The title hydrated salt features a dense array of hydrogen bonds, forming a three-dimensional network.
Keywords: crystal structure, hydrogen bonding, hydrated salt
Abstract
In the title hydrated salt, C5H5Cl2N2
+·C6H4NO3
−·H2O, the pyridine N atom of the cation is protonated and an intramolecular O—H⋯O hydrogen bond is observed in the anion, which generates an S(6) ring. The crystal packing features N—H⋯N, O—H⋯O, N—H⋯O, C—H⋯Cl and C—H⋯O hydrogen bonds, which generate a three-dimensional network.
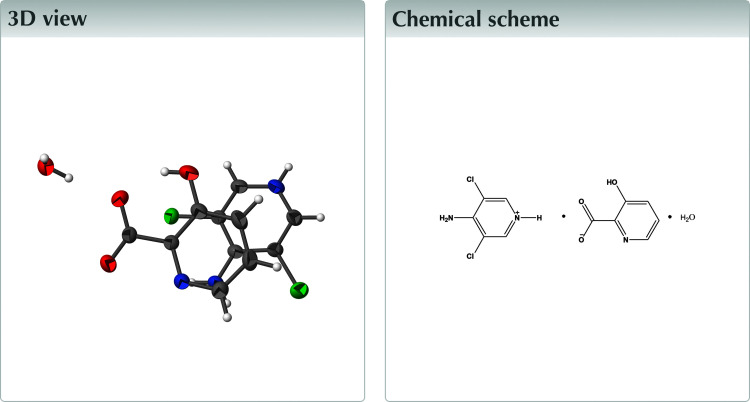
Structure description
4-Aminopyridine and its derivatives are used clinically to treat Lambert–Eaton myasthenic syndrome and multiple sclerosis because they block potassium channels, which prolongs action potentials and increases transmitter release at the neuromuscular junction (Judge & Bever, 2006 ▸). Picolinic acid, which contains N and O donors, has attracted much attention for the design and synthesis of self-assembling systems (e.g., Steiner, 2002 ▸). In this regard, 3-hydroxypicolinic acid is of interest because it can be used as a neutral ligand or, depending on the pH value, as an anionic or cationic ligand. In addition, due to the arrangement of its functional groups, it can act as a monodentate or bidentate ligand, which allows it to form five- or six-membered chelate rings. As part of our work in this area, we now report the synthesis and structure of the title hydrated molecular salt.
The asymmetric unit (Fig. 1 ▸) of the title salt contains a 4-amino-3,5-dichloropyridinium cation, a 3-hydroxy picolinate anion and a water molecule. The pyridinium cation is essentially planar, with a maximum deviation of 0.010 (2) Å for atom C2. A wider than normal angle [C5—N1—C1 = 120.41 (12)°] is subtended at the protonated N1 atom. In the anion, a typical intramolecular O—H⋯O hydrogen bond, which generates an S(6) ring, is seen. In the extended structure, the cations, anions and water molecules are connected by N—H⋯N, O—H⋯O, C—H⋯Cl, N—H⋯O and C—H⋯O hydrogen bonds (Table 1 ▸), forming a three-dimensional network (Figs. 2 ▸ and 3 ▸).
Figure 1.
The molecular structure of the title compound showing 50% displacement ellipsoids. The intramolecular hydrogen bond is shown with dashed lines.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3⋯O2 | 0.82 | 1.79 | 2.5155 (17) | 147 |
| N1—H1⋯N3i | 0.86 | 1.90 | 2.7546 (17) | 171 |
| N2—H2A⋯O1W ii | 0.86 | 2.13 | 2.9414 (17) | 157 |
| N2—H2B⋯O1W iii | 0.86 | 2.05 | 2.8269 (17) | 149 |
| O1W—H1W⋯O2 | 0.85 | 1.90 | 2.7442 (17) | 170 |
| O1W—H2W⋯O1iv | 0.85 | 1.98 | 2.8181 (18) | 170 |
| C5—H5⋯O1i | 0.93 | 2.31 | 2.9864 (18) | 129 |
| C5—H5⋯Cl2v | 0.93 | 2.97 | 3.7363 (16) | 141 |
| C7—H7⋯O3vi | 0.93 | 2.52 | 3.399 (2) | 157 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
 ; (v)
; (v)
 ; (vi)
; (vi)
 .
.
Figure 2.
One-dimensional supramolecular hydrogen-bonded chain mediated by water molecules in the title compound.
Figure 3.
Crystal packing viewed down [100] in the title compound.
