Abstract
SLC25A51 is a member of the mitochondrial carrier family (MCF) but lacks key residues that contribute to the mechanism of other nucleotide MCF transporters. Thus, how SLC25A51 transports NAD+ across the inner mitochondrial membrane remains unclear. To elucidate its mechanism, we use Molecular Dynamics simulations to reconstitute SLC25A51 homology models into lipid bilayers and to generate hypotheses to test. We observe spontaneous binding of cardiolipin phospholipids to three distinct sites on the exterior of SLC25A51's central pore and find that mutation of these sites impairs cardiolipin binding and transporter activity. We also observe that stable formation of the required matrix gate is controlled by a single salt bridge. We identify binding sites in SLC25A51 for NAD+ and show that its selectivity for NAD+ is guided by an electrostatic interaction between the charged nicotinamide ring in the ligand and a negatively charged patch in the pore. In turn, interaction of NAD+ with interior residue E132 guides the ligand to dynamically engage and weaken the salt bridge gate, representing a ligand‐induced initiation of transport.
Keywords: MCART1, mitochondrial carrier family, mitochondrial transport, nicotinamide adenine dinucleotide (NAD), SLC25A51
Subject Categories: Membranes & Trafficking, Metabolism, Organelles
This study reveals how human mitochondrial transporter SLC25A51 distinguishes its cognate ligand, oxidized NAD+, and identifies a requirement for bound membrane phospholipid cardiolipin.
Introduction
SLC25A51 is an essential gene that is ubiquitously expressed across all mammalian cells. Its protein controls the concentration of NAD+ in the mitochondrial matrix by directly importing the oxidized dinucleotide, and thus, mammalian mitochondria depend on SLC25A51 for sustaining NAD+ levels (Girardi et al, 2020; Kory et al, 2020; Luongo et al, 2020). Loss of SLC25A51 resulted in loss of cellular respiration and oxidative flux of the tricarboxylic acid cycle. SLC25A51 further impacted the concentrations of NAD‐derived metabolites and cofactors, including NADH and NADPH (Girardi et al, 2020; Kory et al, 2020; Luongo et al, 2020).
There is little understood, nevertheless, about the underlying mechanisms of SLC25A51 or how the transporter interacts with oxidized NAD+. SLC25A51 is a member of the mitochondrial carrier family (MCF) that localizes to the inner mitochondrial membrane (Girardi et al, 2020; Kory et al, 2020; Luongo et al, 2020; Ziegler et al, 2021). By homology, its structure harbors a pseudo‐tri‐symmetrical pore that is characteristic of the family (Pebay‐Peyroula et al, 2003; Robinson et al, 2008; Ruprecht & Kunji, 2021; Ziegler et al, 2021; Jones et al, 2023; Kang & Chen, 2023). The trice repeated domain comprises two transmembrane helices joined by a short matrix helix parallel to the membrane. Based on cytoplasmic and matrix‐open structures of the related ADP/ATP nucleotide carriers, it is surmised that members of this structurally related family function through an alternating‐access mechanism (Pebay‐Peyroula et al, 2003; Nury et al, 2005; Robinson et al, 2008; Ruprecht et al, 2014; King et al, 2016; Springett et al, 2017; Ruprecht & Kunji, 2021).
SLC25A51 has low conservation (< 20%) in its amino acid sequence compared to its functional homologs in S. cerevisiae and A. thaliana that selectively import mitochondrial NAD+ (Fig EV1) (Todisco et al, 2006; Palmieri et al, 2009; Ziegler et al, 2021). In phylogenetic analyses, SLC25A51 did not cluster with other mammalian nucleotide carriers in the family (Robinson et al, 2008; Palmieri, 2013). Thus, it remains unanswered how SLC25A51 engages and transports its ligand.
Figure EV1. Multiple sequence alignment of representative nucleotide carriers.

-
AAlignment of amino acid sequences for human SLC25A51 (HsSLC25A51), bovine SLC25A4 (BtSLC25A4), fungal ADT (TtADT), yeast Ndt1 (ScNdt1) and Arabidopsis Ndt1 (AtNdt1).
-
BAlignment of amino acid sequences for human SLC25A51 and SLC25A52 and SLC25A51 orthologues from mouse, drosophila, zebrafish, xenopus, and snake.
Data information: Select residues are highlighted by shading (hydrophobic, yellow; hydrophilic, gray; negatively charged, red; positively charged, blue; and proline kink residues, peach).
In this work, we used Molecular Dynamics (MD) simulations to study how SLC25A51 homology models engage the NAD+ ligand under the constraints of thermodynamics and in the context of a lipid environment. We tested our observations in intact cells with mutational analysis, measurements of in situ free mitochondrial NAD+ concentrations using a LigA‐based NAD+ biosensor (Cambronne et al, 2016) and developed a heterologous uptake system to monitor SLC25A51 activity in recombinant E. coli membranes. This study provides the first insight into the binding site of an NAD+ mitochondrial transporter, identifies a mechanism of ligand‐induced transport, and reveals cardiolipin‐binding as a regulator of SLC25A51 activity and consequently mitochondrial NAD+ concentrations.
Results and Discussion
Stable modeling of SLC25A51 in a lipid bilayer
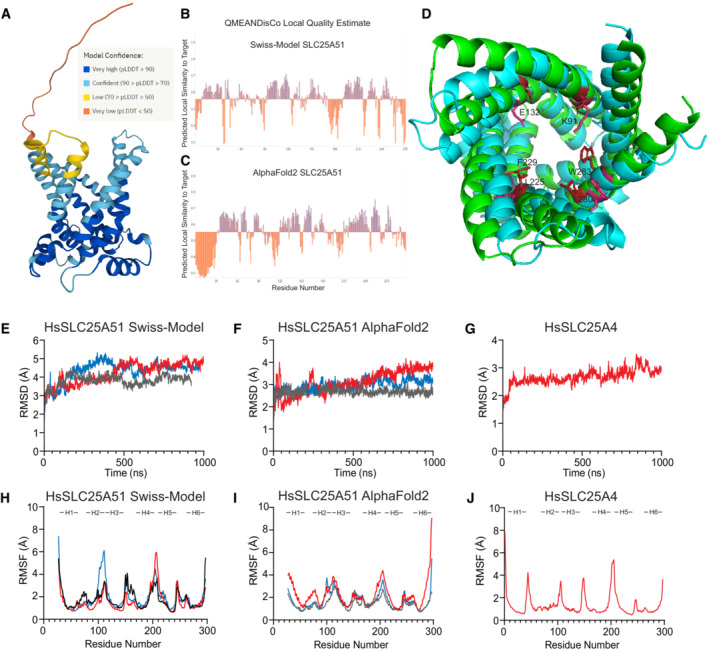
To study how SLC25A51 recognizes NAD+, we first generated apo models of human SLC25A51 in its outward‐facing, cytoplasmic (c‐state) conformation using Swiss‐Model and AlphaFold2 modeling (Waterhouse et al, 2018; Jumper et al, 2021). Both model structures adopted the characteristic MCF fold and were superimposable upon the solved bovine ANT crystal structure (PDB ID: 1OKC) (Pebay‐Peyroula et al, 2003). Using MolProbity, we determined that the quality of the equilibrated AlphaFold2 and Swiss‐Model structures were in the 100th and 97th percentile, respectively (Williams et al, 2018). Model quality was further estimated using pLDDT for AlphaFold2 (Fig EV2A) and QMEANDisCo analysis for both AlphaFold2 and Swiss‐Model with global scores of 0.55 ± 0.05 and 0.56 ± 0.05, respectively (Fig EV2B and C) (Studer et al, 2020; Jumper et al, 2021). A direct comparison revealed an initial difference of 3.5 Å root‐mean‐square deviation (RMSD) in atomic positions between the structures. Closer inspection revealed several residues in the central pore that were hydrophobically buried in one model but exposed to the hydrophilic pore in the other (Fig EV2D). Therefore, we decided to test both models in this study.
