In the crystal, pairs of molecules are linked by N—H⋯N hydrogen bonds, forming
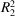 (8) ring motifs. Dimers are connected by N—H⋯O hydrogen bonds, forming layers parallel to the (102) plane. These layers are connected by C—H⋯π and C=O⋯π interactions, consolidating the molecular packing.
(8) ring motifs. Dimers are connected by N—H⋯O hydrogen bonds, forming layers parallel to the (102) plane. These layers are connected by C—H⋯π and C=O⋯π interactions, consolidating the molecular packing.
Keywords: crystal structure, thiazole derivatives, hydrogen bonds, dimers, Hirshfeld surface analysis
Abstract
In the title compound, C6H8N2OS, all atoms except for the methyl H atoms are coplanar, with a maximum deviation of 0.026 (4) Å. In the crystal, pairs of molecules are linked by N—H⋯N hydrogen bonds, forming R 2 2(8) ring motifs. Dimers are connected by N—H⋯O hydrogen bonds, forming layers parallel to the (102) plane. Consolidating the molecular packing, these layers are connected by C—H⋯π interactions between the center of the 1,3-thiazole ring and the H atom of the methyl group attached to it, as well as C=O⋯π interactions between the center of the 1,3-thiazole ring and the O atom of the carboxyl group. According to a Hirshfeld surface study, H⋯H (37.6%), O⋯H/H⋯O (16.8%), S⋯H/H⋯S (15.4%), N⋯H/H⋯N (13.0%) and C⋯H/H⋯C (7.6%) interactions are the most significant contributors to the crystal packing.
1. Chemical context
Heterocyclic aromatic systems are the most important and manifold compounds in organic chemistry (Maharramov et al., 2011b
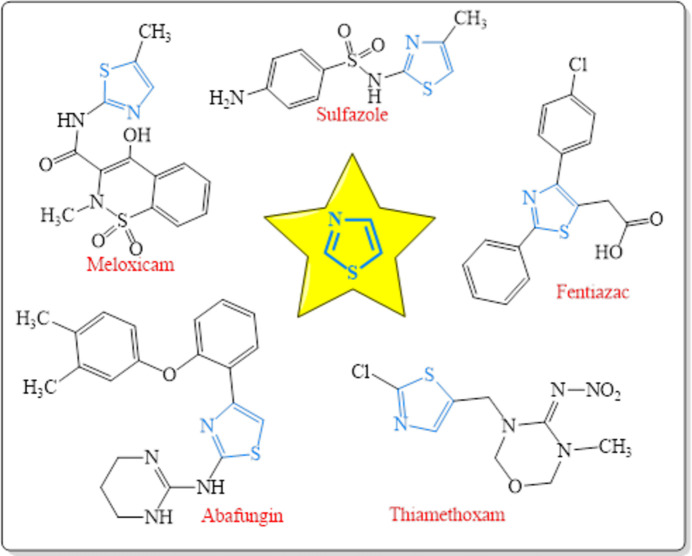
▸; Abdelhamid et al., 2014 ▸). Organic synthesis is developing intensely with newer aromatic heterocyclic compounds that are obtained for diverse medicinal and commercial purposes (Khalilov et al., 2021 ▸). Nowadays, applications of five- and six-membered ring heterocycles have expanded in different branches of chemistry, including sustainable chemistry (Montes et al., 2018 ▸), drug design and development (Tas et al., 2023 ▸) and materials sciences (Yin et al., 2020 ▸). The thiazole core is the most common five-membered heteroaromatic ring system in azole heterocycles (Yadigarov et al., 2009 ▸; Khalilov, 2021 ▸). Thiazoles have potent medicinal applications as it is an essential core scaffold present in many natural (thiamine, penicillin) and synthetic medicinally important compounds (Chhabria et al., 2016 ▸) such as sulfazole, ritonavir, abafungin, fanetizole, meloxicam, fentiazac, nizatidine, thiamethoxam, etc. (Fig. 1 ▸). On the other hand, there have been a variety of significant examples of thiazole derivatives used as target products as well as synthetic intermediates (Akkurt et al., 2018 ▸; Kekeçmuhammed et al., 2022 ▸).