A search of the Cambridge Structural Database (Version 5.43, update November 2022; Groom et al., 2016 ▸) for the 3,5-dichloro-4-amino pyridine fragment with additional substituents yielded hexaaquamagnesium(II) bis(4-amino-3,5,6-trichloro-picolinate) tetrahydrate (CSD refcode BAWGOV; Smith et al., 1981 ▸), [(4-amino-3,5-dichloro-6-fluoropyridin-2-yl)oxy]acetic acid (EZONOY; Park et al., 2016 ▸), sodium picloramate hexahydrate (CURLIM; Smith et al., 2015 ▸), guanidinium 4-amino-3,5,6-trichloropicolinate (GUPICL10; Parthasarathi et al., 1982 ▸), and 6-chloro-3-(trifluoromethoxy)pyridine-2-carboxylic acid (MAFTEU; Manteau et al., 2010 ▸).
Synthesis and crystallization
A hot methanol solution of 3-hydroxy picolinic acid (40 mg) was mixed with a hot aqueous solution of 4-amino 3,5-dichloro pyridine (34 mg). The mixture was cooled slowly and kept at room temperature. After a few days, colourless block shaped crystals were obtained.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C5H5Cl2N2 +·C6H4NO3 −·H2O |
| M r | 320.13 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 296 |
| a, b, c (Å) | 8.4267 (19), 14.084 (3), 10.900 (2) |
| β (°) | 91.953 (8) |
| V (Å3) | 1292.9 (5) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.52 |
| Crystal size (mm) | 0.46 × 0.32 × 0.13 |
| Data collection | |
| Diffractometer | Agilent Xcalibur, Atlas, Gemini |
| Absorption correction | Multi-scan |
| T min, T max | 0.819, 0.937 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 45801, 3296, 2853 |
| R int | 0.038 |
| (sin θ/λ)max (Å−1) | 0.675 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.033, 0.091, 1.03 |
| No. of reflections | 3296 |
| No. of parameters | 185 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.24, −0.35 |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2414314623008210/hb4452sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314623008210/hb4452Isup2.hkl
CCDC reference: 2294939
Additional supporting information: crystallographic information; 3D view; checkCIF report
full crystallographic data
Crystal data
| C5H5Cl2N2+·C6H4NO3−·H2O | F(000) = 656 |
| Mr = 320.13 | Dx = 1.645 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.4267 (19) Å | Cell parameters from 3676 reflections |
| b = 14.084 (3) Å | θ = 2.5–28.8° |
| c = 10.900 (2) Å | µ = 0.52 mm−1 |
| β = 91.953 (8)° | T = 296 K |
| V = 1292.9 (5) Å3 | Plate, colourless |
| Z = 4 | 0.46 × 0.32 × 0.13 mm |
Data collection
| Agilent Xcalibur, Atlas, Gemini diffractometer | 2853 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.038 |
| ω scans | θmax = 28.7°, θmin = 2.4° |
| Absorption correction: multi-scan | h = −11→11 |
| Tmin = 0.819, Tmax = 0.937 | k = −18→18 |
| 45801 measured reflections | l = −14→14 |
| 3296 independent reflections |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.033 | H-atom parameters constrained |
