Abstract
A highly sensitive fluorogenic hexosaminidase substrate, fluorescein di(N-acetyl-β-D-glucosaminide) (FDGlcNAc), was prepared essentially as described previously [Chem. Pharm. Bull. 1993, 41, 314] with some modifications. The fluorescent analog is a substrate for a number of hexosaminidases but here we have focused on the cytoplasmic O-GlcNAcase isoforms. Kinetic analysis using purified O-GlcNAcase and its splice variant (v-O-GlcNAcase) expressed in Escherichia coli suggests that FDGlcNAc is a much more efficient substrate (Km = 84.9 μM) than the conventional substrate, para-nitrophenyl 2-acetamido-2-deoxy-β-D-glucopyranoside (pNP-β-GlcNAc, Km = 1.1 mM) and a previously developed fluorogenic substrate, 4-methylumbelliferyl 2-acetamido-2-deoxy-β-D-glucopyranoside [MUGlcNAc, Km = 0.43 mM; J. Biol. Chem. 2005, 280, 25313] for O-GlcNAcase. The variant O-GlcNAcase, a protein lacking the C-terminal third of the full-length O-GlcNAcase, exhibited a Km of 2.1 mM with respect to FDGlcNAc. This shorter isoform was not previously thought to exhibit O-GlcNAcase activity based on in vitro studies with pNP-β-GlcNAc. However, both O-GlcNAcase isoforms reduced O-GlcNAc protein levels extracted from HeLa and HT-29 cells in vitro, indicating that the splice variant is a bona fide O-GlcNAcase. Fluorescein di-N-acetyl-β-D-galactosaminide (FDGalNAc) is not cleaved by these enzymes, consistent with previous findings that the O-GlcNAcase has substrate specificity toward O-GlcNAc but not O-GalNAc. The enzymatic activity of the shorter isoform of O-GlcNAcase was first detected by using highly sensitive fluorogenic FDGlcNAc substrate. The finding that O-GlcNAcase exists as two distinct isoforms has a number of important implications for the role of O-GlcNAcase in hexosamine signaling.
Keywords: O-GlcNAc, OGT, O-GlcNAcase, v-O-GlcNAcase, FDGlcNAc, MUGlcNAc, pNP-β-GlcNAc
1. Introduction
Many nuclear and cytoplasmic proteins such as transcription factors,1–3 cytoskeletal proteins,4,5 oncogenes,6 kinases,7 phosphatases,8 and chaperones,9–11 are known to be post-translationally modified with O-linked β-N-acetylglucosamine (O-GlcNAc). The O-GlcNAc modification is ubiquitous in all higher eukaryotes that have been examined. In contrast to classical glycosylation, which occurs on membrane and secretory proteins with either N- or O-linkage in a more complex fashion, this modification is added to intracellular proteins through a β-O-linkage. The O-GlcNAc modification may act in a manner analogous to protein phosphorylation in that it is modulated by various extracellular stimuli and growth conditions12 and the turnover rate of the glycan is much faster than that of the polypeptide backbone. This implies that O-GlcNAc is dynamically added to and removed from proteins.13–15 Analogous to kinases and phosphatases, two enzymes are involved in the regulated cycling of O-GlcNAc. Sites modified by O-GlcNAc are often the same or adjacent to those of phosphorylation suggesting a competitive role for O-GlcNAc and phosphorylation in regulating protein function.16–18 Recent evidence suggests that deregulation of cellular O-GlcNAc levels might play a role in type II diabetes,19 cancer,20 and neurological disorders.21 For example, elevation of O-GlcNAc levels by altering one of the enzymes of O-GlcNAc metabolism can give rise to insulin resistance in transgenic mice19 and in tissue culture.22
The enzymes responsible for the cycle of O-GlcNAc addition and removal are the O-GlcNAc transferase (OGT, EC 2.4.1.94) and the O-GlcNAc specific β-N-acetylglucosaminidase known as O-GlcNAcase (EC 3.2.1.52). Both enzymes have been purified, cloned, and characterized.7,23–29 Recently, the crystal structure of the multiple tetratricopeptide repeats (TPR) domain of OGT has been solved and the TPR domain was shown to play a role in dimerization of OGT itself.30 Compared to OGT, very little is known about the O-GlcNAcase or its splice variant. Unlike acidic hexosaminidases in lysosomes, O-GlcNAcase has a neutral pH optimum, nucleocytoplasmic distribution, and substrate specificity toward GlcNAc and not N-acetylgalactosamine (GalNAc).29 Because of these distinct characteristics, O-GlcNAcase is most likely the same enzyme as hexosaminidase C.31,32 The primary sequence of O-GlcNAcase was originally identified as meningioma expressed antigen 5 (MGEA 5), an autoantigen in meningiomas.33 Interestingly, the O-GlcNAcase gene maps to chromosome 10q24 and this locus has been implicated in Alzheimer’s disease.33 Full-length O-GlcNAcase is a 916-amino acid protein having an acidic isoelectric point with a predicted molecular weight of 103 kDa.27,33 The N-terminal and the C-terminal region of O-GlcNAcase has homology to hyaluronidases33,34 and acetyltransferases,35 respectively. The C-terminal of O-GlcNAcase has recently been shown to have histone acetyltransferase (HAT) activity.36 A splice variant of O-GlcNAcase designated as MGEA 5s was also identified.33 This isoform is a 677-amino acid protein that lacks C-terminal third of the full-length O-GlcNAcase (the HAT domain).33,34 The crystal structure of either O-GlcNAcase or v-O-GlcNAcase has not yet been solved. Recently, Vocadlo and co-workers proposed that O-GlcNAcase utilizes a substrate-assisted mechanism of action.37 In that mechanism, 2-acetamido group of the substrate is involved in catalytic hydrolysis via formation and breakdown of a bicyclic oxazoline or oxazolinium ion intermediate leading to the retention of the stereochemistry at anomeric center of the substrate. However, neither residues acting as the catalytic nucleophile or the base, nor the region responsible for either the O-GlcNAcase activity or the substrate binding site has been clearly established. Instead, two opposite hypotheses have been proposed.35,38–40 The N-terminal ‘hyaluronidase’ domain has been proposed to be the catalytic site for O-GlcNAc removal based on the fact that both isoforms of O-GlcNAcase have the same amino terminus and this N-terminal region has limited homology with other glycosidases.38 O-GlcNAcase is categorized into the family 84 of glycoside hydrolases (GH) as defined by the carbohydrate-active enzymes (CAZy) database. In support of the proposal that the N-terminal region is responsible for the catalytic O-GlcNAcase activity, Rigden et al.39 demonstrated that the common conserved N-terminus region in the glycoside hydrolase (GH) family 84 is likely to conform to the catalytic triosephosphate isomerase (TIM) barrel fold. Whereas, the opposite proposal that the O-GlcNAcase catalytic activity resides in the C-terminal acetyltransferase domain was based on the observation40 that O-GlcNAcase enzymatic activity has never been detected in the splice variant of O-GlcNAcase that lacks the putative histone acetyltransferase domain.36 To date, the functional role of the splice variant is still unclear.
The standard in vitro assay for measuring O-GlcNAcase uses pNP-β-GlcNAc as a substrate.29 Enzyme activity is monitored spectrophotometrically by measuring the absorbance at 400 nm of a p-nitrophenolate ion that is enzymatically released from pNP-β-GlcNAc.29,41 However, this assay is relatively insensitive because the substrate undergoes only a modest change in absorbance upon cleavage.
To facilitate the study of O-GlcNAcase isoforms, a highly sensitive method is necessary. Because fluorescence is highly sensitive, a great number of fluorogenic substrates have been widely used in a variety of biological areas.37,42–46 To date, a number of hexosaminidases have been examined by using fluorescent analogues and the sensitivities and usefulness of fluorogenic assays have been demonstrated.37,42–46 For example, fluorescein di-β-D-galactoside (FDG) has been used to measure the degree of transfection of the β-galactosidase reporter gene.43 The sensitivity of this fluorometric assay was found to be 50-fold higher than that of the spectrophotometric assay of b-galactosidase with o-nitrophenyl-β-D-galactopyranoside.44 Vocadlo and co-workers used a series of 4-methylumbelliferyl 2-N-acetyl-2-deoxy-β-D-gluocopyranosides (MUGlcNAc) as fluorogenic substrates for O-GlcNAcase in elucidating the catalytic mechanism of O-GlcNAcase.37 The fluorescent moiety in MUGlcNAc is 4-methylumbelliferone. However, it has been reported that the sensitivity of the assay using FDG is more than 10-fold higher than that of the assay with 4-methylumbelliferyl-β-D-galactoside.45 In other words, the sensitivity of the assay was higher when fluorescein was chosen as a fluorophore than when 4-methylumbelliferone was chosen. Therefore, we have selected fluorescein as the backbone of our substrate to achieve high sensitivity of the assay and prepared fluorescein, di-N-acetyl-β-D-glucosaminide (FDGlcNAc). The synthesis of FDGlcNAc was carried out according to the procedure described by Kasai et al.47 with some modifications. In this article, we evaluate the usefulness of FDGlcNAc as a new fluorogenic substrate for the study of both O-GlcNAcase isoforms by measuring their kinetic parameters. This fluorogenic substrate becomes fluorescent only upon cleavage by O-GlcNAcase. Due to the improved sensitivity of this substrate, we report that shorter splice variant is an active O-GlcNAcase (Scheme 1).
