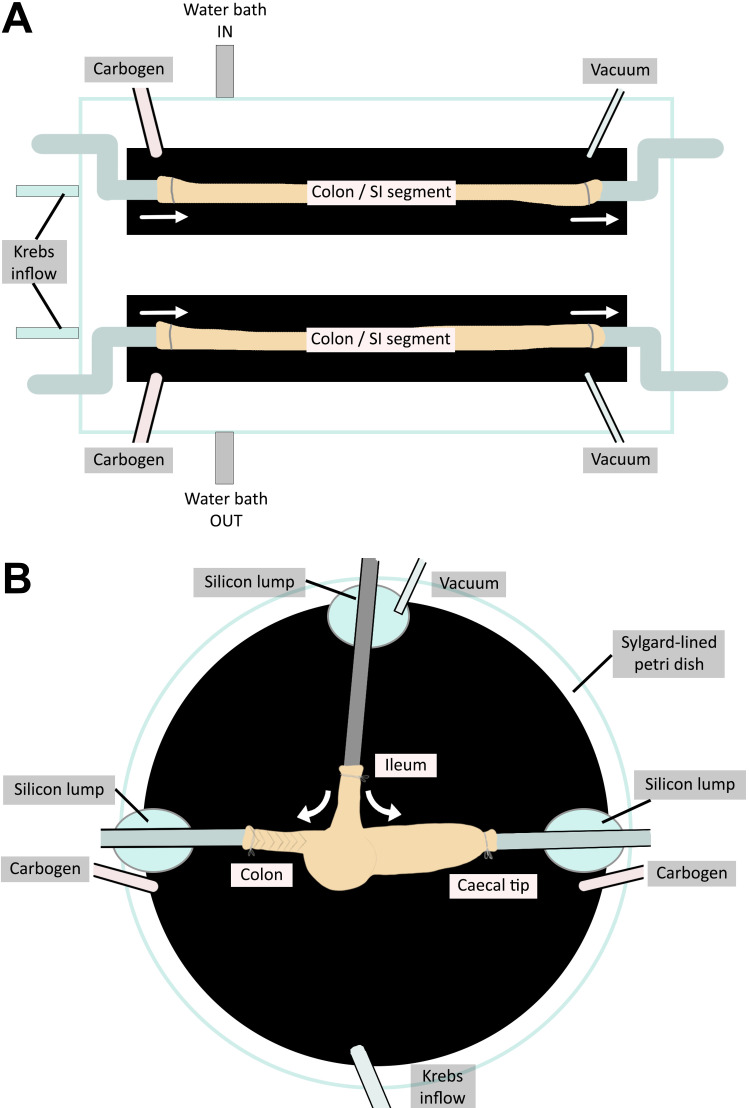
The organ bath used for the analysis of jejunum, ileum, and colon motility (Figure 1A) was made to order by the Physics Department, The University of Melbourne, Victoria, Australia (specifications as described in Swaminathan et al., 2016). The base of the organ bath is lined with black silicone-based elastomer (Sylgard), and the top surfaces of the chamber walls are sealed with a layer of silicone (Multi-purpose Sealant 732; Dow Corning). The presence of silicone ensures that the bath is watertight and provides a base to pin tubing firmly in place on the setup.
Set up the organ bath by attaching the inlet and outlet tubes using pins embedded into the silicone of the bath. The required tubes include one front pressure (FP) and one back pressure (BP) cannula, a carbogen inflow tube, and a vacuum suction tube. The Krebs solution inflow tube is pre-inserted inside the dual chambered organ bath, and silicone is applied to each inlet to avoid water leaks. Ensure that the Krebs solution inflow tube is a minimum of 45 cm in length to connect to the inflow reservoir.
Attach the Masterflex L/S® precision pump tubing (or an equivalent rubber hose) with an outer diameter of 15 mm to the inlet and outlet pipes of the organ bath and connect the opposite end of the tube to the external water bath to allow heated distilled water to flow inside the dual chambers.
-
For caecum motility (Figure 1B), a 14 cm diameter glass Petri dish (with the base coated with black Sylgard) is used as an organ bath; edges of the Petri dish are also lined with silicone.
Set up the organ bath by attaching the tubes with steel insect pins. Tubes required include one FP cannula, two BP cannulas, a carbogen inflow tube, a vacuum suction tube, and a Krebs solution inflow tube. All FP and BP cannulas require a short (2–3 mm length) rubber sleeve at the cannulating tip. The cannulating tip should be cut at an angle (approximately 45°) for easier cannulation.
