Abstract
The burden of Hospital care-associated infections (HCAIs) is becoming a global concern. This is compounded by the emergence of virulent and high-risk bacterial strains such as “ESKAPE” pathogens – (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species), especially within Intensive care units (ICUs) that house high-risk and immunocompromised patients. In this review, we discuss the contributions of AMR pathogens to the increasing burden of HCAIs and provide insights into AMR mechanisms, with a particular focus on last-resort antibiotics like polymyxins. We extensively discuss how structural modifications of surface-membrane lipopolysaccharides and cationic interactions influence and inform AMR, and subsequent severity of HCAIs. We highlight some bacterial phenotypic survival mechanisms against polymyxins. Lastly, we discuss the emergence of plasmid-mediated resistance as a phenomenon making mitigation of AMR difficult, especially within the ICUs. This review provides a balanced perspective on the burden of HCAIs, associated pathogens, implication of AMR and factors influencing emerging AMR mechanisms.
Keywords: Hospital care-associated infections, Nosocomial infections, Antimicrobial resistance, ESKAPE pathogens, AMR mechanisms, Polymyxins
1. Introduction
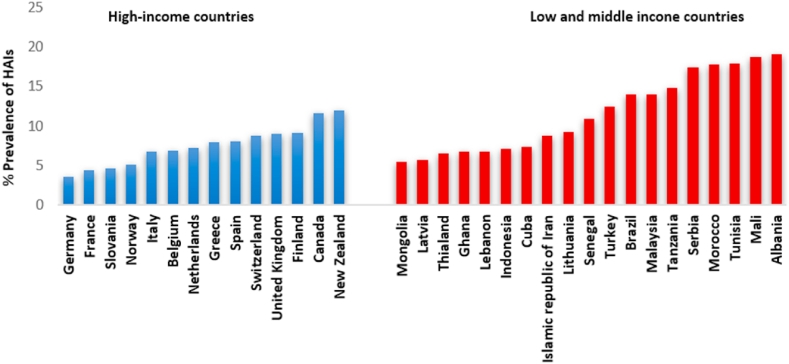
Hospital care-associated infections (HCAI) are major health safety issues worldwide and are defined as infections acquired while receiving treatment for medical or surgical conditions, which were not present during time of admission [1]. HCAIs include hospital-acquired (nosocomial), long-term care-associated, outpatient care-associated and home care-associated infections [2]. Information on the burden of HCAIs outside of the hospital setting is limited due to the onerous process in gathering reliable data on infection, prevention and control practices [3]. However, data on hospital settings points to hospital-acquired infections (HAIs) as the most frequent health challenge within the hospital setting [4]. Hospital-acquired infections (HAIs) occur globally in developed and developing countries with high morbidity and mortality [5]. For example, in the USA and Europe, HAIs are among the leading cause of death [6]. Additionally, HAIs result in prolonged hospital stay, increased microbial resistance to antimicrobials and elevated financial burden on the patient, family and the economy. HAIs from high-income countries show incidence rates ranging from 3.5% to 12% (Fig. 1) and are especially prevalent within intensive care units (ICU) with patient rate of infection about 40% [7,8]. These incidences varied from body sites and determined by the underlying condition of the patient following exposure to medical interventions and hospital environment [7]. High-risk population individuals include patients admitted to ICUs, burn wound and transplant patients, and neonates. The most frequent types of HAIs reported include central line-associated blood stream infections, surgical site infections (SSIs), and ventilator-associated pneumonia, as well as catheter-associated urinary tract infections as the most predominant pathologies [5]. There is an urgency to understand the evolving dimensions of AMR globally, especially in Africa with the recent emergence of ‘Global Priority Pathogens’ in association with HAIs. Therefore, this mini review highlights relevant pathogens majorly implicated in HAIs, prevalence in developed and developing countries. It also provided insights into possible AMR mechanisms employed by Gram-negative ESKAPE pathogens and re-emerging HAI pathogens such as Citrobacter spp. and Proteus spp., majorly to last resort antibiotic polymyxins. Additionally, some of the strategies to mitigate AMR and HAIs in ICUs and the general hospital environment are highlighted.
Fig. 1.
Prevalence of health care-associated infection in high and low/middle income countries by prevalence of HAIs, 1995–2020. 123.
2. Methods
2.1. Literature search strategy and extraction
Search terms “Antimicrobial Resistance”, “Antimicrobial Resistance Mechanisms”, “Global Burden of Hospital Acquired or Nosocomial Infections”, “ESKAPE and Pathogens Implicated in HAIs”, “Polymyxins Antimicrobial Resistance Mechanisms” “Plasmid-mediated Mobile Colistin Resistance” “Infection Prevention and Control of AMR” were used by at least two independent reviewers to conduct literature search. The search was conducted broadly on the global relevance of the highlighted keywords with special focus on their implications in Africa. Pubmed (National Library Medicine), Google Scholar, Scopus, Medline and Web of Science databases were used for literature search. Literature mapping and ranking performed with visual tools like Connected Papers (https://www.connectedpapers.com/), Open Knowledge Maps (https://openknowledgemaps.org/) and LitMap (https://www.litmaps.com/). Referenced database was built with Mendeley (Version 1.19.8) and information generated where processed and represented as figures, graphs and tables; other relevant data processed into paragraphs with headings and subheadings.
2.2. Pathogens Implicated in HAIs
Gram-negative bacteria are commonly implicated in HAIs, contributing as much as 87% of reported cases [9]; however, Staphylococcus aureus is the predominant Gram-positive strain [7]. Pseudomonas aeruginosa, Acinetobacter baumannii and Enterobacteriaceae are the most predominant Gram-negative strains within Europe and Asia [7,10,11]. A. baumannii is the most prevalent pathogen causing ventilator-associated pneumonia (VAP) and catheter-associated bloodstream infections (CAB) among high-risk populations, especially the immunocompromised in the ICUs [12]. In Africa, Klebsiella spp., S. aureus, Acinetobacter spp., and E.coli are the predominant pathogens [13] causing HAIs. High levels of HAI-related methicillin-resistant S. aureus (MRSA) are reported globally [12]. Carbapenem-resistant and Extended spectrum β-lactamases Enterobacteriaceae have been associated with paediatric HAIs [14].
2.3. Burden of antibiotic resistant bacteria and HAIs
Antibiotic resistant bacteria have contributed to the burden of HAIs globally with increasing health-risks especially in developing countries. Their consistent emergence and evolution have made many conventional antibiotics ineffective [15], where there is often only a limited set of last-resort antibiotics as the main treatment option for multidrug resistant (MDR) HAIs [16]. In 2017, the WHO listed twelve antibiotic resistant priority pathogens (Fig. 2.) requiring urgent attention for the development of new antibiotics [17]. The critical group included several high-risk pathogens, namely MDR A. baumannii, carbapenem-resistant Pseudomonas aeruginosa and Enterobacteriaceae. These pathogens exhibit both multidrug resistance and high levels of virulence, especially the notorious ‘ESKAPE’ (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species) pathogens [18,19]. These pathogens have been implicated in HAIs in both developed and developing countries, with a tendency to ‘escape’ lethal doses of antibiotics [20,21].
Fig. 2.
WHO Pathogen Priority list of antibiotic resistant bacteria; figure designed by the authors as adapted from WHO [22].
ESKAPE pathogens represent a group of Gram-positive and -negative bacterial pathogens with high-risk public health implications [20]. Methicillin and vancomycin–resistant S. aureus (MRSA-VRSA) as well as vancomycin-resistant E. faecium (VRE) (the Gram-positive bacteria of the ESKAPE group) have drawn global attention; however, infections caused by Gram-negative (“KAPE”) pathogens are likewise extremely critical causing high morbidity and mortality [23,24]. These pathogens are ubiquitous as they are not restricted to clinical inpatients only, but thrive in diverse environments including water, soil, poultry and air. This environmental diversity promotes unrestricted shedding, sharing and spread of antibiotic resistance genes (ARGs) aided by mobile genetic elements such as plasmids and integrons [25].
2.3.1. Acinetobacter baumannii
Acinetobacter spp., are widely distributed within hospital environments [26]. Acinetobacter spp are non-fermentative, non-motile Gram-negative coccobacilli which represent some notable nosocomial pathogens that rapidly develop resistance to multiple antibiotics [27]. An important member of the genus is A. baumannii, which colonises the skin, infects the bloodstream, urinary tracts and other soft tissues, especially in the immunocompromised individuals, accounting for 20% of ICU-associated infections globally [7]. A. baumannii is becoming MDR globally (to chloramphenicol, first and second-generation cephalosporins and aminopenicillins) with some hospitals reporting pan drug-resistant strains [28]. Also, it exhibits diverse resistance mechanisms such as production of β-lactamases, modifying aminoglycoside target sites, multidrug efflux pumps and antibiotic impermeability [29]. Combination therapy of minocycline/tigecycline and polymyxin is the treatment option since the emergence of A. baumannii strains resistant against carbapenems (meropenem, doripenem, imipenem) [27,30].