Figure EV2. Quality estimation using root mean square deviation and root mean square fluctuation of the simulations.
-
AModel confidence scores for AlphaFold2 structure of HsSLC25A51.
-
B, CQMEANDisCo Local Quality Estimate shows higher confidence for transmembrane helices and lower confidence for flexible loops.
-
DAlignment of Swiss‐Model (Cyan) and AlphaFold2 (Green) structures. Key residues that were initially differentially positioned in the binding site are highlighted.
-
ERMSD of backbone atoms over time with the Apo Swiss‐Model (amino acids 27–297), n = 3.
-
FRMSD of backbone atoms over time with the Apo AlphaFold2 (amino acids 27–297), n = 3.
-
GRMSD of backbone atoms over time with Apo SLC25A4, n = 1.
-
HRMSF of each residue with Apo Swiss‐Model, n = 3.
-
IRMSF of each residue with Apo AlphaFold2, n = 3.
-
JRMSF of each residue with Apo SLC25A4, n = 1.
Source data are available online for this figure.
Each of the apo models was individually embedded in a 86 × 86 Å2 swatch of lipid bilayer using Charmm‐GUI that recapitulated the composition of phospholipid components in mammalian inner mitochondrial membrane (POPC:POPE:CL = 2:3:2) (Comte et al, 1976; Jo et al, 2008; Wu et al, 2014; Mejia & Hatch, 2015; Lee et al, 2016). The system was hydrated and modeled with an ionic concentration of 150 mM KCl. Each model was analyzed in triplicate, and each replicate was ~1 μs in duration (Table EV1). We observed that the mean RMSD over time among the replicates for both stabilized c‐state models were 4.1 ± 0.3 and 2.9 ± 0.2 Å for the Swiss‐Model and AlphaFold2 models, respectively (Fig EV2E and F, Table EV1). In parallel, as a control for the approach, we generated a homology model for c‐state human SLC25A4 (ANT1, ADP/ATP carrier) based on the solved bovine structure (PDB ID: 1OKC) (Pebay‐Peyroula et al, 2003). The model for SLC25A4 (RMSD 2.8 Å) displayed a similar stability compared to the SLC25A51 c‐state models over the course of the simulations (Fig EV2G). We further calculated root‐mean‐square fluctuation (RMSF) values for all c‐state models, and as expected, observed less flexibility in transmembrane regions compared to loop regions (Fig EV2H–J). Together, the data indicated that the experimental model systems were stable.
Cardiolipin is required for SLC25A51 activity
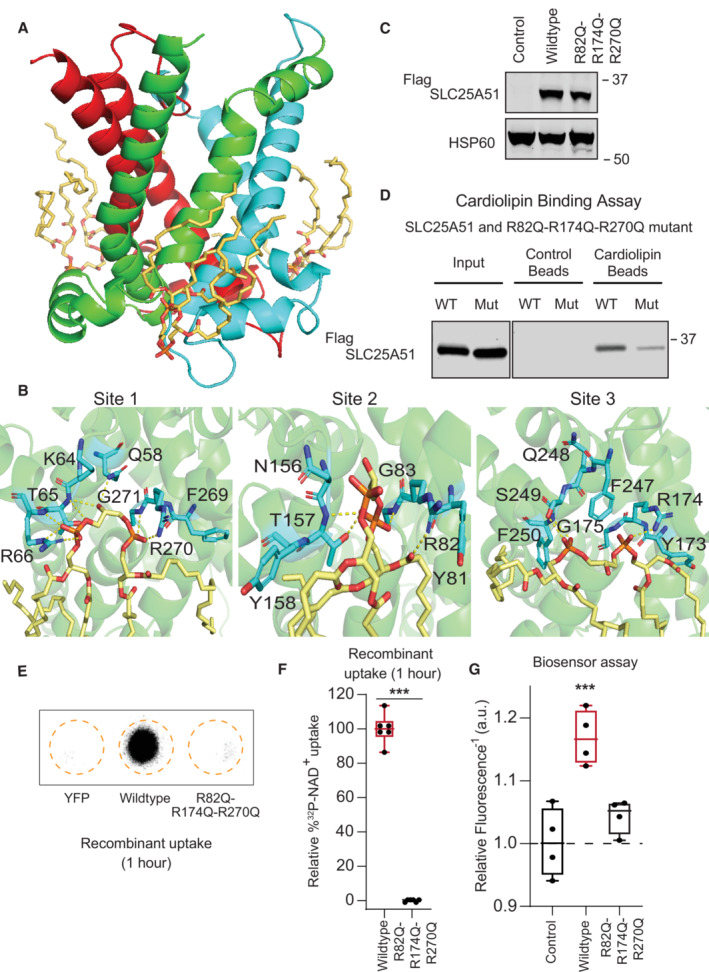
With both the AlphaFold2 and Swiss‐Model models, we observed spontaneous recruitment of cardiolipin molecules to three sites on SLC25A51 (Fig 1A, Movie EV1). Notably, all lipids were initially positioned at random and cardiolipin molecules were not docked to SLC25A51. We observed cardiolipin molecules arriving from different initial positions, displacing other types of phospholipids, and consistently interacting at the same three distinct sites on SLC25A51.
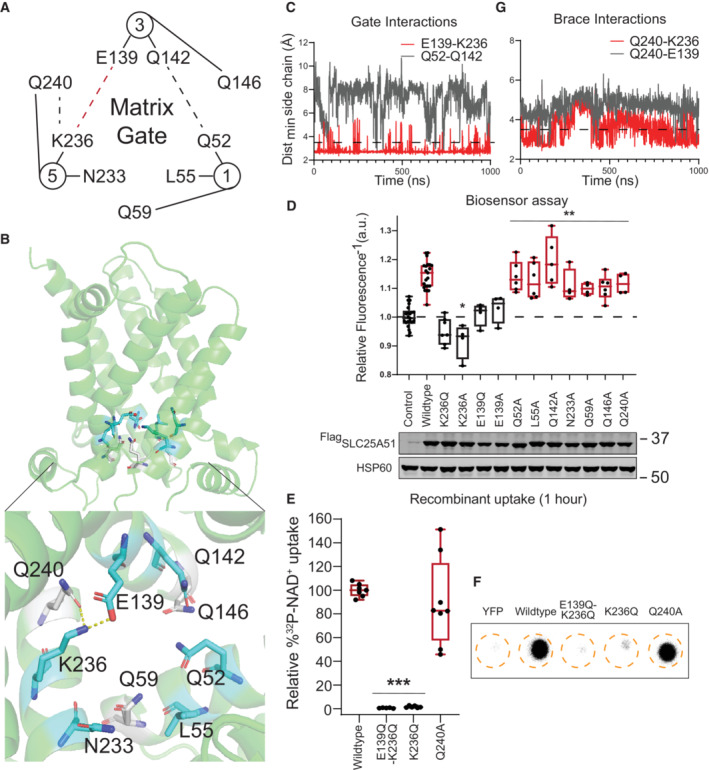
Figure 1. Cardiolipin binding impacts SLC25A51 function.
