Abstract
Effective therapies for hepatocellular carcinoma (HCC) are urgently needed, as it is a type of cancer resistant to chemotherapy. Recent evidence showed that PF-429242, a membrane-bound transcription factor site-1 protease (MBTPS1) inhibitor, exhibited anticancer activities against glioblastomas, renal cell carcinoma, and pancreatic cancer. However, its anticancer activity against HCC has yet to be investigated. In this study, we found that PF-429242 induced autophagy-dependent cell death in HCC cells. RNA-sequencing analysis indicated that the primary effect of PF-429242 was inhibition of the sterol regulatory element-binding protein (SREBP) signaling pathway. However, overexpression of SREBP proteins did not efficiently rescue PF-429242-induced autophagy and cell death. Mechanistically, PF-429242 induced forkhead box protein O1 (FOXO1)-dependent autophagic cell death. Additionally, PF-429242 caused FOXO1-independent upregulation of insulin-like growth factor-binding protein 1 (IGFBP1), ultimately leading to autophagy-independent cell death. The in vivo anticancer activity of PF-429242 against HCC cells was demonstrated in a tumor xenograft mouse model. Therefore, PF-429242 is a potential anticancer agent to treat HCC by triggering FOXO1-dependent autophagic cell death and IGFBP1-mediated anti-survival signaling in parallel.
Keywords: Autophagy, FOXO1, hepatocellular carcinoma, IGFBP1, PF-429242
Introduction
Liver cancer is the fifth and seventh leading cause of cancer deaths, respectively, in men and women worldwide [1]. The most common type of primary liver cancer is hepatocellular carcinoma (HCC), which accounts for 75%~85% of cases [2,3]. Several treatment options are available for HCC, including surgery, liver transplantation, local ablation techniques like ethanol injection or radiofrequency thermal ablation, transarterial chemoembolization/radioembolization, and systemic pharmacological treatments [3-5]. However, advanced or metastatic HCC is a chemo-resistant and refractory tumor type with no reliable treatment options [6]. Although some molecular-targeted agents like sorafenib, regorafenib, lenvatinib, and cabozantinib have been approved for treating advanced HCC, they only slightly improve median overall survival [7-10]. Recently, immunotherapies such as immune checkpoint inhibitors, adoptive cell therapy, engineered cytokines, and therapeutic cancer vaccines have been explored as treatment options for advanced HCC [11,12]. Despite these advances, treatment of advanced HCC remains challenging, and novel therapeutic strategies are needed.
Autophagy is a process that allows cells to break down and recycle their components, often in response to nutrient deprivation or other stressors. During autophagy, cells form double-membraned autophagic vesicles (autophagosomes) to sequester intracellular materials like damaged organelles, protein aggregates, and pathogens, which are then delivered to lysosomes for degradation [13]. Autophagy helps maintain cellular homeostasis, provides an alternative energy source, and allows cells to survive stress by eliminating damaged organelles, misfolded proteins, and pathogens [14]. It can also contribute to type II programmed cell death or autophagic cell death [15,16]. However, relationships of pro-survival autophagy with autophagic cell death and their shared molecular mechanisms are not fully understood [17,18]. Accordingly, the roles of autophagy in cancer progression and responses to treatment are controversial. They may depend on the type, stage, and genetic context of the cancer and the tumor microenvironment. Autophagy can either facilitate drug resistance or contribute to the destruction of cancer cells through autophagic cell death in response to cancer therapies. Therefore, understanding the role of autophagy in cancer treatment and modulating its activity could improve the effectiveness of cancer therapies [19-21].
Sterol regulatory element-binding proteins (SREBPs) are transcription factors that have a basic-helix-loop-helix leucine zipper structure. Among three SREBP isoforms, SREBP1a and SREBP2 are predominantly expressed in cultured cells and respectively activate fatty acid and cholesterol synthesis. SREBP1c is mainly expressed in the liver and is regulated by nutrient and energy statuses. SREBPs sense nutrition levels and regulate lipid metabolism genes. When sterols are present, SREBPs become inactive and are kept in the endoplasmic reticulum by associating with SREBP-cleavage-activating protein (SCAP) and insulin-induced genes 1 and 2 (INSIG1/2). However, in cells with low levels of sterols, SCAP and INSIGs separate, allowing the SREBP-SCAP complex to move to the Golgi apparatus. Here, it is cleaved into a water-soluble N-terminal domain by site-1 protease (S1P) and site-2 protease (S2P), which are respectively encoded by the membrane-bound transcription factor site-1 protease (MBTPS1) and MBTPS2 genes. Mature SREBPs are subsequently taken into nuclei, where they bind to specific sterol regulatory elements and upregulate sterol biosynthesis genes. Sterols then limit additional sterol synthesis by inhibiting SREBP cleavage, forming a negative feedback loop [22,23]. Because of their prominent involvement in lipid metabolism, components of the SREBP signaling pathway have been explored as potential metabolic targets for cancer therapy [22].
Class O forkhead box (FOXO) transcription factors are essential autophagy regulators [24]. Among the four family members (FOXO1, FOXO3, FOXO4, and FOXO6), FOXO3 was first identified as a transcriptional regulator of autophagy genes [25-27]. Later, FOXO1 was also shown to be a transcriptional autophagy regulator [28-30]. FOXO1 can also induce autophagy in a transcription-independent manner. Cytosolic acetylated FOXO1 directly binds to autophagy related 7 (ATG7) to stimulate autophagy [31]. Transcriptional initiation of autophagy by FOXO transcription factors is regulated by their post-translational modifications and subsequent nuclear/cytosolic shuttling. For example, adenosine monophosphate-activated protein kinase (AMPK)-mediated phosphorylation and protein arginine N-methyltransferase 6 (PRMT6)-mediated methylation favor their nuclear translocation, whereas AKT-mediated phosphorylation results in cytoplasmic retention [24].
Insulin-like growth factor (IGF)-binding protein 1 (IGFBP1) belongs to the family of IGF-binding proteins (IGFBP1~6) that modulate the availability of unbound IGFs (IGF1 and IGF2) for interactions with IGF receptors (IGF1R and IGF2R). Physiologically, IGFs mediate the effects of growth hormones, promote cell proliferation and differentiation, and inhibit apoptosis by activating multiple intracellular signaling pathways, including the phosphoinositide 3-kinase (PI3K)/AKT and mitogen-activated protein kinase (MAPK). Thus, IGFs and IGFRs are oncogenic in cancer [32,33]. By binding with IGFs and preventing their interaction with receptors, IGFBP1 can suppress IGF-driven oncogenic signaling.
PF-429242 is the most commonly used experimental MBTPS1 inhibitor [34]. However, its anticancer activity has rarely been investigated. This study identified that PF-429242 exhibits in vitro and in vivo anticancer activities against HCC through the parallel induction of FOXO1-dependent autophagic cell death and FOXO1-independent IGFBP1-mediated anti-survival signaling.
Material and methods
Chemicals and reagents
PF-429242 (#A11230) and AS1842856 (#A15871) were purchased from Adooq Bioscience (Irvine, CA, USA). Doxorubicin (#D-4000) was purchased from LC Laboratories (Woburn, MA, USA). Spautin-1 (#SML0440), chloroquine diphosphate salt (#C6628), and carbenoxolone disodium salt (#C4790) were purchased from Merck (Darmstadt, Germany). ZVAD-FMK (#A1902) was purchased from APExBIO Technology (Houston, TX, USA). Minimum essential medium (MEM; #10-010-CM) and Matrigel (#354248) were purchased from Corning (Corning, NY, USA). L-Glutamine (#25030081), Earle’s balanced salt solution (EBSS; #14155063), sodium bicarbonate (#11360070), non-essential amino acids (NEAA) solution (#11140050), sodium pyruvate (#11360070), fetal bovine serum (FBS; #10437028), alamarBlue cell viability reagent (#DAL1100), M-PER mammalian protein extraction reagent (#78501), Premo autophagy tandem sensor RFP-GFP-LC3B kit (#P36239), Hoechst 33342 solution (#62249), and Lipofectamine RNAiMAX and 3000 transfection reagents (#13778150 and #L3000015, respectively) were purchased from ThermoFisher Scientific (Waltham, MA, USA). Bio-Rad protein assay (#5000006) and iScript cDNA synthesis kit (#1708891) were purchased from Bio-Rad Laboratories (Hercules, CA, USA). Western Lightning ECL Pro reagent (#PK-NEL122) was purchased from PerkinElmer (Boston, MA, USA). GENEzol TriRNA Pure kit (#GZX100) was purchased from Geneaid (New Taipei City, Taiwan). IQ2 SYBR Green Fast qPCR System Master Mix (#DBU-006) was purchased from Bio-Genesis (Taipei, Taiwan).
Cell culture
PLC/PRF/5 (PLC5; #60223) and HepG2 (#RM60025) cells were purchased from the Bioresource Collection and Research Center (Hsinchu City, Taiwan) and authenticated by short tandem repeat profiling. Cells were cultured in 90% MEM with 2 mM L-glutamine and EBSS adjusted to contain 1.5 g/L sodium bicarbonate, 0.1 mM NEAA, 1.0 mM sodium pyruvate, and 10% FBS. All cell lines were incubated in a 37°C humidified incubator and routinely tested to ensure they were free of mycoplasma contamination.
Cell viability assay
PLC5 and HepG2 cells were trypsinized into single-cell suspension and spread in a 96-well plate. After 24 h, cells were treated with drugs for indicated time intervals. At the end of the experiment, alamarBlue reagent was added to each well. After 1~3 h of incubation, fluorescence (excitation at 530~560 nm and emission at 590 nm) was detected using a Varioskan Flash Multimode Reader (ThermoFisher Scientific). Cell viability was calculated after subtraction of the background intensity and normalization with control fluorescence.
Autophagy flux assay
Stages of the autophagic process were detected using a Premo autophagy tandem sensor RFP-GFP-LC3B kit following the manufacturer’s instructions. Briefly, cells were spread on a µ-Slide 8-well chamber slide (#80826; ibidi GmbH; Martinsried, Germany) and allowed to adhere overnight and grow to 70% confluency. Cells were infected with Premo autophagy tandem sensor RFP-GFP-LC3B for 24 h and then exposed to 20 μM PF-429242 for another 24 h. The red fluorescent protein (RFP) and green fluorescent protein (GFP) fluorescence was observed under a fluorescence microscope to illustrate the distributions of autophagosomes (yellow dots in fusion images) and autolysosomes (red dots in fusion images). Cell nuclei were stained with Hoechst 33342.
RNA-sequencing (RNA-Seq) and bioinformatics analysis
PLC5 and HepG2 cells were treated with or without 20 μM PF-429242 for 24 h, and samples were sent to Biotools (New TaipeiCity, Taiwan) for RNA-Seq using the Illumina NovaSeq 6000 platform (San Diego, CA, USA) to generate 150-bp paired-end reads. DEGseq was used to identify the differentially expressed genes (DEGs) with |fold change| > 2 and an adjusted p value of < 0.005. Overlapping DEGs between PLC5 and HepG2 cells were visualized using the VENNY 2.1 online tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html). A heatmap was generated using the Heatmapper online tool [35] (http://www.heatmapper.ca/). A pathway enrichment analysis was performed using the STRING online database [36] (https://string-db.org/) and gene set enrichment analysis (GSEA) software [37,38]. RNA-Seq data (GSE228257) were deposited in the NCBI GEO database.