2.3.2. Pseudomonas aeruginosa
Pseudomonas aeruginosa is ubiquitously present within the normal intestinal flora of humans and widely distributed in the environment, including commonly hospital ICUs [31]. Its broad environmental distribution is based on its metabolic versatility as a pathogen that adapts and survives diverse environmental conditions (broad temperature ranges, salinity, actinomycin, etc) [32]. P. aeruginosa is an opportunistic pathogen that rarely causes infections in healthy individuals; however, it readily causes community and hospital-acquired infections in patients with weakened immune systems. P. aeruginosa has been implicated in surgical site infections, burn and eye injuries, skin/soft tissue, non-healing diabetic wounds, UTIs, bloodstream infections (BSIs) and pneumonia [31]. It is the fourth most isolated nosocomial pathogen, second major cause of VAP, the third most common Gram-negative pathogen implicated in BSIs and a major concern in patients with cystic fibrosis [33,34]. Inherently resistant to a number of antibiotics and antiseptics; P. aeruginosa exhibits diverse AMR mechanisms to many classes of antibiotics [35], including β-lactams, fluoroquinolones, aminoglycosides and third-generation cephalosporins. Historically, carbapenems were the antibiotics of choice for MDR P. aeruginosa; however, with emerging resistance, a combination of polymyxin and anti-pseudomonal agents (piperacillin/tazobactam, imipenem, aztreonam, ceftazidime, cefepime) are currently most effective in treating MDR P. aeruginosa infections [36].
2.3.3. Enterobacteriaceae
Enterobacteriaceae contain both opportunistic and “professional” pathogens which cause diverse infections, including UTIs, BSI, enteric infections, and hospital-acquired pneumonia. The notable pathogens include species of Klebsiella, Enterobacter, Escherichia coli, and Citrobacter [37]. The global challenge associated with Enterobacteriaceae is the production of ESBL for AMR [37]. ESBL hydrolyse most beta lactam antibiotics, including cephalosporins. These genes are present on plasmids that also harbour resistance to aminoglycosides, sulphonamides and cross-resistance to fluoroquinolones, typically making carbapenem the drug of choice for treatment of ESBL-producing pathogens [38]. However, the increased use of carbapenem has resulted in the development of Carbapenem-resistant Enterobacteriaceae (CRE) [39]. Currently, the treatment of infections from MDR Enterobacteriaceae is challenging due to limited treatment options that include some aminoglycosides, tigecycline and polymyxins [40].
2.3.4. Escherichia coli
Escherichia coli is the most common Gram-negative bacterium presenting both a clinical and epidemiological challenge [41]. It is naturally a commensal of the intestinal tract; however, several strains are specialized pathogens of humans and animals [42]. The commensal form inhabits the gastrointestinal tract of humans aiding in digestion [43], whiles the pathogenic forms cause infections resulting in two million deaths annually [44]. Pathogenic E. coli strains are subdivided into several pathotypes causing infections with three main clinical syndromes; UTIs, meningitis/sepsis and enteric diseases [45]. E. coli infections are commonly treated with ciprofloxacin, levofloxacin, fosfomycin and fluoroquinolones; however, resistance to multiple antibiotics have been reported [46]. Resistance to fluoroquinolones and the emergence of ESBLs are of major concerns in treatment [47]. Carbapenems are considered the drug of choice for MDR E. coli infections; however, resistance to carbapenems is also emerging [48].
2.3.5. Klebsiella pneumoniae
Klebsiella is second to E. coli as the most common member of the Enterobacteriaceae. It is responsible for community and hospital-acquired infections (respiratory tract infections, cardiovascular, bacteraemia and UTI [49]. K. pneumoniae causes HAIs, and is frequently isolated in immunocompromised patients with pneumonia, as well as neonatal infections [50]. K. pneumoniae is second to E. coli as the most frequent cause of hospital-acquired BSI globally [51]. It is mainly an opportunistic pathogen; however, hypervirulence (typically associated with hypercapsulation) [52] and resistance to antibiotics has emerged [53,54]. Carbapenem-resistant K. pneumoniae are emerging globally, causing high mortality rates [55], mostly due to acquisition of the namesake Klebsiella pneumoniae carbapenemases (KPC). MDR K. pneumoniae can thus be resistant to all beta-lactams, aminoglycosides, fluoroquinolones. Typically, polymyxin B and colistin E in combination with fosfomycin/tigecycline and some aminoglycosides are used as last resort treatment options [56].
2.4. Enterobacter species
Enterobacter spp. are facultative anaerobic bacteria that are natural commensals of the human gut microbiota [56], but also opportunistic pathogens, typically in the immunocompromised. Twenty-two species of Enterobacter have been identified with E. aerogenes and E. cloacae as the most frequently reported human pathogens. E. cloacae has been implicated in hospital-acquired sepsis, pneumonias, UTIs and postsurgical wound infections [57]. Most isolates of Enterobacter are susceptible to fluoroquinolones, trimethoprim, aminoglycosides and some β-lactams, however intrinsic resistance has emerged to ampicillin, cephalothin and first-generation cephalosporins [58,59] due to the possession of an AmpC type beta lactamase, often in conjunction with porin mutations [60]. The use of extensive broad-spectrum antibiotics has facilitated the development of resistant Enterobacter strains, particularly ESBL-producers in conjunction with multiple other resistance genes circulating globally [58]. Fourth-generation cephalosporins and carbapenems remain effective for treating infections, although there are reports of resistance to these antibiotics [58,61]. In addition, species of Enterobacter easily acquire antibiotic resistance mechanisms hence often restricting treatment options to tigecycline and colistin [57].
2.4.1. Citrobacter spp.
Citrobacter species are motile, non-spore forming bacilli diversely distributed in the soil, water, intestinal tracts of humans and animals as commensals [62,63]. There are 13 species of Citrobacter with Citrobacter freundii as the most commonly isolated [63]. As opportunistic pathogens, they are associated with UTIs, meningitis, septicemia, intestinal infections in neonates and immunocompromised individuals [64]. The different species of Citrobacter show different antimicrobial susceptibility profiles, with C. freundii displaying inherent resistance to ampicillin, carbenicillin and some quinolones [65,66]. Clinical species of Citrobacter are often reported to harbor ESBLs [67] and plasmid-mediated quinolone resistant markers [68]. C. freundii often displays resistance to piperacillin [69], third-generation cephalosporins [70], monobactams and some carbapenems [71]. MDR isolates of C. freundii resistant against quinolones, aminoglycosides, tetracycline and sulfonamides mediated by plasmids have been reported [69,72]. Based on limited therapeutic options for treating current MDR Citrobacter infections, selecting the appropriate antibiotic is imperative. The current treatment recommendation is with meropenems as first choice and fluroquinolones (moxifloxacin, ciprofloxacin) as alternatives [65].
2.5. Polymyxins and AMR bacteria implicated in HAIs
Polymyxins are a group of antibiotics of clinical importance in treating Gram-negative bacterial infections. They are natural products first isolated from Bacillus polymyxa in 1947 [73,74]. Polymyxin use declined in the 1970s due to toxicity concerns and the availability of effective and broader spectrum alternative antibiotics [75]. The rise of MDR pathogens and lack of new antibiotics has necessitated reconsideration of the therapeutic use of polymyxin B and colistin E [75]. Polymyxins are active against P. aeruginosa, A. baumannii and some Enterobacteriaceae; however, Proteus spp., and Serratia marcescens are intrinsically resistant [76]. Polymyxins disrupt the outer membrane integrity of bacterial cells in a poorly-understood way [77,78]. The current toxicity reports of polymyxins show less incidence of nephrotoxicity [79,80]. Following reduction in clinical use, polymyxins were reserved for management of cystic fibrosis, ear and eye infections [81]. However, due to the recent emergence of MDR pathogens, polymyxins B and E have resurfaced as a single dosage and in combinations with other antibiotics (meropenem, imipenem/cilastatin, ampicillin/sulbactam) for clinical use [76].
2.6. Structure and mechanism of action
The polymyxins are five structurally different compounds (A, B, C, D and E), with polymyxin B/E in clinical use [79]. The physicochemical properties of polymyxins are similar to cationic antimicrobial peptides such as defensins of the eukaryotic innate immunity [82]. Polymyxins are non-ribosomal, cyclic lipopeptides and contain a mixture of D and l-amino acids as a general characteristic of most peptide antibiotics. They possess a heptapeptide ring of amino acids and a fatty acid attached to a tripeptide side chain through an amide bond [83]. The amino acid component of polymyxin B includes d-phenylalanine, l-threonine and six 2, 4-diaminobutyric acid residues. Colistin has d-leucine in place of d-phenylalanine. This mixture of lipophilic and hydrophilic groups confers an amphiphilic nature to polymyxins, giving them their bactericidal activity [84].