-
Using suture, tie loose knots around the FP and BP cannulas.
Note: Perform this step before the organ bath is filled with 1× Krebs solution because dry thread is easier to handle.
If working with a pre-made organ bath, switch on the external water bath. To ensure that the temperature of 1× Krebs solution in the organ bath is maintained between 33 and 36 °C, the temperature of the external water bath should be between 50 and 60 °C.
If working with the custom-built caecum organ bath, place the Sylgard-lined Petri dish atop a heat block and set to desired temperature (i.e., between 70 and 90 °C, depending on the environmental temperature) to ensure that the 1× Krebs solution temperature is maintained between 33 and 36 °C.
Set up the Krebs solution inflow bottle by fastening it to the retort stand. Connect the in-flow tube attached to the organ bath to the Krebs solution bottle and twist the 3-way stopcock to allow Krebs solution to flow. Ensure that Krebs solution is flowing into the organ bath (flow rate of 8 mL/min or 500 mL/h) by observing bubbles in the capillary tube of the bottle stopper.
Ensure that the Krebs solution in the organ bath is constantly perfused with carbogen gas.
Switch on the vacuum pump and ensure that vacuum tubes are positioned such that the organ bath does not overflow or leak.
Using a 10 mL syringe, flush 1× Krebs solution into the FP and BP inflow valves to clear any debris potentially blocking the FP and BP tubes. Blockages in these tubes will affect the intraluminal pressure.
Pin a laminated printout of a 2 cm ruler onto the Sylgard-lined base of the intestinal or caecal organ bath and ensure that it is visible in the video frame. This will ensure that the gut length and width can be calculated when the user generates a spatiotemporal heatmap.
-
-
Cannulating intestinal preparations
Transfer the dissected gut segment to the organ bath. The organ bath should now be filled with warmed 1× Krebs solution. When handling the tissue, take care to hold it by the remaining mesentery and avoid touching the gut tissue, as this will affect motility.
-
Small intestine and colon:
Using fine forceps, cannulate the proximal end to the FP inflow tube and tie a knot above the rubber sleeve. This will ensure the preparation does not slip off. Tie a second knot to secure the preparation.
Twist the FP stopcock to allow Krebs solution to flow through the lumen and gently push content to clear the lumen.
Cannulate the distal end to the BP outlet tube and tie a knot above the rubber sleeve. This will ensure the preparation does not slip off. Tie a second knot to secure the preparation.
Once cannulated, carefully remove remaining mesentery using fine dissecting scissors to ensure the edges of the intestinal segment are clearly visible during video recording.
Push the stopper of the capillary rod in the FP reservoir to create a pressure seal. Adjust the height so that the meniscus height in the capillary rod is at 6 cm.
Determine the meniscus height in FP and BP tubes by subtracting the height of the FP or BP meniscus from the height of the organ bath.
-
Cannulating the caecum:
Using fine forceps, cannulate the ileum onto the left cannula that connects to the FP. Tighten the knot above the rubber sleeve to ensure that the preparation does not slip off. Tie a second knot to secure the preparation.
Using fine forceps, carefully grab hold of the caecal tip and snip the tip off (1–2 mm) using fine spring scissors.
Slowly twist open the FP stopcock and allow Krebs solution to flow through the cannula. The FP will activate contractions in the caecum, which will push the caecal contents out through the hole at the caecal tip or colon. To obtain a cleaner preparation (which allows for clearer videos downstream), gently exert a higher FP by using a small 5 mL syringe with a 20 μL pipette tip attached (cut at a 45° angle). Try to clean out the caecum as best as possible; however, avoid exerting too much pressure on the tissue as this will affect motility.
Once the body of the caecum is cleaned out, cannulate the preparation at the caecal tip by tightening the knot on the BP (caecal tip) cannula. Tie a second knot to secure the preparation. Use pins to pin down the cannula in position on the Sylgard-coated surface of the organ bath.
If there is still content in the proximal colon, gently exert pressure from the FP to flush out contents via the colon.
Once the proximal colon is cleaned, cannulate the colon via the BP (colon) cannula. Tighten the knot and tie a second knot to secure the preparation. Use pins to secure the position of the cannula.
Ensure that no blockages are present in each of the cannulas or the caecal preparation by gently exerting pressure via the FP cannula and observing a rising meniscus in the BP tubes.