Western blotting
Cells were lysed using the M-PER mammalian protein extraction reagent. The protein concentration in the lysate was measured with a Bio-Rad protein assay, and equal amounts of protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were transferred to a nitrocellulose membrane, then incubated with a primary antibody overnight at 4°C. After washing with Tris-buffered saline with 0.1% Tween 20 (TBST), the membrane was incubated with a horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h. The membrane was exposed to an enhanced chemiluminescence (ECL) reagent to produce a chemiluminescent signal. The following antibodies were used in this study. SREBP1 (#557036) and SREBP2 (#557037) antibodies were purchased from BD Biosciences (Woburn, MA, USA). MAP1LC3B (#2775), PARP (#9542), cleaved caspase-3 (#9661), phospho-Ser256-FOXO1 (#9461), FOXO1 (#2880), and Tubulin (#2148) antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). The IGFBP1 antibody (#GTX129006) was purchased from GeneTex (HsinchuCity, Taiwan). The DEPP1 antibody (#NBP2-38367) was purchased from Novus Biologicals (Coralville, IA, USA). Acetyl-Lys262/Lys265/Lys274-FOXO1 (#A17406) and Ki67 (#A11390) antibodies were purchased from ABclonal Technology (Woburn, MA, USA). MAP1LC3B (#18725-1-AP) and GAPDH (#60004-1-Ig) antibodies were purchased from Proteintech (Chicago, IL, USA). The anti-Flag antibody (#F3165) was purchased from Merck.
Real-time quantitative polymerase chain reaction (qPCR)
Total RNA was extracted using a GENEzol TriRNA Pure kit, according to the manufacturer’s instructions. First-strand complementary (c)DNA was synthesized using an iScript cDNA synthesis kit. SYBR Green-based PCR amplification was performed on a Roche LightCycler 480 System (Indianapolis, IN, USA) using the following primer pairs: human MBTPS1 (forward 5’-TCCAATTGCTTGGATGACAG-3’ and reverse 5’-TCCAGAACCTTGGAGTACCG-3’), human MBTPS2 (forward 5’-ACCCGTCAATCAACTGACCT-3’ and reverse 5’-TGCCAGATACCTGCACAAAA-3’), human IGFBP1 (forward 5’-CTGCGTGCAGGAGTCTGA-3’ and reverse 5’-CCCAAAGGATGGAATGATCC-3’), human DEPP1 (forward 5’-ATACGTCCTGTGGTGGCATTG-3’ and reverse 5’-CCTGATTCCCGTTCCCTGAT-3’), human MAP1LC3B (forward 5’-AACGGGCTGTGTGAGAAAAC-3’ and reverse 5’-AGTGAGGACTTTGGGTGTGG-3’), human SQSTM1 (forward 5’-CATCGGAGGATCCGAGTGTG-3’ and reverse 5’-TTCTTTTCCCTCCGTGCTCC-3’), and human 18S ribosomal (r)RNA (forward 5’-CGGCGACGACCCATTCGAAC-3’ and reverse 5’-GAATCGAACCCTGATTCCCCGTC-3’). Fold changes in gene expressions were calculated using the comparative cycle threshold method.
Transient transfection
Lipofectamine RNAiMAX and 3000 transfection reagents were used to transiently transfect the following siRNAs and plasmids into cells, respectively, according to the manufacturer’s instructions. si-MBTPS1 (#sc-36496), si-MBTPS2 (#sc-41652), si-IGFBP1 (#sc-39584), si-DEPP1 (#sc-90781), si-FOXO1 (#sc-35382), and control siRNA (#sc-37007) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). pcDNA3.1-2xFLAG-SREBP-1a (Addgene plasmid #26801; http://n2t.net/addgene:26801; RRID: Addgene_26801), pcDNA3.1-2xFLAG-SREBP-1c (Addgene plasmid #26802; http://n2t.net/addgene:26802; RRID: Addgene_26802), and pcDNA3.1-2xFLAG-SREBP-2 (Addgene plasmid #26807; http://n2t.net/addgene:26807; RRID: Addgene_26807) were gifts from Timothy Osborne [39]. At 24 h after transfection, cells were used for further experiments.
Animal xenograft model
Animal studies were approved by the Institutional Animal Care and Use Committee at Mackay Memorial Hospital (MMS-A-S-110-13 and MMS-A-S-111-06) and Taipei Medical University (LAC-2020-0007 and LAC-2021-0421). HepG2 (107) cells mixed with Matrigel (1:1) were subcutaneously injected into 6~8-week-old male NPG (NOD-Prkdcscid IL2rgnull) mice (BioLASCO, Taipei, Taiwan) on the first experimental day (day 0). After 12 days, mice were randomly divided into vehicle (n = 5) and PF-429242 (n = 5) groups. PF-429242 (20 mg/kg) or the vehicle solvent control (8% DMSO in sterile PBS) was intravenously administered to mice twice weekly for 4 weeks. Tumor length and width and mice body weight were measured twice weekly. The tumor volume was calculated by the formula: 0.5 × length × width2. Blinding in group allocation and outcome assessment was not performed in this animal study due to practical limitations and the nature of the interventions used. However, efforts were made to minimize bias and ensure that data were objectively collected and analyzed.
Immunohistochemistry (IHC) staining
After fixation with 10% formaldehyde at room temperature for at least 24 h, HepG2 tumor specimens from xenograft assay were subjected to trimming, dehydration, paraffin embedding, and sectioning. Immunohistochemistry (IHC) was conducted using the primary antibodies (1:100 dilution) for Ki67, IGFBP1, and LC3B. Sections were incubated with 3,3’-diaminobenzidine (DAB) and counterstained with hematoxylin.
Statistical analysis
All experiments were performed independently, with at least three replicates. Results are shown as the mean ± standard error of the mean (SEM) or mean ± standard deviation (SD). Data were assessed for normality using the Kolmogorov-Smirnov test. If the data were normally distributed, an unpaired two-tailed Student’s t-test was used for statistical analysis. If the data were not normally distributed, a two-tailed Mann-Whitney test was used. A p value of < 0.05 was considered statistically significant.
Results
Effect of the MBTPS1 inhibitor, PF-429242, on HCC cells
Recently, the MBTPS1 inhibitor, PF-429242, was found to exhibit in vitro or in vivo anticancer activity against glioblastomas, renal cell carcinoma, and pancreatic cancer [40-42]. Our previous study showed that PF-429242 enhanced the in vitro anticancer activity of GSK343, an enhancer of zeste homolog 2 (EZH2) inhibitor, in HCC cells [43]. In this study, we further explored the anticancer cancer activity of PF-429242 against HCC. We found that PF-429242 dose-dependently inhibited cell viability in two HCC cell lines, PLC5 and HepG2 (Figure 1A). To confirm that PF-429242 indeed inhibited the MBTPS1/SREBP signaling pathway, the proteolytic processing of SREBP1 and SREBP2 was detected by Western blotting. As shown in Figure 1B, the pro-forms (precursors) of SREBP1 and SREBP2 were reduced by PF-429242. Interestingly, PLC5 and HepG2 cells tended to have higher levels of mature SREBP2, which PF-429242 significantly inhibited. These results imply that SREBP2 may be the primary target of PF-429242 in HCC cells. To further characterize the inhibitory effect of PF-429242 on the MBTPS1/SREBP signaling pathway, an RNA-Seq analysis of PF-429242-treated PLC5 and HepG2 cells was performed. There were 5 and 16 commonly upregulated and downregulated genes in the two cell lines (Figure 1C), as visualized in a heatmap (Figure 1D). The pathway enrichment of these genes was analyzed using the STRING database (https://string-db.org/) [36]. We found that PF-429242-downregulated genes were related to lipid metabolism (Figure 1E), which confirmed inhibition of the MBTPS1/SREBP signaling pathway by PF-429242. In addition, GSEA identified that cholesterol homeostasis was the most inhibited cancer hallmark in PF-4294242-treated HCC cells (Figure 1F; Table 1). Consistent with Figure 1B, SREBF2, which encodes the SREBP2 protein, was commonly downregulated by PF-429242 in HCC cells (Figure 1D and 1E). Therefore, these results confirmed inhibition of the MBTPS1/SREBP signaling pathway and cell viability by PF-429242 in HCC cells.
Figure 1.
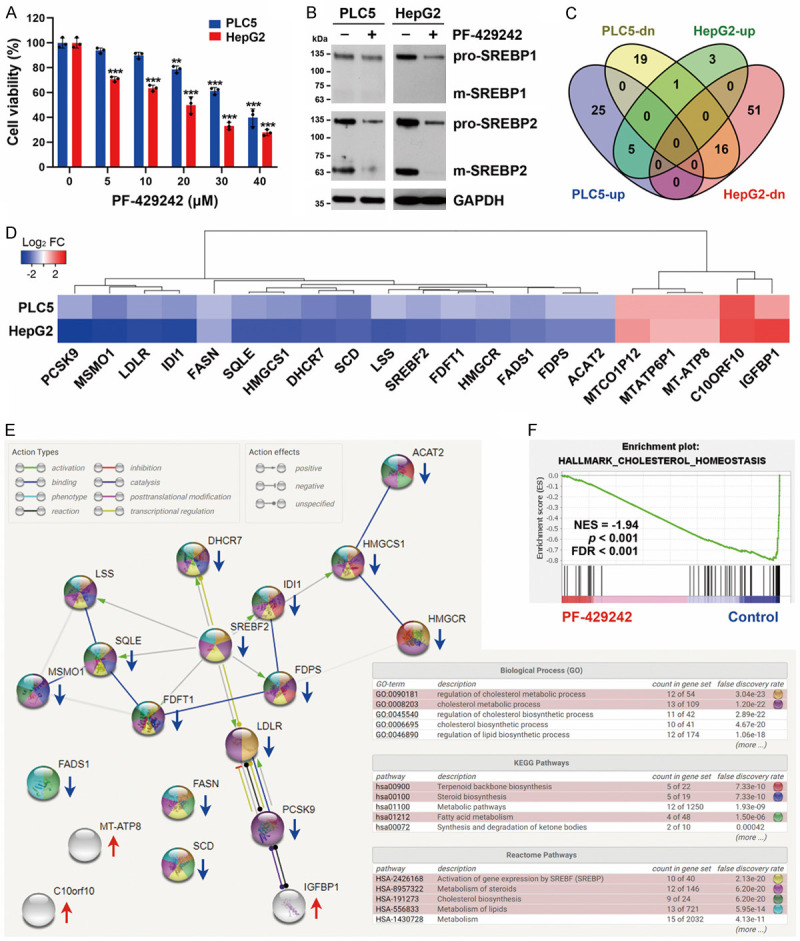
In vitro effect of PF-429242 on hepatocellular carcinoma (HCC) cells. A. PLC5 and HepG2 cells were treated with various doses of PF-429242 for 72 h, and then cell viability was determined by Alamar Blue staining. Error bars are the mean ± SD (n = 3). ** and *** indicate a significant difference (P < 0.01 and P < 0.001, respectively) between PF-429242-treated and control cells. B. PLC5 and HepG2 cells were treated with 20 μM PF-429242 for 24 h. Protein expressions were determined by Western blotting. Pro- and m- respectively indicate the pro-forms (precursors) and mature forms of SREBP. C-E. PLC5 and HepG2 cells were treated with 20 μM PF-429242 for 24 h, and gene expressions were analyzed by RNA-Seq. C. A Venn diagram shows upregulated, downregulated, and overlapping gene numbers in the two HCC cell lines. D. A heatmap is used to visualize the overlapping genes and their fold changes. E. Pathway enrichments of commonly upregulated and downregulated genes were analyzed using the STRING database. MTATP6P1 and MTCO1P12 were excluded because they were pseudogenes. F. A gene set enrichment analysis (GSEA) was performed to visualize the enrichment of “cholesterol homeostasis” cancer hallmark in PF-429242-treated HCC cells.