2.7. Polymyxin interactions with LPS-divalent cations
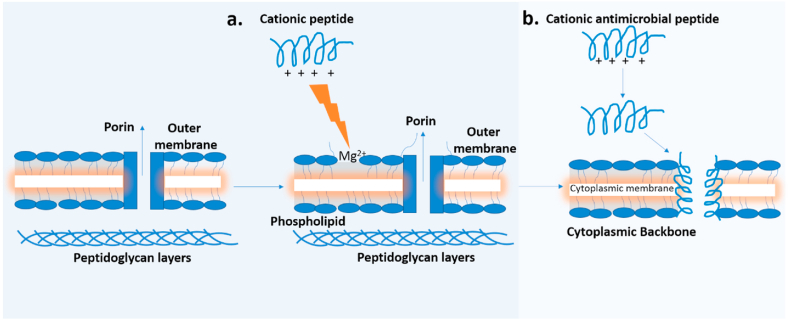
Polymyxin B interacts with the LPS of the outer and potentially LPS precursors in the inner membrane [77] of Gram-negative bacteria by competitively displacing divalent cations (Ca2+and Mg2+) from the negatively charged phosphate group of lipid A. This ultimately results in disruption of the barrier function of the outer membrane [77], as well as depolarization of the inner membrane [85]. This process is described as a self-promoted uptake pathway, where the break in membrane results in passage of various molecules including the cationic peptides (polymyxin). The interaction of the divalent cations on the polypeptide, whose binding affinity is three orders greater than the affinity of the native cations, competitively displaces cations on the binding surface of the LPS. The bulky nature of the polypeptide also disrupts the normal barrier property of the outer membrane (Fig. 3.) [86]. The divalent cations displacement weakened the outer membrane resulting in membrane leakage/breakage. Subsequently, the fatty acid chain of the peptide is directed towards the interior of the cell membrane allowing the heptapeptide ring of amino acids to form an inward channel to further disrupts the membrane integrity leading to bacterial cell death [86,87]. The high transmembrane potential, high negatively charged lipids, lack of cationic lipids and cholesterol generated enhances polymyxins selectivity for bacterial pathogens to eukaryotic hosts [86].
Fig. 3.
The self-promoted uptake of cationic peptides across the outer membrane of Gram-negative bacteria, (a.) The cationic peptide competitively displaces Mg2+ from the LPS causing outer membrane disruption and uptake of antibiotic; (b.) Mechanism of bacterial killing by cationic peptides: the positively charged peptides bind to the negatively charged LPS leading to thinning of the bilayer (cytoplasmic membrane). Membrane potential generated inserts peptide into the membrane to form channels leading to leakage of cytoplasmic molecules and cell death. Figure adapted from (1) [86].
2.8. Bacterial mechanisms of resistance to polymyxins
Resistance to polymyxin has been reported in a number of bacterial pathogens, especially MDR bacteria that used to be sensitive, but became polymyxin resistant after their reintroduction [88]. This emerging resistance is attributed to the increased and inappropriate use of polymyxins globally [89,90]. Investigating polymyxin resistance has led to the detection of diverse mechanisms, including modification of LPS, loss of LPS, efflux pump activity, capsule formation and overexpression of outer membrane protein (OprH) [82]. These resistance mechanisms are either intrinsic (such as in Proteus spp., Serratia spp., Morganella morganii, and Burkholderia spp.), mutational, acquired or adaptive [91] (Table 1).
Table 1.
Mechanisms of polymyxin resistance.
| Bacteria | Polymyxin Mechanisms of Resistance | References |
|---|---|---|
| Klebsiella pneumoniae |
|
[[82], [92], [93]] |
| Acinetobacter baumannii |
|
[92] |
| Pseudomonas aeruginosa |
|
[[82], [93]] |
| Enterobacter |
|
[[94], [95], [96]] |
| E. coli |
|
[[74], [97], [98], [99]] |
| Proteus |
|
[[100], [101]] |
| Citrobacter |
|
[[82], [95]] |
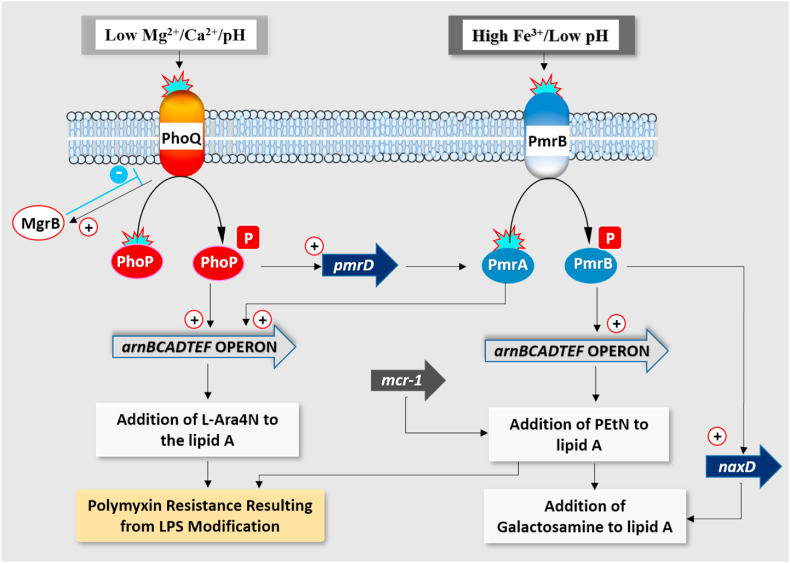
The majority of the resistance mechanisms target the lipid A of LPS, the initial site of action of polymyxin [74]. Two major two-component systems (TCS) (PhoPQ and PmrAB) play crucial roles in regulating gene expressions for lipid A modification in Gram-negative bacteria [94]. In some Enterobacteriaceae, the LPS is targeted by modification of lipid A moiety with 4-amino-4-deoxy-l-arabinose (L-Ara4N) or phosphoethanolamine (PEtN) [77]. In environments with low Mg2+ and Ca2+, PhoQ is activated leading to activation and phosphorylation of cytoplasmic phoP, promoting transcription of the pmrD gene. The pmrD product activates PmrAB at a posttranscriptional level ensuring phosphorylation of PmrA to mediate synthesis or incorporation of L-Ara4N into the lipid A moiety of LPS [102] (Fig. 4.). In some Enterobacteriaceae, like E. cloacae, the L-Ara4N modification is exclusively controlled by the PhoPQ system [103].
Fig. 4.
Schematic overview of mechanisms of LPS modification involved in polymyxin resistance in Gram-negative bacilli (Adapted from Ezadi et al., 2019). [104].
2.9. Role of the mgrB gene in bacterial resistance to polymyxins
MgrB [105] is a small regulatory transmembrane protein with 47 amino acids whose function inhibits the kinase activity of PhoQ or stimulates its phosphatase [106] (Fig. 3b). MgrB thus represses PhoQ activity post-translationally and inactivation of MgrB therefore results in high baseline activation of the PhoPQ signalling pathway. Consequently, PhoPQ-controlled OM modifications are also induced, resulting in high Polymyxin resistance. Mutations in MgrB have been observed in clinical isolates displaying colistin resistance, implicating the mgrB gene in clinical treatment failure [107,108]. Mutations include frameshifts, deletion of gene segments, amino acid substitutions, nonsense mutations and inactivation; the most frequently reported is by transposition of insertional sequences [105]. MgrB is conserved particularly in Klebsiella pneumoniae [109], E. coli [106] and Enterobacter spp [110].
2.10. Plasmid-mediated polymyxin resistance
Resistance to polymyxin was historically linked to chromosomal-related mechanisms with no reports on horizontal gene transfer. However, a plasmid-mediated colistin resistance gene encoding a PEtN transferase was detected in E. coli strains from animal sources in China [111]. The modification with PEtN is not a novel mechanism of resistance; rather, the transferability of the gene among bacteria especially MDR strains, represents a challenge for polymyxin treatment. This plasmid-borne mobile colistin resistance gene (mcr-1) has been reported in more than 30 countries [112], in species of Klebsiella, Enterobacter, Citrobacter, Proteus, Salmonella and E. coli [113]. Currently, new mcr-like genes have emerged with mcr-2 [114], mcr-3 [115], mcr-4 [116], mcr-5 [117], mcr-6 (3) [118], mcr-7 [119], mcr-8 [120], mcr-9 [121] and mcr-10 [122] currently in circulation. These genes have variations compared to the ancestral mcr-1, with about 30 SNPs in mcr-3 [123]. Mcr-2, 3, 4, 5, 6, 7, 8, 9 and 10 share 81%, 32.5%, 34%, 36.1%, 83%,35%, 31%, 36% and 29.31% amino acid sequence similarity with MCR-1 respectively [122,124,125]. The presence of mcr genes on conjugative plasmids replicon such as IncI2, IncHI2, IncX4 and IncF have been confirmed [113,125]. The origin of the mcr-1 gene is unclear; however, the chromosomal region in a Moraxella porci displays significant homology to the mcr-1 structure [118]. The gene is also flanked by insertion sequence (ISApl1) with the transposon Tn6330 as the key element mediating the translocation of mcr-1 into various plasmids [126]. Although mcr genes were initially mainly reported as plasmid-mediated, the ISApl1 and the mcr-1 cassette has been found on the chromosome of E. coli suggesting its integration into the bacterial chromosome [127,128]. The mcr-1 is currently the predominant gene circulating globally with prevalence in the environment (22%), animals (11%) and humans (2.5%) [128] necessitating investigations into its epidemiology and the resistance mechanisms to improve clinical treatment.