Scheme 1.
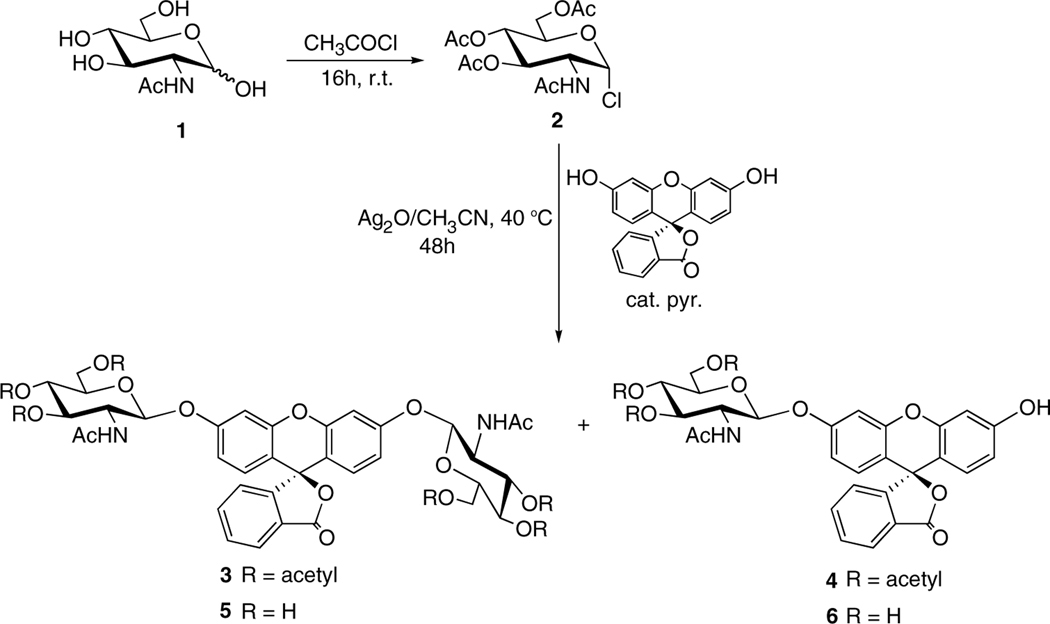
Synthesis of fluorescein di(N-acetyl-β-D-glucosaminide) (FDGlcNAc). The synthesis of FDGlcNAc was carried out essentially according to the procedure described by Kasai et al.47 with some modifications; see details in the Experimental.
2. Results and discussion
The O-GlcNAc specific β-N-acetylglucosaminidase (O-GlcNAcase) is involved in O-GlcNAc removal, playing a key role in O-GlcNAc metabolism. For example, O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino-N-phenylcarbamate (PUGNAC), a potent inhibitor of the O-GlcNAcase, blocks O-GlcNAc cycling, and induces insulin resistance.22,48–51 As mentioned before, the human MGEA5 gene produces alternatively spliced transcripts resulting in two isoforms. One is a full-length enzyme with a unique C-terminal region considered as a putative histone acetyltransferase domain36 and the other is a shorter enzyme that does not have this unique C-terminal region. To date, active O-GlcNAcase has been recognized in only the longer isoform. The report52 that a single nucleotide polymorphism in the MGEA5 gene encoding O-GlcNAcase is associated with diabetes mellitus and age of onset in the Mexican–American population highlights the importance of the physiological role of O-GlcNAcase. Growing interest in O-GlcNAcase stimulated the need for a highly sensitive enzymatic assay method. A series of 4-methylumbelliferyl 2-deoxy-2-N-fluoroacetyl β-D-glucopyranoside substrates for O-GlcNAcase were recently prepared by Vocadlo and co-workers.37 Because a fluorescein-conjugated substrate has higher sensitivity than the corresponding methylumbelliferyl substrate,45 fluorescein was selected as the aglycone of the synthetic fluorogenic substrate.
O-GlcNAcase exhibits both stereo and substrate specificities. This enzyme catalyzes the hydrolysis of β-O-linked pNP-GlcNAc, but not α-O-linked pNP-GlcNAc. Neither β-O-linked pNP-GalNAc, nor para-nitrophenyl gluocopyranoaminide was cleaved by the enzyme (data not shown). Therefore, fluorescein conjugated to two O-GlcNAc moieties via b-linkages, 5, was designed as a substrate for O-GlcNAcase.
The synthesis of 2′,7′-dichlorofluorescein, di-N-acetyl-β-D-glucosaminide and its use as a substrate for the colorimetric rate-assay of urinary N-acetyl β-D-gluocosaminidase (NAGase, EC 3.2.1.30) was originally reported by Kasai et al.47 In the conditions they developed for glycosidation, acetonitrile was used as a solvent and either Et3N or silver oxide (Ag2O) was used as a catalyst. In the optimal reaction system, two GlcNAc moieties were conjugated into a dichloro derivative of fluorescein in a moderate yield.47 However, when fluorescein was used as the aglycone, FDGlcNAc, which is the product of glycosylation of two hydroxyl groups in fluorescein, was not formed, possibly due to the weak reactivity of the secondary hydroxyl group in fluorescein. To overcome this obstacle, a small amount of base was used to increase the reactivity of the hydroxyl group in fluorescein. Thus, FDGlcNAc was successfully synthesized in moderate yield by treatment of fluorescein with a 3-fold molar quantity of 2-acetamido-3,4,6-tri-O-acetyl-2-deoxy-α-D-glucopyranosyl chloride (2) as a glycosyl donor using acetonitrile as the solvent and silver oxide (Ag2O) as a catalyst in the presence of small amounts of pyridine at 40 °C for 48 h. The two hydrophilic GlcNAc moieties of FDGlcNAc provide enhanced solubility (>100 mM) compared to that of the conventional pNP-GlcNAc (<10 mM).
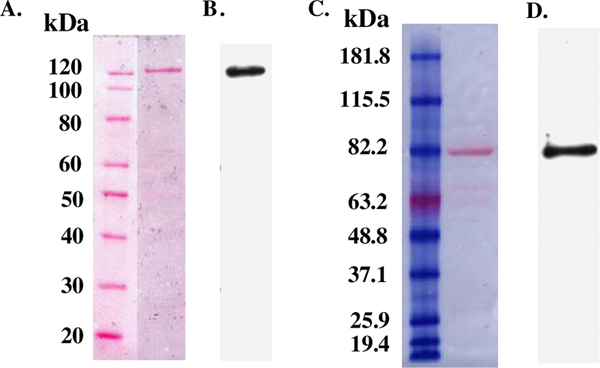
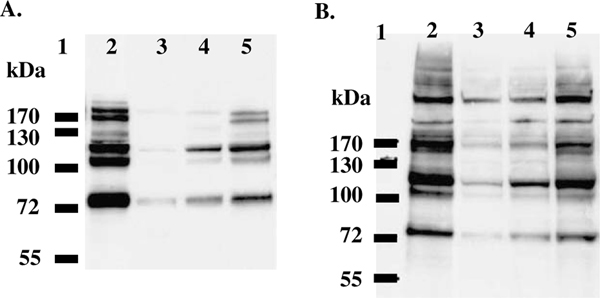
For the purpose of affinity purification and immunological identification, six consecutive histidine residues were recombinantly fused to the O-GlcNAcase, and its variant, at the N-terminus. These recombinant O-GlcNAcase and v-O-GlcNAcase proteins were expressed in Escherichia coli and purified on a nickel column via the His6-tag. To obtain relatively pure protein, an optimized washing protocol was employed (see Methods). The v-O-GlcNAcase eluted with a lower concentration (60 mM) of imidazole as compared to full-length O-GlcNAcase (300 mM). The flow-through fractions containing protein were assayed with two substrates, pNP-β-GlcNAc and FDGlcNAc. The fractions containing enzymatic activity were concentrated with Microcon centrifugal filter devices (Microcon YM-100 and YM-50 for O-GlcNAcase and v-O-GlcNAcase, respectively) and the concentrated enzymes were reconstituted in 20 mM Tris–HCl, pH 7.5, 1 mM DTT aqueous solution. Their purity was assessed by Ponceau staining and immunoblotting (Fig. 1). Recombinant His6-tagged-O-GlcNAcase and -v-O-GlcNAcase migrated at their apparent molecular weight of ~130 and ~80 kDa, respectively. Additionally the modified washing protocol was essential to produce sufficiently pure His6-tagged enzymes.
Figure 1.
Isolation of O-GlcNAcase and v-O-GlcNAcase. The purity of O-GlcNAcase and v-O-GlcNAcase was confirmed by Ponceau S staining of the nitrocellulose membranes (A, C) and Western blot analysis using mouse HRP conjugated 6X His tag antibody [AD1.1.10] (B, D), respectively.