Remove external FP exertion and allow the FP and BP solutions to stabilise to 4 cm height. The meniscus in the FP capillary tube and BP tubes may bob up and down as the caecum contracts but will stabilise at 4 cm during the quiescence period between contraction complexes.
If the caecum floats or bobs around as it contracts, use a small insect pin to gently pin it down to the Sylgard-coated organ bath surface by the mesentery. Ensure that the entire caecal preparation is submerged beneath 1× Krebs solution in the organ bath.
Check organ bath temperature, FP and BP readings, bubbling carbogen, and position of vacuum suction prior to starting video recording.
-
Video recording
Open VirtualDub video capture software. Select File followed by Capture AVI… and enter a file name under File and then Set capture file.
To ensure that the videos do not use up a large amount of storage space, compress the video files by selecting Compression under the Video tab, followed by Xvid MPEG-4 Codec.
Uncheck Enable audio capture under the Audio tab to disable audio recording.
Set desired stop conditions by changing the settings under the Capture tab.
Place a covering (i.e., cardboard box) over the organ bath to reduce glare and flickering from the reflection of the bath solution occurring during the video recording.
Adjust the brightness, contrast, exposure, and gain settings of the video using the webcam driver software (e.g., Logitech driver) to further reduce glare or noisy signals, which may interfere with the recording.
In VirtualDub, start video recording by selecting Capture video under the Capture tab.
Record 15 min videos according to the experimental protocol (e.g., 2 × 15 min video recordings during tissue equilibrium and 4 × 15 min video recordings for control, drug, and washout, i.e., generating a total of 14 video files per experiment). It is recommended to capture 15 min videos to enable periodical assessment of the tissue and experimental setup during the intervals between recordings; however, recording duration can be extended for specific experimental protocols as required. Video files showcasing gastrointestinal motility can be viewed in these published articles: Swaminathan et al. (2016); Hosie et al. (2019); Lee et al. (2023).
At the beginning of each video, record the temperature of the organ bath using a thermometer probe and measure the height of FP and BP meniscus. Record information in a table.
-
Repeat video recording in various conditions
In this example, the nitric oxide synthase (NOS) inhibitor, NOLA 100 mM, was diluted 1:9 with 1× Krebs solution and perfused in the organ bath system to investigate changes in gut motility. NOS is the rate-limiting enzyme in the production of nitric oxide (NO). The main role of NO is to relax the GI tract by inhibiting contractions (Toda and Herman, 2005). Therefore, when NOLA is applied, a reduction in inhibitory tone is expected to result in increased gut constriction and frequency of contractions.
Replace the Krebs solution inflow bottle with a second inflow bottle containing 100 μM NOLA in Krebs solution and repeat steps D5–D7 to visualise motility patterns in the presence of NOLA.
Once recordings in the presence of NOLA are completed, replace with regular Krebs solution, and repeat steps D5–D7 to visualise motility patterns during washout of the drug (NOLA) from the organ bath system.
-
Cleaning up
Remove the intestinal preparation by snipping the suture with dissecting scissors. Discard the tissue appropriately. Please note: although it is not recommended to use tissue samples following motility experimentation for histological analysis, there is potential to use tissue samples following experimentation to assess for neuronal populations via immunofluorescence techniques.
Using a 50 mL syringe, pass distilled water through all cannulas and Krebs solution inflow tube; repeat this three times per tube.
Use the vacuum suction tube to remove the distilled water from the organ bath.
Finally, use a 50 mL syringe to push out any residual water in the tubing.
-
Obtaining spatiotemporal heatmaps
Open MATLAB and select Set Path. Add the GutMap folder to the MATLAB path and save.
To generate a spatiotemporal map, type GutMap at the MATLAB command prompt and select Edge Detection (for video files) from the pop-up window.