Table 1.
Gene set enrichment analysis (GSEA) for hallmarks commonly enriched in PF-429242-treated PLC5 and HepG2 cells
| Hallmark | No. of Genes in Pathway | No. of Pathway Genes Differentially Expressed (% of Total) | NES | p Value | FDR |
|---|---|---|---|---|---|
| CHOLESTEROL_HOMEOSTASIS | 74 | 30 (41%) | -1.95 | < 0.001 | < 0.001 |
| E2F_TARGETS | 200 | 164 (82%) | -1.81 | < 0.001 | < 0.001 |
| G2M_CHECKPOINT | 200 | 134 (67%) | -1.72 | < 0.001 | < 0.001 |
| MITOTIC_SPINDLE | 199 | 128 (64%) | -1.64 | < 0.001 | 0.001 |
| MYC_TARGETS_V1 | 200 | 149 (75%) | -1.59 | < 0.001 | 0.002 |
| OXIDATIVE_PHOSPHORYLATION | 200 | 161 (81%) | -1.56 | < 0.001 | 0.003 |
| PROTEIN_SECRETION | 96 | 76 (79%) | -1.53 | 0.001 | 0.004 |
| DNA_REPAIR | 150 | 103 (69%) | -1.51 | < 0.001 | 0.006 |
| FATTY_ACID_METABOLISM | 158 | 102 (65%) | -1.46 | 0.001 | 0.015 |
| MTORC1_SIGNALING | 200 | 111 (56%) | -1.45 | 0.002 | 0.018 |
Top-10 enriched hallmarks were shown. NES, normalized enrichment score; FDR, false-discovery rate.
PF-429242 induced autophagic cell death
To investigate how PF-429242 inhibits cell viability in HCC cells, cell apoptosis was evaluated by examining cleavage of poly (ADP ribose) polymerase (PARP) and caspase-3. However, PF-429242 did not significantly induce PARP or caspase-3 cleavage (Figure 2A). To confirm this observation, doxorubicin, a chemotherapeutic drug, was used as a positive control. The in vitro anticancer activities of doxorubicin in PLC5 and HepG2 cells were confirmed in Figure 2B. Unlike PF-429242, doxorubicin at 0.25~1 and 2.5 μM respectively induced the cleavages of PARP and caspase-3 in PLC5 and HepG2 cells (Figure 2A). These results indicated that PF-429242 did not induce apoptosis of HCC cells. Interestingly, PF-429242 dose- and time-dependently induced the accumulation of LC3B-II, a marker of autophagy [44] (Figure 2A, 2C). Both increased autophagosome formation and impaired autophagosome-lysosome fusion can induce LC3B-II accumulation [44]. To discriminate between these two possibilities, autophagic flux (LC3B flux) was analyzed by treating PLC5 and HepG2 cells with early-(spautin-1) and late-stage (chloroquine) inhibitors of autophagy. Spautin-1 blocks autophagy initiation by inhibiting ubiquitin-specific peptidase 10 (USP10) and USP13, leading to degradation of vacuolar protein sorting 34 (VPS34)-PI3K complexes [45]. On the other hand, chloroquine reduces the autophagic flux by hindering the fusion of autophagosomes and lysosomes [46]. PF-429242-induced LC3B-II accumulation was reduced by spautin-1 and was enhanced by chloroquine (Figure 2D), suggesting that PF-429242 indeed induces autophagy. To visualize the maturation of autophagosomes in cells, the tandem RFP-GFP-LC3B sensor was transduced into PLC5 and HepG2 cells, and cells were observed under a fluorescence microscope. By using GFP, which is sensitive to acid, in combination with RFP, which is not, it was possible to observe the transition from an autophagosome (which has a neutral pH) to an autolysosome (which has an acidic pH) by tracking the loss of GFP fluorescence and observing only RFP fluorescence [47]. As shown in Figure 2E, PF-429242 increased numbers of autophagosomes (yellowish-green) and autolysosomes (red). To understand the impacts of autophagy and apoptosis on the anticancer activity of PF-429242, the effects of the drug were studied in PLC5 and HepG2 cells in the presence and absence of inhibitors of autophagy (spautin-1) and apoptosis (ZVAD-FMK). As shown in Figure 2F, spautin-1, but not ZVAD-FMK, significantly rescued cells from PF-4294242-induced cytotoxicity, suggesting that PF-429242 induces autophagic cell death.
Figure 2.
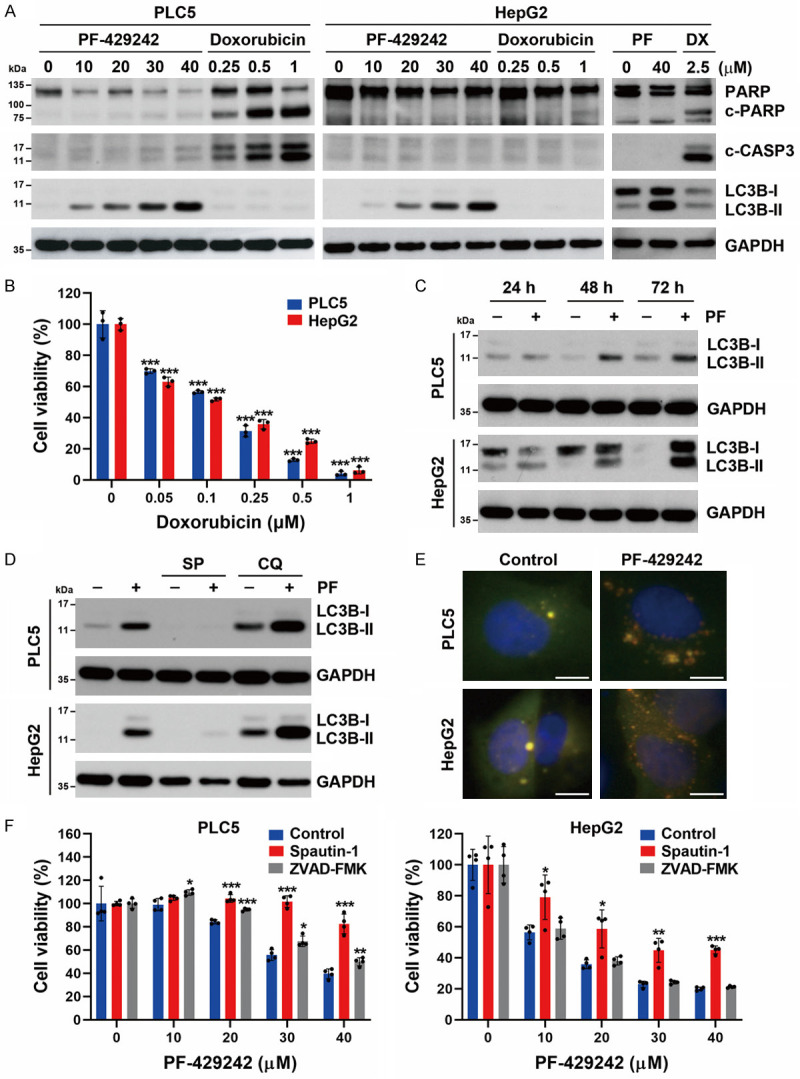
Effects of PF-429242 on apoptosis and autophagy in hepatocellular carcinoma (HCC) cells. A. PLC5 and HepG2 cells were treated with indicated doses of PF-429242 (PF) and doxorubicin (DX) for 48 h. Protein expressions were determined by Western blotting. B. PLC5 and HepG2 cells were treated with various doses of doxorubicin for 72 h, and then cell viability was determined by Alamar Blue staining. Error bars are the mean ± SD (n = 3). *** indicate a significant difference (P < 0.001) between doxorubicin-treated and control cells. C. PLC5 and HepG2 cells were treated with 20 μM PF-429242 for 24, 48, and 72 h. Protein expressions were determined by Western blotting. D. PLC5 and HepG2 cells were treated with 20 μM PF-429242 in the absence or presence of 5 μM spautin-1 (SP) or 30 μM chloroquine (CQ; 6 h before cell harvest) for 24 h. Protein expressions were determined by Western blotting. E. PLC5 and HepG2 cells were transduced with viral particles carrying the RFP-GFP-LC3B tandem sensor for 24 h, followed by treatment with 20 μM PF-429242 for 24 h. RFP and GFP fluorescence was observed under a fluorescence microscope. Cell nuclei were stained with Hoechst 33342. Scale bar, 10 μm. F. PLC5 and HepG2 cells were treated with various doses of PF-429242 with or without 1 μM spautin-1 or 20 μM ZVAD-FMK for 72 h. Cell viability was determined by Alamar Blue staining. Error bars are the mean ± SD (n = 4). *, **, and *** indicate a significant difference (P < 0.05, P < 0.01, and P < 0.001, respectively) in PF-429242-treated cells with or without spautin-1 or ZVAD-FMK.
Inhibition of the MBTPS1/SREBP pathway does not fully contribute to PF-429242-induced autophagy and cell death
It was reported that inhibition of MBTPS1/2 by chemical inhibitors (including PF-429242) and siRNAs can increase the autophagic flux [48,49]. To determine if suppression of MBTPS1 was the cause of the autophagy triggered by PF-429242 in HCC cells, siRNAs specific for MBTPS1 and MBTPS2 (for comparison) were used to reduce their messenger (m)RNA expressions (Figure 3A). Knockdown of MBTPS1, but not MBTPS2, induced a slight accumulation of LC3B-II only when combined with chloroquine, which was incomparable with the effect of PF-429242 (Figure 3B). In addition, knockdown of MBTPS1 or MBTPS2 did not alter cell viability in PLC5 or HepG2 cells (Figure 3C). Therefore, inhibition of MBTPS1 cannot fully explain the PF-429242-induced cell death of HCC cells.
Figure 3.