Neither O-GlcNAcase nor v-O-GlcNAcase cleaved β-O-glycosidic linkage of FDGalNAc. This result is consistent with previous findings that O-GlcNAcase has substrate specificity for O-GlcNAc but not O-GalNAc.
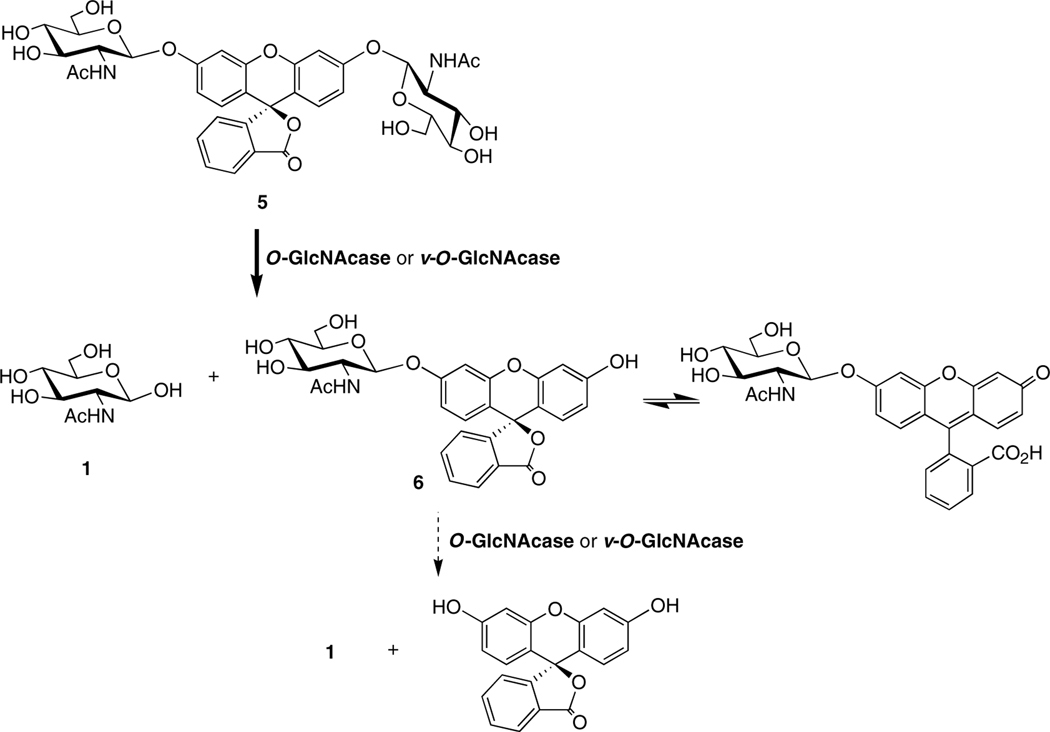
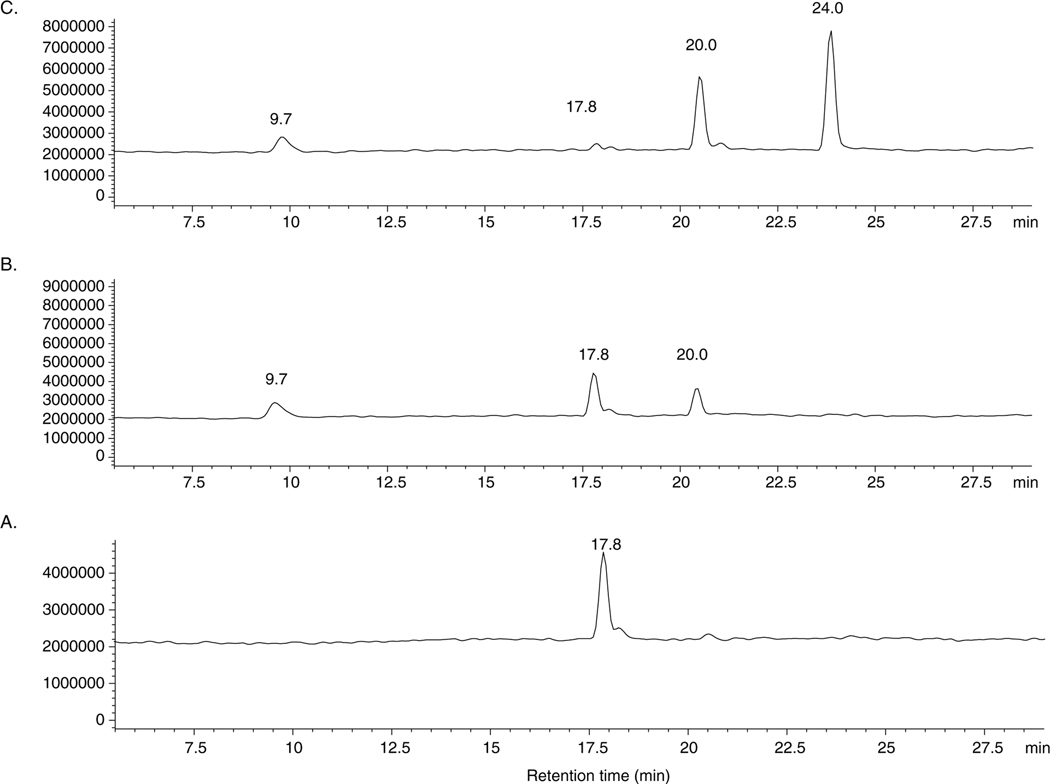
FDGlcNAc is nonfluorescent until cleavage of the β-D-O-glycosidic bond by either O-GlcNAcase or v-O-GlcNAcase. Spontaneous hydrolysis of FDGlcNAc was not observed, demonstrating that FDGlcNAc is as stable in aqueous solution (data not shown) as other substrates such as pNP-β-GlcNAc and MUGlcNAc. Enzymatic hydrolysis produces fluorescent fluorescein mono(N-acetyl-β-D-gluocosaminide) (FMGlcNAc), which is released from FDGlcNAc, tautomerizes to the quinoid form and generates fluorescence (Scheme 2). FMGlcNAc is then further hydrolyzed by the action of enzyme giving rise to fluorescein at longer time (Scheme 2). The products generated by enzymatic hydrolysis of FDGlcNAc were examined by LC–ESI (Fig. 2). A single peak representing FDGlcNAc (tR = 17.8 min) confirms that it is the only species present prior to addition of enzyme (Fig. 2A). All of the enzymatic assays performed were stopped when less than 5% of FDGlcNAc was utilized. As expected, during the time frame of our kinetic assay, FMGlcNAc (tR = 20.0 min) was the only fluorescent species generated by the enzymatic hydrolysis and fluorescence was due to the accumulation of FMGlcNAc alone (Fig. 2B). When 2-fold more enzyme is used and incubation time is extended, the fluorescein (tR = 24.0 min) released upon cleavage of the second N-acetyl-β-D-glucosaminyl group from further hydrolysis of FMGlcNAc was detected (Fig. 2C). However, during the timeframe of our assay for measuring initial velocities of enzymatic hydrolysis with varying concentrations of FDGlcNAc and predetermined amounts of enzyme, only one of the N-acetyl-β-D-gluocosaminyl groups was cleaved from the substrate producing fluorescent FMGlcNAc. Given these data, we used FMGlcNAc as the standard for kinetics of the O-GlcNAcase enzymatic reaction. When the fluorometer was set to have excitation and emission wavelengths at 485 and 535 nm, respectively, with the 4 mm diameter of circular aperture and 1 s of counting time, a plot of increasing concentrations of FMGlcNAc up to 12 μM versus their fluorescence intensities (relative fluorescence units, RFU) resulted in a linear correlation. From this plot the calculated molar fluorescence constant of FMGlcNAc was 119,820 units/μM.
Scheme 2.
Enzymatic hydrolysis reaction of FDGlcNAc (5) by either O-GlcNAcase or v-O-GlcNAcase. FMGlcNAc accumulated at the initial stage of enzymatic hydrolysis. However, fluorescein can be formed under conditions of higher enzyme concentration or longer reaction times.
Figure 2.
Enzymatic hydrolysis of FDGlcNAc examined by LC–ESI. LC conditions are as follows: flow rate, 0.2 mL/min; isocratic elution of H2O/CH3CN (v/v), 97:3 for 5 min, then a gradient of H2O/CH3CN (v/v), from 97:3 to 2:98 for 40 min. (A) A chromatogram of FDGlcNAc (tR = 17.8 min) in the absence of O-GlcNAcase. (B) New peaks arising after a 10 min incubation with O-GlcNAcase (2 μL) at 37 °C are due to GlcNAc (tR = 9.7 min) and FMGlcNAc (tR = 20.0 min), respectively. (C) A chromatogram of FDGlcNAc incubated with O-GlcNAcase (4 μL) at 37 °C for 7 h. Fluorescein is detected and has a retention time of 24 min.
