Abstract
Glioblastoma multiforme (GBM) is the most aggressive type of brain tumor, with an extremely poor prognosis due to resistance to standard-of-care treatments. Strong evidence suggests that the small population of glioma stem cells (GSCs) contributes to the aggressiveness of GBM. One of the mechanisms that promote GSC progression is the dysregulation of membrane transporters, which mediate the influx and efflux of substances to maintain cellular homeostasis. Here, we investigated the role of multidrug and toxin extrusion transporter gene SLC47A1 in GSCs. Results show that SLC47A1 is highly expressed in GSCs compared to non-stem cell glioma cells, and non-tumor cells. Additionally, in-silico analysis of public datasets showed that high SLC47A1 expression is linked to malignancy and a poor prognosis in glioma patients. Further, SLC47A1 expression is correlated with important biological processes and signaling pathways that support tumor growth. Meanwhile, silencing SLC47A1 by short-hairpin RNA (shRNA) influenced cell viability and self-renewal activity in GSCs. Interestingly, SLC47A1 shRNA knockdown or pharmacological inhibition potentiates the effect of temozolomide (TMZ) in GSC cells. The findings suggest that SLC47A1 could serve as a useful therapeutic target for gliomas.
Keywords: SLC47A1, solute transporter, GBM, glioma stem cell, temozolomide
Introduction
One of the deadliest forms of cancer in humans is Glioblastoma multiforme (GBM), a grade IV brain tumor, classified by the World Health Organization (WHO) [1]. Despite the use of standard treatments such as surgery, radiation, and chemotherapy, the survival rate for patients with GBM remains very poor, lasting between 8 and 15 months upon diagnosis [2]. GBM displays significant cellular heterogeneity, with small populations of tumor-initiating glioma stem cells (GSCs) [3]. GSCs are capable to self-renew and differentiate, which promotes tumor growth and recurrence. They can also induce immunosuppression, promote invasion and angiogenesis, and drive resistance to treatment [4]. As such, in-depth research on the molecular processes in GSCs is required to develop effective therapeutic strategies for lowering GBM prognosis and recurrence.
Membrane transporters are crucial for moving a variety of external and endogenous substances across cells and organelles, which enables them to maintain metabolic homeostasis [5]. However, their dysregulation may result in the rapid development of cancers. Membrane transporters frequently serve to feed the increased metabolic demands of the tumors for energy and essential nutrients, as well as to regulate the influx and efflux of chemotherapeutic drugs [6]. Evidence shows that these transporters can promote cancer progression, resistance to chemotherapy, immunosuppression, migration, angiogenesis, and metastasis. Its dysregulation affects a variety of signaling pathways, growth factors, transcription factors, cytokines, and metalloproteinase (MMPs) which are dependent or independent of their primary function [7].
The Solute Carrier Family 47 Member 1 (SLC47A1) which encodes the membrane transport protein, human multidrug and toxin extrusion protein 1 (MATE1), was first described in 2005. MATE1/SLC47A1 is a multidrug efflux pump that functions as an H+/organic cation transporter [8]. The physiological role of MATE1/SLC47A1 is primarily engaged in the elimination of cationic substances, such as endogenous substances, drugs, and toxins, into urine and bile via the kidney and liver, respectively [9]. MATE1/SLC47A1 is confined to the brush-border membrane of the proximal tubules in the kidney. Substances are then pushed from renal cells into the urine by an outward H+ gradient following their absorption by basolateral membrane transporters [10]. High expression of MATE1/SLC47A1 is also detected in the adrenal gland, testis, skeletal muscles, and brain microvessels [11]. Organic cation transporters (OCTs) and other drug efflux transporters from the ABC (ATP-binding cassette) family, including P-glycoprotein (MDR1/ABCB1), breast cancer resistance protein (BCRP/ABCG2) and canalicular multi-specific organic anion transporter (MRP2/ABCC2), work together with MATE1/SLC47A1 [8].
The retention of anticancer drugs in cancer cells can be associated with the expression of MATE1/SLC47A1. For instance, the efficacy of metformin in metastatic colorectal cancer (mCC) with wild-type KRAS is minimal due to the high expression of MATE1 [12]. Also, MATE1/SLC47A1 is downregulated in bone marrow cells of imatinib-nonresponding chronic myeloid leukemia (CML) patients compared to imatinib-responding patients, limiting the drug’s ability to function intracellularly. This finding indicates that MATE1/SLC47A1 mediates imatinib cellular uptake, and thus its expression determines imatinib therapeutic efficacy [13]. Meanwhile, SLC47A1/MATE1 can be a predictor of cancer cell chemosensitivity to the hybrid agent such as platinum-acridine compounds. Treatment of HTC-166 colon cancer cells with epigenetic drugs (EPZ-6438 and EED226) upregulates the expression of SLC47A1/MATE1 enhances the cellular uptake and sensitizes the effect of platinum-acridine compound 1 [14]. On the contrary, the overexpression of the Hedgehog receptor Patched (Ptch1) in adrenocortical carcinoma cells is associated with treatment resistance. Accordingly, overexpression of Ptch1 upregulates the expression of MATE1/SLC47A1 which may be involved in chemotherapy resistance, EMT, and cell proliferation [15]. Recently, it was found that the PPARA transactivates flippase SLC47A1 to suppress ferroptosis induction through lipid remodeling in pancreatic ductal adenocarcinoma cells. Knockdown of SLC47A1 sensitizes cells to ferroptosis in an ACSL4-SOAT1-axis dependent manner to generate polyunsaturated fatty acid-containing (PUFA) cholesterol esters [16]. Although SLC47A1 has been extensively investigated in its role as a membrane transporter, its significance in gliomas remains unclear.
Here, we found that SLC47A1 is highly expressed in glioma stem cells compared to non-stem cell glioma cell lines and non-tumor cells. We also show that high expression of SLC47A1 is correlated to malignancy and poor prognosis in glioma patients. Interestingly, by inhibiting SLC47A1 expression, it reduces the sphere-forming ability and potentiates the effect of TMZ in GSCs.
Materials and methods
Cell culture and reagents
An astrocyte medium (ScienCell Research Laboratories, USA) with 10% fetal bovine serum (FBS; Gibco, USA), 1% astrocyte growth supplement (AGS; ScienCell Research Laboratories, USA), and 1% penicillin-streptomycin (P/S; Welgene, South Korea) was used to maintain normal human astrocytes (NHA). A172, A1207, U87MG, and LN229 non-stem cell glioma cell lines were grown in Dulbecco’s modified media (DMEM/F12; Welgene, South Korea), with 10% FBS and 1% P/S. Glioma stem cells (GSC11, GSC20, GSC23, and GSC267) obtained from the University of Texas MD Anderson Cancer Center [17] were maintained in a Neurobasal Media (NBE) composed of DMEM/F12 medium enriched with 2% B27 (Gibco, USA), 1% P/S solution, basic fibroblast growth factor (bFGF; 20 ng/ml; R&D Systems, USA) and epidermal growth factor (EGF; 20 ng/ml; R&D Systems, USA). All cells were incubated at 37°C with 5% CO2.
Quantitative reverse transcription-PCR (RT-qPCR)
Total RNA was extracted using the RiboEx reagent (GeneAll, South Korea) and purified using the HybridR Kit (GeneAll, South Korea), according to the manufacturer’s protocol. To convert 500 ng of total RNA to complementary DNA (cDNA), the RevertAidTM First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA) was utilized. The quantitative Real-Time PCR (qPCR) was done on a qTOWER3 Real-Time Thermal Cycler (Analytik-Jena, USA) with TB Green® Premix Ex TaqTM (Tli RNaseH Plus; Takara Korea Biomedical Inc., Korea). The cycle threshold (Ct) values were calculated from the qPCR results using the 2-ΔΔCt method. Primer sequences used for qPCR are the following: 18S loading control - Forward (F) 5’-CAGCCACCCGAGATTGAGCA-’3 & Reverse (R) 5’-TAGTAGCGACGGGCGGTGTG-3’; SLC47A1 - (F) 5’-TGCCTGTGACACCCTCATCT-’3 & (R) 5’-GTCTGGGTAAGCCTGGACA-3’; CD15 - (F) 5’-TTGGGACCTCCTAGTTCCAC-’3 & (R) 5’-TGTAAGGAAGCCACATTGGA-3’; CD133 - (F) 5’-CAGGTAAGAACCCGGATCAA-’3 & (R) 5’-TCAGATCTGTGAACGCCTTG-3’; GFAP - (F) 5’-GGAACATCGTGGTGAAGACC-’3 & (R) 5’-AGAGGCGGAGCAACTATCCT-3’; S100β - (F) 5’-TCAAAGAGCAGGAGGTTGTG-’3 & (R) 5’-TCGTGGCAGGCAGTAGTAAC-3’; Tubb3 - (F) 5’-AGTGTGAAAACTGCGACTGC-’3 & (R) 5’-ACGACGCTGAAGGTGTTCAT-’3.