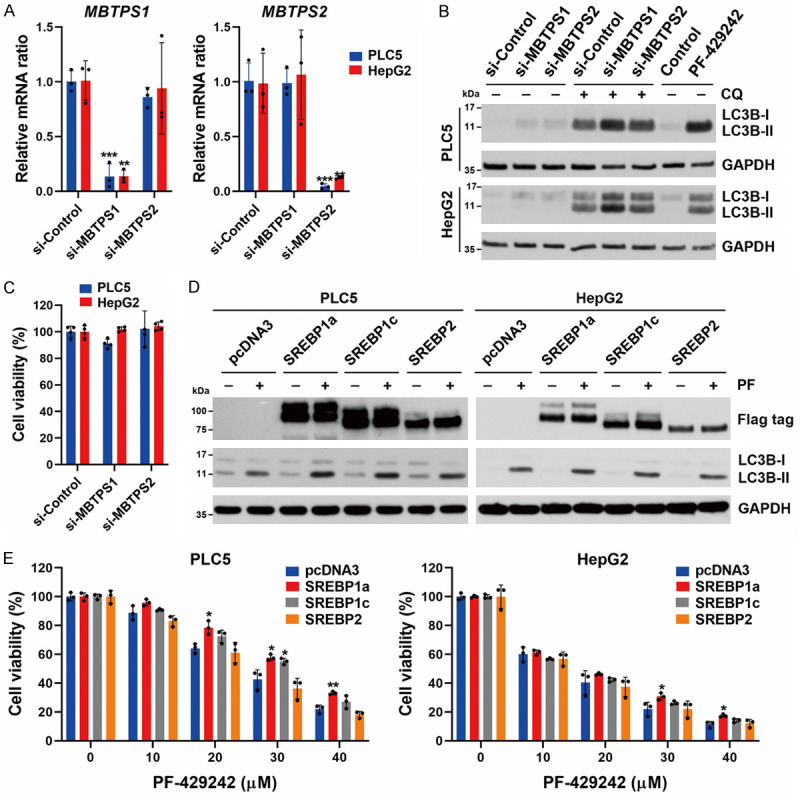
Role of the MBTPS1 and SREBP signaling pathway in PF-429242-induced autophagy and cell death in hepatocellular carcinoma (HCC) cells. A. PLC5 and HepG2 cells were transfected with siRNAs specific for MBTPS1 and MBTPS2 for 48 h, and then the mRNA levels of MBTPS1 and MBTPS2 were determined by a real-time qPCR. Error bars are the mean ± SD (n = 3). *, **, and *** indicate a significant difference (P < 0.05, P < 0.01, and P < 0.001, respectively) compared to si-Control-transfected cells. B. HepG2 and PLC5 cells were transfected with siRNAs specific for MBTPS1 (S1P) and MBTPS2 (S2P) with or without 20 μM chloroquine (CQ; 6 h before cell harvest) for 48 h, or treated with 20 μM PF-429242 for 24 h. Protein expressions were determined by Western blotting. C. PLC5 and HepG2 cells were transfected with siRNAs specific for MBTPS1 and MBTPS2 for 72 h, and then cell viability was determined by Alamar Blue staining. Error bars are the mean ± SD (n = 4). D. HepG2 and PLC5 cells were transfected with plasmids encoding Flag-tagged SREBP1a, SREBP1c, and SREBP2 for 48 h, and then treated with 20 μM PF-429242 for 24 h. Protein expressions were determined by Western blotting. E. SREBP1/2- transfected PLC5 and HepG2 cells were seeded in a 96-well plate, and then treated with indicated doses of PF-429242 for 72 h. Cell viability was determined by Alamar Blue staining. Error bars are the mean ± SD (n = 3). * and ** indicate a significant difference (P < 0.05 and P < 0.01, respectively) between SREBP1/2- and pcDNA3-transfected cells.
Because PF-429242 inhibits the proteolytic processing of SREBPs that enter the nucleus to regulate gene expressions, we investigated whether overexpression of SREBPs could rescue PF-429242-induced autophagy and cell death. PLC5 and HepG2 cells were transfected with plasmids encoding Flag-tagged mature forms of SREBP1a, SREBP1c, or SREBP2 and then were treated with PF-429242. However, overexpression of these SREBP proteins did not affect PF-429242-induced LC3B-II accumulation (Figure 3D), suggesting that PF-429242-induced autophagy in HCC cells is not dependent on inhibiting SREBP processing. In addition, we observed that overexpressions of SREBP1a/c, but not SREBP2, only slightly rescued PF-429242-induced cytotoxicity (Figure 3E). Thus, inhibition of the MBTPS1/SREBP signaling pathway is not central to PF-429242-induced autophagy and cell death in HCC cells.
PF-429242 induces IGFBP1 expression to promote cell death in an autophagy-independent manner
According to the RNA-Seq analysis (Figure 1D), PF-429242 upregulated three genes (C10ORF10, IGFBP1, and MTATP8) and two pseudogenes (MTATP6P1 and MTCO1P12). After a comprehensive literature review, we found that C10ORF10 (also known as DEPP1) was recently identified as an autophagy regulator [50,51]. We hypothesized that DEPP1 upregulation might be responsible for PF-429242-induced autophagy in HCC cells. A real-time qPCR and Western blotting were performed to confirm the induction of DEPP1 and IGFBP1 mRNA and protein expressions (Figure 4A, 4B). Consistent with results of RNA-Seq (Figure 1D), PF-429242 did not induce autophagy-related gene expression, such as MAP1LC3B and SQSTM1 (Figure 4A). To investigate whether DEPP1 and IGFBP1 were responsible for PF-429242-induced autophagy in HCC cells, siRNAs specific to DEPP1 and IGFBP1 were used. However, knockdown of DEPP1 and IGFBP1 did not suppress PF-429242-induced LC3B-II accumulation (Figure 4C, 4D). Interestingly, knockdown of IGFBP1, but not DEPP1, rescued PF-429242-induced cytotoxicity (Figure 4E). Therefore, PF-429242 may induce IGFBP1 expression to promote cell death in an autophagy-independent manner.
Figure 4.
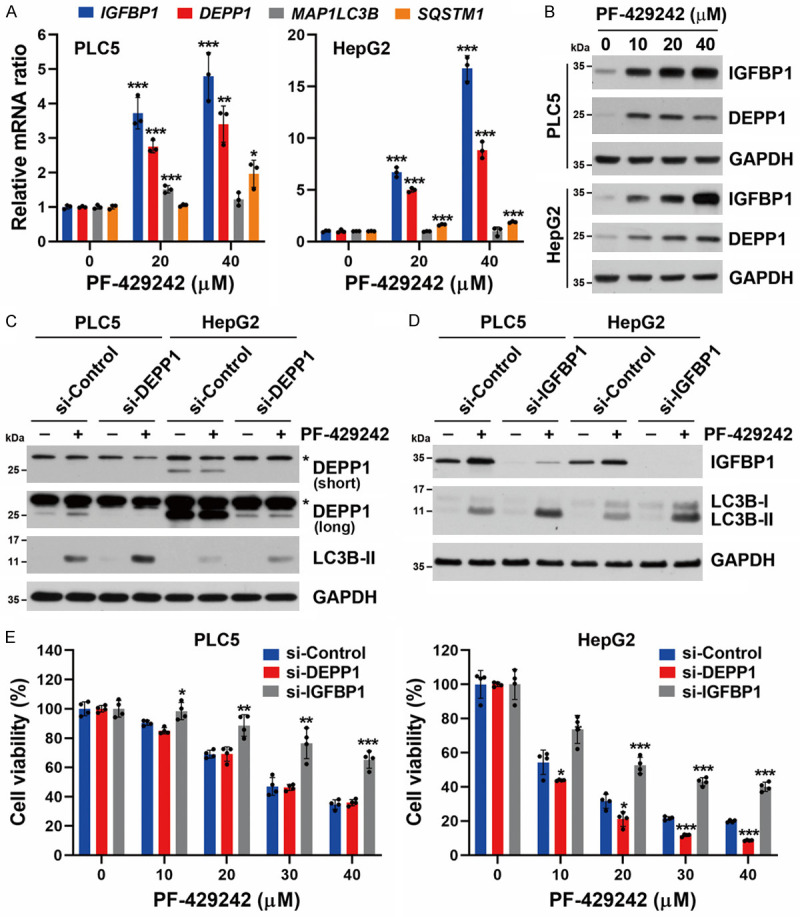
Roles of IGFBP1 and DEPP1 in PF-429242-induced autophagic cell death in hepatocellular carcinoma (HCC) cells. A. PLC5 and HepG2 cells were treated with 20 and 40 μM for PF-429242 for 24 h, and then mRNA levels of the IGFBP1, DEPP1 (C10orf10), MAP1LC3B, and SQSTM1 (p62) genes were determined by a real-time qPCR. Error bars are the mean ± SD (n = 3). *, **, and *** indicate a significant difference (P < 0.05, P < 0.01, and P < 0.001, respectively) compared to untreated control cells. B. PLC5 and HepG2 cells were treated with 10, 20, and 40 μM PF-429242 for 24 h. Protein expressions were determined by Western blotting. C. PLC5 and HepG2 cells were transfected with siRNA specific for DEPP1 for 24 h, then treated with 20 μM PF-429242 for another 24 h. Protein expressions were determined by Western blotting. For DEPP1 expression, both short- and long-exposure images are shown. An asterisk (*) indicates non-specific bands. D. PLC5 and HepG2 cells were transfected with siRNA specific for IGFBP1 for 24 h, then treated with 20 μM PF-429242 for another 24 h. Protein expressions were determined by Western blotting. E. si-DEPP1/si-IGFBP1-transfected PLC5 and HepG2 cells were seeded in a 96-well plate and treated with the indicated doses of PF-429242 for 72 h. Cell viability was determined by Alamar Blue staining. Error bars are the mean ± SD (n = 4). *, **, and *** indicate a significant difference (P < 0.05, P < 0.01, and P < 0.001, respectively) between si-DEPP1/si-IGFBP1- and si-Control-transfected cells.
PF-429242-induced autophagic cell death is mediated by FOXO1
Because both DEPP1 and IGFBP1 were identified as transcriptional targets of FOXO1 and FOXO3 [52-55], we further investigated whether FOXO1 or FOXO3 participates in PF-429242-induced autophagy and cell death. As shown in Figure 5A, the FOXO1 inhibitor, AS1842856, inhibited PF-429242-induced DEPP1, but not IGFBP1, expression. Importantly, PF-429242-induced LC3B-II accumulation was suppressed by AS1842856, suggesting that PF-429242 induces FOXO1-dependent autophagy. In contrast, the FOXO3 inhibitor, carbenoxolone, did not inhibit PF-429242-induced DEPP1/IGFBP1 expression or LC3B-II accumulation (Figure 5B). To further confirm the role of FOXO1 in PF-429242-induced autophagy, a siRNA specific to FOXO1 was used. Consistently, FOXO1-knockdown efficiently reduced PF-429242-induced DEPP1 expression and LC3B-II accumulation (Figure 5C). IGFBP1-knockdown did not alter the expression of FOXO1 (Figure 5D). These observations suggested that there is no correlation between FOXO1 and IGFBP1. Furthermore, PF-429242-induced cytotoxicity was also rescued by FOXO1-knockdown (Figure 5E). Therefore, PF-429242 induces autophagic cell death through a FOXO1-dependent pathway.
Figure 5.