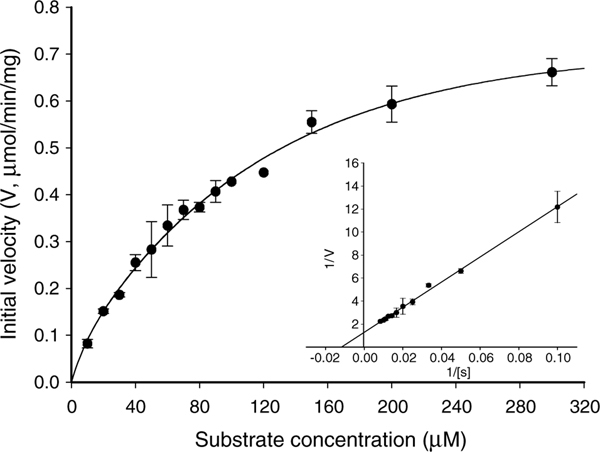
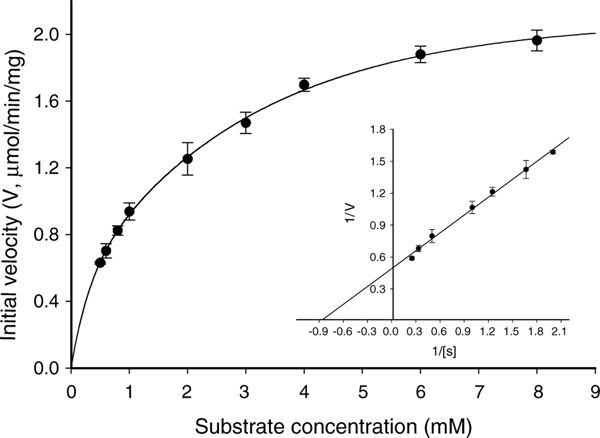
Utilizing FDGlcNAc as a substrate, kinetic parameters including Michaelis constants (Km), specific activities, turnover numbers, and enzymatic efficiencies of the recombinant O-GlcNAcase were obtained through Lineweaver–Burk plots (Fig. 3). Surprisingly, incubation of FDGlcNAc with the v-O-GlcNAcase also produced fluorescence, indicating that this is an active enzyme. The kinetic parameters of the variant enzyme were calculated as well (Fig. 4). For the comparison of these kinetic parameters with FDGlcNAc, enzymes were assayed with pNP-β-GlcNAc (Fig. 5) and the kinetic values of both enzymes for FDGlcNAc and pNP-β-GlcNAc are summarized in Table 1. Using the FDGlcNAc substrate, we established that O-GlcNAcase has a Km of 85 μM and a specific activity of 780 nmol/min/mg with a turnover rate of 1.3/s. Catalytic efficiency (Kcat/Km) of O-GlcNAcase for FDGlcNAc was 1.6 ·× 104 M−1 s−1. On the other hand, O-GlcNAcase exhibited a Km of 1.1 mM and a specific activity of 2035 nmol/min/mg with a turnover number of approximately 3/s and catalytic efficiency of 3.0 ·× 103 M−1 s−1 for pNP-β-GlcNAc.
Figure 3.
Enzyme kinetics of full-length O-GlcNAcase. An activity curve and a Lineweaver–Burk plot were generated for recombinant O-GlcNAcase by varying concentrations of the substrate, FDGlcNAc.
Figure 4.
Enzyme kinetics of variant O-GlcNAcase. An activity curve and a Lineweaver–Burk plot were generated for recombinant v-O-GlcNAcase by varying concentrations of the substrate, FDGlcNAc.
Figure 5.
Enzyme kinetics of O-GlcNAcase. An activity curve and a Lineweaver–Burk plot were generated for recombinant O-GlcNAcase by varying concentrations of the substrate, pNP-β-GlcNAc.
Table 1.
Kinetic parameters of both recombinant O-GlcNAcase isoforms for FDGlcNAc, 5, and pNP-β-GlcNAc
| Kinetic parameter | Substrate | Enzyme | |
|---|---|---|---|
|
|
|||
| O-GlcNAcase | v-O-GlcNAcase | ||
|
| |||
| Km (mM) | FDGGlcNAc | 0.085 ± 0.016 | 2.1 ± 0.2 |
| pNP-β-GlcNAc | 1.1 ± 0.1 | — | |
| Specific activities (nmol/min/mg) | FDGGlcNAc | 780 ± 60 | 1680 ± 165 |
| pNP-β-GlcNAc | 2035 ± 161 | — | |
| Turnover number (Kcat, s−1) | FDGGlcNAc | 1.3 ± 0.1 | 2.1 ± 0.2 |
| pNP-β-GlcNAc | 3.4 ± 0.3 | — | |
| Kcat/Km (efficiency, M−1 s−1) | FDGGlcNAc | 1.6 ·× 104 | 1.0 ·× 103 |
| pNP-β-GlcNAc | 3.1 ·× 103 | — | |
The low Km and high affinity of O-GlcNAcase for FDGlcNAc can be explained by the rigidity and hydrophobicity of the aglycone fluorescein. The respective increase in both the hydrophobicity and rigidity between the three substrates pNP-β-GlcNAc, MUGlcNAc, and FDGlcNAc may reflect an associated decrease in the Km values for these substrates and an increase in apparent affinity. The affinity of O-GlcNAcase for FDGlcNAc (Km = 85 lM) is about 13-fold stronger than it is for pNP-β-GlcNAc (Km = 1.1 mM) and 5-fold stronger than it is for the fluorogenic substrate45 (Km = 430 μM). The most hydrophobic aglycone, fluorescein, is thought to mimic the rigid serine and threonine residues found in proteins. O-GlcNAcaes can access this constrained and hydrophobic substrate better than MUGlcNAc and the least constrained substrate pNP-β-GlcNAc.
While our Km value of O-GlcNAcase for pNP-β-GlcNAc was in good agreement with previous reports,40 the specific activity differed. The specific activity of our recombinant enzyme for pNP-β-GlcNAc was found to be 2035 nmol/min/mg whereas the reported value of their recombinant O-GlcNAcase for the pNP-β-GlcNAc substrate was 652 nmol/min/mg.40 In contrast to the Km, specific activity is not a constant value and depends on the enzymatic assay conditions as well as the purity of enzymes. Wells et al.40 used 50 mM sodium cacodylate, pH 6.5 as an assay buffer for measuring O-GlcNA-case activity whereas O-GlcNAcase activity was assayed in 100 mM citrate phosphate buffer, pH 6.5 in our kinetic experiments. Furthermore, recombinant O-GlcNAcase enzymes have different tags. The cloned human O-GlcNAcase used in our assays has a small tag composed of six histidine amino acids, while recombinant O-GlcNAcase used in their assay has a thioredoxin tag. Thioredoxin is an approximately 10 kDa protein and its presence may reduce O-GlcNAcase catalytic activity. It is likely that a small oligopeptide such as a six consecutive histidine residue does not significantly affect the catalytic activity of O-GlcNAcase. Because the specific activity of our recombinant O-GlcNAcase for pNP-β-GlcNAc is about 3-fold greater than that reported previously,40 other kinetic parameters such as turnover number and catalytic efficiency that are dependent on specific activity value are also 3-fold higher. Interestingly, O-GlcNAcase purified from bovine brain was reported27 to exhibit a specific activity (1840 nmol/min/mg) similar to the specific activity reported here.
Previous reports using pNP-β-GlcNAc (up to 8 mM) as a substrate, did not detect activity of the shorter O-GlcNAcase splice variant.40 However, using FDGlcNAc we found that v-O-GlcNAcase has a Km of 2.1 mM and a specific activity of 1680 nmol/min/mg with a turnover rate of 2.1. The catalytic efficiency of this shorter isoform for FDGlcNAc was determined to be 1.0 ·× 104 M−1 s−1. The activity detected with the shorter O-GlcNAcase was unanticipated. There has been controversy regarding the catalytic region of O-GlcNAcase.35,38–40 As previously mentioned, the C-terminal domain of O-GlcNAcase is homologous to acetyltransferases domain and is absent in v-O-GlcNAcase. Instead, this acetyltransferase domain is replaced by a novel 15 amino acid tail in the variant. Although Hanover et al.38 and Rigden et al.39 proposed that the catalytic domain of O-GlcNAcase is likely to be located in the N-terminal region, there have been no empirical data that support this proposal. On the contrary, the lack of enzymatic activity in the shorter isoform led to such a conclusion that v-O-GlcNAcase is not active, therefore, the O-GlcNAcase catalytic domain resides at the C-terminal end of enzyme.35,40 However, our in vitro kinetic data suggest that v-O-GlcNAcase is quite active, although the Km value of v-O-GlcNAcase for FDGlcNAc was 25-fold higher than that of the full-length O-GlcNAcase.