Lentivirus preparation
SLC47A1 expression was silenced using lentiviral vector producing short-hairpin RNA (shRNA) construct (sh#40 - TRCN0000138440, sh#56 - TRCN0000135356; Sigma-AAldrich, USA). The sh-non-targeting (shNT) lentiviral vector was used as control. The CalPhos Mammalian Transfection Kit (Takara Bio, USA) was used in packaging the lentivirus in 293FT cells. The lentivirus-containing media was harvested and filtered (45 µm) after 72 h transfection. It was then concentrated 100-fold using a Lenti-X concentrator (Takara Bio, USA). Lentivirus production was performed according to the manufacturer’s procedure.
Cell viability assay
GSCs (3 × 105 cells in a laminin-coated 6-well plate) were infected shNT, sh#40, and sh#56 lentivirus. After 48 h, infected cells were reseeded (3000 cells/well; n = 6) in a 96-well plate and exposed to various concentrations of TMZ (Sigma-Aldrich, USA). Similarly, GSCs (3000 cells/well; n = 6) were also seeded in a 96-well plate. After 24 h, cells were co-treated with various doses of SLC47A1 inhibitor (Famotidine; Sigma-Aldrich, USA) and TMZ. Cell viability was assessed after 72 h TMZ exposure by using the alamarBlue® assay, as directed by the manufacturer. A Synergy HTX Multi-Mode Reader (BioTek Instruments Inc., USA) was used to measure the absorbance at a 590 nm wavelength.
In vitro limiting dilution assay
ShRNA-knockdown GSCs were seeded in a 96-well plate at decreasing cell densities (25, 12, 6, 3, and 1 cell(s)/well; n = 30). Cells were supplemented with 10 µl growth media every after 3 days and maintained until 14 days. At the end of the incubation period, the plates were examined under a light microscope for tumor sphere formation. Positive wells were characterized as groups of cells larger than 20 µm in diameter. The frequency of GSCs’ capacity to generate tumorspheres was assessed using the Extreme Limiting Dilution Analysis (ELDA) program [18].
Dataset preparation
The mRNA expression in GSCs under NBE and serum culture conditions were compared using Gene Expression Omnibus (GSE4536) dataset [19]. The dataset was normalized and transformed (Log2 CPM+1) using the Integrated Differential Expression and Pathway analysis (iDEP) version 1.0 [20]. The TCGA_GBMLGG, CGGA, Gravendeel, and Ivy Glioblastoma Atlas Project (Ivy_GAP) datasets were obtained from the Gliovis website [21]. The 434 SLC-related genes were downloaded from the HUGO Gene Nomenclature Committee (HGNC; https://www.genenames.org/).
Bioinformatics analysis
Heatmap and complete linkage clustering analysis were performed Java Tree View software [22]. Volcano plot and log-rank were visualized using GraphPad Prism 8 for Windows (GraphPad Software, USA). Gene ontology (GO) and Kyoto Encyclopedia for Genes and Genome (KEGG) analyses of SLC47A1-related genes were annotated using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics resources [23]. The TCGA_GBMLGG dataset was used to identify clusters (modules) of genes highly correlated with SLC47A1high and SLC47A1low and to relate modules to external sample phenotypes (grade, subtype, IDH status, MGMT status, 1p19q codeletion status, age, and gender). Intramodular hub genes (MM > 0.9 and GS > 0.7) were identified using the Weighted Gene Co-expression Network Analysis (WGCNA) [24]. Univariate and multivariate COX analyses of SLC47A1high and SLC47A1low expression were performed to identify independent variables associated with overall survival (OS) in TCGA_GBMLGG and CGGA datasets using the R package [25]. Kaplan-Meier (KM) and Receiver Operating Characteristics (ROC) and GO and KEGG pathways were plotted using an SR plot (https://www.bioinformatics.com.cn/srplot), an online platform for data analysis and visualization.
RNA-sequencing
GSCs (3 × 105 cells per well) were seeded in a laminin-coated 6-well plate. After 24 h, cells were transfected with either shNT or shSLC47A1 (sh#56) and cultured for 48 h. Cells were resuspended in TRIzol Reagent (Invitrogen; CA, USA) and stored at -80°C before processing and RNA sequencing. RNA extraction, library preparation, and sequencing were outsourced to LAS Co., Ltd. (Gimpo, South Korea). The data that support the findings of this study are openly available in NCBI Repository at http://www.ncbi.nlm.nih.gov/bioproject/1009200, accession number: PRJNA1009200.
Statistical analysis
GraphPad Prism Version 8.0 was used for the statistical analysis. A two-tailed t-test and a one-way ANOVA were used, respectively, to determine the statistical significance between and across groups. Tukey’s multiple comparison tests were then used to compare the results. All data were reported as mean ± Standard Error (SE). A P-value of 0.05 or lower was regarded as statistically significant.
Results
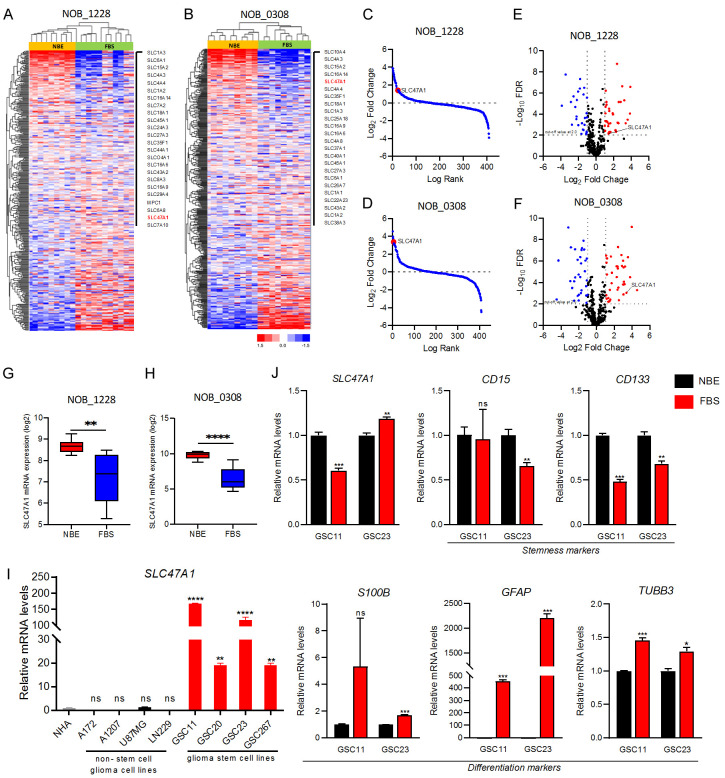
SLC47A1 mRNA is highly expressed in GSCs under NBE culture
To analyze the expression patterns of genes that regulate the stem cell character of GSCs, we compared the mRNA expression profiles of SLC47A1 and other 434 SLC-related genes in GCSs under NBE- and serum-media conditions from the public dataset GSE4536. Heatmap and clustering analysis revealed that SLC47A1 is commonly upregulated in both NOB_1228 and NOB_0308 GSCs under NBE conditions (Figure 1A, 1B). Further, SLC47A1 is one of the top-ranked SLC genes in NBE-cultured GSCs based on a log-rank test (Figure 1C, 1D) and volcano plot with Log2 Fold-change of > 1.5 at P ≤ 0.05 (Figure 1E, 1F). Additionally, we showed that SLC47A1 mRNA expression was considerably higher in both NOB_1228 and NOB_0308 GSCs when cultured in NBE- as compared to serum condition (Figure 1G, 1H). To validate these findings, we next performed RT-qPCR to measure the basal expression levels of SLC47A1 in NHA cells, non-stem cell glioma cell lines (A172, A1207, LN229, and U87MG), and GSC cell lines (GSC11, GSC20, GSC23, and GSC267). Compared to NHA cells and non-stem cell glioma cell lines, the SLC47A1 mRNA level is highly expressed in GSC cell lines (Figure 1I). In the meantime, we also compared the mRNA expression levels of SLC47A1 and stemness (CD15, CD133) and differentiation (S100B, GFAP, TUBB3) markers in GSC11 and GSC23 cells under NBE- and serum media-condition. SLC47A1 expression was observed to be upregulated in GSC11 cells when cultured in NBE condition. Similarly, stemness markers CD15 and CD133 are significantly expressed in GSCs under NBE culture conditions, while differentiation markers S100B, GFAP, and TUBB3 are elevated in GSCs under serum-media conditions (Figure 1J). The results show that SLC47A1’s mRNA expression levels are significantly elevated in GSCs.