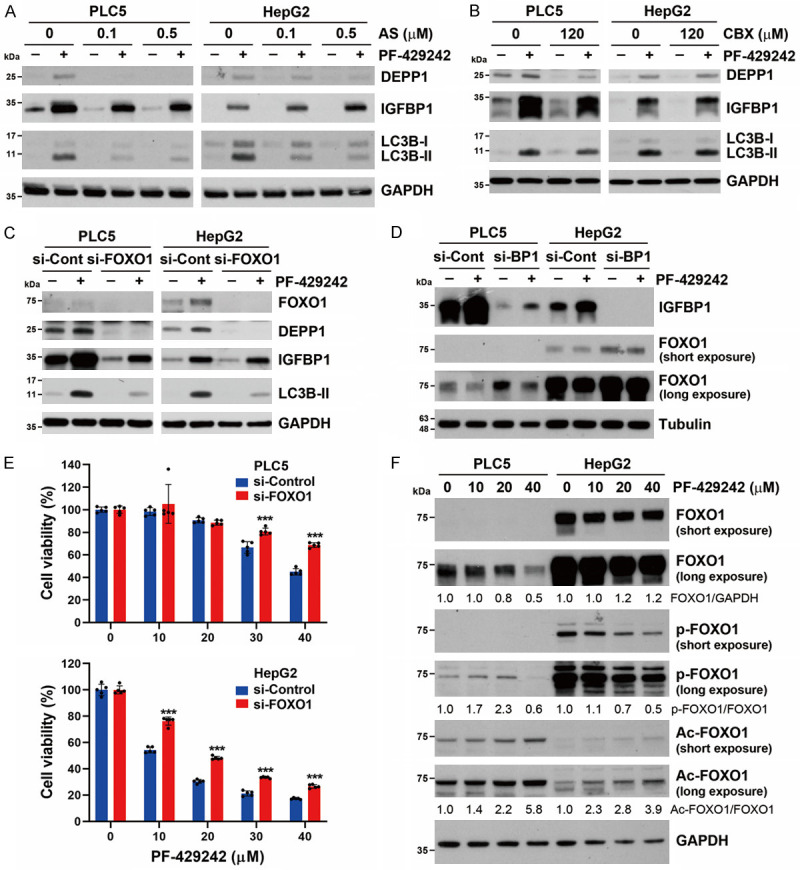
Role of FOXO1 in PF-429242-induced autophagic cell death in hepatocellular carcinoma (HCC) cells. (A) PLC5 and HepG2 cells were treated with 20 μM PF-429242 with or without indicated doses of AS1842856 (AS) for 24 h. Protein expressions were determined by Western blotting. (B) PLC5 and HepG2 cells were treated with 20 μM PF-429242 with or without 120 μM carbenoxolone (CBX) for 24 h. Protein expressions were determined by Western blotting. (C, D) PLC5 and HepG2 cells were transfected with siRNAs specific for FOXO1 in (C) or IGFBP1 (BP1) in (D) for 24 h, and then treated with 20 μM PF-429242 for 24 h. Protein expressions were determined by Western blotting. (E) si-FOXO1-transfected PLC5 and HepG2 cells were seeded in a 96-well plate and then treated with the indicated doses of PF-429242 for 72 h. Cell viability was determined by Alamar Blue staining. Error bars are the mean ± SD (n = 5). *** indicates a significant difference (P < 0.01) between si-FOXO1- and si-Control-transfected cells. (F) PLC5 and HepG2 cells were treated with 10, 20, and 40 μM PF-429242 for 24 h. Protein expressions were determined by Western blotting. Ratios between indicated protein expressions are shown.
FOXO1 was shown to promote autophagy in either a transcription-dependent or -independent manner [28-31]. Because PF-429242 did not significantly induce autophagy gene expression (Figures 1D, 4A), we hypothesized that PF-429242 might induce FOXO1-dependent autophagy in a transcription-independent manner. Post-translational modifications of FOXO1 determine its intracellular localization. Phosphorylation at Ser256 suppresses the transactivation of FOXO1 and promotes its nuclear exclusion [56]. Acetylation of the cytosolic FOXO1 at Lys262, Lys265, and Lys274 stimulates autophagy by directly binding to ATG7 [31]. Thus, the phosphorylation and acetylation of FOXO1 at these residues were examined. As shown in Figure 5F, the effect of PF-429242 on FOXO1 expression was distinct in PLC5 and HepG2 cells. In PLC5 cells, PF-429242 dose-dependently downregulated FOXO1 protein expression. However, the phospho-Ser256 and acetyl-Lys262/265/274 FOXO1 protein levels increased. The endogenous FOXO1 protein level was much higher in PLC5 cells, and it was not downregulated by PF-429242 treatment. However, PF-429242 reduced phospho-Ser256 FOXO1 protein expression. Consistent with that in HepG2 cells, PF-429242 also induced the acetylation of FOXO1. Therefore, PF-429242 may promote autophagy by cytosolic FOXO1 acetylation.
PF-429242 exhibits in vivo anticancer activity in a tumor xenograft mice model
To validate the in vivo anticancer activity of PF-429242, a subcutaneous HepG2 tumor xenograft mouse model was performed. As shown in Figure 6A and 6B, administration of PF-429242 (at a dose of 20 mg/kg) reduced the growth rate of HepG2 tumors in mice without causing significant changes in their body weight. Subsequent IHC staining indicated that PF-429242 reduced the expression of the proliferation marker Ki67 and increased the expression of both IGFBP1 and LC3B in tumors (Figure 6C). These observations supported that PF-429242 exhibited in vivo anticancer activity against HCC cells by inducing autophagic cell death and IGFBP1-dependent anti-survival signaling.
Figure 6.
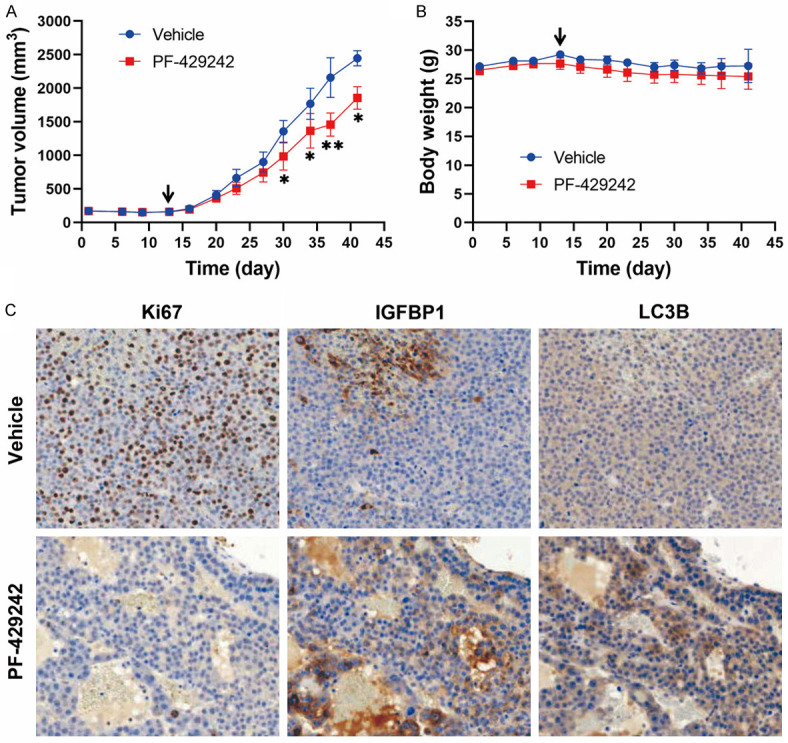
In vivo anticancer activity of PF-429242 in a HepG2 tumor xenograft model. HepG2 xenograft mice received PF-429242 (20 mg/kg) or the vehicle solvent control. Tumor volumes (A) and mice body weights (B) were measured twice weekly. Error bars are the mean ± SEM (n = 5). * and ** indicate a significant difference (P < 0.05 and P < 0.01, respectively) between the vehicle and PF-429242 group at each time point. The arrow indicates the treatment start day. In (C), tumor sections were subjected to IHC staining for the expression of Ki67, IGFBP1, and LC3B. Magnification, 200×.
Discussion
Although the in vitro anticancer activity of PF-429242 was recently demonstrated in glioblastomas, renal cell carcinoma, and pancreatic cancer cells, its in vivo anticancer activity was only validated in renal cell carcinoma [40-42]. Additionally, the anticancer activity of PF-429242 against HCC has not previously been determined. In this study, we validated the in vitro and in vivo anticancer activities of PF-429242 against HCC using a tumor xenograft mouse model. Mechanistically, PF-429242 simultaneously induced FOXO1-dependent autophagic cell death and FOXO1-independent IGFBP1-mediated anti-survival signaling.
Previous studies found that inhibition of MBTPS1/2 by chemical inhibitors (including PF-429242) and siRNAs induce autophagy [48,49]. However, the underlying molecular mechanism has yet to be elucidated. Our results indicated that the knockdown of MBTPS1/2 also induced autophagy in HCC cells, which cannot fully contribute to the effect of PF-429242. Alternatively, PF-429242 induced autophagic cell death in a FOXO1-dependent manner, providing a novel mechanism for PF-429242-induced autophagy.
FOXO1 can regulate autophagy by either promoting the transcription of autophagy regulatory genes or by binding to ATG7 to stimulate autophagy when FOXO1 is acetylated [28-31]. We found that PF-429242 did not significantly induce expression of autophagy-related genes (MAP1LC3B and SQSTM1) or other FOXO1 target genes (except DEPP1). Additionally, PF-429242 increased the level of the acetylated FOXO1 protein. Therefore, we propose that PF-429242 may induce autophagy through the cytosolic FOXO1/ATG7 pathway.
We found that inhibition of autophagy by spautin-1 did not fully rescue PF-429242-induced cytotoxicity, suggesting the presence of autophagy-independent cell death. Additionally, IGFBP1-knockdown rescued PF-429242-induced cell death, but not autophagy. These findings suggest that PF-429242 induces IGFBP1, leading to autophagy-independent cell death. Furthermore, FOXO1-knockdown did not rescue the upregulation of IGFBP1 induced by PF-429242, indicating that FOXO1-dependent autophagic cell death and IGFBP1-dependent anti-survival signaling are two parallel pathways of cell death induced by PF-429242 in HCC cells.
PF-429242 induced IGFBP1 expression to promote cell death in HCC cells, which may have resulted from the effect of IGFBP1 to limit IGF proliferative and survival signaling. However, controversial results exist among cancer types, including HCC [33,57]. IGFBP1 levels in serum and tumor tissues are reduced in HCC, which was associated with poor prognoses of patients [58,59]. The addition of recombinant IGFBP1 inhibited cancer cell invasion and reversed the IGFs’ anti-apoptotic effects in HCC cell lines [58,60]. In addition, IGFBP1 is also a mediator of the anticancer effects of ursolic acid and cannabidiol in HCC cells [61,62]. On the other hand, elevated IGFBP levels in HCC patients’ serum and tumor tissues were also reported by other studies [63,64]. The exact role of IGFBP1 in HCC warrants further investigation.
In conclusion, this study demonstrated the anticancer efficacy of PF-429242 against HCC. We also provide insights into the underlying mechanism of MBTPS1/SREBP inhibition for PF-429242-induced autophagy and cell death in HCC cells. These findings enhance our understanding of PF-429242’s mechanism of action, expanding its potential applications beyond its original target, the MBTPS1/SREBP signaling pathway. The achievements presented here have the potential to advance future application of PF-429242 in cancer therapy.