2.11. Bacterial phenotypic mechanisms of survival against polymyxins
Host organisms utilize diverse defence mechanisms to reduce the burden of infections from bacterial pathogens. They avoid exposure to pathogens (barriers for protection), resist infections (host immunity) or tolerate the presence of the pathogen [129]. However, bacteria alter their lifestyle in response to these host survival mechanisms [130]. These responses include gene expression and protein activity, lifestyle switch from avirulent to more virulent forms within the hosts, biofilm formation/swarming and other adaptive mechanisms [131]. As a particularly well-understood example, bacterial LPS modifications promote bacterial survival against host immunity factors like antimicrobial peptides [132]. The O antigen of the LPS complex has repeating oligosaccharide units that are highly variable immunologically and block the initiation of the complement system of the host innate immune system [133], while the non-repeating lipid A core strengthens the integrity of the outer membrane [133].
3. Discussion
The hospital is characterized as a high health risk environment particularly with reports of HAIs within developed and developing countries [134]. The burden HAIs poses ranges from elevated financial burden, increase in disease severity, high incidence of antimicrobial resistance, morbidity and elevated rates of mortality. Within the hospital setting, bacterial pathogens are associated with nosocomial infections with majority displaying resistance to conventional and last resort antibiotics [4]. Members of ESKAPE pathogens are high-risk pathogens and majorly implicated in HAIs with increased tendency to display multidrug resistance. They are characterized as global priority pathogens as they play critical roles in increasing disease severity by compounding rates of morbidity and mortality.
The multidrug resistance phenotype exhibited by ‘Global Priority Pathogens’ particularly Gram-negative bacteria prompted dependence on two last resort antibiotics carbapenems and polymyxins. Between 2000 and 2010, global use of carbapenems and polymyxins increased by 34% and 14% respectively (4) [135]. However, this emergence of resistance to carbapenems and polymyxins coupled with presence of mobile genetic elements such as plasmids harboring resistant markers challenged treatment options. Gram-negative bacteria developed resistance to polymyxins following reintroduction into clinical and animal use.
Polymyxin resistance mechanisms commonly employed by bacteria were chromosomally or intrinsically mediated with mechanisms such as modification and loss of LPS, efflux pump activity, formation of capsule and overexpression of membrane outer protein. However, recent emergence of circulating plasmid-mediated colistin resistance genes in bacterial strains from animal sources has been reported in over 20 countries with Gram-negative bacteria majorly implicated [112]. The variations in the mobile colistin resistance genes (mcr) predominant in the environment as compared to animals and humans complicates AMR challenges and HAIs. This is further compounded by the transfer of these MDR genes as facilitated by these plasmids.
3.1. Strategies to combat AMR in ICUs
Generally, strategies to combat AMR would majorly involve Infection Prevention/Control (IPCs) and antimicrobial management strategies. By implementing IPCs and antimicrobial stewardship programs in hospital environments and ICUs, the burden AMR poses would be mitigated.
4. Infection Prevention and Control
Emergence of AMR is inevitable as bacteria develop ways to circumvent effectiveness of antibiotics. Administration of antibiotics contributes to selection of antimicrobial resistant bacteria; however, increased complications is linked with prolong antibiotic selection pressure during treatment within the ICU. In this regard, AMR is better prevented and best mitigated by nonpharmacological or Infection Prevention and Control practices (5) [136]. Also, good hygiene practices including hand washing and proper disinfection practices, frequent hospital surveillance of emerging pathogens, establishing effective patient cohorting systems, effective waste disposal protocols/systems and building infrastructural/human capacity and expertise.
5. Antimicrobial management strategies
Proper antibiotic management strategies would involve establishment of good antimicrobial stewardship program in hospitals and use of narrow spectrum antibiotics. Also, effective diagnosis coupled with treatment of infection, shorter course administration of antibiotics and effectively applying pharmacodynamics and kinetics principles in drug administration could help mitigate the rise and spread of resistant bacteria in ICUs and within the hospital environment. Combating antibiotic resistance requires a multifaceted approach that entails international, national and individual level collaborative action to mitigate global spread. Table 2 summarizes a list of strategies to reduce the burden of AMR particularly in hospital setting.
Table 2.
Strategies to combat AMR in ICUs.
| Strategies | Interventions | Relevant global guides and tools/Evidence based study/References |
|---|---|---|
| Nonpharmacological/Infection, Prevention and control |
|
[137], [138], [139] |
|
[140,141] https://apps.who.int/iris/handle/10665/312226. License: CC BY-NC-SA 3.0 IGO; |
|
|
[142] | |
|
WHO Global strategy for containment of AMR, 2001; https://apps.who.int/iris/handle/10665/66860 | |
|
[143,144] | |
|
[143,145] | |
| Antibiotic management |
|
[144] WHO Global strategy for containment of AMR, 2001; https://apps.who.int/iris/handle/10665/66860https://apps.who.int/iris/bitstream/handle/10665/329404/9789241515481-eng.pdf |
|
[145] https://apps.who.int/iris/bitstream/handle/10665/205912/B4691.pdf | |
|
[[146], [147], [148], [149]] https://apps.who.int/iris/bitstream/handle/10665/205912/B4691.pdf | |
|
[150,151] https://apps.who.int/iris/bitstream/handle/10665/205912/B4691.pdf |
6. Conclusion
The burden of hospital acquired infections is increasing globally. The consistent emergence of antimicrobial resistant bacteria, particularly Gram-negative bacteria is contributing to the challenge of HAIs. The plethora of innate resistance mechanisms coupled with acquired genes displayed by Gram-negative bacteria further complicates the efficiency of antibiotics, especially last-resort such as polymyxins. An understanding of the interplay of HAIs and antimicrobial resistant Gram-negative bacteria could inform effective infection control practices to enable implementation of safety protocols.