Figure 1.
SLC47A1 is highly expressed in GSCs and correlates with stemness markers in GSCs cultured in NBE conditions. (A, B) Complex heatmap illustrating the differential expression of 434 solute-carrier related genes in GSCs under NBE and FBS culture condition in GSE4536 dataset; (A) NOB_1228, (B) NOB_0308. (C, D) Log-rank fold-change of SLC47A1 GSCs under NBE and FBS culture condition in GSE4536 dataset; (C) NOB_1228, (D) NOB_0308. (E, F) Volcano plots for identifying genes with significant fold change under the NBE-culture condition compared with the FBS-culture condition in NOB_1228 and NOB_0308 cell lines. Upregulated genes (Log2 FC > 1.5) are presented in red; Downregulated genes (Log2 FC < -1.5) are presented in blue; Not-significant genes are shown in black; (E) NOB_1228, (F) NOB_0308. (G, H) The comparison of SLC47A1 expression in GSCs under NBE and FBS culture conditions according to the GSE4536 dataset; (G), NOB_1228, (H) NOB_0308. Data are means ± SEM (NBE, n = 10 or 11; FBS, n = 11 or 10). (I) RT-qPCR analysis on the SLC47A1 mRNA expression in non-stem cell glioma cell lines (A172, A1207, U87MG, LN229), glioma stem cell lines (GSC11, GSC20, GSC23, GSC267) and normal human astrocyte (NHA). (J) RT-qPCR analysis of SLC47A1, stemness (CD15, CD133), and differentiation (S100B, GFAP, TUBB3) markers in GSC11 and GSC23 cells under NBE and FBS media condition. *P < 0.05, **P < 0.01, ***P < 0.0001, ****P < 0.0001; ns-not significant.
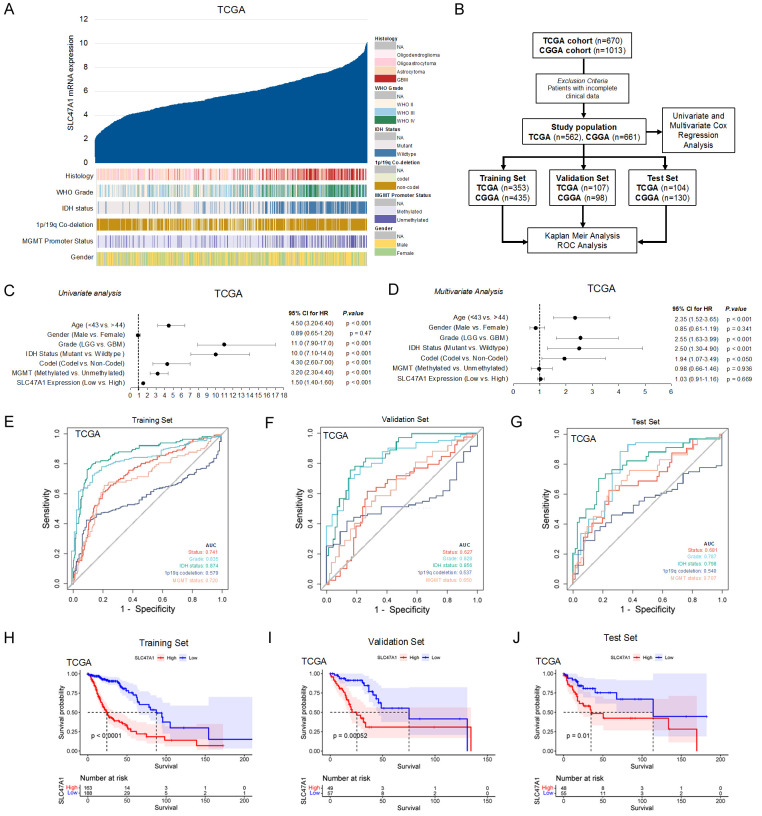
High expression of SLC47A1 is associated with malignancy and poor survival in glioma patients
To understand the clinical significance of SLC47A1, we compared the mRNA expression levels of SLC47A1 in TCGA_GBMLGG and CGGA datasets based on histology, grade, 1p/19q co-deletion status, IDH mutation status, and MGMT promoter methylation (Figures 2A, S1A). Results show that SLC47A1 expression is upregulated in GBM compared to low-grade gliomas. Furthermore, both the TCGA and CGGA datasets demonstrate that the SLC47A1 gene is significantly upregulated in glioma patients with non-codel 1p/19q co-deletion status, IDH wildtype status, and unmethylated MGMT promoter region status. To determine the prognostic value of SLC47A1 expression in glioma patients in the TCGA and CGGA datasets, we conducted our analysis as described in the flow chart (Figure 2B). Univariate analysis revealed that SLC47A1 expression is a high-risk factor (TCGA - HR = 1.50; 95% CI = 1.40-1.60; P < 0.001; CGGA - HR = 1.60; 95% CI = 1.40-1.70; P < 0.001) in glioma patients. Other independent predictive factors were age, gender, WHO grade, IDH status, 1p/19q co-deletion, MGMT status, and primary therapy (Figures 2C, S1B). SLC47A1 was similarly found to be independently associated with overall survival after multivariate analysis (TCGA - HR = 1.03, 95% CI = 0.91-1.16; P = 0.669; CGGA - HR = 1.18, 95% CI = 1.06-1.31; P 0.002). This implies that SLC47A1 might function as an independent prognostic factor for glioma patients (Figures 2D, S1C). Meanwhile, we also assessed the prognostic efficiency of SLC47A1 expression by operating a ROC curve in both TGCA and GCCA datasets (Figures 2E-G, S1D-F). The AUCs (Area under the ROC curve) for survival and SLC47A1 are 0.741, 0.627, and 0.681 for the training set, testing set, and whole set, respectively in the TCGA dataset (Figure 2E-G). Similarly, the AUCs for survival and SLC47A1 expression in CGGA dataset are 0.741, 0.627, and 0.681 for the training set, testing set, and whole set, respectively (Figure S1D-F). Our results indicate a good performance of SLC47A1 expression for survival prediction independent of other clinical factors (Grade, IDH status, 1p/19q codeletion, and, MGMT status). Kaplan-Meir survival curve analysis also shows that increased SLC47A1 expression is associated with poorer patient survival in both TCGA and CGGA datasets (Figures 2H-J, S1G-I). ROC and Kaplan-Meir survival analyses on SLC47A1 expression of TCGA and CGGA whole sets are shown in Figure S1J, S1K. Together, our findings suggest that elevated SLC47A1 expression may be a predictive factor that can influence the prognosis and survival of glioma patients.
Figure 2.
High expression of SLC47A1 is associated with malignancy and poor survival in glioma patients. (A) Correlation between SLC47A1 mRNA expression and glioma histological features in TCGA dataset (n = 670). (B) Flow chart showing the patient population in the TCGA and CGGA cohorts. (C, D) Univariate (C) and multivariate (D) Cox regression analyses to evaluate the correlation of SLC47A1 expression with prognosis of glioma patients in the TCGA cohorts. (E, F) ROC curve and the risk score distribution stratified by SLC47A1 expression levels in patient survival status, grade, 1p/19q codeletion, and MGMT promoter methylation in the TCGA Training set (E), Validation set (F), and Test set (G). (H-J) Kaplan-Meier analysis for SLC47A1 high and low expressions in the TCGA Training set (H), Validation set (I), and Test set (J).