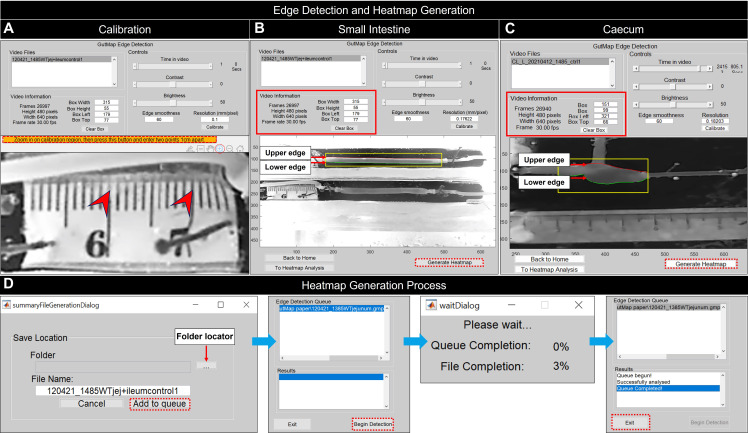
Load .AVI format video recordings and select Calibrate to define video resolution and the length of the gut preparation. This action will open a new window, including an image of the tissue preparation alongside a physical ruler placed within the organ bath. In this window, use the magnify symbol to zoom in. Use the cursor to select two points 1 cm apart on the ruler that is visible in the window. From these points, GutMap will calculate a pixel/mm calibration metric specific to the current video file, ensuring a spatiotemporal heatmap with accurate dimensions (Figure 2A).
Using the mouse, draw a rectangle around the region of interest of the gut tissue. A red contour line will define the upper edge and a green contour line will delineate the lower edge of the gut preparation in the image (Figure 2B, 2C).
Use the time slider function to view a selection of frames and ensure gut tissue is adequately contoured prior to proceeding to the heatmap generation step. If there are issues, adjust brightness, contrast, and edge smoothness to enhance edge detection accuracy.
Click Generate Heatmap. This will prompt the user to name the output file. Select a directory to save the file, and then click Add to queue to process the video.
Repeat steps G3–G6 to edge detect additional videos. Set box dimensions to match between videos if desired. Select Begin Detection to edge-detect the videos listed in the queue (Figure 2D).
Figure 1. Organ baths to assess motility in (A) small intestine (SI) and colon preparations and (B) caecum.
(A) The colon/small intestinal organ bath has two chambers enabling two tissue preparations to be assayed simultaneously. (B) The caecum organ bath is a Petri dish that allows for the three-way cannulation of the caecum preparation. The temperature of setup (A) is regulated by an external water bath. The caecum setup (B) is placed on a heat block to ensure the temperature is kept constant throughout video recording. White arrows indicate the flow of Krebs solution and therefore the pressure exerted on the gut preparations.
Figure 2. GutMap edge detection control panel.
(A) To calibrate the resolution of the current video file, select Calibrate. This will open the calibration figure where the user will be directed to zoom in using the magnifying tool and click on the orange bar. The user selects two points 1 cm apart on the ruler. The calibration figure will then close, and the calibrated resolution value will appear in the Edge Detection Control Panel. Edge detection contours for (B) small intestine and (C) caecum. The selected region of interest contains the upper and lower edges of the gut segment. A red contour delineates the upper edge of the gut, and the green contour identifies the lower edge of the gut. Specifications of the region selected appear in the Video Information area, which can be used to match region-of-interest dimensions between videos. When the gut tissue is adequately contoured, Generate Heatmap is selected by the user. The user is then prompted for the output file name (D) and is next prompted to add the file to the Edge Detection Queue to begin edge detection. For batch processing of multiple files, see instructions in text. A Wait dialogue box appears and indicates the status of the heatmap generation to the user. Once this process is complete, the user must exit.
Data analysis
-
Heatmap analysis
Click To Heatmap Analysis on the Edge Detection Control Panel to access the Heatmap analysis control panel.
Set the minimum and maximum colour display values. This will display contractions and dilations in warm and cool toned colours, respectively. Click Lock Color Range to set the colour range. It is important to keep the colour range constant throughout a dataset to enable comparison between samples and experiments.
-
Cross-sectional heatmap analysis
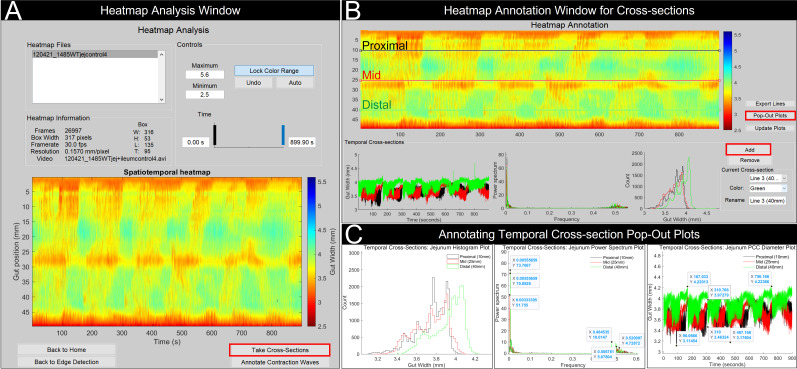
To analyse cross-sections of the spatiotemporal heatmap, select the Take Cross-Sections button (Figure 3A).