To further confirm that the shorter O-GlcNAcase is active, a mixed population of substrates from both HT-29 and HeLa cell extracts were incubated with either O-GlcNAcase or v-O-GlcNAcase as described under Experimental. As shown in Figure 6A, treatment of HT-29 cell extracts with either O-GlcNAcase (lane 3 and 4) or its shorter isoform (lane 5) decreased the level of O-GlcNAc-modified proteins compared to that of cell extracts treated with no enzyme (lane 2). Given the differences in enzymatic kinetics between the two enzymes (Table 1), it was not surprising that v-O-GlcNAcase did not lower the levels of O-GlcNAc as well as did full-length O-GlcNAcase. Similar results were obtained from HeLa cell extracts (Fig. 6B). These findings suggest that both the full-length and variant O-GlcNAcases are active against cellular O-GlcNAc-modified substrates confirming our earlier in vitro data using FDGlcNAc. Based on these data, v-O-GlcNAcase activity should be detected using much higher concentrations of pNP-β-GlcNAc. Given the fact that this shorter isoform is a bona fide O-GlcNAcase and does not possess the C-terminal histone acetyl transferase domain, it is likely that the O-GlcNAcase catalytic domain is located in the common N-terminal domain that has homology to Mu toxin.38 Some evidence suggests that the full-length O-GlcNAcase might be a bifunctional enzyme capable of both catalyzing the hydrolysis of b-linked GlcNAc and catalyzing acetylation of histones.36 Because there has been no tool to measure the enzymatic activity in v-O-GlcNAcase, it is not clear what function this shorter isoform serves in a cellular context. One hypothesis is that it serves as a regulatory tool in concert with the full-length of O-GlcNAcase.40
Figure 6.
Enzymatic activity of O-GlcNAcase and v-O-GlcNAcase on mixed substrates. Western blot analysis of HT-29 (A) and HeLa (B) cell extracts treated with either O-GlcNAcase or v-O-GlcNAcase. Lane 1, protein marker; lane 2, cell extracts without enzyme (control); lanes 3 and 4, cell extracts treated with 7 and 5 μg of O-GlcNAcase, respectively; and lane 5, cell extracts treated with 5 μg of v-O-GlcNAcase.
The nature of FDGlcNAc made it an extremely sensitive substrate for O-GlcNAcase enabling the detection of enzymatic activity in v-O-GlcNAcase for the first time. In conclusion, FDGlcNAc has several advantages for the study of both O-GlcNAcase isoforms over the conventional substrate, pNP-β-GlcNAc and the precedent fluorogenic substrate, MUGlcNAc,37 in terms of sensitivity of the assay and the affinity for the enzyme. In addition, a high-throughput analysis is possible using a multiwell plate format with this new substrate. To dissect the role of each O-GlcNAcase isoform, it is necessary to find specific inhibitors of full-length O-GlcNAcase over its shorter isoform or vice versa. The high-throughput assay with this highly sensitive fluorogenic substrate, FDGlcNAc will accelerate the discovery of novel O-GlcNAcase inhibitors. The highly sensitive substrate FDGlcNAc made it possible to confirm that the v-O-GlcNAcase as an active enzyme. The functional role of each O-GlcNAcase isoform is currently under active investigation. Although FDGlcNAc is cell permeable, its use in intact cells as an imaging agent for the in vivo analysis of O-GlcNAcase is limited due to the lack of enzyme specificity between O-GlcNAcase and lysosomal hexosaminidases. We are currently developing fluorogenic substrates, which are specific for only O-GlcNAcase but not lysosomal hexosaminidases.
3. Experimental
3.1. General methods
All chemicals were of reagent grade and purchased from Sigma Chemical Co. unless otherwise noted. Deuterated DMSO was from Cambridge Isotope Laboratories. FDGalNAc was purchased from Marker Gene Technologies, Inc. Acetonitrile was dried over 4 Å molecular sieves overnight prior to use. The Inertsil ODS-2 column was obtained from GL Science Inc. 1H NMR and 13C NMR spectra were recorded at 300 and 75 MHz, respectively, with a Varian Mercury 300 spectrometer. Chemical shifts are reported in parts per million (δ) and referenced to either the residual signal of the solvent used (DMSO: 1H δ 2.50) or internal tetramethylsilane. Coupling constants, J, are reported in Hertz. Analytical thin-layer chromatography (TLC) was conducted on Silica Gel 60-F254 (E. Merck) with detection by UV light and/or dipping the plates in a cerium sulfate–ammonium molybdate solution followed by heating. Flash column chromatography was carried out on Merck Kiesel gel 60 (SiO2, 230–400 mesh). High performance liquid chromatography (HPLC) was performed on an Inertsil ODS-2 column (10 mm i.d. ×· 250 mm) using a Hewlett Packard Series 1100 multisolvent delivery system with a multiple wavelength detector (flow rate, 2 mL/min; detection, 254, 280 nm; temperature, ambient). Fast atom bombardment (FAB+) MS was obtained at the National Institutes of Health Mass Spectrometry Laboratory. High resolution-electrospray ionization (HR-ESI) mass spectrum was obtained on a Waters LCT Premier time-of-flight (TOF) mass spectrometer. Combined liquid chromatography and electrospray ionization (LC–ESI) mass spectra were obtained on Hewlett Packard Series 1100 MSD equipped with HPLC performed on an AtlantisTM dC18 column (2.1 mm i.d. ·× 100 mm) at the National Institutes of Health Mass Spectrometry Laboratory. Elemental analyses were obtained at Atlantic Microlab, Inc. Fluorometry measurements were obtained at wavelengths of 485 and 535 nm for fluorescence excitation and emission, respectively, with Wallac 1420 multilabel counter (Perkin–Elmer Life Sciences). Absorbance was measured at 400 nm with a UV 160U spectrophotometer (Shimadzu).
3.2. 3′,6′-Bis[[3,4,6-tri-O-acetyl-2-(acetylamino)-2-deoxy-β-D-glucopyranosyl]oxy]-spiro[isobenzofuran-1(3H), 90-[9H]-xanthen]-3-one (3)
Fluorescein (1.0 g, 3 mmol) was added to a solution of N-acetyl 2-deoxy-3,4,6-tri-O-acetyl-a-D-glucopyranosyl chloride53 (2, 3.3 g, 9.0 mmol) in dry CH3CN. To this mixture were, subsequently, added silver oxide (Ag2O, 2.09 g, 9.0 mmol) and five drops of pyridine. The reaction solution was stirred for 2 days at 40 °C, after which the remaining solid residue was removed by filtration and the filtrate was concentrated to yield a dark brown syrup. Flash column silica chromatography of the resulting residue washing with a mixture of hexane/EtOAc (2:1), then eluting with a gradient of CHCl3/CH3CN from 1:1 to 1:2 (v/v) offered the desired product 3 as a white solid (1.19 g, 1.2 mmol, 40%): Rf = 0.12 (EtOAc); 1H NMR (300 MHz, DMSO-d6): δ 8.08 (app. dd, 2H, J = 8.7, 2.7 Hz, arom H), 8.03 (d, 2H, J = 7.2 Hz, NHCOCH3), 7.80 (dt, 1H, J = 7.6, 1.5 Hz, arom H), 7.74 (dt, 1H, J = 7.2, 1.1 Hz, arom H), 7.31 (d, 1H, J = 7.6 Hz, arom H), 7.03 (dd, 2H, J = 5.3, 2.0 Hz, xanthene H), 6.81–6.72 (m, 4H, xanthene H), 5.44 (d, 2H, J = 8.4 Hz, GlcNAc H-1), 5.24 (app. dt, 2H, J = 9.7, 3.0 Hz, GlcNAc H-3), 4.94 (t, 2H, J = 9.5 Hz, GlcNAc H-4), 4.25–3.96 (m, 8H, GlcNAc H-5, H-6a, H-6b, H-2), 2.06, 2.03 (s, each 3H, OCOCH3), 2.01, 1.96 (s, each 6H, OCOCH3), 1.78, 1.77 (s, each 3H, NHCOCH3); 13C NMR (75 MHz, DMSO-d6): δ 170.02, 169.98, 169.69, 169.54, 169.34, 168.51 (9C@O), 158.28, 158.22, 152.39, 151.42, 151.39 (5C), 135.84 (arom CH), 130.38 (arom CH), 129.34 (xanthene, 2CH), 125.65 (1C), 124.86 (arom CH), 124.00 (arom CH), 113.86 (xanthene, 2CH), 112.81, 112.87 (2C), 103.44 (xanthene, 2CH), 97.50 (2CH, C-1), 81.58 (xanthene, spiro C), 72.24 (2CH, C-3), 70.96 (2CH, C-5), 68.33 (2CH, C-4), 61.64 (2CH, C-6), 53.05 (2CH, C-2), 20.34, 20.44, 20.48 (6CH3, COCH3), 22.62 (2CH3, NHCOCH3). Anal. Calcd for C48H50N2O21·H2O: C, 57.14; H, 5.19; N, 2.78. Found: C, 56.81; H, 5.18; N, 2.89.