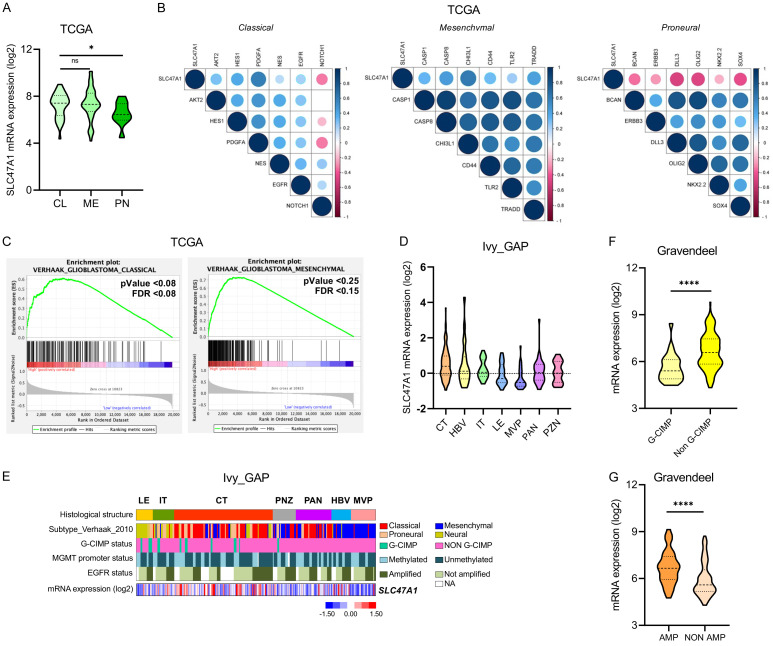
SLC47A1 is overexpressed in classical and mesenchymal GBM subtypes and upregulated in cellular tumors
We then used the TCGA dataset to evaluate SLC47A1 preferential expression in GBM subtypes. The expression of SLC47A1 is significantly higher in the classical and mesenchymal subtypes compared to the proneural subtype (Figure 3A). Furthermore, SLC47A1 has a strong association with classical and mesenchymal markers, according to Pearson’s correlation analysis (Figure 3B). Hence, Gene-set Enrichment Analysis (GSEA) back-upped these findings (Figure 3C). We then next used the Ivy-GAP dataset to determine the GBM anatomical region that favors SLC47A1 expression. Results show that SLC47A1 expression is significantly upregulated in the cellular tumor (CT) region (Figure 3D, 3E). Meanwhile, the Gravendeel dataset was used to assess the mRNA expression of SLC47A1 with CIMP and EGFR status (Figure 3F, 3G). Results show that SLC47A1 is highly expressed in non-G-CIMP and EGFR amplification samples. Collectively, these findings indicate that upregulation of SLC47A1 mRNA expression is associated with classical and mesenchymal GBM subtypes and preferentially expressed in the tumor’s CT region.
Figure 3.
SLC47A1 is upregulated in classical and mesenchymal GBM subtypes and enriched in the central tumor region. (A) Comparison of SLC47A1 mRNA levels among the groups with GBM subtypes in the TCGA database (CL - Classical, ME - Mesenchymal, PN - Proneural). (B) The correlation analysis between SLC47A1 expression with classical, mesenchymal, proneural GSC markers in the TCGA dataset. (C) GSEA analysis of SLC47A1 expression with glioma subtypes in TCGA dataset. (D) Comparison of SLC47A1 mRNA expression in different GBM regions using the IvyGAP dataset (LE, n = 19; IT, n = 24; CT, n = 111; PNZ, n = 26; PAN, n = 40; HBV, n = 22; MVP, n = 28). LE - leading edge; IT - infiltration tumor; CT - cellular tumor; PNZ - perinecrotic zone; PAN - pseudopalisading cells around necrosis; HBV - hyperplastic blood vessels; MVP - microvascular proliferation. (E) Heatmap of SLC47A1 expression signatures in IvyGAP dataset. (F, G) The expression of SLC47A1 in the Gravendeel dataset based on (E) CIMP status and (F) EGFR amplification status. *P < 0.05 **P < 0.01;***P < 0.001, ****P < 0.0001, ns - not significant.
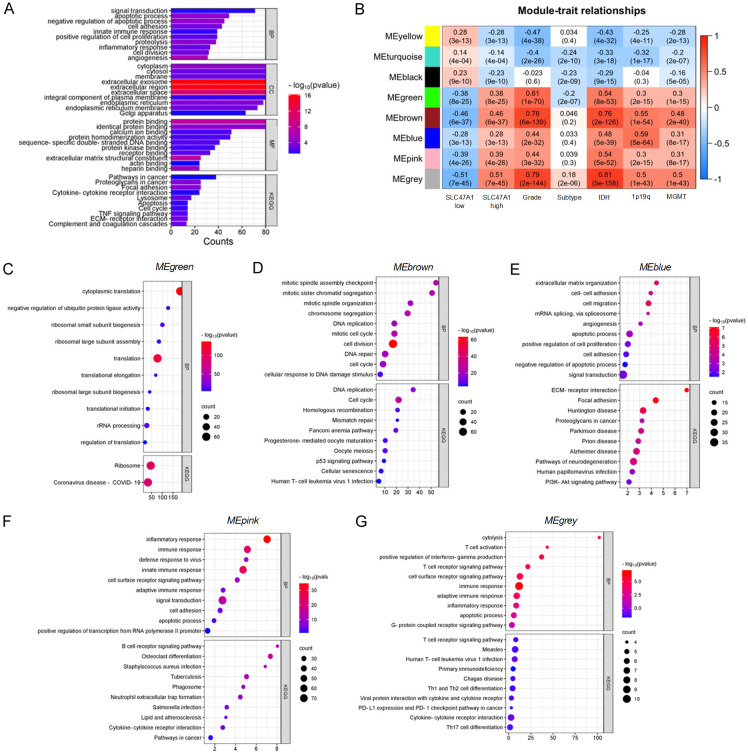
SLC47A1 expression is associated with biological processes and pathways that support tumor growth
We next performed correlation analysis on the TCGA_GBMLGG dataset to gain a better understanding of the biological processes and signaling pathways associated with SLC47A1 expression. 1299 genes are significantly correlated with SLC47A1 expression (Pearson’s R ≥ 0.40). Using DAVID functional annotation, we found that these genes are related to biological processes like fibril organization, immune response, signal transduction, integrin-mediated signaling, angiogenesis, extracellular matrix organization, positive regulation of cell migration, and negative regulation of the apoptotic process. Meanwhile, KEGG pathway analysis shows that these genes are involved in ECM-receptor interaction, TNF signaling pathways, cytokine-cytokine interaction, and focal adhesion (Figure 4A). To further determine the role of high SLC47A1 expression in glioma, we performed WGCNA analysis using the TCGA_GBMLGG dataset. The findings reveal a total of 2297 protein-coding genes grouped into 9 modules that are related to the levels of SLC47A1 expression. Among these modules, 5 modules (MEgreen, MEbrown, MEblue, MEpink, MEgrey) are positively correlated with high SLC47A1 expression, and phenotypes high-grade glioma, classical subtype, IDH-wildtype, 1p/19q non-codeletion, and unmethylated MGMT promoter region (R = 0.3-0.6; P < 0.0001) (Figure 4B). Interestingly, these modules contain gene sets that are related to different biological processes such as translation, cell cycle, cell proliferation, adhesion, angiogenesis, migration and apoptosis, and immune and inflammatory response (Figure 4C-G). KEGG pathways analysis also revealed that these gene sets are associated with p53, focal adhesion, PI3K-Akt, ECM-receptor interaction, and cytokine-cytokine interaction (Figure 4C-G). Clearly, we show that high expression of SLC47A1 not only serve as a solute transporter but also influences various biological processes and signaling pathways that are important in glioma progression.
Figure 4.
High expression of SLC47A1 influences important biological processes and pathways in glioma. (A) DAVID functional annotation of genes associated with SLC47A1 in TCGA_GBMLGG using Pearson R correlation (R > 0.4). (B) Module-trait-relationship between high and low SLC47A1 with glioma WHO grade, subtype, IDH, 1p19q codeletion, and MGMT promoter methylation status analyzed by WGCNA analysis. (C-G) DAVID functional enrichment analysis of biological processes (BP) and Kyoto Encyclopedia of Genes and Genomes pathways (KEGG) in (C) Green, (D) Brown, (E) Blue, (F) Pink, and (G) Grey Module genes.