-
ACardiolipin (wheat) bound to SLC25A51 at the junctions of its repeated domains, highlighted in red, green, and blue.
-
BRepresentative poses depicting identified interactions at sites 1–3. Yellow dashes indicate interactions < 3.5 Å. At site 2, the binding of a single cardiolipin phosphate was observed.
-
CAnti‐Flag Western blot for transiently transfected FlagSLC25A51 variants and empty vector control in HeLa cells; HSP60, loading control.
-
DIn vitro pull down of FlagSLC25A51 variants using cardiolipin‐bound or control beads and detected with anti‐Flag Western blots.
-
ERepresentative phosphorimage of 32P‐NAD+ taken up after 1 h in recombinant E. coli cells expressing indicated SLC25A51 variants (amino acids 29–297) or negative control protein YFP.
-
FQuantitation of 32P‐NAD+ uptake after 1 h in E. coli cells expressing wildtype SLC25A51 (amino acids 29–297, red) and the indicated mutant. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, n = 6 biological replicates, unpaired two‐sided t‐test ***P < 0.001.
-
GFree mitochondrial NAD+ levels measured using a ratiometric biosensor in HeLa cells co‐expressing empty vector control or FlagSLC25A51 variants. Measurements were taken at 24 h post‐transfection; the dashed line indicates the baseline defined by the empty vector control, and red denotes data equivalent to wildtype. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, n = 4 biological replicates, ANOVA < 0.01 with post‐hoc Dunnett's Test compared to empty vector control, ***P < 0.001.
Source data are available online for this figure.
Cardiolipin binding was mediated by polar interactions between its phosphates and exterior‐facing residues on the transporter at Binding Site 1 (T65, R66, R270, and G271), Binding Site 2 (N156, T157, R82, and G83), and Binding Site 3 (S249, R174, G175) (Fig 1B). The observed sites correlated in position with cardiolipin binding motifs [F/Y/W] X G and [F/Y/W] [K/R] G identified from ADP/ATP transporter studies (Pebay‐Peyroula et al, 2003; Nury et al, 2005; Ruprecht et al, 2014, 2019; Crichton et al, 2015; Duncan et al, 2018; Senoo et al, 2020; preprint: Senoo et al, 2023). All binding sites were on the exterior of the central pore facing the matrix leaflet and positioned at the junctions between the three repeated protein domains that comprised SLC25A51's pore (Fig 1A and B). Each cardiolipin molecule spanned an even‐numbered helix from one domain and the matrix helix of the neighboring domain. At sites 1 and 3, cardiolipin bridged adjacent domains through extensive engagement of each of its phosphates (Fig 1B). At Site 2, a single phosphate of the cardiolipin molecule bridged the domains in 3 out of 4 replicates (Fig 1B). This suggested at least two binding modalities for cardiolipin and that cardiolipin may engage SLC25A51 asymmetrically. The dynamic and asymmetric recruitment of cardiolipin may be needed for matching the asymmetric shape of NAD+ or to create flexibility in the pore SLC25A51; binding sites with distinct roles have been recently described in the ADP/ATP carrier (Mao et al, 2021; Yi et al, 2022; preprint: Senoo et al, 2023).
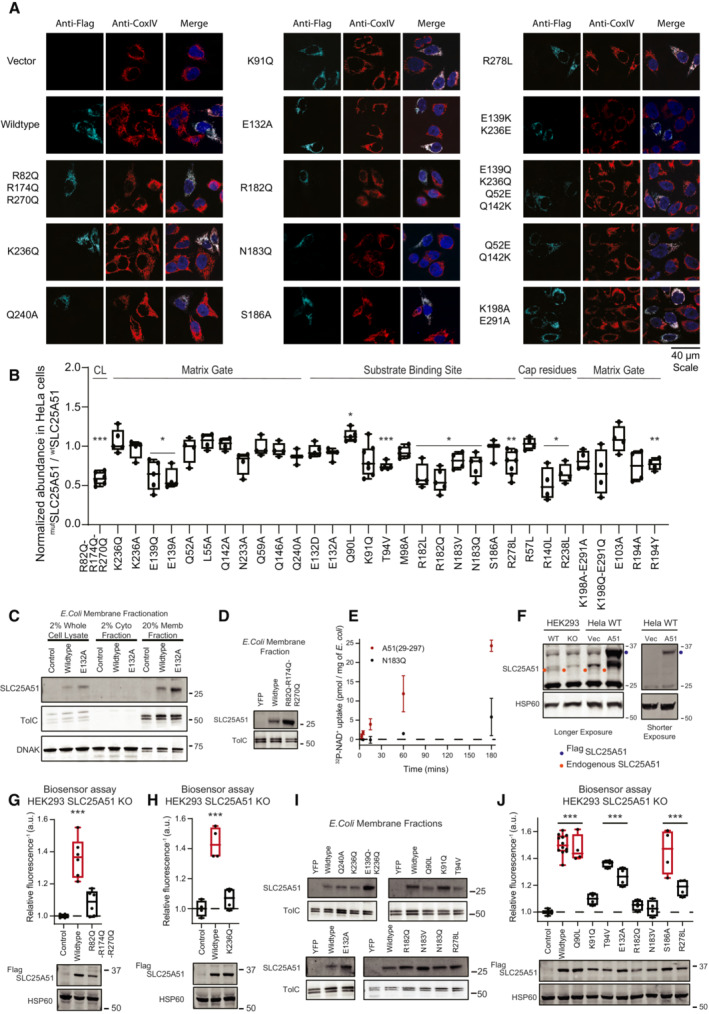
We introduced point mutations R82Q, R174Q, and R270Q at conserved residues to blunt the predicted electrostatic interactions at each binding site (Fig EV1B). These sites were exterior to the transporter's pore, distinct from the active site, and not obviously involved in interhelical interactions. Flag‐SLC25A51 and Flag‐SLC25A51R82Q‐R174Q‐R270Q were detectable via Western blot and each co‐localized with mitochondrial inner membrane protein Cox IV using immunofluorescence assays (Figs 1C and EV3A and B). We compared binding of immunoprecipitated Flag‐SLC25A51 and Flag‐SLC25A51R82Q‐R174Q‐R270Q to cardiolipin‐conjugated beads in vitro and found that the mutations diminished binding (Fig 1D).
Figure EV3. Localization, expression, and function of SLC25A51 mutants.
-
AImmunofluorescence co‐localization of CoxIV and FlagSLC25A51 variants transiently transfected in HeLa cells. Anti‐Flag is shown in cyan, anti‐CoxIV in red, and DAPI in blue.