-
Click Add and place a line at a specific position on the heatmap. Three temporal plots will appear below the spatiotemporal heatmap displaying gut width data over time, a frequency power spectrum, and a histogram of contractility frequency (Figure 3B).
The gut width temporal plot displays a range of peaks; maximum peaks represent resting gut diameter, and minimum peaks represent constricted gut diameter.
The frequency temporal plot displays frequency power; lower frequency peaks represent neurogenic input, and higher frequency peaks represent myogenic input.
The histogram plot displays the frequency of contractility.
Temporal plots can be annotated by selecting Pop-Out Plots from the Heatmap Annotation Window to create individual figure windows of the temporal plots. Use the magnification tools to zoom in or out of the temporal plots, and hover over the peaks to place xy-coordinates (Figure 3C).
-
Cross-sectional heatmap analysis: gut width over time temporal plot
Place xy-coordinates on the gut width vs. time temporal plot to obtain the maximal resting gut diameter, maximal contracted gut diameter, duration of contractile complexes, and quiescent period between contractile complexes.
-
Cross-sectional heatmap analysis: power spectrum frequency temporal plot
Place data tips on peaks to obtain myogenic and neurogenic input measures.
-
Cross-sectional heatmap analysis: histogram temporal plot
The histogram plots contractile activity from the selected cross-section line.
Click Export Lines to export data values from the cross-section line in a text file format.
-
Annotation of contraction waves
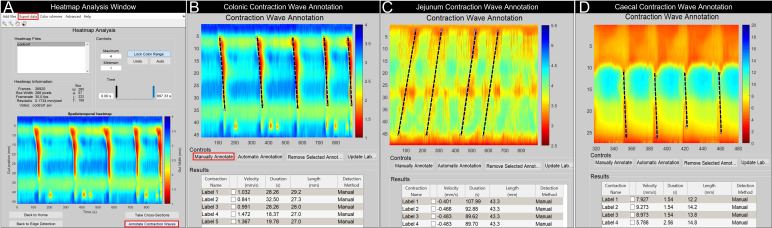
To analyse contractions, select Annotate Contraction Waves from the heatmap analysis control panel (Figure 4A).
Use the magnification tools in the Annotate Contraction Waves window to zoom in to the contractions to be analysed.
Select Manually Annotate to draw a line from the start to the end of the contraction (see Figure 4B–4D for examples).
Metrics of the annotated line (velocity, duration of contraction, and length of the annotated contraction) will appear in the table below the heatmap. Note: Contraction Name can be modified.
Select the Update Label button to make changes to the annotated contraction. The results table will update accordingly.
To remove an annotation, select the individual check box within the results table. This will make the selected annotation appear as a red line on the heatmap. Then, click Remove Selected Annotation.
To export the results, return to the heatmap analysis control panel and select Export data (Figure 4A).
Figure 3. GutMap cross-section analysis.
(A) In the Heatmap Analysis Window, lock colour range and select the Take Cross-Sections button. This will open a new window (B) where the user can add cross-sectional lines to specific positions on the spatiotemporal heatmap. Output is provided from these cross-sectional lines including gut width, power spectra, and histogram plots of contractile activity. (C) By selecting Pop-Out Plots, these temporal cross-sections can be annotated for further analysis.
Figure 4. GutMap contraction wave annotation windows.
(A) In the Heatmap Analysis Window, lock the colour range and select Annotate Contraction Waves. A new window will open, allowing the user to manually annotate the contractions or groups of contractions. Annotated examples are shown for (B) colonic, (C) jejunal, and (D) caecal heatmaps [annotated region depicts caecal contractions propagating from the colon-caecal junction (upper red region of map) towards the caecal tip]. The Contraction Wave Annotation tool can be used to measure velocity, duration, and length of the contraction by selecting Manually Annotate and drawing a line following the slope of the contraction or contractile cluster. The data obtained in the results table can be exported by selecting Export Data in the Heatmap Analysis Window.