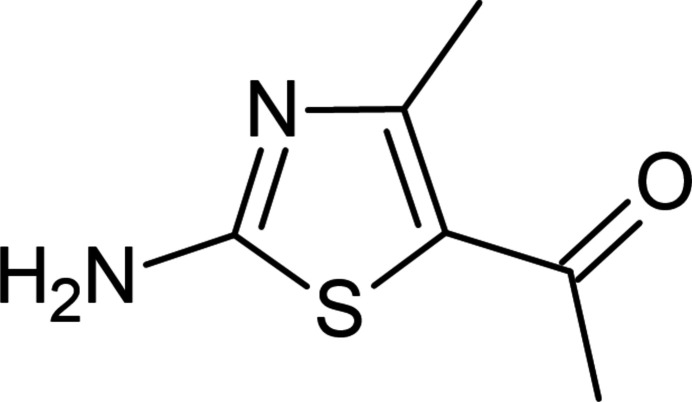
Figure 1.
Some marketed drugs containing the thiazole moiety.
In a continuation of our investigations of heterocyclic systems with biological activity and in the framework of ongoing structural studies (Maharramov et al., 2011a ▸; Askerov et al., 2020 ▸; Karimli et al., 2023 ▸), we report here the crystal structure and Hirshfeld surface analysis of the title compound, 1-(2-amino-4-methyl-1,3-thiazol-5-yl)ethan-1-one.
2. Structural commentary
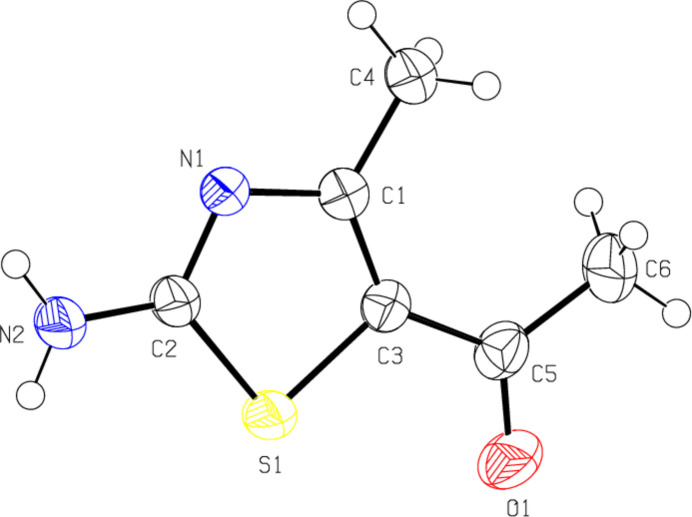
In the title compound, Fig. 2 ▸, all atoms except for the methyl H atoms are coplanar, with a maximum deviation of 0.026 (4) Å for C6. The geometric parameters of the title compound are normal and comparable to those of related compounds listed in the Database survey section.
Figure 2.
The molecular structure of the title compound, showing the atom labeling and displacement ellipsoids drawn at the 50% probability level.
3. Supramolecular features and Hirshfeld surface analysis
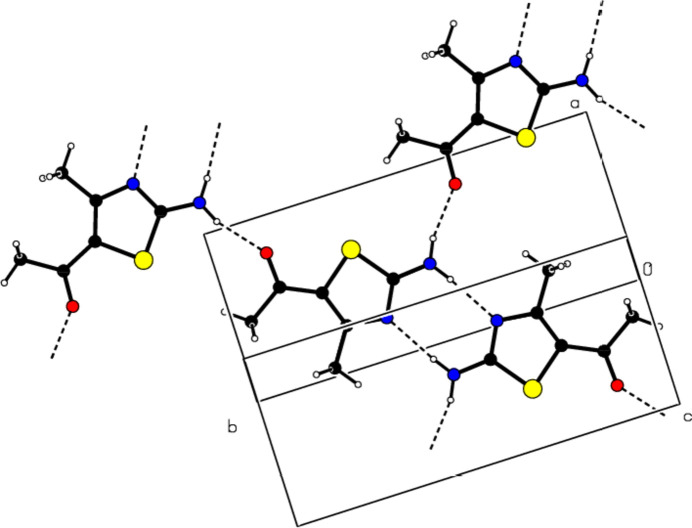
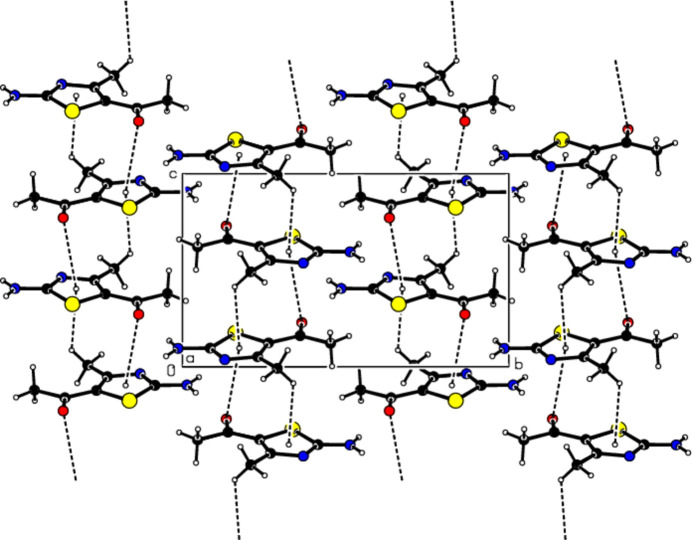
In the crystal, pairs of molecules are linked by N—H⋯N hydrogen bonds, forming
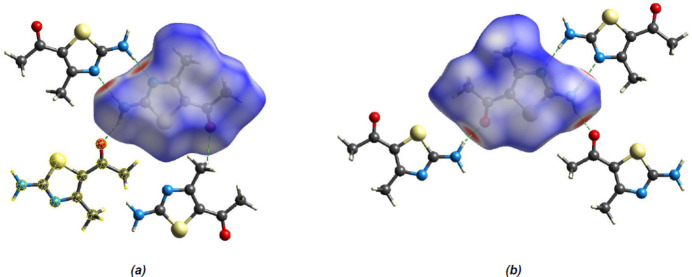