-
BRelative abundance of FlagSLC25A51 mutants over wildtype from HeLa whole cell lysates normalized from Western blot data (24 h post‐transfection). Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, n = 4–7 biological replicates. One‐sample two‐sided t‐test for significant difference from 1, *P < 0.05, **P < 0.01, ***P < 0.001.
-
CMembrane enrichment of E. coli cells expressing YFP control, wildtype SLC25A51 (amino acids 29–297) or the indicated mutant detected with anti‐SLC25A51 Western blot; anti‐TolC (membrane) and anti‐DNAK (cytoplasmic), fractionation markers.
-
DMembrane expression of wildtype SLC25A51 (amino acids 29–297), indicated variants, or YFP control in E. coli cells detected with anti‐SLC25A51 Western blot; TolC, loading control.
-
ERadiolabeled NAD+ uptake with time by E. coli cells expressing wildtype SLC25A51 (amino acids 29–297) or its N183Q mutant. Data are mean ± SD and n = 3–4 biological replicates.
-
FAnti‐SLC25A51 Western blot of HEK293 wildtype, HEK293 SLC25A51 KO and HeLa cells transiently transfected with FlagSLC25A51 or empty vector control; HSP60, loading control. Red dots, endogenous SLC25A51; green dots, ectopically expressed FlagSLC25A51.
-
G, HFree mitochondrial NAD+ levels measured using a ratiometric biosensor in HEK293 SLC25A51 KO cells co‐expressing empty vector control, wildtype FlagSLC25A51, and indicated mutants. Measurements were taken at 48 h post‐transfection; the dashed line indicates the baseline defined by the empty vector control, and red denotes data equivalent to wildtype. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, n = 4–6 biological replicates, ANOVA < 0.0001 with post‐hoc Dunnett's Test compared to empty vector control ***P < 0.001. (bottom) Protein expression from HEK293 SLC25A51 KO cells transiently transfected with empty vector control, wildtype FlagSLC25A51 and the indicated variants detected using anti‐Flag Western blot; HSP60, loading control.
-
IMembrane expression of wildtype SLC25A51 (amino acids 29–297), indicated variants or YFP control in E. coli cells using anti‐SLC25A51 Western blot; TolC, loading control.
-
JFree mitochondrial NAD+ levels measured using a ratiometric biosensor in HEK293 SLC25A51 KO cells expressing empty vector control (n = 10 biological replicates), wildtype FlagSLC25A51 (n = 12 biological replicates), and indicated mutants (n = 4 biological replicates). Measurements were taken at 48 h post‐transfection; the dashed line indicates the baseline defined by the empty vector control, and red denotes data equivalent to wildtype. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, ANOVA < 0.0001 with post‐hoc Dunnett's Test compared to empty vector control ***P < 0.001. (bottom) Protein expression from HEK293 SLC25A51 KO cells transiently transfected with empty vector control, wildtype FlagSLC25A51 and the indicated variants using anti‐Flag Western blot; HSP60, loading control.
Source data are available online for this figure.
To determine whether cardiolipin binding was required for SLC25A51 activity, we monitored SLC25A51‐dependent uptake of 32P‐NAD+ (Eller et al, 2023) by adopting an assay previously used to characterize mitochondrial ADP/ATP transporter activity in bacterial cells (Haferkamp et al, 2002; Ravaud et al, 2012; Mifsud et al, 2013). The induced expression of SLC25A51 variants in E. coli cells resulted in their localization to the membrane fraction (Fig EV3C and D), reported to contain 5–10% cardiolipin (Dowhan, 1997). We observed a time‐dependent internalization of 32P‐NAD+ upon induction of wildtype SLC25A51 (amino acids 29–297) but not with control cells expressing a predicted binding pocket mutant of SLC25A51 (Fig EV3E). Mutation of cardiolipin binding sites resulted in significantly less uptake compared to wildtype after 1 h (Fig 1E and F).
To determine whether cardiolipin binding impacted SLC25A51 activity in intact cells, we expressed a genetically encoded single fluorescent‐protein ratiometric sensor for free NAD+ in the mitochondria of HeLa cells (Cambronne et al, 2016; Eller et al, 2019; Luongo et al, 2020). Figure 1G depicts the inverted fluorescence intensity of the sensor that is a readout of relative NAD+ concentrations. Acute ectopic expression of FlagSLC25A51 resulted in increased steady‐state levels of free mitochondrial NAD+ compared to empty vector, demonstrating that this assay could measure the activity of SLC25A51 of importing NAD+ in intact and respiring cells (Fig 1G). Flag‐SLC25A51R82Q‐R174Q‐R270Q was unable to efficiently increase mitochondrial NAD+ concentrations indicating that cardiolipin binding was required for SLC25A51 function (Fig 1G). We observed a similar loss of activity compared to wildtype when Flag‐SLC25A51R82Q‐R174Q‐R270Q was expressed in HEK293 SLC25A51 Knockout (KO) line (Fig EV3F and G).
The matrix gate is comprised of a single salt bridge interaction
To study the control of SLC25A51 opening to the matrix, we examined all possible inter‐helical interactions at the position where gated salt‐bridge networks have been identified in the related MCF carriers (Nelson et al, 1998; Ruprecht & Kunji, 2021; Ziegler et al, 2021). In SLC25A51, there were only two potential interhelical interactions: a putative hydrogen bond between Q52 and Q142, and a putative ionic salt‐bridge between E139 and K236 (Fig 2A). Time evolution of each interaction over 1,000 ns in both AlphaFold2 and Swiss‐Model models indicated that the hydrogen bond (Q52‐Q142) was not formed (combined mean percent occupancy, m.p.o. 2% ± 0.7%) while the salt bridge interaction (E139‐K236) was stable (combined m.p.o., 93% ± 3%) (Fig 2B and C).
Figure 2. A single salt bridge stabilizes the NAD+‐accessible, outward facing conformation.
-
AGraphical representation of the matrix gate with dash lines indicating possible polar non‐covalent interactions. Interactions were either indicated (red) or unsupported (gray) by data.
-
BRelative position of the matrix gate in a cartoon representation of SLC25A51; tested glutamine brace residues (white) and interactions (dashed lines).
-
CTime evolution of distance between indicated side chains for putative gate interactions. Interaction cutoff of 3.5 Å is shown as a dashed line.
-
DFree mitochondrial NAD+ levels measured using a ratiometric biosensor in HeLa cells expressing empty vector control (n = 25 biological replicates), wildtype FlagSLC25A51 (n = 25 biological replicates), and indicated mutants (n = 4–6 biological replicates). Measurements were taken at 24 h post‐transfection; the dashed line indicates the baseline defined by the empty vector control, and red denotes data equivalent to wildtype. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, ANOVA P < 0.0001 with post‐hoc Dunnett's test compared to empty vector control, *P < 0.05, **P < 0.01. (bottom) Protein expression from HeLa cells transiently transfected with empty vector control, wildtype FlagSLC25A51 and the indicated variants using anti‐Flag Western blot; HSP60, loading control.
-
EQuantitation of uptake of 32P‐NAD+ after 1 h by E. coli cells expressing wildtype SLC25A51 (amino acids 29–297) and the indicated mutants; wildtype activity and equivalent, red. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, n = 5–8 biological replicates, ANOVA P < 0.0001 with post‐hoc Dunnett's test compared to wildtype, ***P < 0.001.