Validation of protocol
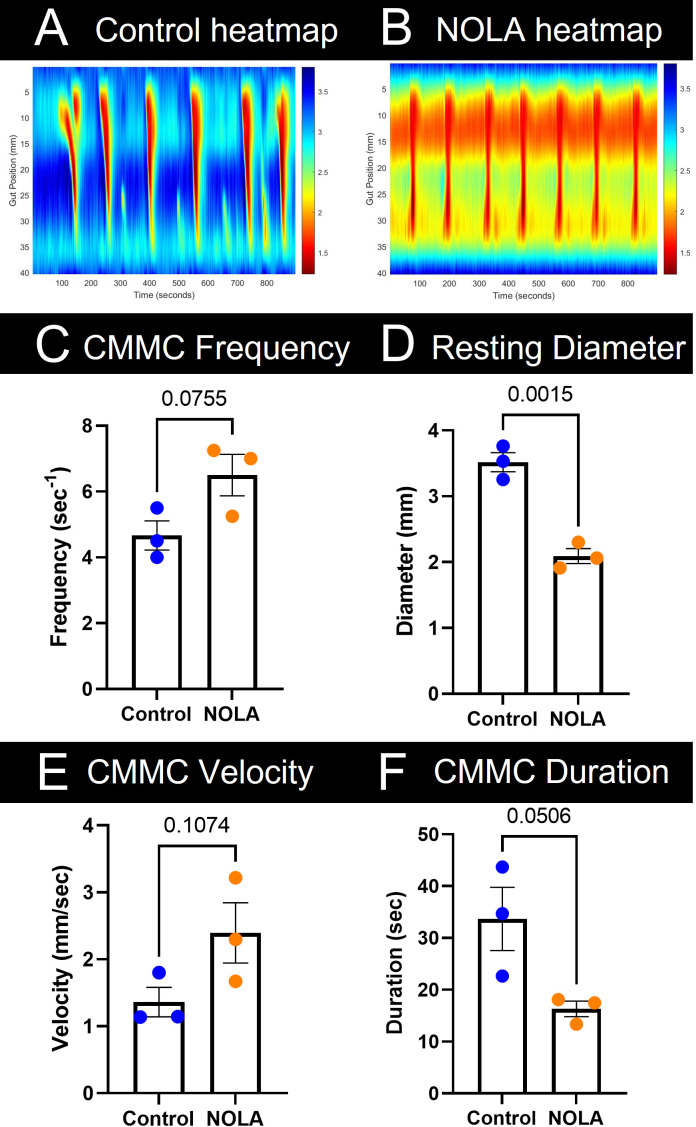
To compare control and experimental conditions (i.e., motility patterns in the presence of NOLA) data, data tips from each heatmap (four control and four NOLA heatmaps) were averaged. Averaged values can then be inputted to GraphPad Prism version 9.5.0 (or alternative statistical software), where an unpaired t-test is performed to compare various parameters (frequency, diameter, velocity, and duration). This analysis process can be applied to various gastrointestinal tissue segments (i.e., small intestine, caecum, and colon). For example, here we show colonic motility data obtained from n = 3 female mice during baseline and NOLA conditions (Figure 5). We observed a reduction in colonic diameter (constriction) at rest upon NOLA treatment; however, there were no changes to frequency, velocity, or duration of colon migrating motor complexes.
Figure 5. Analysis of colon migrating motor complexes (CMMCs) in n = 3 female mice.
Representative colonic heatmaps in (A) control and (B) NOLA conditions display contractile activity in warm tones and relaxation in cool tones. (C) CMMCs that propagated for greater than 50% of the length of the colon were counted and compared between control and NOLA groups. (D) Data tips placed at the peaks of the temporal cross-section plot were averaged to obtain the maximal resting diameter between control and NOLA groups. (E) and (F) The Manually Annotate tool was used to annotate the slope of the CMMCs propagating for greater than 50% of the length of the colon. Velocity and duration results are automatically outputted and then averaged and compared between control and NOLA groups using an Unpaired student t-test.
Acknowledgments
All authors approved the manuscript for submission. ELH-Y was supported by an ARC Future Fellowship (FT160100126) and an RMIT Vice Chancellor’s Senior Research Fellowship. TAS and CYQL received the RMIT Research Stipend Scholarship (RRSS). This protocol describes a significantly enhanced approach based on our previously published methods protocol: Swaminathan et al. (2016).
Competing interests
The authors declare no conflict of interest.