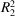 (8) ring motifs (Bernstein et al., 1995 ▸; Table 1 ▸, Fig. 3 ▸). Dimers are connected by N—H⋯O hydrogen bonds, forming layers parallel to the (102) plane (Table 1 ▸, Fig. 4 ▸). Consolidating the molecular packing, these layers are connected by C—H⋯π interactions between the center of the 1,3-thiazole ring and the H atom of the methyl group attached to it, as well as C=O⋯π interactions between the center of the 1,3-thiazole ring and the O atom of the carboxyl group (Table 1 ▸, Figs. 5 ▸ and 6 ▸).
(8) ring motifs (Bernstein et al., 1995 ▸; Table 1 ▸, Fig. 3 ▸). Dimers are connected by N—H⋯O hydrogen bonds, forming layers parallel to the (102) plane (Table 1 ▸, Fig. 4 ▸). Consolidating the molecular packing, these layers are connected by C—H⋯π interactions between the center of the 1,3-thiazole ring and the H atom of the methyl group attached to it, as well as C=O⋯π interactions between the center of the 1,3-thiazole ring and the O atom of the carboxyl group (Table 1 ▸, Figs. 5 ▸ and 6 ▸).
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 is the centroid of the (N1/S1/C1–C3) 1,3-thiazole ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H1A⋯N1i | 0.86 | 2.11 | 2.963 (4) | 175 |
| N2—H1B⋯O1ii | 0.86 | 2.02 | 2.835 (4) | 158 |
| C4—H4B⋯Cg1iii | 0.96 | 2.89 | 3.603 (4) | 132 |
Symmetry codes: (i)
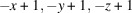 ; (ii)
; (ii)
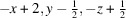 ; (iii)
; (iii)