-
FRepresentative phosphorimage of 32P‐NAD+ taken up after 1 h in recombinant E. coli cells expressing indicated SLC25A51 variants (amino acids 29–297) or negative control protein YFP.
-
GTime evolution of distance between indicated side chains of putative brace interactions. Interaction cutoff at 3.5 Å is shown as a dashed line.
Source data are available online for this figure.
We used the mitochondrial NAD+ sensor assay and an E. coli uptake assay to test whether these interactions were required for SLC25A51 activity. Loss of the gate is predicted to favor the inward‐facing, matrix (m‐state) conformation, which would not be amenable to NAD+ import. Expression of SLC25A51 variants in HeLa cells with an ablated salt bridge (K236Q, K236A, E139Q, or E139A point mutations) were not able to increase steady‐state levels of free mitochondrial NAD+ levels compared to empty vector control (Fig 2D). We also tested the K236Q mutation in HEK293 SLC25A51 KO cells and observed no significant mitochondrial NAD+ increase (Fig EV3H). This indicated that formation of the E139‐K236 salt bridge was required for SLC25A51 activity. In line with the simulation results, disruption of the putative hydrogen bond with Q52A or Q142A did not have any major impact on SLC25A51 activity (Fig 2D). Furthermore, conserved residues in the consensus matrix gate motif (PX[D/E]XX[K/R]XXQ)—including L55 and N233 whose counterparts were identified as critical in the yeast and plant mitochondrial carriers (Miniero et al, 2022)—were not required for SLC25A51 activity in cells (Fig 2D). We further confirmed the necessity of the salt bridge using uptake assays and found that introduction of either E139Q‐K236Q or K236Q mutations resulted in loss of activity (Figs 2E and F, and EV3I). Together, the modeling and the biochemical assays indicated that a single interhelical salt‐bridge between E139 and K236 formed the matrix gate.
Glutamine braces are not required for SLC25A51 activity
In many MCF carriers, glutamine braces play important roles in regulating the matrix gate (Ruprecht & Kunji, 2021). We observed an analogously positioned glutamine residue Q240 in SLC25A51 to putatively brace the identified E139‐K236 salt‐bridge (Fig 2A and B). Modeling analyses were used to test the stability of either a Q240‐E139 or Q240‐K236 interaction. We did not observe consistent interactions in either model (Q240‐E139 combined m.p.o. of 5% ± 3%; Q240‐K236 combined m.p.o of 45% ± 16%) (Fig 2G). In agreement, mutation of Q240, or analogous glutamine residues Q59 and Q146 in adjacent domains, minimally impaired SLC25A51 activity (Figs 2D–F and EV3I). Together, our data demonstrated that the identified salt‐bridge gate in SLC25A51 can function without stabilization from glutamine braces, distinctly unique from many other MCF carriers (Ruprecht & Kunji, 2021).
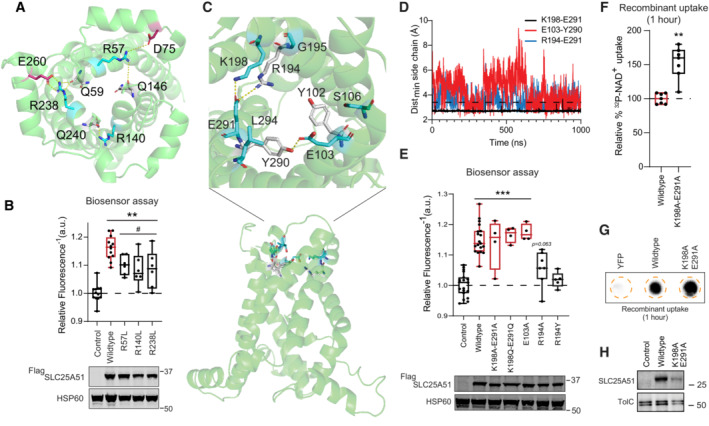
Arginine cap residues contribute to regulating SLC25A51 activity
Cap residues at the matrix opening of the transporter have been proposed to help stabilize the c‐state of MCF carriers by neutralizing c‐terminal negative dipoles of converging odd‐numbered helices or by forming stabilizing ionic interactions with matrix helices (Pierri et al, 2014; Yi et al, 2019). While modeling SLC25A51, we observed three arginine residues, R57, R238, and R140, positioned to putatively serve as cap residues (Fig EV4A). Mutation of each of the three cap residues (R57L, R140L and R238L) partially impaired SLC25A51 activity (Fig EV4B). In agreement, we observed stable interactions over time between R140 and the backbone of residue Q240 (m.p.o 95% ± 3% AlphaFold2 model, m.p.o. 80% ± 16% Swiss‐Model), as well as between R238 and the backbone of residue Q59 (m.p.o. 70% ± 10%, Swiss‐Model).
Figure EV4. The role of cap residues and cytoplasmic gate for SLC25A51 function.
-
ARelative positions of cap residues and their interactions (dashed lines) in a cartoon representation of SLC25A51.
-
BFree mitochondrial NAD+ levels measured using a ratiometric biosensor in HeLa cells expressing empty vector control (n = 12 biological replicates), wildtype FlagSLC25A51 (n = 12 biological replicates), and indicated mutants (n = 6 biological replicates). Measurements were taken at 24 h post‐transfection; the dashed line indicates the baseline defined by the empty vector control, and red denotes data equivalent to wildtype. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, ANOVA P < 0.0001, post‐hoc Dunnett's test compared to empty vector control (**) and compared to wildtype FlagSLC25A51 (#), **P < 0.01 and # P < 0.05. (bottom) Protein expression from HeLa cells transiently transfected with empty vector control, wildtype FlagSLC25A51 and the indicated variants using anti‐Flag Western blot; HSP60, loading control.
-
CRelative position of the cytoplasmic gate in a cartoon representation of SLC25A51.
-
DTime evolution of distance between indicated side chains for putative gate interactions. Interaction cutoff at 3.5 Å is shown as a dashed horizontal line.
-
EFree mitochondrial NAD+ levels measured using a ratiometric biosensor in HeLa cells expressing empty vector control (n = 19 biological replicates), wildtype FlagSLC25A51 (n = 19 biological replicates), and indicated mutants (n = 4–7 biological replicates). Measurements taken at 24 h post‐transfection; empty vector control baseline is shown as dashed line and wildtype activity and equivalent is in red color. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, ANOVA P < 0.0001, post‐hoc Dunnett's test compared to empty vector control, ***P < 0.001. (bottom) Protein expression from HeLa cells transiently transfected with empty vector control, wildtype FlagSLC25A51 and the indicated variants using anti‐Flag Western blot; HSP60, loading control.
-
FQuantitation of uptake of 32P‐NAD+ after 1 h by E. coli cells expressing wildtype SLC25A51 (amino acids 29–297) and the indicated mutant; wildtype activity and equivalent is in red color and the wildtype baseline is shown as dashed line. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, n = 7 biological replicates, unpaired t‐test ***P < 0.001.
-
GRepresentative phosphorimage of retained 32P‐NAD+ after 1 h by E. coli cells expressing wildtype SLC25A51 (amino acids 29–297), indicated variant, or YFP control.