Financial disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Monegro A.F., Muppidi V., Regunath H. StatPearls. StatPearls Publishing; 2020. Hospital Acquired Infections.http://www.ncbi.nlm.nih.gov/pubmed/28722887 [PubMed] [Google Scholar]
- 2.Horan T.C., Andrus M., Dudeck M.A. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am. J. Infect. Control. 2008;36(5):309–332. doi: 10.1016/j.ajic.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Kung H.-C., Hoyert D.L., Xu J., Murphy S.L. Natl. Vital Stat. Rep. 2005;56 http://www.cdc.gov/nchs/deaths.htm Number 10, (April 24, 2008) (Vol. 56. [PubMed] [Google Scholar]
- 4.Peleg A.Y., Hooper D.C. Hospital-acquired infections due to gram-negative bacteria. N. Engl. J. Med. 2010;362(19):1804–1813. doi: 10.1056/nejmra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan H.A., Baig F.K., Mehboob R. Hainan Medical University; 2017. Nosocomial Infections: Epidemiology, Prevention, Control and Surveillance. Asian Pacific Journal of Tropical Biomedicine. May 1. [DOI] [Google Scholar]
- 6.McFee R.B. Nosocomial or hospital-acquired infections: an overview. Disease-a-Month: DM. 2009;55(7):422–438. doi: 10.1016/j.disamonth.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vincent J.L., Rello J., Marshall J., Silva E., Anzueto A., Martin C.D.…Reinhart K. International study of the prevalence and outcomes of infection in intensive care units. JAMA, J. Am. Med. Assoc. 2009;302(21):2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 8.Nejad S.B., Allegranzi B., Syed S.B., Ellisc B., Pittetd D. Bulletin of the World Health Organization; 2011, October. Infections liées aux soins de santé en afrique: Une étude systématique. [DOI] [Google Scholar]
- 9.Sydnor E.R.M., Perl T.M. Hospital epidemiology and infection control in acute-care settings. Clin. Microbiol. Rev. 2011;24(1):141–173. doi: 10.1128/CMR.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Who . WHO Report; 2014. Antimicrobial Resistance: Global Report on Surveillance. :9789241564748, April 2014. [Google Scholar]
- 11.Saleem Z., Godman B., Hassali M.A., Hashmi F.K., Azhar F., Rehman I.U. Taylor and Francis Ltd; 2019. Point Prevalence Surveys of Health-Care-Associated Infections: a Systematic Review. Pathogens and Global Health. May 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allegranzi B., Nejad S.B., Combescure C., Graafmans W., Attar H., Donaldson L., Pittet D. Burden of endemic health-care-associated infection in developing countries: systematic review and meta-analysis. Lancet. 2011;377(9761):228–241. doi: 10.1016/S0140-6736(10)61458-4. [DOI] [PubMed] [Google Scholar]
- 13.Irek E.O., Amupitan A.A., Obadare T.O., Aboderin A.O. AOSIS OpenJournals Publishing AOSIS (Pty) Ltd; 2018. A Systematic Review of Healthcare-Associated Infections in Africa: an Antimicrobial Resistance Perspective. African Journal of Laboratory Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flokas M.E., Detsis M., Alevizakos M., Mylonakis E. Prevalence of ESBL-producing Enterobacteriaceae in paediatric urinary tract infections: a systematic review and meta-analysis. J. Infect. 2016;73(6):547–557. doi: 10.1016/J.JINF.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Auer G.K., Oliver P.M., Rajendram M., Lin T.-Y., Yao Q., Jensen G.J., Weibel D.B. Bacterial swarming reduces Proteus mirabilis and Vibrio parahaemolyticus cell stiffness and increases β-lactam susceptibility. mBio. 2019;10(5) doi: 10.1128/MBIO.00210-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frieri M., Kumar K., Boutin A. Antibiotic resistance. Journal of Infection and Public Health. 2017;10(4):369–378. doi: 10.1016/J.JIPH.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Tacconelli E., Carrara E., Savoldi A., Kattula D., Burkert F. 2015. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics.http://www.cdc.gov/drugresistance/threat-report-2013/ Retrieved from. [Google Scholar]
- 18.Agaba P., Tumukunde J., Tindimwebwa J.V.B., Kwizera A. Nosocomial bacterial infections and their antimicrobial susceptibility patterns among patients in Ugandan intensive care units : a cross sectional study. BMC Res. Notes. 2017:1–12. doi: 10.1186/s13104-017-2695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monegro A.F., Regunath H. StatPearls. StatPearls Publishing; 2018. Hospital acquired infections. [PubMed] [Google Scholar]
- 20.Rice L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008;197(8):1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 21.Founou R.C., Founou L.L., Essack S.Y. Public Library of Science; 2017, December 1. Clinical and Economic Impact of Antibiotic Resistance in Developing Countries: A Systematic Review and Meta-Analysis. PLoS ONE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO Microbial fact sheets. World health organization guidelines for drinking. WHO Report, page accessed in 2019. 2004:221–296. http://scholar.google.com/scholar?hl=en&btnG=Search&q=intitle:Microbial+fact+sheets#0 [Google Scholar]
- 23.Tacconelli E., Carrara E., Savoldi A., et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018 Mar;18(3):318–327. doi: 10.1016/s1473-3099(17)30753-3. PMID: 29276051. [DOI] [PubMed] [Google Scholar]
- 24.Breijyeh Z., Jubeh B., Karaman R. Resistance of gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules. 2020;25(6) doi: 10.3390/MOLECULES25061340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Wintersdorff C.J.H., Penders J., van Niekerk J.M., Mills N.D., Majumder S., van Alphen L.B.…Wolffs P.F.G. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 2016;0(FEB):173. doi: 10.3389/FMICB.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin M.-F., Lan C.-Y. Antimicrobial resistance in Acinetobacter baumannii: from bench to bedside. World Journal of Clinical Cases : WJCC. 2014;2(12):787. doi: 10.12998/WJCC.V2.I12.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee C.-R., Lee J.H., Park M., Park K.S., Bae I.K., Kim Y.B.…Lee S.H. Biology of acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 2017;7(MAR):55. doi: 10.3389/FCIMB.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falagas M.E., Makris G.C., Dimopoulos G., Matthaiou D.K. Heteroresistance: a concern of increasing clinical significance? Clin. Microbiol. Infect. 2008;14(2):101–104. doi: 10.1111/j.1469-0691.2007.01912.x. [DOI] [PubMed] [Google Scholar]
- 29.Matar G.M., Sun Yoon S., University Y., Korea Ravi Jhaveri S., Lee C.-R., Hun Lee J.…Hee Lee S. Biology of acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Frontiers in Cellular and Infection Microbiology | Www.Frontiersin.Org. 2017;7:55. doi: 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doi Y., Murray G.L., Peleg A.Y. Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 2015;36(1):85–98. doi: 10.1055/s-0034-1398388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Driscoll J.A., Brody S.L., Kollef M.H. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2012:351–368. doi: 10.2165/00003495-200767030-00003. 2007 67:3, 67(3) [DOI] [PubMed] [Google Scholar]
- 32.Diggle S.P., Whiteley M. Microbe Profile: Pseudomonas aeruginosa: opportunistic pathogen and lab rat. Microbiology. 2020;166(1):30. doi: 10.1099/MIC.0.000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elborn J.S. Cystic fibrosis. Lancet. 2016;388(10059):2519–2531. doi: 10.1016/S0140-6736(16)00576-6. [DOI] [PubMed] [Google Scholar]
- 34.Campos JC. de M., Antunes L.C., Ferreira R.B. 2020. Global Priority Pathogens: Virulence, Antimicrobial Resistance and Prospective Treatment Options.https://www.futuremedicine.com/doi/abs/10.2217/fmb-2019-0333 Jun 4 [cited 2021 Sep 14];15(8). Available from: [DOI] [PubMed] [Google Scholar]
- 35.Pachori P., Gothalwal R., Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes & Diseases. 2019;6(2):109. doi: 10.1016/J.GENDIS.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bassetti M., Vena A., Croxatto A., Righi E., Guery B. How to manage Pseudomonas aeruginosa infections. Drugs Context (US) 2018;7 doi: 10.7573/DIC.212527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paterson D.L. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Infect. Control. 2006;34(5):S20–S28. doi: 10.1016/J.AJIC.2006.05.238. [DOI] [PubMed] [Google Scholar]
- 38.Pitout J.D., Laupland K.B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 2008;8(3):159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 39.Rodríguez-Baño J., Gutiérrez-Gutiérrez B., Machuca I., Pascual A. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Rev. 2018;31(2) doi: 10.1128/CMR.00079-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morrill H.J., Pogue J.M., Kaye K.S., LaPlante K.L. Treatment options for carbapenem-resistant Enterobacteriaceae infections. Open Forum Infect. Dis. 2015;2(2) doi: 10.1093/OFID/OFV050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paitan Y. Current trends in antimicrobial resistance of Escherichia coli. Curr. Top. Microbiol. Immunol. 2018;416:181–211. doi: 10.1007/82_2018_110. [DOI] [PubMed] [Google Scholar]