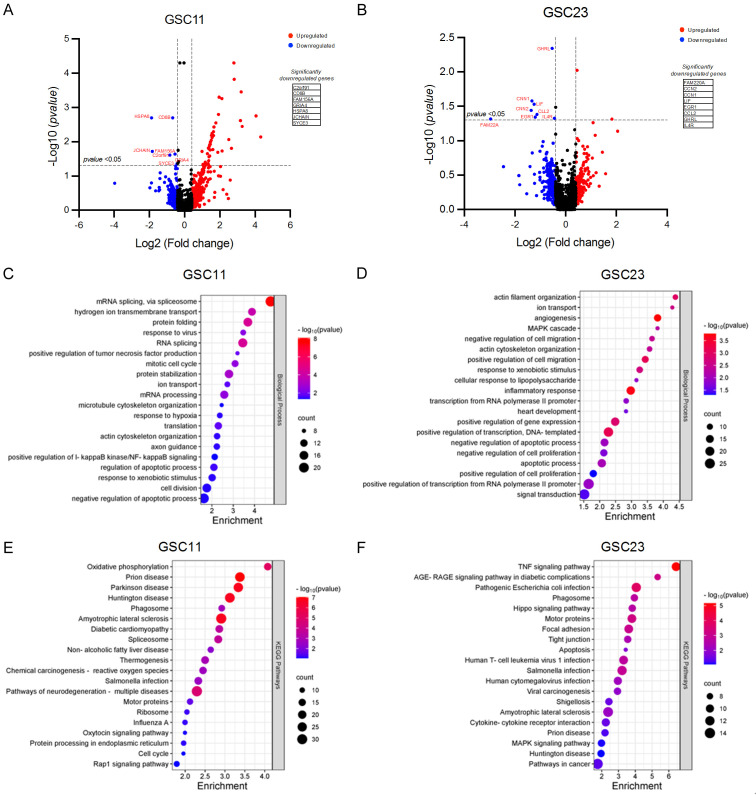
RNA-Seq investigation of SLC47A1 inhibition in GSCs
To further explore the downstream mechanism of SLC47A1 in GSCs, RNA sequencing analysis was performed in SLC47A1 knockdown and non-targeting control GSC11 and GSC23 cells. There are 7 (C2orf91, CD8B, FAM156A, GRIA4, HSPA8, JCHAIN, SYCE3) and 8 (FAM220A, CCN2, CCN1, LIF, EGR1, CCL2, GHRL, IL4R) significantly downregulated genes in SLC47A1 knockdown GSC11 and GSC23 cells, respectively (Figure 5A, 5B). To investigate the biological functions of the downregulated genes (Log2 Fold-Change < -0.4) in this study, we conducted functional annotation analysis by gene ontology (Figure 5C, 5D). The most significantly enriched biological processes in knockdown cells are related to SLC47A1’s primary functions such as hydrogen ion transmembrane transport, ion transport, and response to xenobiotic stimulus. Interestingly, we also found that genes related to mitotic cell cycle, cell division, response to hypoxia, negative regulation of the apoptotic process, angiogenesis, positive regulation of cell migration, inflammatory response, signal transduction, and positive regulation of cell proliferation are downregulated in SLC47A1 knockdown GSCs. Meanwhile, the KEGG pathway analysis was performed to identify the potential signaling pathways (Figure 5E, 5F). The results indicate that genes associated with pathways including oxidative phosphorylation, chemical carcinogenesis, Rap1 signaling, cell cycle, TNF signaling, Hippo signaling, focal adhesion, apoptosis, cytokine-cytokine interaction, and MAPK signaling pathways are downregulated in SLC47A1 knockdown GSCs. The association of these biological processes and pathways based on RNA-seq results corroborates with our findings in the in-silico analysis in Figure 4. Notably, analyzing the associated biological processes and pathways in the significantly downregulated genes in SLC47A1 knockdown cells is related to immune response (Figure S2E, S2F). Meanwhile, the upregulated genes upon SLC47A1 knockdown are associated with biological processes (Figure S2A, S2B) such as regulation of the apoptotic process, negative regulation of cell proliferation, cell differentiation, negative regulation of cell migration and lipid metabolic process, and pathways (Figure S2C, S2D) such as TNF, NFKB, TGFB, p53, FOXO, MAPK, and amino acid metabolism pathways. These results suggest that SLC47A1 is not only involved in transmembrane transport as its primary function but also involved in various biological processes and pathways that are important to the progression of gliomas.
Figure 5.
RNA sequencing analysis between SLC47A1 knockdown and non-target control GSC11 and GSC23 cells. (A, B) Volcano plot of the differentially expressed genes in the GSC11 (A) and GSC23 (B) knockdown cells. The red color indicates differentially expressed genes with high expression, while the blue color indicates low expression. (C, D) Gene Ontology analysis on the differentially expressed genes between SLC47A1 knockdown and non-target control GSC11 (C) and GSC23 (D) cells. (E, F) KEGG pathway analysis on the differentially expressed genes between SLC47A1 knockdown and non-target control GSC11 (E) and GSC23 (F) cells.
SLC47A1 knockdown suppresses self-renewal activity in GSC
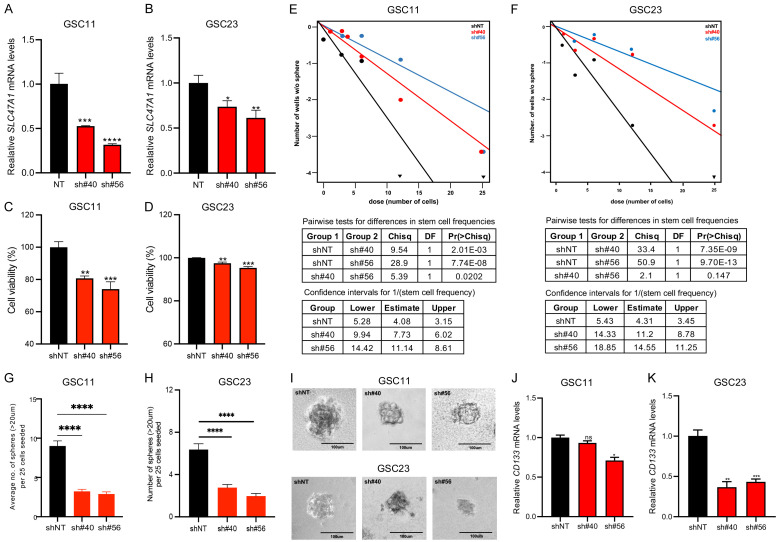
To investigate the role of SLC47A1 in GSCs, we used a shRNA lentivirus to knockdown of SLC47A1 in the GSC11 and GSC23 cell lines. We verified the shRNA knockdown efficiency using RT-qPCR (Figure 6A, 6B). Next, we investigated the effect of SLC47A1 knockdown on the viability of GSC11 and GSC23 cells. Cell viability in both GSC11 and GSC23 cells is slightly reduced (10-20%) after knockdown when compared to control cells (Figure 6C, 6D). Meanwhile, SLC47A1 knockdown inhibited GSC tumorsphere formation ability when compared to nontargeting shRNA-treated cells (Figure 6E-I). We also looked at the expression of the stemness surface marker CD133. CD133 expression was likewise reduced in SLC47A1 knockdown cells compared to controls (Figure 6J, 6K). These results imply that SLC47A1 may also play a role in GSC self-renewal property.
Figure 6.
SLC47A1 knockdown suppresses stemness in GSC. (A, B) RT-qPCR analysis showing SLC47A1 knockdown after transfection with shRNA in GSC11 (A) and GSC23 (B). Data are means ± SEM (n = 3). (C, D) Cell viability of GSC11 (C) and GSC23 (D) after 96 h post-transfection of SLC47A1 shRNA. Data are means ± SEM (n = 6). (E-I), Effects of SLC47A1 knockdown on neurosphere formation in GSC11 and GSC23. Scale bars representative image is 100 μm. (J, K) mRNA expression levels of CD133 after shRNA knockdown of SLC47A1 in GSC11 (J) and GSC23 (K). Data are means ± SEM (n = 3). *P < 0.05 **P < 0.01;***P < 0.001, ****P < 0.0001, ns - not significant.