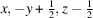 .
.
Figure 3.
Partial view of the N—H⋯N and N—H⋯O bonds in the (102) plane of the title compound.
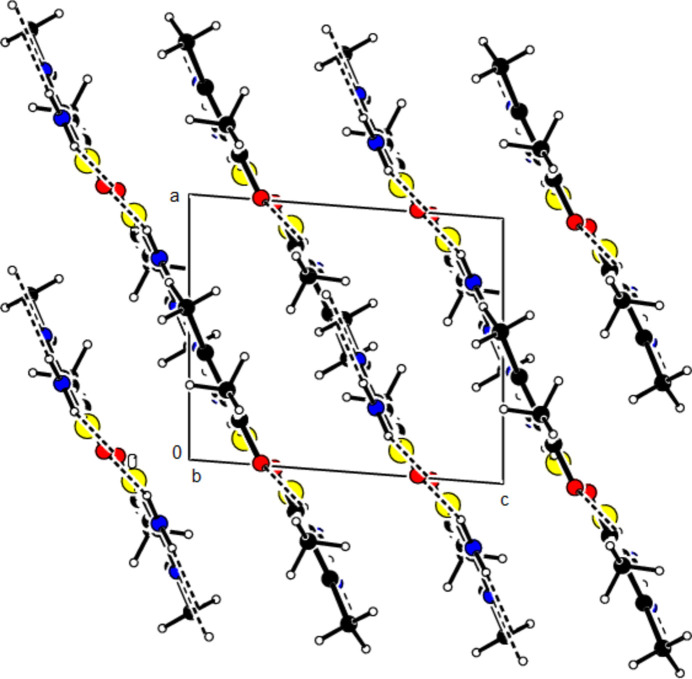
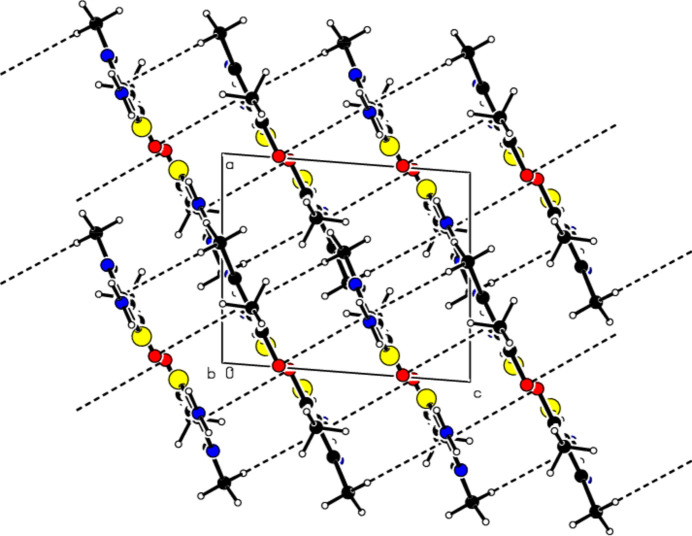
Figure 4.
View of the packing of the title compound along the b-axis.
Figure 5.
View of the C—H⋯π and C=O⋯π interactions of the title compound down the a axis.
Figure 6.
View of the C—H⋯π and C=O⋯π interactions of the title compound down the b axis.
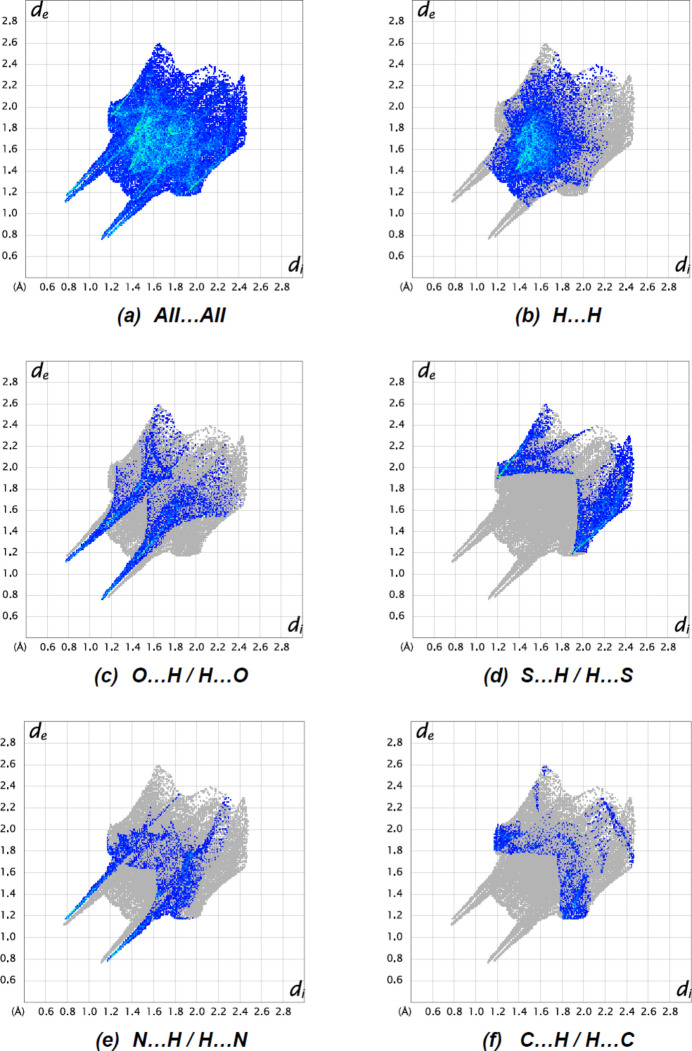
Crystal Explorer 17.5 (Spackman et al., 2021 ▸) was used to generate Hirshfeld surfaces and two-dimensional fingerprint plots in order to quantify the intermolecular interactions in the crystal. The Hirshfeld surfaces were mapped over d norm in the range −0.5624 (red) to 0.9850 (blue) a.u. (Fig. 7 ▸). The interactions given in Table 2 ▸ play a key role in the molecular packing of the title compound. The most important interatomic contact is H⋯H as it makes the highest contribution to the crystal packing (37.6%, Fig. 8 ▸ b). Other major contributors are O⋯H/H⋯O (16.8%, Fig. 8 ▸ c), S⋯H/H⋯S (15.4%, Fig. 8 ▸ d), N⋯H/H⋯N (13.0%, Fig. 8 ▸ e) and C⋯H/H⋯C (7.6%, Fig. 8 ▸ f) interactions. Other, smaller contributions are made by S⋯C/C⋯S (2.7%), C⋯O/O⋯C (2.6%), C⋯C (1.8%), N⋯C/C⋯N (1.5%), S⋯O/O⋯S (0.8%), S⋯N/N⋯S (0.1%) and O⋯N/N⋯O (0.1%) interactions.