- 42.Elsas J. D. van, Semenov A.V., Costa R., Trevors J.T. Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 2011;5(2):173. doi: 10.1038/ISMEJ.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berthe T., Ratajczak M., Clermont O., Denamur E., Petit F. Evidence for coexistence of distinct Escherichia coli populations in various aquatic environments and their survival in estuary water. Appl. Environ. Microbiol. 2013;79(15):4684. doi: 10.1128/AEM.00698-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaper J.B., Nataro J.P., Mobley H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004:123–140. doi: 10.1038/nrmicro818. 2004 2:2, 2(2) [DOI] [PubMed] [Google Scholar]
- 45.Mainil J. Escherichia coli virulence factors. Vet. Immunol. Immunopathol. 2013;152(1–2):2–12. doi: 10.1016/J.VETIMM.2012.09.032. [DOI] [PubMed] [Google Scholar]
- 46.Ramírez-Castillo F.Y., Moreno-Flores A.C., Avelar-González F.J., Márquez-Díaz F., Harel J., Guerrero-Barrera A.L. An evaluation of multidrug-resistant Escherichia coli isolates in urinary tract infections from Aguascalientes, Mexico: cross-sectional study. Ann. Clin. Microbiol. Antimicrob. 2018;17(1):34. doi: 10.1186/S12941-018-0286-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicolle L.E. Urinary tract infection. Crit. Care Clin. 2013;29(3):699–715. doi: 10.1016/J.CCC.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 48.Hidron A.I., Edwards J.R., Patel J., Horan T.C., Sievert D.M., Pollock D.A., Facilities N.H.S.N.T., H. S. N P.N. Antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2006–2007. Infect. Control Hosp. Epidemiol. 2008;29(11):996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 49.Lee D., Oh J.Y., Sum S., Park H.-M. Prevalence and antimicrobial resistance of Klebsiella species isolated from clinically ill companion animals. J. Vet. Sci. 2021;22(2):1–13. doi: 10.4142/JVS.2021.22.E17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paczosa M.K., Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. : MMBR (Microbiol. Mol. Biol. Rev.) 2016;80(3):629. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reid C.B., Steele L., Pasquill K., Parfitt E.C., Laupland K.B. Occurrence and determinants of Klebsiella species bloodstream infection in the western interior of British Columbia, Canada. BMC Infect. Dis. 2019;19(1):1–7. doi: 10.1186/S12879-019-4706-8. 2019 19:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shon A.S., Bajwa R.P., Russo T.A. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bengoechea J.A., Sa Pessoa J. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2019;43(2):123–144. doi: 10.1093/FEMSRE/FUY043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flores-Valdez M., Ares M.A., Rosales-Reyes R., Torres J., Girón J.A., Weimer B.C.…Cruz M. A. D. la. Whole genome sequencing of pediatric Klebsiella pneumoniae strains reveals important insights into their virulence-associated traits. Front. Microbiol. 2021;12 doi: 10.3389/FMICB.2021.711577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su S., Zhang J., Zhao Y., Yu L., Wang Y., Wang Y.…Zhang X. Outbreak of KPC-2 carbapenem-resistant Klebsiella pneumoniae ST76 and carbapenem-resistant K2 hypervirulent Klebsiella pneumoniae ST375 strains in northeast China: molecular and virulent characteristics. BMC Infect. Dis. 2020;20(1) doi: 10.1186/S12879-020-05143-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee C.R., Lee J.H., Park K.S., Kim Y.B., Jeong B.C., Lee S.H. Frontiers Media S.A; 2016. Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Frontiers in Microbiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mezzatesta M.L., Gona F., Stefani S. Enterobacter cloacae complex. clinical impact and emerging antibiotic resistance. 2012;7(7):887–902. doi: 10.2217/FMB.12.61. [DOI] [PubMed] [Google Scholar]
- 58.Davin-Regli A., Lavigne J.-P., Pagès J.-M. Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin. Microbiol. Rev. 2019;32(4) doi: 10.1128/CMR.00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanders W.E., Sanders C.C. Enterobacter spp. Pathogens Poised To Flourish at the Turn of the Century. 1997;10(2):220–241. doi: 10.1128/cmr.10.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacoby G.A. 2009. AmpC Β-Lactamases. Clin Microbiol Rev [Internet] Jan[cited 2022 Jul 6];22(1):161–82. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harris P.N.A., Wei J.Y., Shen A.W., Abdile A.A., Paynter S., Huxley R.R.…Paterson D.L. Carbapenems versus alternative antibiotics for the treatment of bloodstream infections caused by Enterobacter, Citrobacter or Serratia species: a systematic review with meta-analysis. J. Antimicrob. Chemother. 2016;71(2):296–306. doi: 10.1093/JAC/DKV346. [DOI] [PubMed] [Google Scholar]
- 62.Ranjan K.P., Ranjan N. Citrobacter: an emerging health care associated urinary pathogen. Urol. Ann. 2013;5(4):313. [PMC free article] [PubMed] [Google Scholar]
- 63.Nayar Ritu, Shukla A.I. Epidemiology, prevalence and identification of citobacter spesies in clinical spesimen in a tertiary care hospital in India. International Journal of Scientific and Research Publications. 2014;4(4):2250–3153. [Google Scholar]
- 64.Liu L., Lan R., Liu L., Wang Y., Zhang Y., Wang Y., et al. 2017. Antimicrobial Resistance and Cytotoxicity of Citrobacter Spp. In Maanshan Anhui Province, China.http://journal.frontiersin.org/article/10.3389/fmicb.2017.01357/full [cited 2020 Jul 18];8(JUL):1357. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kus J.V., Burrows L.L. Infections due to Citrobacter and Enterobacter. XPharm: The Comprehensive Pharmacology Reference. 2007;1–12 doi: 10.1016/B978-008055232-3.60868-2. [DOI] [Google Scholar]
- 66.Liu L.H., Wang N.Y., Wu A.Y.J., Lin C.C., Lee C.M., Liu C.P. Citrobacter freundii bacteremia: risk factors of mortality and prevalence of resistance genes. J. Microbiol. Immunol. Infect. 2018;51(4):565–572. doi: 10.1016/J.JMII.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 67.Moland E.S., Hanson N.D., Black J.A., Hossain A., Song W., Thomson K.S. Prevalence of newer β-lactamases in gram-negative clinical isolates collected in the United States from 2001 to 2002. J. Clin. Microbiol. 2006;44(9):3318. doi: 10.1128/JCM.00756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shao Y., Xiong Z., Li X., Hu L., Shen J., Li T., Hu F., Chen S. Prevalence of plasmid-mediated quinolone resistance determinants in Citrobacter freundii isolates from Anhui province, PR China. J. Med. Microbiol. 2011;60(Pt 12):1801–1805. doi: 10.1099/jmm.0.034082-0. [DOI] [PubMed] [Google Scholar]
- 69.Khorasani G., Salehifar E., Eslami G. Profile of microorganisms and antimicrobial resistance at a tertiary care referral burn centre in Iran: emergence of Citrobacter freundii as a common microorganism. Burns. 2008;34(7):947–952. doi: 10.1016/J.BURNS.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 70.Jones R.N., Jenkins S.G., Hoban D.J., Pfaller M.A., Ramphal R. In vitro efficacy of six cephalosporins tested against Enterobacteriaceae isolated at 38 North American medical centres participating in the SENTRY Antimicrobial Surveillance Program, 1997–1998. Int. J. Antimicrob. Agents. 2000;15(2):111–118. doi: 10.1016/S0924-8579(00)00152-7. [DOI] [PubMed] [Google Scholar]
- 71.Gaibani P., Ambretti S., Farruggia P., Bua G., Berlingeri A., Tamburini M.V.…Sambri V. Outbreak of Citrobacter freundii carrying VIM-1 in an Italian Hospital, identified during the carbapenemases screening actions, June 2012. Int. J. Infect. Dis. 2013;17(9):e714–e717. doi: 10.1016/J.IJID.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 72.Liu L., Qin L., Hao S., Lan R., Xu B., Guo Y.…Zhao C. Lineage, antimicrobial resistance and virulence of Citrobacter spp. Pathogens. 2020;9(3) doi: 10.3390/PATHOGENS9030195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stansly P.G., Schlosser M.E. Studies on polymyxin: isolation and identification of Bacillus polymyxa and differentiation of polymyxin from certain known antibiotics. J. Bacteriol. 1947;54(5):549–556. doi: 10.1128/JB.54.5.549-556.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trimble M.J., Mlynárčik P., Kolář M., Hancock R.E.W. Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harbor Perspectives in Medicine. 2016;6(10) doi: 10.1101/CSHPERSPECT.A025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bergen P.J., Landersdorfer C.B., Lee H.J., Li J., Nation R.L. ‘Old’ antibiotics for emerging multidrug-resistant bacteria. Curr. Opin. Infect. Dis. 2012;25(6):626. doi: 10.1097/QCO.0B013E328358AFE5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Falagas M.E., Rafailidis P.I., Matthaiou D.K. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist. Updates. 2010;13(4–5):132–138. doi: 10.1016/J.DRUP.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 77.Nation R.L., Li J. Colistin in the 21st century. Curr. Opin. Infect. Dis. 2009;22(6):535. doi: 10.1097/QCO.0B013E328332E672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sabnis A., Hagart K.L., Klöckner A., Becce M., Evans L.E., Furniss R., Mavridou D.A., Murphy R., Stevens M.M., Davies J.C., Larrouy-Maumus G.J., Clarke T.B., Edwards A.M. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. Elife. 2021;10 doi: 10.7554/eLife.65836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Falagas M.E., Kasiakou S.K. Toxicity of polymyxins: a systematic review of the evidence from old and recent studies. Crit. Care. 2006;10(1):R27. doi: 10.1186/CC3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Phe K., Lee Y., McDaneld P.M., Prasad N., Yin T., Figueroa D.A.…Tam V.H. In vitro assessment and multicenter cohort study of comparative nephrotoxicity rates associated with colistimethate versus polymyxin b therapy. Antimicrob. Agents Chemother. 2014;58(5):2740–2746. doi: 10.1128/AAC.02476-13/SUPPL_FILE/ZAC005142831SO1.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reed M.D., Stern R.C., O'Riordan M.A., Blumer J.L. The pharmacokinetics of colistin in patients with cystic fibrosis. J. Clin. Pharmacol. 2001;41(6):645–654. doi: 10.1177/00912700122010537. [DOI] [PubMed] [Google Scholar]