Inhibition of SLC47A1 potentiates the effect of TMZ in GSC
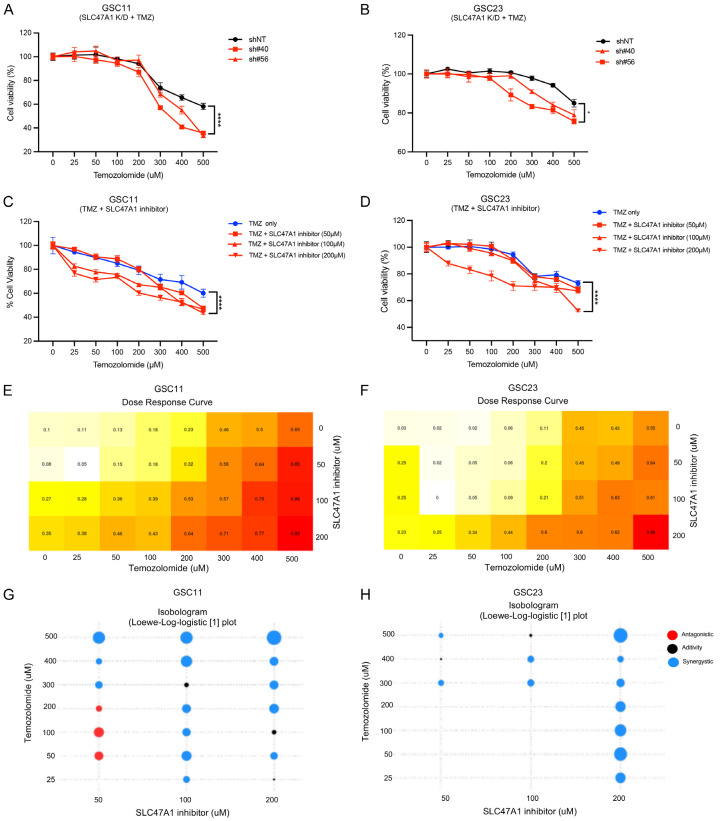
We next assessed the effect of the chemotherapeutic drug TMZ in GSCs when SLC47A1 is inhibited by shRNA knockdown or using Famotidine, a potential MATE1/SLC47A1 inhibitor [26]. SLC47A1 knockdown and TMZ co-treatment significantly reduced cell viability in a dose-dependent manner relative to control (Figure 7A, 7B). On the other hand, treatment of Famotidine alone did not cause a significant decrease in cell viability of GSC11 and GSC23 cells (Figure S3A, S3B). Interestingly, co-treatment with TMZ and a SLC47A1 inhibitor reduced cell viability significantly in a dose-dependent manner when compared to the control. (Figure 7C, 7D). The synergistic anticancer effect of TMZ and SLC47A1 inhibitor co-treatment in GSC11 and GSC23 cells was further confirmed by SiCoDEA [27], a standalone web tool for evaluating and visualizing data on multi-drug combination response (Figure 7E-H). These findings show that inhibition of SLC47A1 can potentiate the effect of TMZ in GSC cells.
Figure 7.
Inhibition of SLC47A1 potentiates the effect of TMZ in GSC. (A) Dose-response viability (%) of SLC47A1 K/D GSC11 (A) and GSC23 (B) cells treated with various concentrations of TMZ (25, 50, 100, 200, 300, 400, 500 µM) assessed by alamarBlue assay after 72 h exposure. (C, D) Cell viability (%) of GSC11 (C) and GSC23 (D) after 72 h co-treatment of increasing doses of TMZ (25, 50, 100, 200, 300, 400, 500 µM) and Famotidine (SLC47A1 inhibitor; 50, 100, 200 µM). (E-H) Dose-response curve and Isobologram (Loewe-log logistic [1] model) of TMZ and SLC47A1 inhibitor co-treatment in GSC11 and GSC23 cells. *P < 0.05 **P < 0.01;***P < 0.001, ****P < 0.0001, ns - not significant.
Discussion
Membrane transporters are required for the movement of hormones, neurotransmitters, xenobiotics, and essential metabolites such as sugars, amino acids, and nucleosides across cell or organelle membranes [5]. However, because tumor cells require more energy and nutrients to maintain internal homeostasis, the expression of membrane transporters is often dysregulated [28]. Here, we investigated the role of multidrug and toxin extrusion transporter gene SLC47A1 in GSC. The SLC47A1 gene, which encodes the multidrug and toxin extrusion protein 1 (MATE1), regulates the coupled organic cation efflux in various tissues including the kidney, liver, adrenal gland, skeletal muscle, testis, and in the first-trimester placenta [8]. Our data show that SLC47A1 is one of the differently expressed membrane transporters in GSCs in the GSE4536 dataset. According to reports, NBE-cultured GSCs maintain their capacity for self-renewal in a manner comparable to that of regular NSCs. GSCs cultured on serum-containing media, on the other hand, lose their ability to self-renew, differentiate, and exhibit gene expression profiles that are not similar to NSCs or the initial GBMs from which they were derived, and are neither clonogenic nor tumorigenic [19]. RT-qPCR validation on the SLC47A1 mRNA expression is significantly higher in GSC cell lines compared to non-stem cell gliomas and NHA cells. This implies that SLC47A1 expression may aid in tumorigenesis and GSC self-renewal ability.
Accordingly, some studies show that high SLC47A1 expression is a good prognostic factor for endometrial, lung, renal and pancreatic cancers [29,30]. On the contrary, it can also influence poor outcomes in metastatic colorectal and non-small cell lung cancers [31,32]. Here, we also show that SLC47A1 is significantly expressed in high-grade gliomas. Additionally, patients with gliomas that exhibit such high expression have a poor outcome. These findings imply that SLC47A1 may serve as a marker for the prognosis of GBM patients. We also investigated the SLC47A1 expression patterns in the various subtypes of GBM to better understand the significance of SLC47A1 expression in GBM [33,34]. We found that SLC47A1 is upregulated in both classical and mesenchymal subtypes. The classical subtype has abnormal alterations such as Chr.7 amplification, Chr.10 loss, inactivation of the RB pathway, and a localized 9p21.3 homozygous deletion. Furthermore, the classical subtype has high levels of expression of Sonic hedgehog pathways, Notch signaling pathways, and the neural precursor and stem cell marker NES [35]. In a recent study, the Hedgehog receptor Patched (Ptch1) is overexpressed in adrenocortical carcinoma which influences the upregulation of SLC47A1 [15]. On the other hand, the classical subtype has a high level of EGFR amplification and is infrequent with other GBM subtypes [35]. In the present study, we found that SLC47A1 is increased in GBM with EGFR amplification using the Gravendeel dataset. Meanwhile, we also found that SLC47A1 is highly expressed in the mesenchymal GBM subtype. Extensive necrosis and inflammation, increased expression of TNF superfamily and NF-B pathway genes, loss of the tumor suppressor genes p53, NF1, and PTEN, and overexpression of interstitial and angiogenesis genes are all characteristics of the mesenchymal subtype. Mesenchymal subtypes have the lowest prognosis of all GBM subtypes. Recently, it was found that VEGF-A, VEGF-B, ANG1, and ANG24 genes are highly expressed in the mesenchymal subtype [35].
In this study, in-silico analysis revealed that high expression of SLC47A1 is related to various biological processes like translation, cell cycle, cell proliferation, adhesion, angiogenesis, migration and apoptosis, and immune and inflammatory response. KEGG pathways associated with high SLC47A1 expression include p53, focal adhesion, PI3K-Akt, ECM-receptor interaction, and cytokine-cytokine interaction. There are specific predictive core genes in the cell cycle, the JAK-STAT pathway, and DNA repair in the classical subtype. Meanwhile, certain prognostic genes in the mesenchymal subtype are linked to the PI3K/Akt, MAPK, ERK, and Wnt pathways, as well as the transition of mesenchymal cells [35]. Taken together, we show that high SLC47A1 expression is associated with biological processes and signaling pathways that are important for glioma progression and malignancy. Our findings in the in-silico analysis corroborate our RNA-seq analysis. However, further in vitro and in vivo studies are required to confirm the function of SLC47A1 and how it affects certain biological processes and signaling pathways that are crucial for the advancement of gliomas.
We also found that SLC47A1 could influence self-renewal ability in GSCs. We show that the sphere-forming ability and the expression of the stemness marker, CD133, are reduced when SLC47A1 expression is inhibited. Several studies show that expression of MATE1/SLC47A1 could support cancer stem cells and promote chemoresistance. For example, bone marrow stem cells (CD34+) of CML patients have higher expression of influx (SLC22A) and efflux transporters (ABCB1, ABCG2, ABCC1, SLC47A1, SLC47A2) could result in infectivity of imatinib treatment [36]. Similarly, in breast cancer stem cells, overexpression of extrusion transporters (MATE1 and PMAT) confers resistance to metformin treatment [37]. The co-expression of MATE1/SLC47A1 with ABC efflux transporters increases cell detoxification and excretory mechanism [8]. GSCs are more resistant to chemotherapy and radiation than non-stem glioma cells. A lack of or abnormal DNA repairs mechanisms, such as an increase in MGMT which repairs TMZ-induced DNA damage, and overexpression of anti-apoptotic signals are examples of resistance mechanisms in GBM. In addition, lower cellular accumulation of chemotherapeutics is caused by enhanced ABC efflux transporter expression [38].