| wR(F2) = 0.091 | w = 1/[σ2(Fo2) + (0.0465P)2 + 0.3959P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max < 0.001 |
| 3296 reflections | Δρmax = 0.24 e Å−3 |
| 185 parameters | Δρmin = −0.35 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. The water H atoms were located in a difference Fourier map and allowed to refine freely. The remaining H atoms were positioned geometrically (C—H = 0.93 and N—H = 0.86 Å) and were refined using a riding model, with Uiso(H) = 1.2 Ueq(C). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.85851 (4) | 0.68420 (3) | 0.15096 (4) | 0.04683 (12) | |
| Cl2 | 0.89478 (5) | 0.30311 (2) | 0.10521 (4) | 0.04737 (12) | |
| O1 | 0.42463 (12) | 0.67099 (7) | 0.27823 (11) | 0.0439 (3) | |
| O2 | 0.61861 (13) | 0.71788 (7) | 0.40818 (11) | 0.0459 (3) | |
| O3 | 0.82471 (14) | 0.60173 (8) | 0.48694 (13) | 0.0535 (3) | |
| H3 | 0.780642 | 0.653220 | 0.477230 | 0.080* | |
| N3 | 0.50360 (13) | 0.48436 (8) | 0.31752 (10) | 0.0321 (2) | |
| N1 | 1.19792 (14) | 0.49866 (8) | 0.22147 (11) | 0.0371 (3) | |
| H1 | 1.294531 | 0.500301 | 0.249479 | 0.045* | |
| N2 | 0.73770 (13) | 0.49223 (8) | 0.08094 (11) | 0.0355 (3) | |
| H2A | 0.685153 | 0.544264 | 0.072057 | 0.043* | |
| H2B | 0.694906 | 0.439155 | 0.059109 | 0.043* | |
| C10 | 0.59379 (14) | 0.55395 (9) | 0.36845 (11) | 0.0288 (3) | |
| C11 | 0.53872 (15) | 0.65512 (9) | 0.34965 (13) | 0.0326 (3) | |
| C3 | 0.88438 (14) | 0.49383 (9) | 0.12820 (11) | 0.0291 (3) | |
| C4 | 0.96128 (15) | 0.57881 (9) | 0.16585 (12) | 0.0321 (3) | |
| C6 | 0.73510 (16) | 0.53325 (10) | 0.43364 (13) | 0.0353 (3) | |
| C2 | 0.97853 (16) | 0.41096 (9) | 0.14404 (12) | 0.0325 (3) | |
| C5 | 1.11412 (16) | 0.57934 (10) | 0.21103 (13) | 0.0362 (3) | |
| H5 | 1.160941 | 0.636508 | 0.234961 | 0.043* | |
| C1 | 1.13192 (17) | 0.41529 (10) | 0.18845 (13) | 0.0373 (3) | |
| H1A | 1.191375 | 0.359836 | 0.195919 | 0.045* | |
| C9 | 0.55198 (17) | 0.39478 (10) | 0.32671 (14) | 0.0388 (3) | |
| H9 | 0.488901 | 0.347163 | 0.291583 | 0.047* | |
| C8 | 0.69351 (18) | 0.37002 (10) | 0.38699 (15) | 0.0428 (3) | |
| H8 | 0.725723 | 0.306886 | 0.390273 | 0.051* | |
| C7 | 0.78549 (17) | 0.43927 (11) | 0.44166 (15) | 0.0431 (3) | |
| H7 | 0.880028 | 0.423758 | 0.483433 | 0.052* | |
| O1W | 0.48614 (13) | 0.86692 (8) | 0.53048 (11) | 0.0441 (3) | |
| H1W | 0.518086 | 0.816660 | 0.495934 | 0.066* | |
| H2W | 0.462944 | 0.849278 | 0.602250 | 0.066* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0388 (2) | 0.03527 (19) | 0.0658 (3) | 0.00513 (13) | −0.00686 (17) | −0.01076 (16) |
| Cl2 | 0.0497 (2) | 0.03070 (18) | 0.0608 (3) | −0.00776 (14) | −0.01108 (18) | 0.00405 (15) |
| O1 | 0.0385 (5) | 0.0339 (5) | 0.0585 (7) | 0.0032 (4) | −0.0120 (5) | 0.0042 (5) |
| O2 | 0.0429 (6) | 0.0320 (5) | 0.0621 (7) | −0.0008 (4) | −0.0073 (5) | −0.0116 (5) |
| O3 | 0.0416 (6) | 0.0449 (6) | 0.0723 (8) | −0.0023 (5) | −0.0249 (6) | −0.0072 (6) |
| N3 | 0.0294 (5) | 0.0297 (5) | 0.0371 (6) | 0.0000 (4) | −0.0023 (4) | −0.0008 (4) |