Ethical considerations
All animal handling and euthanasia procedures were performed according to strict guidelines as approved by the RMIT Animal Ethics Committee (AEC#: 21268).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1.Browning K. N. and Travagli R. A.(2014). Central Nervous System Control of Gastrointestinal Motility and Secretion and Modulation of Gastrointestinal Functions. Compr. Physiol.: 1339–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cersosimo M. G., Raina G. B., Pecci C., Pellene A., Calandra C. R., Gutiérrez C., Micheli F. E. and Benarroch E. E.(2013). Gastrointestinal manifestations in Parkinson’s disease: prevalence and occurrence before motor symptoms. J. Neurol. 260(5): 1332-1338. [DOI] [PubMed] [Google Scholar]
- 3.Costa M., Dodds K. N., Wiklendt L., Spencer N. J., Brookes S. J. H. and Dinning P. G.(2013). Neurogenic and myogenic motor activity in the colon of the guinea pig, mouse, rabbit, and rat. Am. J. Physiol. Gastrointest. Liver Physiol. 305(10): G749-G759. [DOI] [PubMed] [Google Scholar]
- 4.Costa M., Wiklendt L., Simpson P., Spencer N. J., Brookes S. J. and Dinning P. G.(2015). Neuromechanical factors involved in the formation and propulsion of fecal pellets in the guinea-pig colon. Neurogastroenterol. Motil. 27(10): 1466-1477. [DOI] [PubMed] [Google Scholar]
- 5.Furness J. B.(2012). The enteric nervous system and neurogastroenterology. Nat. Rev. Gastroenterol. Hepatol. 9(5): 286-294. [DOI] [PubMed] [Google Scholar]
- 6.Giancola F., Torresan F., Repossi R., Bianco F., Latorre R., Ioannou A., Guarino M., Volta U., Clavenzani P., Mazzoni M., et al.(2017). Downregulation of neuronal vasoactive intestinal polypeptide in Parkinson's disease and chronic constipation. Neurogastroenterol. Motil. 29(5): e12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorrindo P., Williams K. C., Lee E. B., Walker L. S., McGrew S. G. and Levitt P.(2012). Gastrointestinal Dysfunction in Autism: Parental Report, Clinical Evaluation, and Associated Factors. Autism Res. 5(2): 101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gwynne R. M. and Bornstein J. C.(2007). Mechanisms underlying nutrient-induced segmentation in isolated guinea pig small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 292(4): G1162-G1172. [DOI] [PubMed] [Google Scholar]
- 9.Gwynne R. M., Thomas E. A., Goh S. M., Sjövall H. and Bornstein J. C.(2004). Segmentation induced by intraluminal fatty acid in isolated guinea-pig duodenum and jejunum. J. Physiol. 556(2): 557-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hennig G. W., Costa M., Chen B. N. and Brookes S. J. H.(1999). Quantitative analysis of peristalsis in the guinea‐pig small intestine using spatio‐temporal maps. J. Physiol. 517(2): 575-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosie S., Ellis M., Swaminathan M., Ramalhosa F., Seger G. O., Balasuriya G. K., Gillberg C., Råstam M., Churilov L., McKeown S. J., et al.(2019). Gastrointestinal dysfunction in patients and mice expressing the autism‐associated R451C mutation in neuroligin‐3. Autism Res. 12(7): 1043-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hulls C., Lentle R. G., de Loubens C., Janssen P. W. M., Chambers P. and Stafford K. J.(2012). Spatiotemporal mapping of ex vivo motility in the caecum of the rabbit. J Comp. Physiol. B 182(2): 287-297. [DOI] [PubMed] [Google Scholar]
- 13.Hulls C., Lentle R. G., Reynolds G. W., Janssen P. W. M., Chambers P. and de Loubens C.(2016). Ex vivo motility in the base of the rabbit caecum and its associated structures: an electrophysiological and spatiotemporal analysis. J. Physiol. Biochem. 72(1): 45-57. [DOI] [PubMed] [Google Scholar]
- 14.Janssen P. W. M., Lentle R. G., Hulls C., Ravindran V. and Amerah A. M.(2009). Spatiotemporal mapping of the motility of the isolated chicken caecum. J Comp. Physiol. B 179(5): 593-604. [DOI] [PubMed] [Google Scholar]
- 15.Kupsky W. J., Grimes M. M., Sweeting J., Bertsch R. and Cote L. J.(1987). Parkinson's disease and megacolon: Concentric hyaline inclusions(Lewy bodies) in enteric ganglion cells. Neurology 37(7): 1253-1253. [DOI] [PubMed] [Google Scholar]