Acknowledgements
This work was supported by the National Science and Technology Council [grant numbers 109-2314-B-038-040, 110-2314-B-195-022, 111-2314-B-195-021, 112-2314-B-195-017, and 111-2314-B-038-105-MY3]; the Mackay Memorial Hospital [grant numbers MMH111-15 and MMH112-13]; the Ministry of Education [grant number DP2-111-21121-01-C-01-05]; and the Health and Welfare Surcharge of Tobacco Products from the Ministry of Health and Welfare [grant number MOHW111-TDU-B-221-014013]. The authors also express their gratitude for the financial support provided by the “TMU Research Center of Cancer Translational Medicine” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan. The authors would like to thank the Office of Research and Development at Taipei Medical University for the help in English editing.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 4.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PubMed] [Google Scholar]
- 6.Roberts LR, Gores GJ. Hepatocellular carcinoma: molecular pathways and new therapeutic targets. Semin Liver Dis. 2005;25:212–225. doi: 10.1055/s-2005-871200. [DOI] [PubMed] [Google Scholar]
- 7.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Haussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J SHARP Investigators Study Group. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 8.Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, Gerolami R, Masi G, Ross PJ, Song T, Bronowicki JP, Ollivier-Hourmand I, Kudo M, Cheng AL, Llovet JM, Finn RS, LeBerre MA, Baumhauer A, Meinhardt G, Han G RESORCE Investigators. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 9.Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, Baron A, Park JW, Han G, Jassem J, Blanc JF, Vogel A, Komov D, Evans TRJ, Lopez C, Dutcus C, Guo M, Saito K, Kraljevic S, Tamai T, Ren M, Cheng AL. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391:1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 10.Abou-Alfa GK, Meyer T, Cheng AL, El-Khoueiry AB, Rimassa L, Ryoo BY, Cicin I, Merle P, Chen Y, Park JW, Blanc JF, Bolondi L, Klumpen HJ, Chan SL, Zagonel V, Pressiani T, Ryu MH, Venook AP, Hessel C, Borgman-Hagey AE, Schwab G, Kelley RK. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N Engl J Med. 2018;379:54–63. doi: 10.1056/NEJMoa1717002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z, Liu X, Liang J, Liu Y, Hou X, Zhang M, Li Y, Jiang X. Immunotherapy for hepatocellular carcinoma: current status and future prospects. Front Immunol. 2021;12:765101. doi: 10.3389/fimmu.2021.765101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, Pikarsky E, Zhu AX, Finn RS. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19:151–172. doi: 10.1038/s41571-021-00573-2. [DOI] [PubMed] [Google Scholar]
- 13.Kirkin V, Rogov VV. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol Cell. 2019;76:268–285. doi: 10.1016/j.molcel.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Anding AL, Baehrecke EH. Cleaning house: selective autophagy of organelles. Dev Cell. 2017;41:10–22. doi: 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, Annicchiarico-Petruzzelli M, Antonov AV, Arama E, Baehrecke EH, Barlev NA, Bazan NG, Bernassola F, Bertrand MJM, Bianchi K, Blagosklonny MV, Blomgren K, Borner C, Boya P, Brenner C, Campanella M, Candi E, Carmona-Gutierrez D, Cecconi F, Chan FK, Chandel NS, Cheng EH, Chipuk JE, Cidlowski JA, Ciechanover A, Cohen GM, Conrad M, Cubillos-Ruiz JR, Czabotar PE, D’Angiolella V, Dawson TM, Dawson VL, De Laurenzi V, De Maria R, Debatin KM, DeBerardinis RJ, Deshmukh M, Di Daniele N, Di Virgilio F, Dixit VM, Dixon SJ, Duckett CS, Dynlacht BD, El-Deiry WS, Elrod JW, Fimia GM, Fulda S, Garcia-Saez AJ, Garg AD, Garrido C, Gavathiotis E, Golstein P, Gottlieb E, Green DR, Greene LA, Gronemeyer H, Gross A, Hajnoczky G, Hardwick JM, Harris IS, Hengartner MO, Hetz C, Ichijo H, Jaattela M, Joseph B, Jost PJ, Juin PP, Kaiser WJ, Karin M, Kaufmann T, Kepp O, Kimchi A, Kitsis RN, Klionsky DJ, Knight RA, Kumar S, Lee SW, Lemasters JJ, Levine B, Linkermann A, Lipton SA, Lockshin RA, Lopez-Otin C, Lowe SW, Luedde T, Lugli E, MacFarlane M, Madeo F, Malewicz M, Malorni W, Manic G, Marine JC, Martin SJ, Martinou JC, Medema JP, Mehlen P, Meier P, Melino S, Miao EA, Molkentin JD, Moll UM, Munoz-Pinedo C, Nagata S, Nunez G, Oberst A, Oren M, Overholtzer M, Pagano M, Panaretakis T, Pasparakis M, Penninger JM, Pereira DM, Pervaiz S, Peter ME, Piacentini M, Pinton P, Prehn JHM, Puthalakath H, Rabinovich GA, Rehm M, Rizzuto R, Rodrigues CMP, Rubinsztein DC, Rudel T, Ryan KM, Sayan E, Scorrano L, Shao F, Shi Y, Silke J, Simon HU, Sistigu A, Stockwell BR, Strasser A, Szabadkai G, Tait SWG, Tang D, Tavernarakis N, Thorburn A, Tsujimoto Y, Turk B, Vanden Berghe T, Vandenabeele P, Vander Heiden MG, Villunger A, Virgin HW, Vousden KH, Vucic D, Wagner EF, Walczak H, Wallach D, Wang Y, Wells JA, Wood W, Yuan J, Zakeri Z, Zhivotovsky B, Zitvogel L, Melino G, Kroemer G. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang D, Kang R, Berghe TV, Vandenabeele P, Kroemer G. The molecular machinery of regulated cell death. Cell Res. 2019;29:347–364. doi: 10.1038/s41422-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doherty J, Baehrecke EH. Life, death and autophagy. Nat Cell Biol. 2018;20:1110–1117. doi: 10.1038/s41556-018-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denton D, Kumar S. Autophagy-dependent cell death. Cell Death Differ. 2019;26:605–616. doi: 10.1038/s41418-018-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganzleben I, Neurath MF, Becker C. Autophagy in cancer therapy-molecular mechanisms and current clinical advances. Cancers (Basel) 2021;13:5575. doi: 10.3390/cancers13215575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020;19:12. doi: 10.1186/s12943-020-1138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulcahy Levy JM, Thorburn A. Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 2020;27:843–857. doi: 10.1038/s41418-019-0474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng X, Li J, Guo D. SCAP/SREBPs are central players in lipid metabolism and novel metabolic targets in cancer therapy. Curr Top Med Chem. 2018;18:484–493. doi: 10.2174/1568026618666180523104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimano H, Sato R. SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat Rev Endocrinol. 2017;13:710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- 24.Cheng Z. The FoxO-autophagy axis in health and disease. Trends Endocrinol Metab. 2019;30:658–671. doi: 10.1016/j.tem.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez AM, Csibi A, Raibon A, Cornille K, Gay S, Bernardi H, Candau R. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J Cell Biochem. 2012;113:695–710. doi: 10.1002/jcb.23399. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Liu HY, Han J, Cao SY, Hong T, Zhuo D, Shi J, Liu Z, Cao W. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287:39107–39114. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu P, Das M, Reilly J, Davis RJ. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 2011;25:310–322. doi: 10.1101/gad.1984311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, Zhu WG. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 32.Furstenberger G, Senn HJ. Insulin-like growth factors and cancer. Lancet Oncol. 2002;3:298–302. doi: 10.1016/s1470-2045(02)00731-3. [DOI] [PubMed] [Google Scholar]
- 33.Lin YW, Weng XF, Huang BL, Guo HP, Xu YW, Peng YH. IGFBP-1 in cancer: expression, molecular mechanisms, and potential clinical implications. Am J Transl Res. 2021;13:813–832. [PMC free article] [PubMed] [Google Scholar]
- 34.Hawkins JL, Robbins MD, Warren LC, Xia D, Petras SF, Valentine JJ, Varghese AH, Wang IK, Subashi TA, Shelly LD, Hay BA, Landschulz KT, Geoghegan KF, Harwood HJ Jr. Pharmacologic inhibition of site 1 protease activity inhibits sterol regulatory element-binding protein processing and reduces lipogenic enzyme gene expression and lipid synthesis in cultured cells and experimental animals. J Pharmacol Exp Ther. 2008;326:801–808. doi: 10.1124/jpet.108.139626. [DOI] [PubMed] [Google Scholar]
- 35.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 38.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toth JI, Datta S, Athanikar JN, Freedman LP, Osborne TF. Selective coactivator interactions in gene activation by SREBP-1a and -1c. Mol Cell Biol. 2004;24:8288–8300. doi: 10.1128/MCB.24.18.8288-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caruana BT, Skoric A, Brown AJ, Lutze-Mann LH. Site-1 protease, a novel metabolic target for glioblastoma. Biochem Biophys Res Commun. 2017;490:760–766. doi: 10.1016/j.bbrc.2017.06.114. [DOI] [PubMed] [Google Scholar]
- 41.Siqingaowa, Sekar S, Gopalakrishnan V, Taghibiglou C. Sterol regulatory element-binding protein 1 inhibitors decrease pancreatic cancer cell viability and proliferation. Biochem Biophys Res Commun. 2017;488:136–140. doi: 10.1016/j.bbrc.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 42.Wang TB, Geng M, Jin H, Tang AG, Sun H, Zhou LZ, Chen BH, Shen G, Sun Q. SREBP1 site 1 protease inhibitor PF-429242 suppresses renal cell carcinoma cell growth. Cell Death Dis. 2021;12:717. doi: 10.1038/s41419-021-03999-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang PM, Hong YH, Hsu KC, Liu TP. p38alpha/S1P/SREBP2 activation by the SAM-competitive EZH2 inhibitor GSK343 limits its anticancer activity but creates a druggable vulnerability in hepatocellular carcinoma. Am J Cancer Res. 2019;9:2120–2139. [PMC free article] [PubMed] [Google Scholar]