Figure 7.
(a) Front and (b) back sides of the three-dimensional Hirshfeld surface of the title compound mapped over d norm, with a fixed color scale of −0.5624 to 0.9850 a.u.
Table 2. Summary of short interatomic contacts (Å) in the title compound.
| O1⋯H4A | 2.69 | 1 + x, y, z |
| O1⋯H1B | 2.02 | 2 − x,
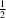 + y,
+ y,
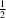 − z
− z
|
| C1⋯H4B | 3.09 |
x,
 − y, −
− y, −
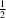 + z
+ z
|
| H1A⋯N1 | 2.11 | 1 − x, 1 − y, 1 − z |
| N2⋯H6B | 2.89 | 1 − x, −
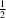 + y,
+ y,
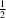 − z
− z
|
Figure 8.
The two-dimensional fingerprint plots of the title compound, showing (a) all interactions, and delineated into (b) H⋯H, (c) O⋯H/H⋯O, (d) S⋯H/H⋯S, (e) N⋯H/H⋯N and (f) C⋯H/H⋯C interactions. [d e and d i represent the distances from a point on the Hirshfeld surface to the nearest atoms outside (external) and inside (internal) the surface, respectively].
4. Database survey
A search of the Cambridge Structural Database (CSD, Version 5.43, last update November 2022; Groom et al., 2016 ▸) for the central five-membered ring 1,3-thiazole yielded five compounds related to the title compound, viz. CSD refcodes IXAMAV (Kennedy et al., 2004a ▸), ABEGAQ (Kennedy et al., 2004b ▸), FEFKUY (Hazra et al., 2012 ▸), DUTZEY (Chen & Xu, 2010 ▸) and LAMQOJ (Fait et al., 2021 ▸).
In the crystal of IXAMAV, the supramolecular network is based upon N—H⋯N hydrogen-bonded centrosymmetric dimers linked by N—H⋯O contacts. ABEGAQ forms a supramolecular network based on N—H⋯N hydrogen-bonded centrosymmetric dimers that are linked in turn by N—H⋯O contacts. In the crystal of FEFKUY, an interplay of O—H⋯N and C—H⋯O hydrogen bonds connects the molecules to form C(6)
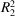 (8) polymeric chains, which are further linked via weak C—H⋯O hydrogen bonds into a two-dimensional supramolecular framework. The crystal structure of DUTZEY involves intermolecular N—H⋯N hydrogen bonds. In the crystal of LAMQOJ, weak C—H⋯N hydrogen bonds build up a wavy layer of molecules in the (011) plane. The layers are stacked in the [100] direction by weak π–π stacking interactions between the 1,3-thiazole rings.
(8) polymeric chains, which are further linked via weak C—H⋯O hydrogen bonds into a two-dimensional supramolecular framework. The crystal structure of DUTZEY involves intermolecular N—H⋯N hydrogen bonds. In the crystal of LAMQOJ, weak C—H⋯N hydrogen bonds build up a wavy layer of molecules in the (011) plane. The layers are stacked in the [100] direction by weak π–π stacking interactions between the 1,3-thiazole rings.
5. Synthesis and crystallization
The title compound was synthesized using a reported procedure (Donald et al., 2012 ▸), and colorless crystals were obtained upon recrystallization from an ethanol/water (3:1) solution at room temperature.
6. Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. All H atoms were placed in calculated positions (C—H = 0.96 Å and N—H = 0.86 Å) and refined as riding with U iso(H) = 1.2U eq(N) for the NH2 group and 1.5U eq(C) for CH3 groups.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | C6H8N2OS |
| M r | 156.20 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 296 |
| a, b, c (Å) | 6.7445 (15), 13.498 (3), 8.010 (2) |
| β (°) | 94.421 (7) |
| V (Å3) | 727.1 (3) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.37 |
| Crystal size (mm) | 0.60 × 0.45 × 0.35 |
| Data collection | |
| Diffractometer | Bruker APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸ |
| T min, T max | 0.649, 0.745 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 14701, 1492, 940 |
| R int | 0.144 |
| (sin θ/λ)max (Å−1) | 0.626 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.050, 0.142, 1.04 |
| No. of reflections | 1492 |
| No. of parameters | 93 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.29, −0.28 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989023007181/vm2288sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989023007181/vm2288Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989023007181/vm2288Isup3.cml
CCDC reference: 2288949
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
This study was supported by Baku State University, Erciyes University, Tribhuvan University and the Universidad de Antofagasta. Authors’ contributions are as follows. Conceptualization, EZH, KAA and AMM; methodology, EZH, IB and MA; investigation, EZH and IB; writing (original draft), MA and AB; writing (review and editing of the manuscript), MA and EZH; visualization, MA, RMR and IB; funding acquisition, EZH, AB and IB; resources, AB, IB and MA; supervision, MA and AMM.
supplementary crystallographic information
Crystal data
| C6H8N2OS | F(000) = 328 |
| Mr = 156.20 | Dx = 1.427 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 6.7445 (15) Å | Cell parameters from 2051 reflections |
| b = 13.498 (3) Å | θ = 3.0–26.4° |
| c = 8.010 (2) Å | µ = 0.37 mm−1 |
| β = 94.421 (7)° | T = 296 K |
| V = 727.1 (3) Å3 | Prism, colourless |
| Z = 4 | 0.60 × 0.45 × 0.35 mm |
Data collection
| Bruker APEXII CCD diffractometer | 940 reflections with I > 2σ(I) |
| φ and ω scans | Rint = 0.144 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015 | θmax = 26.4°, θmin = 3.0° |
| Tmin = 0.649, Tmax = 0.745 | h = −8→8 |
| 14701 measured reflections | k = −16→16 |
| 1492 independent reflections | l = −10→10 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.050 | H-atom parameters constrained |
| wR(F2) = 0.142 | w = 1/[σ2(Fo2) + (0.0616P)2 + 0.4474P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.04 | (Δ/σ)max < 0.001 |
| 1492 reflections | Δρmax = 0.29 e Å−3 |
| 93 parameters | Δρmin = −0.28 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.5920 (5) | 0.7303 (2) | 0.4486 (4) | 0.0354 (8) | |
| C2 | 0.7302 (5) | 0.5848 (2) | 0.4036 (4) | 0.0356 (8) | |
| C3 | 0.7583 (5) | 0.7636 (2) | 0.3769 (4) | 0.0349 (8) | |
| C4 | 0.4274 (5) | 0.7905 (3) | 0.5084 (5) | 0.0446 (9) | |
| H4A | 0.356539 | 0.822440 | 0.414759 | 0.067* | |
| H4B | 0.480953 | 0.839710 | 0.585759 | 0.067* | |
| H4C | 0.338231 | 0.748209 | 0.563299 | 0.067* | |
| C5 | 0.8348 (5) | 0.8605 (3) | 0.3393 (4) | 0.0408 (9) | |