In the present study, we also found that shRNA knockdown or pharmacological inhibition of SLC47A1/MATE1 and co-treatment of chemotherapeutic drug TMZ has reduced the viability of GSCs. Recently, it was found that inhibition of SLC47A1 inhibited the proliferation and migration ability of uveal melanoma cells (MUM-2B) in vitro [39]. Meanwhile, Cimetidine and Famotidine are both histamine H2 receptor antagonists [40] and are potential inhibitors for SLC47A1 [11]. In a recent study, famotidine can serve as a radioprotector for rectal mucosa in prostate cancer patients treated with radiotherapy [41]. A previous study found that cimetidine can act as an anti-adhesive and an anti-migratory agent for glioma cells, and could complement the cytotoxic TMZ to combat both proliferating and migrating GBM cells. In vivo, analysis revealed that combined cimetidine and TMZ treatment significantly increase survival in the orthotopic xerograph model [42]. Here, we show that inhibiting SLC47A1, either by shRNA or pharmacological inhibitor, could potentiate the effect of TMZ in GSCs. Hence, further studies are needed to elucidate the molecular mechanism of how SLC47A1 expression can influence tumorigenicity and treatment resistance in GSCs.
The expression and activation of SLC47A1 are regulated by several transcription factors including Sp1, AP-1, HNF4a, NRF2, and PPAR [43,44]. These transcription factors are involved in the regulation of several important oncogenes and tumor suppressors, as well as genes involved in fundamental cellular activities such as proliferation, differentiation, the DNA damage response, apoptosis, tumor invasion, senescence, dedifferentiation, aerobic glycolysis, inflammation and angiogenesis [45-49]. On the other hand, over 980 single nucleotide polymorphisms (SNPs) had been identified in the SLC47A1. These polymorphic sites influence the loss of function or decrease the transcription and transport activity of SLC47A1 [50]. Similarly, the transport activity of SLC47A1/MATE1 is dependent on the environmental pH [44]. MATE1 exchanges its substrates against the proton gradient. Consequently, substrate uptake is driven by an in-to-out proton gradient with a maximum uptake activity at an extracellular pH of 8.5 [51]. Acidification of the extracellular environment improves the export function of MATE1, while extracellular alkalinization or intracellular acidification increases the uptake of substrates [52]. Hence, further studies are required to confirm the clinical relevance of the various transcription factors, polymorphism, and environmental pH that influence the regulation and function of SLC47A1 genes in gliomagenesis.
In summary, our study suggests that SLC47A1 is a potential prognostic biomarker to characterize gliomas. Importantly, SLC47A1 influences the stemness properties in GSCs and molecular and pharmacological inhibition of SLC47A1 can sensitize the effect of the chemotherapeutic drug TMZ. Understanding the molecular function of SLC47A1 in GSCs could be an avenue to improve or develop more efficient treatment strategies for GBM.
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science, and Technology (No. NRF-2022R1I1A3070961, and NRF-2022R1A2C1011742).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Wu W, Klockow JL, Zhang M, Lafortune F, Chang E, Jin L, Wu Y, Daldrup-Link HE. Glioblastoma multiforme (GBM): an overview of current therapies and mechanisms of resistance. Pharmacol Res. 2021;171:105780. doi: 10.1016/j.phrs.2021.105780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King JL, Benhabbour SR. Glioblastoma multiforme-a look at the past and a glance at the future. Pharmaceutics. 2021;13:1053. doi: 10.3390/pharmaceutics13071053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan K, Yang K, Rich JN. The evolving landscape of glioblastoma stem cells. Curr Opin Neurol. 2013;26:701–707. doi: 10.1097/WCO.0000000000000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alves ALV, Gomes INF, Carloni AC, Rosa MN, da Silva LS, Evangelista AF, Reis RM, Silva VAO. Role of glioblastoma stem cells in cancer therapeutic resistance: a perspective on antineoplastic agents from natural sources and chemical derivatives. Stem Cell Res Ther. 2021;12:206. doi: 10.1186/s13287-021-02231-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q, Shu Y. Role of solute carriers in response to anticancer drugs. Mol Cell Ther. 2014;2:15. doi: 10.1186/2052-8426-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nyquist MD, Prasad B, Mostaghel EA. Harnessing solute carrier transporters for precision oncology. Molecules. 2017;22:539. doi: 10.3390/molecules22040539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rashid K, Ahmad A, Liang L, Liu M, Cui Y, Liu T. Solute carriers as potential oncodrivers or suppressors: their key functions in malignant tumor formation. Drug Discov Today. 2021;26:1689–1701. doi: 10.1016/j.drudis.2021.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Staud F, Cerveny L, Ahmadimoghaddam D, Ceckova M. Multidrug and toxin extrusion proteins (MATE/SLC47); role in pharmacokinetics. Int J Biochem Cell Biol. 2013;45:2007–11. doi: 10.1016/j.biocel.2013.06.022. [DOI] [PubMed] [Google Scholar]
- 9.Motohashi H, Inui KI. Multidrug and toxin extrusion family SLC47: physiological, pharmacokinetic and toxicokinetic importance of MATE1 and MATE2-K. Mol Aspects Med. 2013;34:661–668. doi: 10.1016/j.mam.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Liu X, Wang Y, Zhou N, Peng J, Gong L, Ren J, Luo C, Luo X, Jiang H, Chen K, Zheng M. Combinatorial pharmacophore modeling of multidrug and toxin extrusion transporter 1 inhibitors: a theoretical perspective for understanding multiple inhibitory mechanisms. Sci Rep. 2015;5:13684. doi: 10.1038/srep13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wittwer MB, Zur AA, Khuri N, Kido Y, Kosaka A, Zhang X, Morrissey KM, Sali A, Huang Y, Giacomini KM. Discovery of potent, selective multidrug and toxin extrusion transporter 1 (MATE1, SLC47A1) inhibitors through prescription drug profiling and computational modeling. J Med Chem. 2013;56:781–795. doi: 10.1021/jm301302s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie J, Xia L, Xiang W, He W, Yin H, Wang F, Gao T, Qi W, Yang Z, Yang X, Zhou T, Gao G. Metformin selectively inhibits metastatic colorectal cancer with the KRAS mutation by intracellular accumulation through silencing MATE1. Proc Natl Acad Sci U S A. 2020;117:13012–13022. doi: 10.1073/pnas.1918845117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrach S, Schmidt-Lauber C, Pap T, Pavenstädt H, Schlatter E, Schmidt E, Berdel WE, Schulze U, Edemir B, Jeromin S, Haferlach T, Ciarimboli G, Bertrand J. MATE1 regulates cellular uptake and sensitivity to imatinib in CML patients. Blood Cancer J. 2016;6:e470. doi: 10.1038/bcj.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao X, Watkins NH, Brown-Harding H, Bierbach U. A membrane transporter determines the spectrum of activity of a potent platinum-acridine hybrid anticancer agent. Sci Rep. 2020;10:15201. doi: 10.1038/s41598-020-72099-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feliz Morel ÁJ, Hasanovic A, Morin A, Prunier C, Magnone V, Lebrigand K, Aouad A, Cogoluegnes S, Favier J, Pasquier C, Mus-Veteau I. Persistent properties of a subpopulation of cancer cells overexpressing the hedgehog receptor patched. Pharmaceutics. 2022;14:988. doi: 10.3390/pharmaceutics14050988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Z, Liu J, Long F, Kang R, Kroemer G, Tang D, Yang M. The lipid flippase SLC47A1 blocks metabolic vulnerability to ferroptosis. Nat Commun. 2022;13:7965. doi: 10.1038/s41467-022-35707-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, Conroy S, Long L, Lelic N, Wang S, Gumin J, Raj D, Kodama Y, Raghunathan A, Olar A, Joshi K, Pelloski CE, Heimberger A, Kim SH, Cahill DP, Rao G, Den Dunnen WFA, Boddeke HWGM, Phillips HS, Nakano I, Lang FF, Colman H, Sulman EP, Aldape K. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 20.Ge SX, Son EW, Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics. 2018;19:534. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowman RL, Wang Q, Carro A, Verhaak RG, Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017;19:139–141. doi: 10.1093/neuonc/now247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 23.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 24.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shalabh Univariate, bivariate and multivariate statistics using R: quantitative tools for data analysis and data science. J R Stat Soc Ser A Stat Soc. 2022;185:736–737. [Google Scholar]