- 44.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi R, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NY, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai XY, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ET, Banhegyi G, Bartholomew CR, Bassham DC, Bast RC Jr, Batoko H, Bay BH, Beau I, Bechet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin JP, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, Brunetti-Pierri N, Brunk UT, Bulman DE, Bultman SJ, Bultynck G, Burbulla LF, Bursch W, Butchar JP, Buzgariu W, Bydlowski SP, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calabretta B, Calvo-Garrido J, Camougrand N, Campanella M, Campos-Salinas J, Candi E, Cao L, Caplan AB, Carding SR, Cardoso SM, Carew JS, Carlin CR, Carmignac V, Carneiro LA, Carra S, Caruso RA, Casari G, Casas C, Castino R, Cebollero E, Cecconi F, Celli J, Chaachouay H, Chae HJ, Chai CY, Chan DC, Chan EY, Chang RC, Che CM, Chen CC, Chen GC, Chen GQ, Chen M, Chen Q, Chen SS, Chen W, Chen X, Chen X, Chen X, Chen YG, Chen Y, Chen Y, Chen YJ, Chen Z, Cheng A, Cheng CH, Cheng Y, Cheong H, Cheong JH, Cherry S, Chess-Williams R, Cheung ZH, Chevet E, Chiang HL, Chiarelli R, Chiba T, Chin LS, Chiou SH, Chisari FV, Cho CH, Cho DH, Choi AM, Choi D, Choi KS, Choi ME, Chouaib S, Choubey D, Choubey V, Chu CT, Chuang TH, Chueh SH, Chun T, Chwae YJ, Chye ML, Ciarcia R, Ciriolo MR, Clague MJ, Clark RS, Clarke PG, Clarke R, Codogno P, Coller HA, Colombo MI, Comincini S, Condello M, Condorelli F, Cookson MR, Coombs GH, Coppens I, Corbalan R, Cossart P, Costelli P, Costes S, Coto-Montes A, Couve E, Coxon FP, Cregg JM, Crespo JL, Cronje MJ, Cuervo AM, Cullen JJ, Czaja MJ, D’Amelio M, Darfeuille-Michaud A, Davids LM, Davies FE, De Felici M, de Groot JF, de Haan CA, De Martino L, De Milito A, De Tata V, Debnath J, Degterev A, Dehay B, Delbridge LM, Demarchi F, Deng YZ, Dengjel J, Dent P, Denton D, Deretic V, Desai SD, Devenish RJ, Di Gioacchino M, Di Paolo G, Di Pietro C, Diaz-Araya G, Diaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dikic I, Dinesh-Kumar SP, Ding WX, Distelhorst CW, Diwan A, Djavaheri-Mergny M, Dokudovskaya S, Dong Z, Dorsey FC, Dosenko V, Dowling JJ, Doxsey S, Dreux M, Drew ME, Duan Q, Duchosal MA, Duff K, Dugail I, Durbeej M, Duszenko M, Edelstein CL, Edinger AL, Egea G, Eichinger L, Eissa NT, Ekmekcioglu S, El-Deiry WS, Elazar Z, Elgendy M, Ellerby LM, Eng KE, Engelbrecht AM, Engelender S, Erenpreisa J, Escalante R, Esclatine A, Eskelinen EL, Espert L, Espina V, Fan H, Fan J, Fan QW, Fan Z, Fang S, Fang Y, Fanto M, Fanzani A, Farkas T, Farre JC, Faure M, Fechheimer M, Feng CG, Feng J, Feng Q, Feng Y, Fesus L, Feuer R, Figueiredo-Pereira ME, Fimia GM, Fingar DC, Finkbeiner S, Finkel T, Finley KD, Fiorito F, Fisher EA, Fisher PB, Flajolet M, Florez-McClure ML, Florio S, Fon EA, Fornai F, Fortunato F, Fotedar R, Fowler DH, Fox HS, Franco R, Frankel LB, Fransen M, Fuentes JM, Fueyo J, Fujii J, Fujisaki K, Fujita E, Fukuda M, Furukawa RH, Gaestel M, Gailly P, Gajewska M, Galliot B, Galy V, Ganesh S, Ganetzky B, Ganley IG, Gao FB, Gao GF, Gao J, Garcia L, Garcia-Manero G, Garcia-Marcos M, Garmyn M, Gartel AL, Gatti E, Gautel M, Gawriluk TR, Gegg ME, Geng J, Germain M, Gestwicki JE, Gewirtz DA, Ghavami S, Ghosh P, Giammarioli AM, Giatromanolaki AN, Gibson SB, Gilkerson RW, Ginger ML, Ginsberg HN, Golab J, Goligorsky MS, Golstein P, Gomez-Manzano C, Goncu E, Gongora C, Gonzalez CD, Gonzalez R, Gonzalez-Estevez C, Gonzalez-Polo RA, Gonzalez-Rey E, Gorbunov NV, Gorski S, Goruppi S, Gottlieb RA, Gozuacik D, Granato GE, Grant GD, Green KN, Gregorc A, Gros F, Grose C, Grunt TW, Gual P, Guan JL, Guan KL, Guichard SM, Gukovskaya AS, Gukovsky I, Gunst J, Gustafsson AB, Halayko AJ, Hale AN, Halonen SK, Hamasaki M, Han F, Han T, Hancock MK, Hansen M, Harada H, Harada M, Hardt SE, Harper JW, Harris AL, Harris J, Harris SD, Hashimoto M, Haspel JA, Hayashi S, Hazelhurst LA, He C, He YW, Hebert MJ, Heidenreich KA, Helfrich MH, Helgason GV, Henske EP, Herman B, Herman PK, Hetz C, Hilfiker S, Hill JA, Hocking LJ, Hofman P, Hofmann TG, Hohfeld J, Holyoake TL, Hong MH, Hood DA, Hotamisligil GS, Houwerzijl EJ, Hoyer-Hansen M, Hu B, Hu CA, Hu HM, Hua Y, Huang C, Huang J, Huang S, Huang WP, Huber TB, Huh WK, Hung TH, Hupp TR, Hur GM, Hurley JB, Hussain SN, Hussey PJ, Hwang JJ, Hwang S, Ichihara A, Ilkhanizadeh S, Inoki K, Into T, Iovane V, Iovanna JL, Ip NY, Isaka Y, Ishida H, Isidoro C, Isobe K, Iwasaki A, Izquierdo M, Izumi Y, Jaakkola PM, Jaattela M, Jackson GR, Jackson WT, Janji B, Jendrach M, Jeon JH, Jeung EB, Jiang H, Jiang H, Jiang JX, Jiang M, Jiang Q, Jiang X, Jiang X, Jimenez A, Jin M, Jin S, Joe CO, Johansen T, Johnson DE, Johnson GV, Jones NL, Joseph B, Joseph SK, Joubert AM, Juhasz G, Juillerat-Jeanneret L, Jung CH, Jung YK, Kaarniranta K, Kaasik A, Kabuta T, Kadowaki M, Kagedal K, Kamada Y, Kaminskyy VO, Kampinga HH, Kanamori H, Kang C, Kang KB, Kang KI, Kang R, Kang YA, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kanthasamy A, Karantza V, Kaushal GP, Kaushik S, Kawazoe Y, Ke PY, Kehrl JH, Kelekar A, Kerkhoff C, Kessel DH, Khalil H, Kiel JA, Kiger AA, Kihara A, Kim DR, Kim DH, Kim DH, Kim EK, Kim HR, Kim JS, Kim JH, Kim JC, Kim JK, Kim PK, Kim SW, Kim YS, Kim Y, Kimchi A, Kimmelman AC, King JS, Kinsella TJ, Kirkin V, Kirshenbaum LA, Kitamoto K, Kitazato K, Klein L, Klimecki WT, Klucken J, Knecht E, Ko BC, Koch JC, Koga H, Koh JY, Koh YH, Koike M, Komatsu M, Kominami E, Kong HJ, Kong WJ, Korolchuk VI, Kotake Y, Koukourakis MI, Kouri Flores JB, Kovacs AL, Kraft C, Krainc D, Kramer H, Kretz-Remy C, Krichevsky AM, Kroemer G, Kruger R, Krut O, Ktistakis NT, Kuan CY, Kucharczyk R, Kumar A, Kumar R, Kumar S, Kundu M, Kung HJ, Kurz T, Kwon HJ, La Spada AR, Lafont F, Lamark T, Landry J, Lane JD, Lapaquette P, Laporte JF, Laszlo L, Lavandero S, Lavoie JN, Layfield R, Lazo PA, Le W, Le Cam L, Ledbetter DJ, Lee AJ, Lee BW, Lee GM, Lee J, Lee JH, Lee M, Lee MS, Lee SH, Leeuwenburgh C, Legembre P, Legouis R, Lehmann M, Lei HY, Lei QY, Leib DA, Leiro J, Lemasters JJ, Lemoine A, Lesniak MS, Lev D, Levenson VV, Levine B, Levy E, Li F, Li JL, Li L, Li S, Li W, Li XJ, Li YB, Li YP, Liang C, Liang Q, Liao YF, Liberski PP, Lieberman A, Lim HJ, Lim KL, Lim K, Lin CF, Lin FC, Lin J, Lin JD, Lin K, Lin WW, Lin WC, Lin YL, Linden R, Lingor P, Lippincott-Schwartz J, Lisanti MP, Liton PB, Liu B, Liu CF, Liu K, Liu L, Liu QA, Liu W, Liu YC, Liu Y, Lockshin RA, Lok CN, Lonial S, Loos B, Lopez-Berestein G, Lopez-Otin C, Lossi L, Lotze MT, Low P, Lu B, Lu B, Lu B, Lu Z, Luciano F, Lukacs NW, Lund AH, Lynch-Day MA, Ma Y, Macian F, MacKeigan JP, Macleod KF, Madeo F, Maiuri L, Maiuri MC, Malagoli D, Malicdan MC, Malorni W, Man N, Mandelkow EM, Manon S, Manov I, Mao K, Mao X, Mao Z, Marambaud P, Marazziti D, Marcel YL, Marchbank K, Marchetti P, Marciniak SJ, Marcondes M, Mardi M, Marfe G, Marino G, Markaki M, Marten MR, Martin SJ, Martinand-Mari C, Martinet W, Martinez-Vicente M, Masini M, Matarrese P, Matsuo S, Matteoni R, Mayer A, Mazure NM, McConkey DJ, McConnell MJ, McDermott C, McDonald C, McInerney GM, McKenna SL, McLaughlin B, McLean PJ, McMaster CR, McQuibban GA, Meijer AJ, Meisler MH, Melendez A, Melia TJ, Melino G, Mena MA, Menendez JA, Menna-Barreto RF, Menon MB, Menzies FM, Mercer CA, Merighi A, Merry DE, Meschini S, Meyer CG, Meyer TF, Miao CY, Miao JY, Michels PA, Michiels C, Mijaljica D, Milojkovic A, Minucci S, Miracco C, Miranti CK, Mitroulis I, Miyazawa K, Mizushima N, Mograbi B, Mohseni S, Molero X, Mollereau B, Mollinedo F, Momoi T, Monastyrska I, Monick MM, Monteiro MJ, Moore MN, Mora R, Moreau K, Moreira PI, Moriyasu Y, Moscat J, Mostowy S, Mottram JC, Motyl T, Moussa CE, Muller S, Muller S, Munger K, Munz C, Murphy LO, Murphy ME, Musaro A, Mysorekar I, Nagata E, Nagata K, Nahimana A, Nair U, Nakagawa T, Nakahira K, Nakano H, Nakatogawa H, Nanjundan M, Naqvi NI, Narendra DP, Narita M, Navarro M, Nawrocki ST, Nazarko TY, Nemchenko A, Netea MG, Neufeld TP, Ney PA, Nezis IP, Nguyen HP, Nie D, Nishino I, Nislow C, Nixon RA, Noda T, Noegel AA, Nogalska A, Noguchi S, Notterpek L, Novak I, Nozaki T, Nukina N, Nurnberger T, Nyfeler B, Obara K, Oberley TD, Oddo S, Ogawa M, Ohashi T, Okamoto K, Oleinick NL, Oliver FJ, Olsen LJ, Olsson S, Opota O, Osborne TF, Ostrander GK, Otsu K, Ou JH, Ouimet M, Overholtzer M, Ozpolat B, Paganetti P, Pagnini U, Pallet N, Palmer GE, Palumbo C, Pan T, Panaretakis T, Pandey UB, Papackova Z, Papassideri I, Paris I, Park J, Park OK, Parys JB, Parzych KR, Patschan S, Patterson C, Pattingre S, Pawelek JM, Peng J, Perlmutter DH, Perrotta I, Perry G, Pervaiz S, Peter M, Peters GJ, Petersen M, Petrovski G, Phang JM, Piacentini M, Pierre P, Pierrefite-Carle V, Pierron G, Pinkas-Kramarski R, Piras A, Piri N, Platanias LC, Poggeler S, Poirot M, Poletti A, Pous C, Pozuelo-Rubio M, Praetorius-Ibba M, Prasad A, Prescott M, Priault M, Produit-Zengaffinen N, Progulske-Fox A, Proikas-Cezanne T, Przedborski S, Przyklenk K, Puertollano R, Puyal J, Qian SB, Qin L, Qin ZH, Quaggin SE, Raben N, Rabinowich