-
HMembrane expression of wildtype SLC25A51 (amino acids 29–297), its indicated variant or YFP control in E. coli cells using anti‐SLC25A51 Western blot; TolC, loading control.
Source data are available online for this figure.
NAD + import can occur independently of a stable cytoplasmic gate
SLC25A51 is predicted to function via an alternating‐access mechanism similarly to other MCF family members (Robinson et al, 2008; Ziegler et al, 2021; Cimadamore‐Werthein et al, 2023). We thus generated an m‐state model (MolProbity 94th percentile)—using Swiss‐Homology Modeling and the solved structure of T. thermophila ADP/ATP carrier (PDB ID: 6GCI) (Ruprecht et al, 2019)—to help identify the cytoplasmic gate. Performed in triplicate (~1 μs/replicate, Table EV1), we observed consistent and stable formation of a putative ionic gate between K198 and E291 (m.p.o. 95% ± 5%) in an analogous plane to the identified cytoplasmic gate in the ADP/ATP carrier (Fig EV4C and D). This suggested that SLC25A51 has single salt bridge gates on each side of its pore. Mutation of the cytoplasmic gate is expected to favor the c‐state conformation. Interestingly, introduction of double mutations K198Q‐E291Q or K198A‐E291A did not impair SLC25A51 activity (Fig EV4E–H) unlike previously observed for ADP/ATP carrier (King et al, 2016; Ruprecht et al, 2019). This indicated that a c‐state conformation was sufficient for importing NAD+ and that a stable m‐state was not required for continuous uptake of NAD+. This capacity for uniport was previously predicted based on relative strengths of the gates (Robinson et al, 2008).
Putative tyrosine brace interactions were also observed but with lowered percent occupancy (E103‐Y290 m.p.o. 68% ± 24%; R194‐E291 m.p.o. 63% ± 8.5%) (Fig EV4C and D). In agreement with the idea that a stable m‐state was not essential, many of the disruptive mutations had minimal effect on SLC25A51 activity (Fig EV4E). The exception was that R194A and R194Y blocked SLC25A51 activity; however, mutation of its binding partner E291 did not result in any impairment, so any contribution from R194 was likely independent of its brace function. Together, the data indicated that cytoplasmic gate and brace features were not critical to SLC25A51's import function.
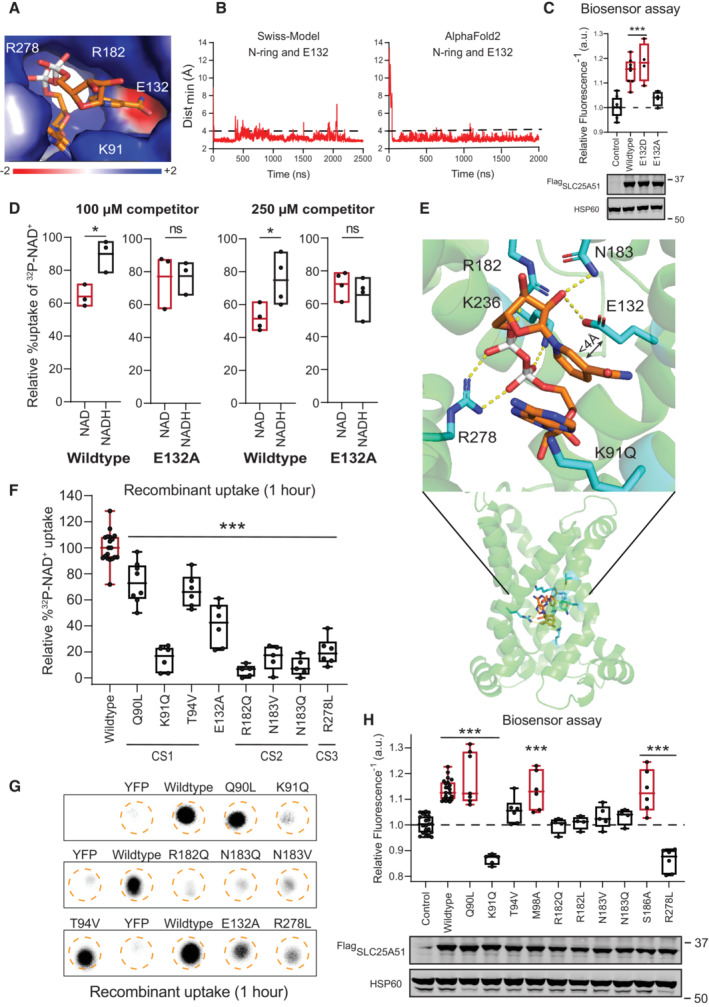
NAD + is orientated by charge to bind at three conserved contact points
To study how SLC25A51 engages its ligand, we unbiasedly modeled the binding of NAD+ onto both c‐state models. We used the Autodock 4.2 algorithm (Morris et al, 2009; Forli et al, 2016) to generate a series of poses ordered by calculated binding energy scores. We chose the poses with the lowest binding energy scores from each model to continue with analyses using MD simulations.
With both models (Table EV1 Simulations 10–18), we observed a prompt (within 100 ns) and consistent positioning of the nicotinamide ring (Fig 3A and B, Movies EV2 and EV3). The central ligand‐binding pore of SLC25A51 is characterized by positive charges arising from inward facing residues K91, R182, and R278 on the even‐numbered helices, and a distinct negatively charged region from acidic residue E132 on helix 3 (Fig 3A). We observed in both models that the positively‐charged nicotinamide ring of oxidized NAD+ consistently oriented away from the basic residues in the binding site and toward E132 (Fig 3A and B, Movies EV2 and EV3). Ablation of E132's charge impaired SLC25A51 activity, but a E132D variant retained function (Figs 3C and F, and EV3I and J). E132 is conserved across SLC25A51 orthologs (Fig EV1B).
Figure 3. E132 and conserved contact sites comprise a selective NAD+ binding site.
-
AElectrostatic environment of the substrate binding site (APBS as Pymol plugin). Representative positioning of NAD+ from simulation 17 at 1,230 ns (Table EV1).
-
BMinimal distance over time between the nicotinamide ring and E132 in simulations 17 (Swiss‐Model) and 12 (AlphaFold2) (Table EV1). Interaction cutoff at 4 Å shown as a dashed line.
-
CFree mitochondrial NAD+ levels measured using a ratiometric biosensor in HeLa cells co‐expressing empty vector control (n = 8 biological replicates), wildtype FlagSLC25A51 (n = 8 biological replicates), and indicated mutants (n = 4 biological replicates). Measurements were taken at 24 h post‐transfection; the dashed line indicates the baseline defined by the empty vector control, and red denotes data equivalent to wildtype. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, ANOVA P < 0.0001 with post‐hoc Dunnett's test compared to empty vector control ***P < 0.001. (bottom) Protein expression from HeLa cells transiently transfected with empty vector control, wildtype FlagSLC25A51 and the indicated variants using anti‐Flag Western blot; HSP60, loading control.
-
DRelative uptake of 32P‐NAD+ after 1 h by E. coli cells expressing wildtype SLC25A51 (amino acids 29–297) or the E132A mutant when competed by 100 μM (n = 3 biological replicates) and 250 μM (n = 4 biological replicates) unlabeled NAD+ (red) or NADH (black). Data is shown as floating bars with central line at mean and the box represents the min and max value, unpaired two‐sided t‐test *P < 0.05.