- 16.Lee C. Y. Q., Balasuriya G. K., Herath M., Franks A. E., and Hill-Yardin E. L.(2023). Impaired cecal motility and secretion alongside expansion of gut-associated lymphoid tissue in the Nlgn3R451C mouse model of autism. Sci. Rep. 13: 12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee C. Y. Q., Franks A. E. and Hill-Yardin E. L.(2020). Autism-associated synaptic mutations impact the gut-brain axis in mice. Brain Behav. Immun. 88: 275-282. [DOI] [PubMed] [Google Scholar]
- 18.Leembruggen, A. J. L., Balasuriya, G. K., Zhang, J., Schokman, S., Swiderski, K., Bornstein, J. C., Nithianantharajah, J. and Hill‐Yardin, E. L.(2020). Colonic dilation and altered ex vivo gastrointestinal motility in the neuroligin‐3 knockout mouse. Autism Res. 13(5): 691-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., Hao M. M., Van den Haute C., Baekelandt V., Boesmans W. and Vanden Berghe P.(2019). Regional complexity in enteric neuron wiring reflects diversity of motility patterns in the mouse large intestine. eLife 8: e42914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyster D. J., Bywater R. A. and Taylor G. S.(1995). Neurogenic control of myoelectric complexes in the mouse isolated colon. Gastroenterology 108(5): 1371-1378. [DOI] [PubMed] [Google Scholar]
- 21.McElhanon B. O., McCracken C., Karpen S. and Sharp W. G.(2014). Gastrointestinal Symptoms in Autism Spectrum Disorder: A Meta-analysis. Pediatrics 133(5): 872-883. [DOI] [PubMed] [Google Scholar]
- 22.Neal K. B., Parry L. J. and Bornstein J. C.(2009). Strain-specific genetics, anatomy and function of enteric neural serotonergic pathways in inbred mice. J. Physiol. 587(3): 567-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts R. R., Murphy J. F., Young H. M. and Bornstein J. C.(2007). Development of colonic motility in the neonatal mouse-studies using spatiotemporal maps. Am. J. Physiol. Gastrointest. Liver Physiol. 292(3): G930-G938. [DOI] [PubMed] [Google Scholar]
- 24.K. Schulze‐Delrieu, Brown B., Lange W., T. Custer‐Hagen, Lu C., Shirazi S. and Lepsien G.(1996). Volume shifts, unfolding and rolling of haustra in the isolated guinea pig caecum. Neurogastroenterol. Motil. 8(3): 217-225. [DOI] [PubMed] [Google Scholar]
- 25.Singaram C., Gaumnitz E., Torbey C., Ashraf W., Quigley E., Sengupta A. and Pfeiffer R.(1995). Dopaminergic defect of enteric nervous system in Parkinson's disease patients with chronic constipation. Lancet 346(8979): 861-864. [DOI] [PubMed] [Google Scholar]
- 26.Spencer N. J., Hibberd T. J., Travis L., Wiklendt L., Costa M., Hu H., Brookes S. J., Wattchow D. A., Dinning P. G., Keating D. J., et al.(2018). Identification of a Rhythmic Firing Pattern in the Enteric Nervous System That Generates Rhythmic Electrical Activity in Smooth Muscle. J. Neurosci. 38(24): 5507-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swaminathan M., Hill-Yardin E., Ellis M., Zygorodimos M., Johnston L. A., Gwynne R. M. and Bornstein J. C.(2016). Video Imaging and Spatiotemporal Maps to Analyze Gastrointestinal Motility in Mice. J. Vis. Exp.: e3791/53828-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toda N. and Herman A. G.(2005). Gastrointestinal Function Regulation by Nitrergic Efferent Nerves. Pharmacol. Res. 57(3): 315-338. [DOI] [PubMed] [Google Scholar]
- 29.Tonini M., Costa M., Brookes S. and Humphreys C.(1996). Dissociation of the ascending excitatory reflex from peristalsis in the guinea-pig small intestine. Neuroscience 73(1): 287-297. [DOI] [PubMed] [Google Scholar]
- 30.van Staaveren N., Krumma J., Forsythe P., Kjaer J. B., Kwon I. Y., Mao Y. K., West C., Kunze W. and Harlander-Matauschek A.(2020). Cecal motility and the impact of Lactobacillus in feather pecking laying hens. Sci. Rep. 10(1): e1038/s41598-020-69928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vuong H. E. and Hsiao E. Y.(2017). Emerging Roles for the Gut Microbiome in Autism Spectrum Disorder. Biol. Psychiatry 81(5): 411-423. [DOI] [PMC free article] [PubMed] [Google Scholar]