H, Rabkin SW, Rahman I, Rami A, Ramm G, Randall G, Randow F, Rao VA, Rathmell JC, Ravikumar B, Ray SK, Reed BH, Reed JC, Reggiori F, Regnier-Vigouroux A, Reichert AS, Reiners JJ Jr, Reiter RJ, Ren J, Revuelta JL, Rhodes CJ, Ritis K, Rizzo E, Robbins J, Roberge M, Roca H, Roccheri MC, Rocchi S, Rodemann HP, Rodriguez de Cordoba S, Rohrer B, Roninson IB, Rosen K, Rost-Roszkowska MM, Rouis M, Rouschop KM, Rovetta F, Rubin BP, Rubinsztein DC, Ruckdeschel K, Rucker EB 3rd, Rudich A, Rudolf E, Ruiz-Opazo N, Russo R, Rusten TE, Ryan KM, Ryter SW, Sabatini DM, Sadoshima J, Saha T, Saitoh T, Sakagami H, Sakai Y, Salekdeh GH, Salomoni P, Salvaterra PM, Salvesen G, Salvioli R, Sanchez AM, Sanchez-Alcazar JA, Sanchez-Prieto R, Sandri M, Sankar U, Sansanwal P, Santambrogio L, Saran S, Sarkar S, Sarwal M, Sasakawa C, Sasnauskiene A, Sass M, Sato K, Sato M, Schapira AH, Scharl M, Schatzl HM, Scheper W, Schiaffino S, Schneider C, Schneider ME, Schneider-Stock R, Schoenlein PV, Schorderet DF, Schuller C, Schwartz GK, Scorrano L, Sealy L, Seglen PO, Segura-Aguilar J, Seiliez I, Seleverstov O, Sell C, Seo JB, Separovic D, Setaluri V, Setoguchi T, Settembre C, Shacka JJ, Shanmugam M, Shapiro IM, Shaulian E, Shaw RJ, Shelhamer JH, Shen HM, Shen WC, Sheng ZH, Shi Y, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shintani T, Shirihai OS, Shore GC, Sibirny AA, Sidhu SB, Sikorska B, Silva-Zacarin EC, Simmons A, Simon AK, Simon HU, Simone C, Simonsen A, Sinclair DA, Singh R, Sinha D, Sinicrope FA, Sirko A, Siu PM, Sivridis E, Skop V, Skulachev VP, Slack RS, Smaili SS, Smith DR, Soengas MS, Soldati T, Song X, Sood AK, Soong TW, Sotgia F, Spector SA, Spies CD, Springer W, Srinivasula SM, Stefanis L, Steffan JS, Stendel R, Stenmark H, Stephanou A, Stern ST, Sternberg C, Stork B, Stralfors P, Subauste CS, Sui X, Sulzer D, Sun J, Sun SY, Sun ZJ, Sung JJ, Suzuki K, Suzuki T, Swanson MS, Swanton C, Sweeney ST, Sy LK, Szabadkai G, Tabas I, Taegtmeyer H, Tafani M, Takacs-Vellai K, Takano Y, Takegawa K, Takemura G, Takeshita F, Talbot NJ, Tan KS, Tanaka K, Tanaka K, Tang D, Tang D, Tanida I, Tannous BA, Tavernarakis N, Taylor GS, Taylor GA, Taylor JP, Terada LS, Terman A, Tettamanti G, Thevissen K, Thompson CB, Thorburn A, Thumm M, Tian F, Tian Y, Tocchini-Valentini G, Tolkovsky AM, Tomino Y, Tonges L, Tooze SA, Tournier C, Tower J, Towns R, Trajkovic V, Travassos LH, Tsai TF, Tschan MP, Tsubata T, Tsung A, Turk B, Turner LS, Tyagi SC, Uchiyama Y, Ueno T, Umekawa M, Umemiya-Shirafuji R, Unni VK, Vaccaro MI, Valente EM, Van den Berghe G, van der Klei IJ, van Doorn W, van Dyk LF, van Egmond M, van Grunsven LA, Vandenabeele P, Vandenberghe WP, Vanhorebeek I, Vaquero EC, Velasco G, Vellai T, Vicencio JM, Vierstra RD, Vila M, Vindis C, Viola G, Viscomi MT, Voitsekhovskaja OV, von Haefen C, Votruba M, Wada K, Wade-Martins R, Walker CL, Walsh CM, Walter J, Wan XB, Wang A, Wang C, Wang D, Wang F, Wang F, Wang G, Wang H, Wang HG, Wang HD, Wang J, Wang K, Wang M, Wang RC, Wang X, Wang X, Wang YJ, Wang Y, Wang Z, Wang ZC, Wang Z, Wansink DG, Ward DM, Watada H, Waters SL, Webster P, Wei L, Weihl CC, Weiss WA, Welford SM, Wen LP, Whitehouse CA, Whitton JL, Whitworth AJ, Wileman T, Wiley JW, Wilkinson S, Willbold D, Williams RL, Williamson PR, Wouters BG, Wu C, Wu DC, Wu WK, Wyttenbach A, Xavier RJ, Xi Z, Xia P, Xiao G, Xie Z, Xie Z, Xu DZ, Xu J, Xu L, Xu X, Yamamoto A, Yamamoto A, Yamashina S, Yamashita M, Yan X, Yanagida M, Yang DS, Yang E, Yang JM, Yang SY, Yang W, Yang WY, Yang Z, Yao MC, Yao TP, Yeganeh B, Yen WL, Yin JJ, Yin XM, Yoo OJ, Yoon G, Yoon SY, Yorimitsu T, Yoshikawa Y, Yoshimori T, Yoshimoto K, You HJ, Youle RJ, Younes A, Yu L, Yu L, Yu SW, Yu WH, Yuan ZM, Yue Z, Yun CH, Yuzaki M, Zabirnyk O, Silva-Zacarin E, Zacks D, Zacksenhaus E, Zaffaroni N, Zakeri Z, Zeh HJ 3rd, Zeitlin SO, Zhang H, Zhang HL, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang MY, Zhang XD, Zhao M, Zhao YF, Zhao Y, Zhao ZJ, Zheng X, Zhivotovsky B, Zhong Q, Zhou CZ, Zhu C, Zhu WG, Zhu XF, Zhu X, Zhu Y, Zoladek T, Zong WX, Zorzano A, Zschocke J, Zuckerbraun B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, Jin M, Zhu Z, Wang H, Yu J, Li Y, Hao Y, Choi A, Ke H, Ma D, Yuan J. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ, Coppes RP, Engedal N, Mari M, Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 48.Deng X, Pan X, Cheng C, Liu B, Zhang H, Zhang Y, Xu K. Regulation of SREBP-2 intracellular trafficking improves impaired autophagic flux and alleviates endoplasmic reticulum stress in NAFLD. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:337–350. doi: 10.1016/j.bbalip.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Al-Maskari M, Care MA, Robinson E, Cocco M, Tooze RM, Doody GM. Site-1 protease function is essential for the generation of antibody secreting cells and reprogramming for secretory activity. Sci Rep. 2018;8:14338. doi: 10.1038/s41598-018-32705-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salcher S, Hermann M, Kiechl-Kohlendorfer U, Ausserlechner MJ, Obexer P. C10ORF10/DEPP-mediated ROS accumulation is a critical modulator of FOXO3-induced autophagy. Mol Cancer. 2017;16:95. doi: 10.1186/s12943-017-0661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stepp MW, Folz RJ, Yu J, Zelko IN. The c10orf10 gene product is a new link between oxidative stress and autophagy. Biochim Biophys Acta. 2014;1843:1076–1088. doi: 10.1016/j.bbamcr.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 52.Salcher S, Hagenbuchner J, Geiger K, Seiter MA, Rainer J, Kofler R, Hermann M, Kiechl-Kohlendorfer U, Ausserlechner MJ, Obexer P. C10ORF10/DEPP, a transcriptional target of FOXO3, regulates ROS-sensitivity in human neuroblastoma. Mol Cancer. 2014;13:224. doi: 10.1186/1476-4598-13-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen S, Gai J, Wang Y, Li H. FoxO regulates expression of decidual protein induced by progesterone (DEPP) in human endothelial cells. FEBS Lett. 2011;585:1796–1800. doi: 10.1016/j.febslet.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 54.Nakamura M, Takakura M, Fujii R, Maida Y, Bono Y, Mizumoto Y, Zhang X, Kiyono T, Kyo S. The PRB-dependent FOXO1/IGFBP-1 axis is essential for progestin to inhibit endometrial epithelial growth. Cancer Lett. 2013;336:68–75. doi: 10.1016/j.canlet.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 55.Tang Q, Zheng F, Wu J, Xiao Q, Li L, Hann SS. Combination of solamargine and metformin strengthens IGFBP1 gene expression through inactivation of Stat3 and reciprocal interaction between FOXO3a and SP1. Cell Physiol Biochem. 2017;43:2310–2326. doi: 10.1159/000484383. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, Gan L, Pan H, Guo S, He X, Olson ST, Mesecar A, Adam S, Unterman TG. Phosphorylation of serine 256 suppresses transactivation by FKHR (FOXO1) by multiple mechanisms. Direct and indirect effects on nuclear/cytoplasmic shuttling and DNA binding. J Biol Chem. 2002;277:45276–45284. doi: 10.1074/jbc.M208063200. [DOI] [PubMed] [Google Scholar]
- 57.Adamek A, Kasprzak A. Insulin-like growth factor (IGF) system in liver diseases. Int J Mol Sci. 2018;19:1308. doi: 10.3390/ijms19051308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai B, Ruan B, Wu J, Wang J, Shang R, Sun W, Li X, Dou K, Wang D, Li Y. Insulin-like growth factor binding protein-1 inhibits cancer cell invasion and is associated with poor prognosis in hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:5645–5654. [PMC free article] [PubMed] [Google Scholar]
- 59.Abou-Alfa GK, Capanu M, O’Reilly EM, Ma J, Chou JF, Gansukh B, Shia J, Kalin M, Katz S, Abad L, Reidy-Lagunes DL, Kelsen DP, Chen HX, Saltz LB. A phase II study of cixutumumab (IMC-A12, NSC742460) in advanced hepatocellular carcinoma. J Hepatol. 2014;60:319–324. doi: 10.1016/j.jhep.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nielsen KO, Mirza AH, Kaur S, Jacobsen KS, Winther TN, Glebe D, Pociot F, Hogh B, Storling J. Hepatitis B virus suppresses the secretion of insulin-like growth factor binding protein 1 to facilitate anti-apoptotic IGF-1 effects in HepG2 cells. Exp Cell Res. 2018;370:399–408. doi: 10.1016/j.yexcr.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 61.Shangguan F, Zhou H, Ma N, Wu S, Huang H, Jin G, Wu S, Hong W, Zhuang W, Xia H, Lan L. A novel mechanism of cannabidiol in suppressing hepatocellular carcinoma by inducing GSDME dependent pyroptosis. Front Cell Dev Biol. 2021;9:697832. doi: 10.3389/fcell.2021.697832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang LJ, Tang Q, Wu J, Chen Y, Zheng F, Dai Z, Hann SS. Inter-regulation of IGFBP1 and FOXO3a unveils novel mechanism in ursolic acid-inhibited growth of hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2016;35:59. doi: 10.1186/s13046-016-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hwang DL, Huang SP, Lan WS, Lee PD. Elevated insulin, proinsulin and insulin-like growth factor-binding protein-1 in liver disease. Growth Horm IGF Res. 2003;13:316–321. doi: 10.1016/s1096-6374(03)00042-x. [DOI] [PubMed] [Google Scholar]
- 64.Nedic O, Malenkovic V, Dukanovic B, Baricevic I. Association of elevated IGFBP-1 with increased IGF-II concentration in patients with carcinoma of the liver. Int J Biol Markers. 2008;23:225–230. doi: 10.1177/172460080802300405. [DOI] [PubMed] [Google Scholar]