| N1 | 0.0295 (5) | 0.0419 (6) | 0.0393 (6) | −0.0021 (5) | −0.0075 (5) | 0.0026 (5) |
| N2 | 0.0278 (5) | 0.0345 (6) | 0.0438 (6) | −0.0027 (4) | −0.0051 (5) | −0.0019 (5) |
| C10 | 0.0268 (6) | 0.0282 (6) | 0.0313 (6) | −0.0006 (5) | 0.0004 (5) | −0.0009 (5) |
| C11 | 0.0294 (6) | 0.0290 (6) | 0.0397 (7) | 0.0010 (5) | 0.0023 (5) | −0.0010 (5) |
| C3 | 0.0273 (6) | 0.0342 (6) | 0.0258 (6) | −0.0031 (5) | 0.0007 (5) | 0.0009 (5) |
| C4 | 0.0299 (6) | 0.0327 (6) | 0.0335 (6) | −0.0009 (5) | −0.0010 (5) | −0.0027 (5) |
| C6 | 0.0290 (6) | 0.0371 (7) | 0.0396 (7) | −0.0004 (5) | −0.0039 (5) | 0.0003 (6) |
| C2 | 0.0340 (6) | 0.0304 (6) | 0.0329 (6) | −0.0047 (5) | −0.0028 (5) | 0.0039 (5) |
| C5 | 0.0323 (6) | 0.0387 (7) | 0.0374 (7) | −0.0054 (5) | −0.0035 (5) | −0.0036 (6) |
| C1 | 0.0357 (7) | 0.0363 (7) | 0.0396 (7) | 0.0008 (5) | −0.0048 (6) | 0.0065 (6) |
| C9 | 0.0387 (7) | 0.0292 (6) | 0.0483 (8) | −0.0009 (5) | −0.0021 (6) | −0.0018 (6) |
| C8 | 0.0413 (8) | 0.0320 (7) | 0.0549 (9) | 0.0084 (6) | 0.0016 (7) | 0.0068 (6) |
| C7 | 0.0327 (7) | 0.0436 (8) | 0.0526 (9) | 0.0074 (6) | −0.0066 (6) | 0.0075 (7) |
| O1W | 0.0418 (6) | 0.0358 (5) | 0.0542 (7) | 0.0069 (4) | −0.0077 (5) | −0.0016 (5) |
Geometric parameters (Å, º)
| Cl1—C4 | 1.7233 (14) | C10—C11 | 1.5103 (18) |
| Cl2—C2 | 1.7217 (14) | C3—C4 | 1.4151 (17) |
| O1—C11 | 1.2366 (17) | C3—C2 | 1.4184 (18) |
| O2—C11 | 1.2697 (17) | C4—C5 | 1.3631 (18) |
| O3—C6 | 1.3446 (17) | C6—C7 | 1.392 (2) |
| O3—H3 | 0.8200 | C2—C1 | 1.3661 (19) |
| N3—C9 | 1.3287 (18) | C5—H5 | 0.9300 |
| N3—C10 | 1.3481 (16) | C1—H1A | 0.9300 |
| N1—C5 | 1.3406 (18) | C9—C8 | 1.386 (2) |
| N1—C1 | 1.3430 (18) | C9—H9 | 0.9300 |
| N1—H1 | 0.8600 | C8—C7 | 1.370 (2) |
| N2—C3 | 1.3229 (16) | C8—H8 | 0.9300 |
| N2—H2A | 0.8600 | C7—H7 | 0.9300 |
| N2—H2B | 0.8600 | O1W—H1W | 0.8499 |
| C10—C6 | 1.3965 (18) | O1W—H2W | 0.8500 |
| C6—O3—H3 | 109.5 | O3—C6—C10 | 121.79 (13) |
| C9—N3—C10 | 119.47 (12) | C7—C6—C10 | 118.91 (13) |
| C5—N1—C1 | 120.41 (12) | C1—C2—C3 | 121.66 (12) |
| C5—N1—H1 | 119.8 | C1—C2—Cl2 | 120.10 (11) |
| C1—N1—H1 | 119.8 | C3—C2—Cl2 | 118.24 (10) |
| C3—N2—H2A | 120.0 | N1—C5—C4 | 120.96 (13) |
| C3—N2—H2B | 120.0 | N1—C5—H5 | 119.5 |
| H2A—N2—H2B | 120.0 | C4—C5—H5 | 119.5 |
| N3—C10—C6 | 121.12 (12) | N1—C1—C2 | 120.81 (13) |
| N3—C10—C11 | 117.62 (11) | N1—C1—H1A | 119.6 |
| C6—C10—C11 | 121.24 (11) | C2—C1—H1A | 119.6 |
| O1—C11—O2 | 125.31 (13) | N3—C9—C8 | 122.09 (13) |
| O1—C11—C10 | 118.99 (12) | N3—C9—H9 | 119.0 |
| O2—C11—C10 | 115.68 (12) | C8—C9—H9 | 119.0 |
| N2—C3—C4 | 122.67 (12) | C7—C8—C9 | 119.48 (13) |
| N2—C3—C2 | 123.00 (12) | C7—C8—H8 | 120.3 |
| C4—C3—C2 | 114.33 (11) | C9—C8—H8 | 120.3 |
| C5—C4—C3 | 121.81 (12) | C8—C7—C6 | 118.88 (13) |
| C5—C4—Cl1 | 119.66 (11) | C8—C7—H7 | 120.6 |
| C3—C4—Cl1 | 118.52 (10) | C6—C7—H7 | 120.6 |
| O3—C6—C7 | 119.29 (12) | H1W—O1W—H2W | 104.5 |