3.3. 3′-[[3,4,6-Tri-O-acetyl-2-(acetylamino)-2-deoxy-β-D-glucopyranosyl]oxy]-6′-hydroxyspiro[isobenzofuran-1(3H),90-[9H]-xanthen]-3-one (4)
Silver oxide (Ag2O, 0.28 g, 1.2 mmol) and two drops of pyridine were added to a solution of 2 (0.44 g, 1.2 mmol) and fluorescein (0.2 g, 1.2 mmol) in dry CH3CN. The reaction mixture was stirred for 2 days at 40 °C, after which the remaining solid residue was removed by filtration. The filtrate was concentrated and subjected to flash column silica chromatography. Elution with a slow gradient of CH3Cl/CH3CN from 2:1 to 1:1 (v/v) gave 0.48 g of 4 as a yellow colored powder (0.73 mmol, 60%); Rf = 0.47 (EtOAc); 1H NMR (300 MHz, DMSO-d6): δ 8.10 (app. dd, 1H, J = 9.0, 3.9 Hz, arom H), 8.01 (d, 2H, J = 6.9 Hz, NHCOCH3), 7.79–7.70 (m, 2H, arom H), 7.29 (app. t, 1H, J = 9.0 Hz, arom H), 7.09–7.07 (m, 1H, xanthene H), 6.73–6.69 (m, 3H, xanthene H), 6.60–6.58 (m, 2H, xanthene H), 5.43 (d, 1H, J = 8.7 Hz, GlcNAc H-1), 5.22, 5.21 (t, each 1/2H, J = 9.8 Hz, GlcNAc H-3), 4.92 (t, 1H, J = 9.7 Hz, GlcNAc H-4), 4.27–4.15 (m, 2H, GlcNAc H-5, H-6a), 4.15–3.98 (m, 2H, GlcNAc H-2, H-6b), 2.06, 2.04 (s, each 3/2H, OCOCH3), 2.01, 1.95 (s, each 3H, OCOCH3), 1.77, 1.76 (s, each 3/2H, NHCOCH3); LR LC–ESIMS: m/z 684.2 [M+Na+].
3.4. 3′,6′-Bis[[2-(acetylamino)-2-deoxy-β-D-glucopyranosyl]oxy]-spiro-[isobenzofuran-1(3H),9′-[9H]-xanthen]-3-one (5)
A cooled (—20 °C) solution of 3 (300 mg, 0.30 mmol) in CH3OH (5 mL) was treated dropwise with aqueous NH3 (28%, 1 mL, 7.2 mmol). The reaction mixture was warmed to rt over 2 h, stirred for an additional 1 h at rt, and concentrated in vacuo to yield a pale yellow solid. This crude material was purified by reverse-phase HPLC to give 5 (170 mg, 0.23 mmol, 76.7%) as a white solid: HPLC: tR = 16.8 min (H2O/CH3CN (v/v) from 95:5 to 80:20 for 5 min, 80:20 to 60:40 for 15 min, 60:40 to 35:65 for 25 min, 2 mL/min); 1H NMR (300 MHz, DMSO-d6): δ 8.02 (d, 1H, J = 7.42 Hz, arom H), 7.82–7.71 (m, 4H, 2 ·× arom H, 2 ·× NHCOCH3), 7.30 (d, 1H, J = 7.42 Hz, arom H), 6.96 (app. m, 2H, xanthene H), 6.71 (app. s, 4H, xanthene H), 5.14–5.09 (m, 6H, GlcNAc H-1, OH-3, OH-4), 4.65 (app. q, 2H, J = 4.7 Hz, OH-6), 3.75–3.63 (m, 4H, GlcNAc H-2, H-6a), 3.53–3.15 (m, 8H, GlcNAc H-3, H-4, H-5, H-6b), 1.80, 1.79 (s, 6H, 2 ·× NHCOCH3); 3C NMR (75 MHz, DMSO-d6): δ 169.30 (2NHC=O), 168.57 (C=O), 159.05, 159.01, 152.45, 151.51, 151.46 (5C), 135.80 (arom CH), 130.33 (arom CH), 129.20 (xanthene, 2CH), 125.74 (1C), 124.81 (arom CH), 123.98 (arom CH), 113.77 (xanthene, 2CH), 112.31, 112.28 (2C), 103.27, 103.19 (xanthene, CH), 98.78 (2CH, C-1), 81.87 (xanthene, spiro C), 77.32, 73.91, 70.20 (each 2CH, C-3, C-4, C-5), 60.71 (2CH, C-6), 55.32 (2CH, C-2), 23.04 (2CH3, NHCOCH3); HR FAB MS: Calcd for C36H38N2O15Cs: m/z 871.1327 [M+Cs]+. Found: m/z 871.1300. Anal. Calcd for C36H38N2O15·H2O: C, 57.14; H, 5.33; N, 3.70. Found: C, 56.89; H, 5.45; N, 3.70.
3.5. 3′-[[2-(Acetylamino)-2-deoxy-β-D-glucopyranosyl]oxy]-6′-hydroxyspiro[isobenzofuran-1(3H),9′-[9H]-xanthen]-3-one (6)
O-Deacetylation of 4 (150 mg, 0.23 mmol) with aqueous NH3 was carried out as described for 4. Purification of the crude material by reverse-phase HPLC offered 96 mg of 5 as an orange colored powder (0.18 mmol, 78%): HPLC: tR = 11.3 min (H2O/CH3CN (v/v) from 95:5 to 50:50 for 4 min, 50:50 to 27:73 for 11 min, 27:73 to 20:80 for 1 min, 2 mL/min); 1H NMR (300 MHz, DMSO-d6): δ 8.01 (d, 1H, J = 7.2 Hz, arom H), 7.81–7.69 (m, 3H, 2 ·× arom H, NHCOCH3), 7.28 (app. dd, 1H, J = 7.5, 3.6 Hz arom H), 6.95 (app. s, 1H, xanthene H), 6.71–6.64 (m, 3H, xanthene H), 6.58 (app. s, 2H, xanthene H), 5.20–5.01 (m, 2H, OH), 5.07 (d, 1H, J = 8.7 Hz, GlcNAc H-1), 4.67 (br s, 1H, OH), 3.80–3.16 (m, 6H, GlcNAc H-2, H-3, H-4, H-5, H-6a, H-6b), 1.79 (s, 3H, NHCOCH3); HR-ESIMS: Calcd for C28H25N1O10Na: m/z 558.1376 [M+Na]+. Found: m/z 558.1390.
3.6. Expression and purification of recombinant O-GlcNAcase and v-O-GlcNAcase
Details describing the cloning of pBAD/HisA human O-GlcNAcase (MGEA 5) and its splice variant (MGEA5 s) clones will be reported elsewhere.54 Cultures of TOP 10 cells containing pBAD/HisA expression vector (Invitrogen) encoding the His6-tagged-O-GlcNAcase were grown overnight in Luria–Bertani (LB) broth (Digene) supplemented with ampicillin (50 lμ/mL) at 37 °C at 200 rpm. Induction of O-GlcNAcases was initiated by adding arabinose solution (0.02%) when the OD600 of cultures reached at around 0.5. The cultures were further grown for 4 h at 30 °C at 200 rpm. The cells were harvested by centrifugation at 4000g for 15 min in a Sorvall® RC 5C Plus (DuPont) centrifuge. The pellet was subjected to freeze–thaw and suspended in 1/50 of the original volume in 0.1 mg/mL lysozyme, 20 mM Tris–HCl, pH 7.5, 1 mM DTT, 0.1% Triton X-100, an EDTA-free protease inhibitor cocktail tablet (1 tablet/10 mL). The suspension was incubated at rt for 5 min and sonicated on ice (4 ·× 10 s, setting 3, Misonix Ultrasonic Processor). The supernatant obtained after centrifugation at 20,000g for 15 min was stored in aliquots at −80 °C. Expressed His6-tagged O-GlcNAcase enzyme in the supernatant was purified on His-Trap HP column (Amersham Biosciences) according to the manufacture’s protocol with minor variations. In this protocol, phosphate buffer (pH 7.4) containing 40 mM imidazole was used as a binding buffer and the column loaded with sample was washed successively with the binding buffer (20 mL) and phosphate buffer (pH 7.4) containing 60 mM concentration of imidazole (20 mL). Elution with phosphate buffer (pH 7.4) containing 300 mM imidazole was performed at 1.0 mL/min and the eluate was collected in 1 mL fractions. Fractions were assayed for protein by measuring absorbance at 280 nm. Fractions containing protein were further assayed for O-GlcNAcase activity using the protocol described below in this section. Fractions showing enzyme activity were pooled and concentrated using a Centricon 100 microconcentrator (Amicon). Small amount of concentrated enzyme fraction was then reconstituted in 20 mM Tris–HCl, pH 7.5, 1 mM DTT aqueous solution. Pierce BCA protein assay reagent was used to estimate protein concentration by using bovine serum albumin (BSA) as a standard protein. The purified shorter splice variant of O-GlcNAcase (v-O-GlcNAcase) was prepared by the same protocol described above except using the phosphate buffer containing 10 mM concentration of imidazole as a binding buffer and washing thoroughly with 20 mL of the binding buffer and eluting successively with 60, 100, 300 mM imidazole phosphate buffer (pH 7.4). Enzyme containing fractions were concentrated by using a Centricon 50 microconcentrator (Amicon) after fractions showing enzyme activity were pooled. Purity of the enzymes was confirmed by Ponceau S staining of the membrane (Sigma) and Western blot analysis using anti His-Tag antibody (Abcam) (see Fig. 1).