-
EBound NAD+ (orange) in the ligand binding pocket (cyan) is shown in both side and enlarged views. Non‐covalent interactions < 3.5 Å, dashed yellow lines.
-
FQuantitation of 32P‐NAD+ uptake after 1 h in E. coli cells expressing wildtype SLC25A51 (amino acids 29–297) and indicated mutants; wildtype activity (n = 19 biological replicates) and equivalent, red. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, n = 5–8 biological replicates, ANOVA P < 0.0001 with post‐hoc Dunnett's test compared to wildtype ***P < 0.001.
-
GRepresentative phosphorimage of retained 32P‐NAD+ after 1 h by E. coli cells expressing wildtype SLC25A51 (amino acids 29–297), indicated variants, or YFP control.
-
HFree mitochondrial NAD+ levels measured using a ratiometric biosensor in HeLa cells expressing empty vector control (n = 27 biological replicates), wildtype FlagSLC25A51 (n = 27 biological replicates), and indicated mutants (n = 4–7 biological replicates). Measurements were taken at 24 h post‐transfection; the dashed line indicates the baseline defined by the empty vector control, and red denotes data equivalent to wildtype. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, ANOVA P < 0.0001, post‐hoc Dunnett's test compared to empty vector control ***P < 0.001. (bottom) Protein expression from HeLa cells transiently transfected with empty vector control, wildtype FlagSLC25A51 and the indicated variants detected by anti‐Flag Western blot; HSP60, loading control.
Source data are available online for this figure.
We had previously determined that the presence of an equal concentration of NADH (100 μM) did not significantly interfere with SLC25A51 uptake activity of NAD+ (Fig 3D) (Luongo et al, 2020). A positive charge in the nicotinamide ring structure is a feature that distinguishes the oxidized form of NAD+ from reduced NADH. We hypothesized that electrostatic charges may facilitate an interaction between NAD+ and E132 in the pore of SLC25A51. To test this, we compared the ability for either unlabeled NAD+ or NADH to compete for SLC25A51‐dependent uptake of 32P‐NAD+ either at equivalent (100 μM) or 2.5‐fold higher concentrations (250 μM). With both concentrations, oxidized NAD+ successfully competed and impeded 32P‐NAD+ uptake (Fig 3D). With wildtype SLC25A51, oxidized NAD+ was a significantly better competitor compared to NADH. However, loss of E132 resulted in a loss of discrimination for SLC25A51 between NAD+ and NADH when ligands were present at equal concentrations (Fig 3D). This suggests that E132 contributes to ligand selectivity. The relatively subtle E132 may be enough to create a molecular discrimination based on electrostatic charge, given the much higher concentrations of NAD+ compared to NADH available for import in mitochondria. Free cytosolic NAD+ is approximately 100 to 1,000‐fold more abundant than free NADH (Williamson et al, 1967; Zhang et al, 2002; Hung et al, 2011; Zhao et al, 2011). Cytosolic ratios—which due to non‐discriminating porin channels in the outer membrane—likely represent what is experienced by intracellular SLC25A51.
We observed the nicotinamide ribose (NR) moiety formed consistent contacts (in 8 out of 9 simulations with both Swiss‐Model and AlphaFold2 models) (Table EV1). Borrowing the terminology used in the ADP/ATP binding pocket (Kunji & Robinson, 2006; Robinson & Kunji, 2006; Robinson et al, 2008), we observed that SLC25A51 used residue T94 in contact site 1 and N183 in contact site 2 to stabilize the NR moiety through specific hydrogen bonds with its carboxamide group and ribose (2′ and 3′ OH), respectively (Fig 3E). R278 in contact site 3, R182, and K91 engaged the negative charges from the phosphates of NAD+ (Fig 3E, Movies EV2 and EV3).
We mutated and tested the requirement of conserved residues in the three contact sites using the sensor and uptake assays (Figs 3F–H, EV1B and EV3I and J) (Ruprecht & Kunji, 2021; Ziegler et al, 2021). For contact site 1, we found that SLC25A51 variants with a K91Q mutation—and to a lesser extent T94V—impaired SLC25A51 import with minimal effects on protein stability. Mutating adjacent contact site residues (Q90 and M98) that were not observed to participate in binding of the NR moiety minimally impaired SLC25A51 despite being conserved residues (Figs 3F–H, EV1B and EV3I and J). Similarly, we confirmed that R182 and N183 were required for site 2 but not neighboring residue S186, and R278 was required for site 3 (Figs 3F–H and EV3I and J). The results were consistent with previous docking and mutational analyses (Kory et al, 2020; Ziegler et al, 2021). We observed that all three contact sites in SLC25A51's pore must be engaged to orient the ligand such that it can travel deeper into the pore. This may explain why nicotinamide, or its mononucleotide were unable to efficiently compete with NAD+ for transport (Kory et al, 2020; Luongo et al, 2020).
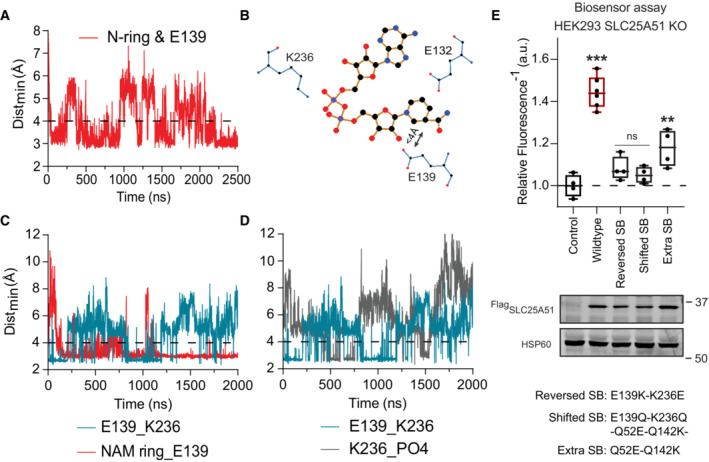
NAD + directly engaged with the gate in SLC25A51
While modeling the binding of NAD+, we unexpectedly observed two simulations wherein the ligand traversed deeper into the pore (Movie EV4 and Table EV1 simulations 16 and 17). To determine whether an extended simulation could provide additional insights, we continued with a pose that originated from the unbiased docking with the Swiss‐Homology model but had scored among lowest binding energy scores (Table EV1 simulations 19–22). This pose positioned the nicotinamide ring deeper in the pocket.
In all four replicate simulations 19–22, we observed channeling of the nicotinamide ring from E132 to E139 (Fig 4A and B, Movie EV4). This was reminiscent of the electrostatic funneling observed with ADP/ATP carriers (Heidkämper et al, 1996; Dehez et al, 2008; Wang & Tajkhorshid, 2008; Krammer et al, 2009; Monné et al, 2013; Mavridou et al, 2022). In both model structures, residue E139 of the salt‐bridge gate was positioned below E132 on the same helix 3. We also observed that as the nicotinamide ring interacted with E139 (85, 48, 69 and 93% occupancy), the phosphates on NAD+ could interact with K236, the other half of the salt bridge. Resultingly, formation of the E139‐K236 salt bridge was negatively correlated with the NAD+ phosphate‐K236 interaction (R = −0.192, −0.045, −0.681 and −0.336 per replicate, P < 0.001), as well as with the nicotinamide ring‐E139 interaction (R = −0.298, −0.053, −0.091 and −0.342 calculated per replicate, P < 0.001) (Fig 4C and D, Movie EV5). Together this disrupted an otherwise stable gate interaction (Fig 4, Movie EV5). An analogous ligand‐initiated mechanism has been proposed for the ADP/ATP carrier involving interference of the gate via the N6 atom of the adenine moiety on the nucleotide or the guanidino NH1 or NH2 of a conserved binding site arginine residue (Kunji & Robinson, 2006).