- 82.Olaitan A.O., Morand S., Rolain J.M. Frontiers Media S.A; 2014, November 26. Mechanisms of Polymyxin Resistance: Acquired and Intrinsic Resistance in Bacteria. Frontiers in Microbiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gallardo-Godoy A., Muldoon C., Becker B., Elliott A.G., Lash L.H., Huang J.X.…Cooper M.A. 2016. Activity and Predicted Nephrotoxicity of Synthetic Antibiotics Based on Polymyxin B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Velkov T., Roberts K.D., Nation R.L., Thompson P.E., Li J. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol. 2013;8(6):711–724. doi: 10.2217/FMB.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Epand R.M., Walker C., Epand R.F., Magarvey N.A. Molecular mechanisms of membrane targeting antibiotics. Biochim. Biophys. Acta. 2016;1858(5):980–987. doi: 10.1016/j.bbamem.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 86.Hancock R.E.W. Peptide antibiotics. Lancet. 1997;349(9049):418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 87.Mohapatra S.S., Dwibedy S.K., Padhy I. Polymyxins, the last-resort antibiotics: mode of action, resistance emergence, and potential solutions. J. Biosci. 2021;(3):1–18. doi: 10.1007/S12038-021-00209-8. 2021 46:3, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bogdanovich T., Adams-Haduch J.M., Tian G.B., Nguyen M.H., Kwak E.J., Muto C.A., Doi Y. Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)–Producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin. Infect. Dis. 2011;53(4):373–376. doi: 10.1093/CID/CIR401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baron S., Hadjadj L., Rolain J.M., Olaitan A.O. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int. J. Antimicrob. Agents. 2016;48(6):583–591. doi: 10.1016/j.ijantimicag.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 90.Srinivas P., Rivard K. Polymyxin resistance in gram-negative pathogens. Curr. Infect. Dis. Rep. 2017;19(11):1–9. doi: 10.1007/S11908-017-0596-3. 2017 19:11. [DOI] [PubMed] [Google Scholar]
- 91.Poirel L., Jayol A., Nordmanna P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017;30(2):557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jeannot K., Bolard A., Plésiat P. Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents. 2017;49(5):526–535. doi: 10.1016/j.ijantimicag.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 93.Ahmed M.A., E.-G E.-S., Zhong L.-L., Shen C., Yang Y., Doi Y., Tian G.-B. Colistin and its role in the Era of antibiotic resistance: an extended review(2000–2019) Emerg. Microb. Infect. 2020;9(1):868. doi: 10.1080/22221751.2020.1754133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang J., Li C., Song J., Velkov T., Wang L., Zhu Y., Li J. Regulating polymyxin resistance in Gram-negative bacteria: roles of two-component systems PhoPQ and PmrAB. 2020;15(6):445–459. doi: 10.2217/FMB-2019-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wand M.E., Mark Sutton J. Mutations in the two component regulator systems PmrAB and PhoPQ give rise to increased colistin resistance in Citrobacter and Enterobacter spp. J. Med. Microbiol. 2020;69(4):521–529. doi: 10.1099/JMM.0.001173/CITE/REFWORKS. [DOI] [PubMed] [Google Scholar]
- 96.Telke A.A., Olaitan A.O., Morand S., Rolain J.M. soxRS induces colistin hetero-resistance in Enterobacter asburiae and Enterobacter cloacae by regulating the acrAB-tolC efflux pump. J. Antimicrob. Chemother. 2017;72(10):2715–2721. doi: 10.1093/JAC/DKX215. [DOI] [PubMed] [Google Scholar]
- 97.Phan M.-D., Nhu N.T.K., Achard M.E.S., Forde B.M., Hong K.W., Chong T.M.…Schembri M.A. Modifications in the pmrB gene are the primary mechanism for the development of chromosomally encoded resistance to polymyxins in uropathogenic Escherichia coli. J. Antimicrob. Chemother. 2017;72(10):2729–2736. doi: 10.1093/JAC/DKX204. [DOI] [PubMed] [Google Scholar]
- 98.Petrou V.I., Herrera C.M., Schultz K.M., Clarke O.B., Vendome J., Tomasek D.…Mancia F. Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation. Science. 2016;351(6273):608–612. doi: 10.1126/SCIENCE.AAD1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Warner D.M., Levy S.B. Different effects of transcriptional regulators MarA, SoxS and Rob on susceptibility of Escherichia coli to cationic antimicrobial peptides (CAMPs): rob-dependent CAMP induction of the marRAB operon. Microbiology. 2010;156(Pt 2):570. doi: 10.1099/MIC.0.033415-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ro′z A., Ro′z R., Alski ˙, Sidorczyk Z., Kotełko K. Potential virulence factors of Proteus bacilli. Microbiol. Mol. Biol. Rev. 1997;61(1):65–89. doi: 10.1128/mmbr.61.1.65-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aquilini E., Merino S., Knirel Y.A., Regué M., Tomás J.M. Functional identification of Proteus mirabilis eptC gene encoding a core lipopolysaccharide phosphoethanolamine transferase. Int. J. Mol. Sci. 2014;15(4):6689. doi: 10.3390/IJMS15046689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Groisman E.A. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 2001;183(6):1835. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kang K.N., Klein D.R., Kazi M.I., Guérin F., Cattoir V., Brodbelt J.S., Boll J.M. Colistin heteroresistance in Enterobacter cloacae is regulated by PhoPQ-dependent 4-amino-4-deoxy-l-arabinose addition to lipid A. Mol. Microbiol. 2019;111:1604–1616. doi: 10.1111/mmi.14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ezadi F., Ardebili A., Mirnejad R. Antimicrobial susceptibility testing for polymyxins: challenges, issues, and recommendations. J. Clin. Microbiol. 2019;57(4) doi: 10.1128/JCM.01390-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cannatelli A., Giani T., D'Andrea M.M., Pilato V. Di, Arena F., Conte V., Rossolini G.M. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 2014;58(10):5696. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lippa A.M., Goulian M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 2009;5(12) doi: 10.1371/JOURNAL.PGEN.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aghapour Z., Gholizadeh P., Ganbarov K., Bialvaei A.Z., Mahmood S.S., Tanomand A.…Kafil H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 2019;12:965. doi: 10.2147/IDR.S199844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Murtha A.N., Kazi M.I., Schargel R.D., Cross T., Fihn C., Cattoir V., Carlson E.E., Boll J.M., Dörr T. High-level carbapenem tolerance requires antibiotic-induced outer membrane modifications. PLoS Pathog. 2022;18(2) doi: 10.1371/journal.ppat.1010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cannatelli A., D'Andrea M.M., Giani T., Di Pilato V., Arena F., Ambretti S.…Rossolini G.M. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 2013;57(11):5521. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wozniak J.E., Chande A.T., Burd E.M., Band V.I., Satola S.W., Farley M.M., Jacob J.T., Jordan I.K., Weiss D.S. Absence of mgrB alleviates negative growth effects of colistin resistance in Enterobacter cloacae. Antibiotics (Basel, Switzerland) 2020;9(11):825. doi: 10.3390/antibiotics9110825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu Y.Y., Wang Y., Walsh T.R., Yi L.X., Zhang R., Spencer J., et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016 Feb 1;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 112.Sun J., Zhang H., Liu Y.-H., Feng Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018;26(9):794–808. doi: 10.1016/J.TIM.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 113.Nang S.C., Li J., Velkov T. The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit. Rev. Microbiol. 2019;45(2):131. doi: 10.1080/1040841X.2018.1492902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xavier B.B., Lammens C., Ruhal R., Kumar-Singh S., Butaye P., Goossens H., Malhotra-Kumar S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2 , in Escherichia coli , Belgium, June 2016. Euro Surveill. 2016;21(27) doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 115.Yin W., Li H., Shen Y., Liu Z., Wang S., Shen Z.…Wang Y. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio. 2017;8(3) doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carattoli A., Villa L., Feudi C., Curcio L., Orsini S., Luppi A.…Magistrali C.F. Novel plasmid-mediated colistin resistance mcr-4 g ene in Salmonella and Escherichia coli , Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017;22(31) doi: 10.2807/1560-7917.ES.2017.22.31.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Borowiak M., Fischer J., Hammerl J.A., Hendriksen R.S., Szabo I., Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 2017;72(12):3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 118.AbuOun M., Stubberfield E.J., Duggett N.A., Kirchner M., Dormer L., Nunez-Garcia J.…Anjum M.F. mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 2017;72(10):2745–2749. doi: 10.1093/JAC/DKX286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yang Y.Q., Li Y.X., Lei C.W., Zhang A.Y., Wang H.N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018;73(7):1791–1795. doi: 10.1093/jac/dky111. [DOI] [PubMed] [Google Scholar]