- 26.Hibma JE, Zur AA, Castro RA, Wittwer MB, Keizer RJ, Yee SW, Goswami S, Stocker SL, Zhang X, Huang Y, Brett CM, Savic RM, Giacomini KM. The effect of famotidine, a MATE1-selective inhibitor, on the pharmacokinetics and pharmacodynamics of metformin. Clin Pharmacokinet. 2016;55:711–721. doi: 10.1007/s40262-015-0346-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spinozzi G, Tini V, Ferrari A, Gionfriddo I, Ranieri R, Milano F, Pierangeli S, Donnini S, Mezzasoma F, Silvestri S, Falini B, Martelli MP. SiCoDEA: a simple, fast and complete app for analyzing the effect of individual drugs and their combinations. Biomolecules. 2022;12:904. doi: 10.3390/biom12070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Gebali S, Bentz S, Hediger MA, Anderle P. Solute carriers (SLCs) in cancer. Mol Aspects Med. 2013;34:719–734. doi: 10.1016/j.mam.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Meng Y, Li Y, Fang D, Huang Y. Identification of solute carrier family genes related to the prognosis and tumor-infiltrating immune cells of pancreatic ductal adenocarcinoma. Ann Transl Med. 2022;10:57. doi: 10.21037/atm-21-6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edemir B. Identification of prognostic organic cation and anion transporters in different cancer entities by in silico analysis. Int J Mol Sci. 2020;21:4491. doi: 10.3390/ijms21124491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian CY, Zheng Y, Wang Y, Chen J, Liu JY, Zhou HH, Yin JY, Liu ZQ. Associations of genetic polymorphisms of the transporters organic cation transporter 2 (OCT2), multidrug and toxin extrusion 1 (MATE1), and ATP-binding cassette subfamily C member 2 (ABCC2) with platinum-based chemotherapy response and toxicity in non-small cell lung cancer patients. Chin J Cancer. 2016;35:85. doi: 10.1186/s40880-016-0145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suenaga M, Schirripa M, Cao S, Zhang W, Yang D, Dadduzio V, Salvatore L, Borelli B, Pietrantonio F, Ning Y, Okazaki S, Berger MD, Miyamoto Y, Gopez R Jr, Barzi A, Yamaguchi T, Loupakis F, Lenz HJ. Potential role of polymorphisms in the transporter genes ENT1 and MATE1/OCT2 in predicting TAS-102 efficacy and toxicity in patients with refractory metastatic colorectal cancer. Eur J Cancer. 2017;86:197–206. doi: 10.1016/j.ejca.2017.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, Barthel F, Cho HJ, Lin YH, Satani N, Martinez-Ledesma E, Zheng S, Chang E, Sauvé CG, Olar A, Lan ZD, Finocchiaro G, Phillips JJ, Berger MS, Gabrusiewicz KR, Wang G, Eskilsson E, Hu J, Mikkelsen T, DePinho RA, Muller F, Heimberger AB, Sulman EP, Nam DH, Verhaak RGW. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32:42–56. e6. doi: 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P, Xia Q, Liu L, Li S, Dong L. Current opinion on molecular characterization for GBM classification in guiding clinical diagnosis, prognosis, and therapy. Front Mol Biosci. 2020;7:562798. doi: 10.3389/fmolb.2020.562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreira-Nunes CFA, Beltrão ACS, Francês LTVM, Sousa RGMA, Silva IT, Silva AL, Silva WA Jr, Lemos JAR. Drug efflux transporters and imatinib mesylate insensitivity in chronic myeloid leukemia. Blood. 2012;120:4424–4424. [Google Scholar]
- 37.Samuel SM, Varghese E, Koklesová L, Líšková A, Kubatka P, Büsselberg D. Counteracting chemoresistance with metformin in breast cancers: targeting cancer stem cells. Cancers (Basel) 2020;12:2482. doi: 10.3390/cancers12092482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wijaya J, Fukuda Y, Schuetz JD. Obstacles to brain tumor therapy: key ABC transporters. Int J Mol Sci. 2017;18:2544. doi: 10.3390/ijms18122544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang W, Yang F, Zhang Y, Fang Q, Lai Y, Lan Y. A newly established cuproptosis-related gene signature for predicting prognosis and immune infiltration in uveal melanoma. Int J Mol Sci. 2023;24:11358. doi: 10.3390/ijms241411358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmadi A, Ebrahimzadeh MA, Ahmad-Ashrafi S, Karami M, Mahdavi MR, Saravi SS. Hepatoprotective, antinociceptive and antioxidant activities of cimetidine, ranitidine and famotidine as histamine H2 receptor antagonists. Fundam Clin Pharmacol. 2011;25:72–79. doi: 10.1111/j.1472-8206.2009.00810.x. [DOI] [PubMed] [Google Scholar]
- 41.Razzaghdoust A, Mozdarani H, Mofid B. Famotidine as a radioprotector for rectal mucosa in prostate cancer patients treated with radiotherapy: phase I/II randomized placebo-controlled trial. Strahlenther Onkol. 2014;190:739–744. doi: 10.1007/s00066-014-0602-8. [DOI] [PubMed] [Google Scholar]
- 42.Lefranc F, James S, Camby I, Gaussin JF, Darro F, Brotchi J, Gabius J, Kiss R. Combined cimetidine and temozolomide, compared with temozolomide alone: significant increases in survival in nude mice bearing U373 human glioblastoma multiforme orthotopic xenografts. J Neurosurg. 2005;102:706–714. doi: 10.3171/jns.2005.102.4.0706. [DOI] [PubMed] [Google Scholar]
- 43.Kajiwara M, Terada T, Ogasawara K, Iwano J, Katsura T, Fukatsu A, Doi T, Inui KI. Identification of multidrug and toxin extrusion (MATE1 and MATE2-K) variants with complete loss of transport activity. J Hum Genet. 2009;54:40–46. doi: 10.1038/jhg.2008.1. [DOI] [PubMed] [Google Scholar]
- 44.Aleksunes LM, Woo V, Joy MS. Multidrug and toxin extrusion proteins. Drug Transporters Wiley. 2022:33–55. [Google Scholar]
- 45.Beishline K, Azizkhan-Clifford J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015;282:224–258. doi: 10.1111/febs.13148. [DOI] [PubMed] [Google Scholar]
- 46.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 47.Lv DD, Zhou LY, Tang H. Hepatocyte nuclear factor 4α and cancer-related cell signaling pathways: a promising insight into cancer treatment. Exp Mol Med. 2021;53:8–18. doi: 10.1038/s12276-020-00551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pouremamali F, Pouremamali A, Dadashpour M, Soozangar N, Jeddi F. An update of Nrf2 activators and inhibitors in cancer prevention/promotion. Cell Commun Signal. 2022;20:100. doi: 10.1186/s12964-022-00906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan Y, Wang M, Yang K, Chi T, Liao Z, Wei P. PPAR-α modulators as current and potential cancer treatments. Front Oncol. 2021;11:599995. doi: 10.3389/fonc.2021.599995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yonezawa A, Inui K. Importance of the multidrug and toxin extrusion MATE/SLC47A family to pharmacokinetics, pharmacodynamics/toxicodynamics and pharmacogenomics. Br J Pharmacol. 2011;164:1817–1825. doi: 10.1111/j.1476-5381.2011.01394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grottker J, Rosenberger A, Burckhardt G, Hagos Y. Interaction of human multidrug and toxin extrusion 1 (MATE1) transporter with antineoplastic agents. Drug Metabol Drug Interact. 2011;26:181–9. doi: 10.1515/DMDI.2011.024. [DOI] [PubMed] [Google Scholar]
- 52.Strobel J, Müller F, Zolk O, Endreß B, König J, Fromm MF, Maas R. Transport of asymmetric dimethylarginine (ADMA) by cationic amino acid transporter 2 (CAT2), organic cation transporter 2 (OCT2) and multidrug and toxin extrusion protein 1 (MATE1) Amino Acids. 2013;45:989–1002. doi: 10.1007/s00726-013-1556-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.