3.7. O-GlcNAcase assay and enzyme kinetics with FDGlcNAc
One international unit (1 U) of O-GlcNAcase is defined as the amount that catalyzes the formation of 1.0 μmol of FMGlcNAc and 1.0 μmol of N-acetyl-D-glucosamine per minute at pH 6.5 at 37 °C. Purified recombinant O-GlcNAcase (0.17 μg, 2 μL) was added to the solution containing the fluorogenic substrate, 5, of which concentration was varied (10–300 μM) in 20 μL of 0.5 M citrate/phosphate buffer, pH 6.5, in a final volume of 100 μL at the start of the incubation at 37 °C. The assays were terminated by adding 900 μL of 0.5 M Na2CO3 solution after 3–5 min incubation. The assay solution (200 μL) was transferred into a 96-well plate and fluorescence was measured at the excitation wavelength of 485 nm (λex = 485 nm) and at the emission wavelength of 535 nm (λem = 535 nm) on the Wallac 1420 fluorometer (Perkin–ElmerTM Life Sciences). Assays were performed in triplicate.
3.8. v-O-GlcNAcase assay and enzyme kinetics with FDGlcNAc
One unit of v-O-GlcNAcase is defined as the amount that catalyzes the formation of 1.0 μmol of FMGlcNAc and 1.0 μmol of N-acetyl-D-gluocosamine per minute at pH 6.5 at 37 °C. The enzymatic reaction was initiated by adding 4 μL of partially purified recombinant v-O-GlcNAcase (1.19 μg) to the solution containing the substrate, 5, which is varied (0.2–20 mM) in 20 μL of 0.5 M citrate/phosphate buffer, pH 6.5, in a final volume of 100 μL. The assays were terminated and quantitated as described for the O-GlcNAcase. Assays were performed in triplicate.
3.9. O-GlcNAcase assay and enzyme kinetics with pNP-β-GlcNAc
One unit of O-GlcNAcase is defined as the amount that hydrolyzes 1.0 μmol of pNP-β-GlcNAc to p-nitrophenol and N-acetyl-D-gluocosamine per minute at pH 6.5 at 37 °C. pNP-β-GlcNAc (14 mM) was prepared in 5% DMSO in water and used as a stock solution. Purified recombinant O-GlcNAcase (0.18 μg) was added to increasing concentrations of pNP-β-GlcNAc (0.3– 8 mM) in 20 μL of 0.5 M citrate/phosphate buffer, pH 6.5, in a final volume of 100 μL at the start of the incubation at 37 °C. The assays were terminated by adding 900 μL of 0.5 M Na2CO3 solution after 5 min incubation. The p-phenolate ion generated from the hydrolysis of pNP-β-GlcNAc by the action of enzyme was spectrophotometrically measured at 400 nm on the UV 160U spectrophotometer (Shimadzu). Assays were performed in triplicate.
3.10. Molar fluorescence coefficient of FMGlcNAc
Standard dilutions made for calculating the molar fluorescence coefficient of FMGlcNAc on the Wallac 1420 fluorometer were as follows: 0.01, 0.05, 0.1, 0.5, 1.0, 2.0, 3.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10, and 12 μM in 20 μL of 0.5 M citrate/phosphate buffer, pH 6.5, 900 μL of 0.5 M Na2CO3 solution in a total volume of 1 mL. The assay solution (200 μL) was transferred into a 96-well plate and fluorescence was measured at λex = 485 nm and at λem = 535 nm on the Wallac 1420 fluorometer with a continuous wave lamp set to 5028 in the stabilized energy mode and with the 4 mm diameter of circular aperture and 1 s of counting time. The plot of the concentrations of the standard versus their corresponding fluorescence intensities was constructed and the line-of-best-fit through the experimental points was obtained. The slope of this straight line is the molar fluorescence coefficient.
The calculated molar fluorescence constant of FMGlcNAc is 119,820 units/μM.
3.11. Molar absorptivity of pNP-β-GlcNAc
pNP-β-GlcNAc (10 mM) was prepared in 5% DMSO in water and used as a stock solution. 3, 7, 10, 20, 30, 40, 50, 65, 80, 100, 110, and 120 μM of pNP-β-GlcNAc were made in 20 μL of 0.5 M citrate/phosphate buffer, pH 6.5, 900 μL of 0.5 M Na2CO3 solution in a total volume of 1 mL. Absorbance of para-phenolate ion in a series of dilution standards was measured at 400 nm at 25 °C on the UV 160U spectrophotometer (Shimadzu). Assays were performed in triplicate. The plot of the concentrations of the standard versus their corresponding absorbance was constructed and the line-of-best-fit through the experimental points was obtained. From the slope of this straight line, calculated molar absorptivity of pNP-β-GlcNAc is 18.8 mM cm−1.
3.12. Substrate specificity
Either 2 μL of purified recombinant O-GlcNAcase (0.17 μg) or 4 μL of partially purified recombinant v-O-GlcNAcase (1.19 μg) was added to the solution containing 30 μM of FDGalNAc (Marker Gene Technologies, Inc.) in 20 μL of 0.5 M citrate/phosphate, pH 6.5, in a final volume of 100 μL and incubated at 37 °C for 30 min. The assays were terminated by adding 900 μL of 0.5 M Na2CO3 solution. The assay solution (200 μL) was transferred into a 96-well plate and fluorescence was measured at λex = 485 nm and at λem = 535 nm on the Wallac 1420 fluorometer.
3.13. Preparation of HT-29 and HeLa cell extracts, enzyme activity assay, and Western blotting
The suspension of 10 mL culture of PUGNAC treated HT-29 cells was centrifuged at 2500g for 10 min. The pellet was suspended in 0.3 mL of lysis buffer containing 20 mM Tris–HCl, pH 8.0, 1 mM DTT, 0.1% Triton X-100, an EDTA-free protease inhibitor cocktail (1 tablet/10 mL). The suspension was incubated at rt for 5 min and sonicated on ice (4 ·× 10 s, setting 3, Misonix Ultrasonic Processor). The supernatant obtained after centrifugation at 20,000g for 15 min was stored in aliquots at −80 °C. PUGNAC treated HeLa cell extracts were prepared using MPER protein extraction reagent (Pierce) according to the manufacturer’s protocol from a 10 mL suspension of cultured cells. Protein (20 μg) was incubated with 0 (control), 7 μg, 5 μg of O-GlcNAcase, and 5 μg of v-O-GlcNAcase in 0.05 M citrate/phosphate buffer, pH 6.5, in a final volume of 22.5 μL at 37 °C for 3 h. Reactions were stopped by adding 7.5 μL of NuPAGE® LDS sample buffer (4·×). Samples were loaded onto a 10% NuPAGE pre-cast gel and proteins were separated by electrophoresis and then transferred to a nitrocellulose membrane for immunoblotting. The blot was probed with O-GlcNAc specific antibody RL2 (mouse IgG1, Affinity Bioreagents, Inc.) at 1:1000 dilution. Peroxidase-conjugated goat anti-mouse IgG (Jackson Immunoresearch Laboratories, Inc.) was used as the secondary antibody. The blot was visualized with enhanced SuperSignal® West Pico Chemiluminescent Substrate (Pierce) (see Fig. 6).
Acknowledgments
This research was financially supported by National Institute of Health Grants (VFJA009594). We thank Mr. Victor Livengood who is an operator of the mass spectrometers in the NIDDK, NIH for obtaining the mass spectral data and allowing us to use LC–ESI instrument for monitoring the time-course of the enzyme reaction.