- 120.Wang X., Wang Y., Zhou Y., Wang Z., Wang Y., Zhang S., Shen Z. Emergence of colistin resistance gene mcr-8 and its variant in raoultella ornithinolytica. Front. Microbiol. 2019;10(FEB):228. doi: 10.3389/fmicb.2019.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Carroll L.M., Gaballa A., Guldimann C., Sullivan G., Henderson L.O., Wiedmann M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype typhimurium isolate. mBio. 2019;10(3) doi: 10.1128/MBIO.00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang C., Feng Y., Liu L., Wei L., Kang M., Zong Z. Identification of novel mobile colistin resistance gene mcr-10. 2020;9(1):508–516. doi: 10.1080/22221751.2020.1732231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hadjadj L., Baron S.A., Olaitan A.O., Morand S., Rolain J.-M. Co-Occurrence of variants of mcr-3 and mcr-8 genes in a Klebsiella pneumoniae isolate from Laos. Front. Microbiol. 2019;0:2720. doi: 10.3389/FMICB.2019.02720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Elbediwi M., Li Y., Paudyal N., Pan H., Li X., Xie S.…Yue M. Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980–2018) Microorganisms. 2019;7(10) doi: 10.3390/MICROORGANISMS7100461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Luo Q., Wang Y., Xiao Y. Prevalence and transmission of mobilized colistin resistance (mcr) gene in bacteria common to animals and humans. Biosafety and Health. 2020;2(2):71–78. doi: 10.1016/j.bsheal.2020.05.001. [DOI] [Google Scholar]
- 126.Snesrud E., He S., Chandler M., Dekker J.P., Hickman A.B., McGann P., Dyda F. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob. Agents Chemother. 2016;60(11):6973. doi: 10.1128/AAC.01457-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zurfluh K., Tasara T., Poirel L., Nordmann P., Stephan R. Draft genome sequence of Escherichia coli S51, a chicken isolate harboring a chromosomally encoded mcr-1 gene. Genome Announc. 2016;4(4) doi: 10.1128/genomeA.00796-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moubareck C.A. Polymyxins and bacterial membranes: a review of antibacterial activity and mechanisms of resistance. Membranes. 2020;10(8):181. doi: 10.3390/MEMBRANES10080181. 2020, Vol. 10, Page 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bufe, B., Teuchert, Y., Schmid, A., Pyrski, M., Pérez-Gómez, A., Eisenbeis, J., … Zufall, F. (n.d.). Bacterial MgrB peptide activates chemoreceptor Fpr3 in mouse accessory olfactory system and drives avoidance behaviour. 10.1038/s41467-019-12842-x. [DOI] [PMC free article] [PubMed]
- 130.Desai S.K., Kenney L.J. Switching Lifestyles Is an in vivo Adaptive Strategy of Bacterial Pathogens. Front Cell Infect Microbiol. 2019;11(9) doi: 10.3389/fcimb.2019.00421. PMID: 31921700; PMCID: PMC6917575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Grobas I., Polin M., Asally M. Swarming bacteria undergo localized dynamic phase transition to form stress-induced biofilms. Elife. 2021;10 doi: 10.7554/ELIFE.62632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gao J., Guo Z. Progress in the synthesis and biological evaluation of lipid A and its derivatives. Med. Res. Rev. 2018;38(2):556. doi: 10.1002/MED.21447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Maldonado R.F., Sá-Correia I., Valvano M.A. Oxford University Press; 2016. Lipopolysaccharide Modification in Gram-Negative Bacteria during Chronic Infection. FEMS Microbiology Reviews. July 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Report on the Burden of Endemic Health Care-Associated Infection Worldwide Clean Care Is Safer Care. 2011. www.who.int [Google Scholar]
- 135.Buckner M.M.C., Ciusa M.L., Piddock L.J.V. FEMS Microbiol Rev [Internet; 2018. Strategies to Combat Antimicrobial Resistance: Anti-plasmid and Plasmid Curing; p. 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kollef M.H., Micek S.T. Strategies to prevent antimicrobial resistance in the intensive care unit. Crit. Care Med. 2005;33(8):1845–1853. doi: 10.1097/01.ccm.0000171849.04952.79. [DOI] [PubMed] [Google Scholar]
- 137.Pratt R.J., Pellowe C., Loveday H.P., Robinson N., Smith G.W., Barrett S., et al. The epic project: developing national evidence-based guidelines for preventing healthcare associated infections. J. Hosp. Infect. 2001 Jan 1;47(SUPPL. 1) doi: 10.1053/jhin.2000.0886. S3–4. [DOI] [PubMed] [Google Scholar]
- 138.Mathur P. Indian J Med Res [Internet; 2011. Hand Hygiene: Back to the Basics of Infection Control. [cited 2023 Aug 16];134(5):611. Available from:/pmc/articles/PMC3249958/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.de Kraker M.E.A., Tartari E., Tomczyk S., Twyman A., Francioli L.C., Cassini A., et al. Implementation of hand hygiene in health-care facilities: results from the WHO Hand Hygiene Self-Assessment Framework global survey 2019. Lancet Infect. Dis. 2022 Jun 1;22(6):835–844. doi: 10.1016/S1473-3099(21)00618-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Riaz L., Yang Q., Sikandar A., Safeer R., Anjum M., Mahmood T., et al. 2020. Antibiotics Use in Hospitals and Their Presence in the Associated Waste.https://link.springer.com/chapter/10.1007/978-3-030-40422-2_2 [cited 2023 Aug 21];27–49. Available from: [Google Scholar]
- 141.Safe Management of Wastes from Health-Care Activities. second ed. 2023. Aug 17]. Available from: https://www.who.int/publications/i/item/9789241548564. [Google Scholar]
- 142.Blomberg B., Mwakagile D.S., Urassa W.K., et al. Surveillance of antimicrobial resistance at a tertiary hospital in Tanzania. BMC Public Health. 2004;4:45. doi: 10.1186/1471-2458-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fitzgibbon J.E., Wallis C.L. Laboratory challenges conducting international clinical research in resource-limited settings. J Acquir Immune Defic Syndr. 2014;1(65 Suppl 1(0 1):S36–S39. doi: 10.1097/QAI.0000000000000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Patel D., Lawson W., Guglielmo B.J. Antimicrobial stewardship programs: interventions and associated outcomes. Expert Rev Anti Infect Ther. 2008;6(2):209–222. doi: 10.1586/14787210.6.2.209. PMID: 18380603. [DOI] [PubMed] [Google Scholar]
- 145.Tenover F.C., Mohammed M.J., Stelling J., O’Brien T., Williams R. Ability of laboratories to detect emerging antimicrobial resistance: proficiency testing and quality control results from the World Health Organization’s external quality assurance system for antimicrobial susceptibility testing. J Clin Microbiol. 2001;39(1):241–250. doi: 10.1128/JCM.39.1.241-250.2001. PMID: 11136778; PMCID: PMC87709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.White A.C., Jr., Atmar R.L., Wilson J., Cate T.R., Stager C.E., Greenberg S.B. Effects of requiring prior authorization for selected antimicrobials: expenditures, susceptibilities, and clinical outcomes. Clin Infect Dis. 1997;25(2):19230–19239. doi: 10.1086/514545. PMID: 9332517. [DOI] [PubMed] [Google Scholar]
- 147.Sbarbaro JA. Can we influence prescribing patterns? Clin Infect Dis. 2001 Sep 15;33(Suppl 3):S240–S244. doi: 10.1086/321856. PMID: 11524726. [DOI] [PubMed] [Google Scholar]
- 148.MacDougall C, Polk RE. Antimicrobial stewardship programs in health care systems. Clin Microbiol Rev. 2005 Oct;18(4):638–656. doi: 10.1128/CMR.18.4.638-656.2005. PMID: 16223951; PMCID: PMC1265911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Giblin TB, Sinkowitz-Cochran RL, Harris PL, Jacobs S, Liberatore K, Palfreyman MA, Harrison EI, Cardo DM. CDC Campaign to Prevent Antimicrobial Resistance Team. Clinicians' perceptions of the problem of antimicrobial resistance in health care facilities. Arch Intern Med. 2004 Aug 9-23;164(15):1662–1668. doi: 10.1001/archinte.164.15.1662. PMID: 15302636. [DOI] [PubMed] [Google Scholar]
- 150.Owens RC Jr. Antimicrobial stewardship: concepts and strategies in the 21st century. Diagn Microbiol Infect Dis. 2008 May;61(1):110–128. doi: 10.1016/j.diagmicrobio.2008.02.012. Epub 2008 Apr 2. PMID: 18384997. [DOI] [PubMed] [Google Scholar]
- 151.Khilnani GC, Zirpe K, Hadda V, Mehta Y, Madan K, Kulkarni A, Mohan A, Dixit S, Guleria R, Bhattacharya P. Guidelines for Antibiotic Prescription in Intensive Care Unit. Indian J Crit Care Med. 2019 Jan;23(Suppl 1):S1–S63. doi: 10.5005/jp-journals-10071-23101. PMID: 31516211; PMCID: PMC6734471. [DOI] [PMC free article] [PubMed] [Google Scholar]