| C6 | 0.7269 (6) | 0.9538 (3) | 0.3790 (5) | 0.0567 (11) | |
| H6A | 0.718170 | 0.958584 | 0.497742 | 0.085* | |
| H6B | 0.595477 | 0.952624 | 0.323674 | 0.085* | |
| H6C | 0.798079 | 1.010036 | 0.340806 | 0.085* | |
| N1 | 0.5761 (4) | 0.62959 (19) | 0.4631 (4) | 0.0359 (7) | |
| N2 | 0.7505 (4) | 0.4865 (2) | 0.4051 (4) | 0.0498 (9) | |
| H1A | 0.661573 | 0.449933 | 0.445684 | 0.060* | |
| H1B | 0.852593 | 0.459698 | 0.365376 | 0.060* | |
| S1 | 0.90402 (13) | 0.66254 (6) | 0.32437 (12) | 0.0412 (3) | |
| O1 | 0.9925 (4) | 0.8657 (2) | 0.2713 (4) | 0.0571 (8) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0349 (18) | 0.0323 (17) | 0.039 (2) | 0.0005 (15) | 0.0045 (15) | −0.0011 (15) |
| C2 | 0.0348 (18) | 0.0287 (16) | 0.044 (2) | 0.0034 (14) | 0.0063 (15) | 0.0031 (15) |
| C3 | 0.0343 (18) | 0.0308 (17) | 0.040 (2) | −0.0023 (14) | 0.0035 (15) | 0.0009 (15) |
| C4 | 0.042 (2) | 0.0320 (18) | 0.061 (2) | 0.0032 (16) | 0.0105 (18) | −0.0005 (17) |
| C5 | 0.0410 (19) | 0.0351 (19) | 0.046 (2) | −0.0044 (15) | 0.0016 (17) | 0.0066 (15) |
| C6 | 0.066 (3) | 0.0313 (19) | 0.075 (3) | 0.0012 (19) | 0.015 (2) | 0.0054 (19) |
| N1 | 0.0318 (14) | 0.0294 (14) | 0.0476 (18) | −0.0007 (11) | 0.0093 (13) | 0.0001 (13) |
| N2 | 0.0450 (18) | 0.0295 (16) | 0.078 (2) | 0.0006 (13) | 0.0263 (17) | 0.0004 (15) |
| S1 | 0.0371 (5) | 0.0341 (5) | 0.0547 (6) | 0.0002 (4) | 0.0172 (4) | 0.0035 (4) |
| O1 | 0.0474 (16) | 0.0460 (16) | 0.080 (2) | −0.0083 (12) | 0.0182 (15) | 0.0148 (14) |
Geometric parameters (Å, º)
| C1—N1 | 1.370 (4) | C4—H4B | 0.9600 |
| C1—C3 | 1.375 (4) | C4—H4C | 0.9600 |
| C1—C4 | 1.484 (5) | C5—O1 | 1.234 (4) |
| C2—N1 | 1.322 (4) | C5—C6 | 1.501 (5) |
| C2—N2 | 1.334 (4) | C6—H6A | 0.9600 |
| C2—S1 | 1.730 (3) | C6—H6B | 0.9600 |
| C3—C5 | 1.446 (5) | C6—H6C | 0.9600 |
| C3—S1 | 1.752 (3) | N2—H1A | 0.8600 |
| C4—H4A | 0.9600 | N2—H1B | 0.8600 |
| N1—C1—C3 | 115.6 (3) | O1—C5—C3 | 118.5 (3) |
| N1—C1—C4 | 116.8 (3) | O1—C5—C6 | 119.6 (3) |
| C3—C1—C4 | 127.6 (3) | C3—C5—C6 | 121.9 (3) |
| N1—C2—N2 | 122.3 (3) | C5—C6—H6A | 109.5 |
| N1—C2—S1 | 115.4 (2) | C5—C6—H6B | 109.5 |
| N2—C2—S1 | 122.3 (2) | H6A—C6—H6B | 109.5 |
| C1—C3—C5 | 134.3 (3) | C5—C6—H6C | 109.5 |
| C1—C3—S1 | 109.7 (2) | H6A—C6—H6C | 109.5 |
| C5—C3—S1 | 116.0 (2) | H6B—C6—H6C | 109.5 |
| C1—C4—H4A | 109.5 | C2—N1—C1 | 110.7 (3) |
| C1—C4—H4B | 109.5 | C2—N2—H1A | 120.0 |
| H4A—C4—H4B | 109.5 | C2—N2—H1B | 120.0 |
| C1—C4—H4C | 109.5 | H1A—N2—H1B | 120.0 |
| H4A—C4—H4C | 109.5 | C2—S1—C3 | 88.60 (15) |
| H4B—C4—H4C | 109.5 | ||
| N1—C1—C3—C5 | −179.3 (4) | N2—C2—N1—C1 | 179.3 (3) |
| C4—C1—C3—C5 | 2.0 (7) | S1—C2—N1—C1 | −0.5 (4) |
| N1—C1—C3—S1 | 0.0 (4) | C3—C1—N1—C2 | 0.3 (5) |
| C4—C1—C3—S1 | −178.7 (3) | C4—C1—N1—C2 | 179.2 (3) |
| C1—C3—C5—O1 | −179.5 (4) | N1—C2—S1—C3 | 0.4 (3) |
| S1—C3—C5—O1 | 1.3 (5) | N2—C2—S1—C3 | −179.4 (3) |
| C1—C3—C5—C6 | −0.1 (7) | C1—C3—S1—C2 | −0.2 (3) |
| S1—C3—C5—C6 | −179.4 (3) | C5—C3—S1—C2 | 179.2 (3) |
Hydrogen-bond geometry (Å, º)
Cg1 is the centroid of the (N1/S1/C1–C3) 1,3-thiazole ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H1A···N1i | 0.86 | 2.11 | 2.963 (4) | 175 |
| N2—H1B···O1ii | 0.86 | 2.02 | 2.835 (4) | 158 |
| C4—H4B···Cg1iii | 0.96 | 2.89 | 3.603 (4) | 132 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+2, y−1/2, −z+1/2; (iii) x, −y+1/2, z−1/2.