Abbreviations:
- FDGlcNAc
fluorescein di-N-acetyl-β-D-glucosaminide
- O-GlcNAcase
a full-length form of peptide O-GlcNAc-β-N-acetyl-glucosaminidase
- v-O-GlcNAcase
a shorter isoform of peptide O-GlcNAc-β-N-acetylglucosaminidase
- PNP
para-nitrophenyl
- O-GlcNAc
O-linked β-N-acetylglucosamine
- FDGalNAc
fluorescein di-N-acetyl-β-D-galactosaminide
- O-GalNAc
O-linked β-N-acetylgalactosamine
- OGT
protein uridine diphospho-N-acetylglucosamine: polypeptide β-N-acetylglucosaminyl transferase
- MUGlcNAc
4-methylumbelliferyl 2-acetamido-2-deoxy-β-D-glucopyranoside
- UDP
uridine diphosphate
- DMSO
dimethyl sulfoxide
- NMR
nuclear magnetic resonance
- TLC
thin-layer chromatography
- FAB MS
fast atom bombardment mass spectroscopy
- LC–ESIMS
combined liquid chromatography and electrospray ionization spectroscopy
- HPLC
high performance liquid chromatography
- PBS
phosphate-buffered saline
- DMEM
Dulbecco’s modified eagle medium
- PUGNAC
O-(2-acetamido-2-deoxy-D-glucopyranosylidene)-amino-N-phenylcarbamate
References
- 1.Jackson SP; Tjian R. Cell 1988, 55, 125–133. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP; Tjian R. Proc. Natl. Acad. Sci. U.S.A 1989, 86, 1781–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly WG; Dahmus ME; Hart GW J. Biol. Chem 1993, 268, 10416–10424. [PubMed] [Google Scholar]
- 4.Holt GW; Haltiwanger RS; Torres C-R; Hart GW J. Biol. Chem 1987, 262, 14847–14850. [PubMed] [Google Scholar]
- 5.Dong L-Y; Xu Z-S; Chevrier MR; Cotter RJ;Cleveland DW; Hart GW J. Biol. Chem 1993, 268, 16679–16687. [PubMed] [Google Scholar]
- 6.Chou T-Y; Dang CV; Hart GW Proc. Natl. Acad. Sci. U.S.A 1995, 92, 4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lubas WA; Hanover JA J. Biol. Chem 2000, 275, 10983–10988. [DOI] [PubMed] [Google Scholar]
- 8.Meikrantz W; Smith DM; Sladicka MM; Schlegel RA J. Cell Sci 1991, 98, 303–307. [DOI] [PubMed] [Google Scholar]
- 9.Walgren JL; Vincent TS; Schey KL; Buse MG Am. J. Physiol.: Endocrinol. Metab 2003, 284, E424–E434. [DOI] [PubMed] [Google Scholar]
- 10.Roquemore EP; Chevrier MR; Cotter RJ; Hart GW Biochemistry 1996, 35, 3578–3586. [DOI] [PubMed] [Google Scholar]
- 11.Lefebvre T; Cieniewsi C; Lemoine J; Guerardel Y;Leroy Y; Zanetta JP; Michalski JC Biochem. J 2001, 360, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kearse KP; Hart GW Proc. Natl. Acad. Sci. U.S.A 1991, 88, 1701–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou CF; Smith AJ; Omary MB J. Biol. Chem 1992, 267, 3901–3906. [PubMed] [Google Scholar]
- 14.Ku NO; Omary MB Exp. Cell Res 1994, 211, 24–35. [DOI] [PubMed] [Google Scholar]
- 15.Favreau C; Worman HJ; Wozniak RW; Frappier T; Courvalin JC Biochemistry 1996, 35, 8035–8044. [DOI] [PubMed] [Google Scholar]
- 16.Cheng X; Cole RN; Zaia J; Hart GW Biochemistry 2000, 39, 11609–11620. [DOI] [PubMed] [Google Scholar]
- 17.Comer FI; Hart GW Biochemistry 2001, 40, 7845–7852. [DOI] [PubMed] [Google Scholar]
- 18.Chou TY; Hart GW; Dang CV J. Biol. Chem 1995, 270, 18961–18965. [DOI] [PubMed] [Google Scholar]
- 19.McClain DA; Lubas WA; Cooksey RC; Hazel M;Parker GJ; Love DC; Hanover JA Proc. Natl. Acad. Sci. U.S.A 2002, 99, 10695–10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw P; Freeman J; Bovey R; Iggo R. Oncogene 1996, 12, 921–930. [PubMed] [Google Scholar]
- 21.Arnold CS; Johnson GV; Cole RN; Dong DL;Lee M; Hart GW J. Biol. Chem 1996, 271, 28741–28744. [DOI] [PubMed] [Google Scholar]
- 22.Vosseller K; Wells L; Lane MD; Hart GW Proc. Natl. Acad. Sci. U.S.A 2002, 99, 5313–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haltiwanger RS; Blomberg MA; Hart GW J. Biol. Chem 1992, 267, 9005–9013. [PubMed] [Google Scholar]
- 24.Hartweck LM; Scott CL; Olszewski NE Genetics 2002, 161, 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lubas WA; Frank DW; Krause M; Hanover JA J. Biol. Chem 1997, 272, 9316–9324. [DOI] [PubMed] [Google Scholar]
- 26.Kreppel LK; Hart GW J. Biol. Chem 1999, 274, 32015–32022. [DOI] [PubMed] [Google Scholar]
- 27.Gao Y; Wells L; Comer FI; Parker GJ; Hart GW J. Biol. Chem 2001, 276, 9838–9845. [DOI] [PubMed] [Google Scholar]
- 28.Wrabl JO; Grishin NV J. Mol. Biol 2001, 314, 365–374. [DOI] [PubMed] [Google Scholar]
- 29.Dong DL; Hart GW J. Biol. Chem 1994, 269, 19321–19330. [PubMed] [Google Scholar]
- 30.Jinek M; Rehwinkel J; Lazarus BD; Izaurralde E;Hanover JA; Conti E. Nat. Struct. Mol. Biol 2004, 11, 1001–1007. [DOI] [PubMed] [Google Scholar]
- 31.Braidman I; Carroll M; Dance N; Robinson D;Poenaru L; Weber A; Dreyfus JC; Overdijk B; Hooghwinkel GJ FEBS Lett. 1974, 41, 181–184. [DOI] [PubMed] [Google Scholar]
- 32.Overdijk B; Van der Kroef WM; Van Steijn GJ;Lisman JJ Biochim. Biophys. Acta 1981, 659, 255–266. [DOI] [PubMed] [Google Scholar]
- 33.Heckel D; Comtesse N; Brass N; Blin N; Zang KD; Meese E. Hum. Mol. Genet 1998, 7, 1859–1872. [DOI] [PubMed] [Google Scholar]
- 34.Comtesse N; Maldener E; Meese E. Biochem. Biophys. Res. Commun 2001, 283, 634–640. [DOI] [PubMed] [Google Scholar]
- 35.Schultz J; Pils B. FEBS Lett. 2002, 529, 179–182. [DOI] [PubMed] [Google Scholar]
- 36.Toleman C; Paterson AJ; Whisenhunt TR; Kudlow JE J. Biol. Chem 2004, 279, 53665–53673. [DOI] [PubMed] [Google Scholar]
- 37.Macauley MS; Whitworth GE; Debowski A; Chin D; Vocadlo DJ J. Biol. Chem 2005, 280, 25313–25322. [DOI] [PubMed] [Google Scholar]
- 38.Hanover JA FASEB J. 2001, 15, 1865–1876. [DOI] [PubMed] [Google Scholar]
- 39.Rigden DJ; Jedrzejas MJ; de Mello LV FEBS Lett. 2003, 544, 103–111. [DOI] [PubMed] [Google Scholar]
- 40.Wells L; Gao Y; Mahoney JA; Voseller K; Chen C; Rosen A; Hart GW J. Biol. Chem 2002, 277, 1755–1761. [DOI] [PubMed] [Google Scholar]
- 41.Glasgow LR; Paulson JA; Hill RL J. Biol. Chem 1977, 252, 8615–8623. [PubMed] [Google Scholar]
- 42.Yegorov AM; Markaryan AN; Vozniy YV;Cherednikova TV; Demcheva MV; Berezin IV Anal. Lett 1988, 21, 193–209. [Google Scholar]
- 43.Rakhmanova VA; MacNonald RC Anal. Biochem 1998, 257, 234–237. [DOI] [PubMed] [Google Scholar]
- 44.MacGregor GR; Nolan GP; Fiering S; Roederer M; Herzenberg LA In Gene Expression in Vivo; Murray, Walker EJ, Eds JM; Humana Press: Clifton, NJ, 1990; pp 217–236. [Google Scholar]
- 45.Young DC; Kingsley SD; Ryan KA; Dutko FJ Anal. Biochem 1993, 215, 24–30. [DOI] [PubMed] [Google Scholar]
- 46.Berg M; Undisz K; Thiericke R; Moore T; Posten CJ Biomol. Screen 2000, 5, 71–76. [DOI] [PubMed] [Google Scholar]
- 47.Kasai K; Uchida R; Yamaji N. Chem. Pharm. Bull 1993, 41, 314–318. [DOI] [PubMed] [Google Scholar]
- 48.Arias EB; Kim J; Cartee GD Diabetes 2004, 53, 921–930. [DOI] [PubMed] [Google Scholar]
- 49.Park SY; Ryu J; Lee W. Exp. Mol. Med 2005, 37, 220–229. [DOI] [PubMed] [Google Scholar]
- 50.Arias EB; Cartee GD Acta Physiol. Scand 2005, 183, 281–289. [DOI] [PubMed] [Google Scholar]
- 51.Perreira M; Kim EJ; Thomas CJ; Hanover JA Bioorg. Med. Chem 2006, 14, 837–846. [DOI] [PubMed] [Google Scholar]
- 52.Lehman DM; Fu DJ; Freeman AB; Hunt KJ;Leach RJ; Johnson-Pais T; Hamlington J; Dyer TD; Arya R; Abboud H; Goring HH; Duggirala R; Blangero J; Konrad RJ; Stern MP Diabetes 2005, 54, 1214–1221. [DOI] [PubMed] [Google Scholar]
- 53.Horton D. In Methods in Carbohydrate Chemistry; Whistler, BeMiller RL, Eds JN; Academic: New York, 1972; pp 282–285. [Google Scholar]
- 54.Kang DO; Lee G; Kim EJ; Hanover JA, inpreparation. [Google Scholar]