| C9—N3—C10—C6 | −1.8 (2) | C4—C3—C2—C1 | −1.97 (19) |
| C9—N3—C10—C11 | 176.65 (12) | N2—C3—C2—Cl2 | −2.54 (18) |
| N3—C10—C11—O1 | −7.84 (19) | C4—C3—C2—Cl2 | 178.22 (10) |
| C6—C10—C11—O1 | 170.60 (13) | C1—N1—C5—C4 | −0.6 (2) |
| N3—C10—C11—O2 | 173.80 (12) | C3—C4—C5—N1 | 0.0 (2) |
| C6—C10—C11—O2 | −7.76 (19) | Cl1—C4—C5—N1 | −178.74 (11) |
| N2—C3—C4—C5 | −178.05 (13) | C5—N1—C1—C2 | −0.2 (2) |
| C2—C3—C4—C5 | 1.20 (19) | C3—C2—C1—N1 | 1.5 (2) |
| N2—C3—C4—Cl1 | 0.74 (18) | Cl2—C2—C1—N1 | −178.65 (11) |
| C2—C3—C4—Cl1 | 179.99 (10) | C10—N3—C9—C8 | −0.2 (2) |
| N3—C10—C6—O3 | −178.70 (13) | N3—C9—C8—C7 | 1.6 (2) |
| C11—C10—C6—O3 | 2.9 (2) | C9—C8—C7—C6 | −0.9 (2) |
| N3—C10—C6—C7 | 2.4 (2) | O3—C6—C7—C8 | −179.94 (15) |
| C11—C10—C6—C7 | −175.97 (13) | C10—C6—C7—C8 | −1.0 (2) |
| N2—C3—C2—C1 | 177.27 (13) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3···O2 | 0.82 | 1.79 | 2.5155 (17) | 147 |
| N1—H1···N3i | 0.86 | 1.90 | 2.7546 (17) | 171 |
| N2—H2A···O1Wii | 0.86 | 2.13 | 2.9414 (17) | 157 |
| N2—H2B···O1Wiii | 0.86 | 2.05 | 2.8269 (17) | 149 |
| O1W—H1W···O2 | 0.85 | 1.90 | 2.7442 (17) | 170 |
| O1W—H2W···O1iv | 0.85 | 1.98 | 2.8181 (18) | 170 |
| C5—H5···O1i | 0.93 | 2.31 | 2.9864 (18) | 129 |
| C5—H5···Cl2v | 0.93 | 2.97 | 3.7363 (16) | 141 |
| C7—H7···O3vi | 0.93 | 2.52 | 3.399 (2) | 157 |
Symmetry codes: (i) x+1, y, z; (ii) x, −y+3/2, z−1/2; (iii) −x+1, y−1/2, −z+1/2; (iv) x, −y+3/2, z+1/2; (v) −x+2, y+1/2, −z+1/2; (vi) −x+2, −y+1, −z+1.
Funding Statement
MH thanks SERB-IRE for financial support (Ref. No. SIR/2022/000011). SJK thanks TANSCHE for financial support (File No. RGP/2019–20/MTWU/HECP-0080).
References
- Agilent (2012). CrysAlis PRO and CrysAlis RED. Agilent Technologies Ltd, Yarnton, England.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Judge, S. & Bever, C. (2006). Pharmacol. Ther. 111, 224–259. [DOI] [PubMed]
- Manteau, B., Genix, P., Brelot, L., Vors, J.-P., Pazenok, S., Giornal, F., Leuenberger, C. & Leroux, F. R. (2010). Eur. J. Org. Chem. 604.
- Park, H., Choi, M. Y., Kwon, E. & Kim, T. H. (2016). Acta Cryst. E72, 1836–1838. [DOI] [PMC free article] [PubMed]
- Parthasarathi, V., Wolfrum, S., Noordik, J. H., Beurskens, P. T., Smith, G., Reilly, E. J. O. & Kennard, C. H. L. (1982). Cryst. Struct. Commun. 11, 1519.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Smith, G. (2015). Acta Cryst. E71, 931–933. [DOI] [PMC free article] [PubMed]
- Smith, G., Reilly, E. J. O. & Kennard, C. H. L. (1981). Cryst. Struct. Commun. 10, 1277.
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Steiner, T. (2002). Angew. Chem. Int. Ed. 41, 48–76.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2414314623008210/hb4452sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314623008210/hb4452Isup2.hkl
CCDC reference: 2294939
Additional supporting information: crystallographic information; 3D view; checkCIF report