References
- Abdelhamid, A. A., Mohamed, S. K., Maharramov, A. M., Khalilov, A. N. & Allahverdiev, M. A. (2014). J. Saudi Chem. Soc. 18, 474–478.
- Akkurt, M., Duruskari, G. S., Toze, F. A. A., Khalilov, A. N. & Huseynova, A. T. (2018). Acta Cryst. E74, 1168–1172. [DOI] [PMC free article] [PubMed]
- Askerov, R. K., Maharramov, A. M., Khalilov, A. N., Akkurt, M., Akobirshoeva, A. A., Osmanov, V. K. & Borisov, A. V. (2020). Acta Cryst. E76, 1007–1011. [DOI] [PMC free article] [PubMed]
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Bruker (2016). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin. USA.
- Chen, X. & Xu, L. (2010). Acta Cryst. E66, o2148. [DOI] [PMC free article] [PubMed]
- Chhabria, M. T., Patel, S., Modi, P. & Brahmkshatriya, P. S. (2016). Curr. Top. Med. Chem. 16, 2841–2862. [DOI] [PubMed]
- Donald, M. B., Rodriguez, K. X., Shay, H., Phuan, P.-W., Verkman, A. S. & Kurth, M. J. (2012). Bioorg. Med. Chem. 20, 5247–5253. [DOI] [PMC free article] [PubMed]
- Fait, M. J. G., Spannenberg, A., Kondratenko, E. V. & Linke, D. (2021). IUCrData, 6, x211332. [DOI] [PMC free article] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hazra, D. K., Mukherjee, M., Helliwell, M. & Mukherjee, A. K. (2012). Acta Cryst. C68, o452–o455. [DOI] [PubMed]
- Karimli, E. G., Khrustalev, V. N., Kurasova, M. N., Akkurt, M., Khalilov, A. N., Bhattarai, A. & Mamedov, İ. G. (2023). Acta Cryst. E79, 474–477. [DOI] [PMC free article] [PubMed]
- Kekeçmuhammed, H., Tapera, M., Tüzün, B., Akkoç, S., Zorlu, Y. & Sarıpınar, E. (2022). ChemistrySelect, 7, e202201502.
- Kennedy, A. R., Khalaf, A. I., Suckling, C. J. & Waigh, R. D. (2004a). Acta Cryst. E60, o1188–o1190.
- Kennedy, A. R., Khalaf, A. I., Suckling, C. J. & Waigh, R. D. (2004b). Acta Cryst. E60, o1510–o1512.
- Khalilov, A. N. (2021). Rev. Roum. Chim. 66, 719–723.
- Khalilov, A. N., Tüzün, B., Taslimi, P., Tas, A., Tuncbilek, Z. & Cakmak, N. K. (2021). J. Mol. Liq. 344, 117761.
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- Maharramov, A. M., Khalilov, A. N., Gurbanov, A. V., Allahverdiyev, M. A. & Ng, S. W. (2011a). Acta Cryst. E67, o721. [DOI] [PMC free article] [PubMed]
- Maharramov, A. M., Khalilov, A. N., Gurbanov, A. V. & Brito, I. (2011b). Acta Cryst. E67, o1307. [DOI] [PMC free article] [PubMed]
- Montes, V., Miñambres, J. F., Khalilov, A. N., Boutonnet, M., Marinas, J. M., Urbano, F. J., Maharramov, A. M. & Marinas, A. (2018). Catal. Today, 306, 89–95.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spackman, P. R., Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Jayatilaka, D. & Spackman, M. A. (2021). J. Appl. Cryst. 54, 1006–1011. [DOI] [PMC free article] [PubMed]
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Tas, A., Tüzün, B., Khalilov, A. N., Taslimi, P., Ağbektas, T. & Cakmak, N. K. (2023). J. Mol. Struct. 1273, 134282.
- Yadigarov, R. R., Khalilov, A. N., Mamedov, I. G., Nagiev, F. N., Magerramov, A. M. & Allakhverdiev, M. A. (2009). Russ. J. Org. Chem. 45, 1856–1858.
- Yin, J., Khalilov, A. N., Muthupandi, P., Ladd, R. & Birman, V. B. (2020). J. Am. Chem. Soc. 142, 60–63. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989023007181/vm2288sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989023007181/vm2288Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989023007181/vm2288Isup3.cml
CCDC reference: 2288949
Additional supporting information: crystallographic information; 3D view; checkCIF report