Figure 4. Substrate induced disruption of the matrix gate.
-
AMinimum distance between the nicotinamide ring and E139 over time, simulation 17 (Table EV1). Interaction cutoff at 4 Å is shown as a dashed line.
-
BTwo‐dimensional projection (Ligplot+) of NAD+ and its interaction with E132 and the E139‐K236 salt bridge.
-
CTime evolutions of the E139‐K236 salt bridge (teal) compared to the nicotinamide ring and E139 interaction (red); simulation 19 (Table EV1). Interaction cutoff at 4 Å is shown as a dashed line.
-
DTime evolutions of the E139‐K236 salt bridge (teal) compared to the phosphates in NAD+ and K236 (gray); simulation 19 (Table EV1). Interaction cutoff at 4 Å is shown as a dashed line.
-
EFree mitochondrial NAD+ levels measured using a ratiometric biosensor in HEK293 SLC25A51 KO cells expressing empty vector control, wildtype FlagSLC25A51, and indicated mutants. Measurements were taken at 48 h post‐transfection; the dashed line indicates the baseline defined by the empty vector control, and red denotes data equivalent to wildtype. Data are shown in box and whisker format, with hinges at 25th and 75th percentiles, whiskers represent min and max and the line is the median, n = 4–6 biological replicates, ANOVA P < 0.0001, post‐hoc Dunnett's test compared to empty vector control **P < 0.01, ***P < 0.001. (bottom) Protein expression from HEK293 SLC25A51 KO cells transiently transfected with empty vector control, wildtype FlagSLC25A51 and the indicated variants detected with anti‐Flag Western blot; HSP60, loading control.
Source data are available online for this figure.
Previously, studies showed that AMP, dUTP, NADH, GDP and GTP can bind the ADP/ATP carrier, suggesting that the dynamics of transport in cells involves an additional differentiation step to discriminate between bound molecules (Mifsud et al, 2013; Majd et al, 2018). To determine the requirement of the alignment between E132 and E139 for activity we reversed the positioning of E139 with its salt bridge (E139K‐K236E) and tested SLC25A51 activity using sensor assays. We also shifted the position of the entire salt bridge so that it was misaligned (E139Q‐K236Q‐Q52E‐Q142K). Activity was lost in the cases where the position of E139 was reversed or shifted, indicating that the alignment of this residue was critical (Fig 4E). We also introduced an additional salt bridge (Q52E‐Q142K), while maintaining the original salt bridge. The presence of an extra constitutive salt‐bridge that did not have an established regulatory mechanism resulted in significant loss of SLC25A51 activity (Fig 4E). Nevertheless, SLC25A51 activity was not fully diminished. The residual activity indicated that the preserved E132/E139 alignment was functional but that its role was partially hampered by the extra salt bridge (Fig 4E). Together, the data suggest that alignment of E132/E139 functions to facilitate gate opening.
With this study, we have gained insight into how the binding pocket of SLC25A51 aids the selective transport of oxidized NAD+, as well as evidence for ligand‐induced gate opening. The preference for NAD+ implies SL25A51 can alter flux through the tricarboxylic acid cycle by controlling the local NAD+/NADH ratio and oxidoreductive reactions. As specific point mutations can partially destabilize SLC25A51, we have sought corroborating data from multiple independent assays. A limitation is that a fully in vitro reconstitution of SLC25A51 activity has yet to be achieved. Future elucidation of the full dynamics of SLC25A51 will accelerate the development of SLC25A51 modulators for targeting disease (Fig EV5).
Figure EV5. Summary of the SLC25A51 residues mutated in this study.
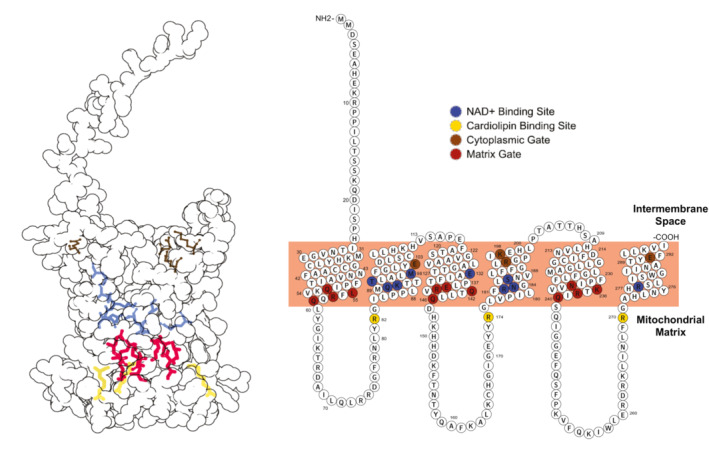
The mutations studied in this work are highlighted in a tertiary structure representation (Left) generated using Protein Imager (Tomasello et al, 2020) and a modified primary structure representation (Right) generated using Protter (Omasits et al, 2014).
Materials and Methods
Multiple sequence alignment
Multiple Sequence Alignment was performed using Cobalt sequence alignment tool (Papadopoulos & Agarwala, 2007) and was viewed and analyzed using Unipro UGENE (Okonechnikov et al, 2012). Figure EV1A depicts the alignment of representative nucleotide carriers HsSLC25A51 (UniProt: Q9H1U9), BtSLC25A4 (UniProt: P02722, PDB ID: 1OKC), TtADT (UniProt: G2QNH0, PDB ID: 6GCI), ScNdt1 (UniProt: P40556) and AtNdt1 (UniProt: O22261). Figure EV1B shows the alignment for the following SLC25A51 homologs and paralogue: human SLC25A51 (UniProt: Q9H1U9), human SLC25A52 (UniProt: Q3SY17), mouse SLC25A51 (UniProt: Q5HZI9), drosophila SLC25A51 (UniProt: Q7K483), zebrafish SLC25A51 (UniProt: Q6PBV5), xenopus SLC25A51 (UniProt: A9UMI0) and snake SLC25A51 (UniProt: A0A6P9B6W2).
SLC25A51 models
The homology models of cytoplasmic open state of apo‐SLC25A51 (amino acids 27–297) and apo‐SLC25A4 were generated using the Swiss‐Model server with bovine SLC25A4 crystal structure as the template (PDB ID: 1OKC) (Pebay‐Peyroula et al, 2003; Waterhouse et al, 2018). Bovine SLC25A4 was chosen as it has a high‐resolution crystal structure available (2.2 Å) and its GMQE score of 0.55 was the highest for SLC25A51 among all the c‐state MCF carrier structures available. The homology model of mitochondrial matrix open state of apo‐SLC25A51 (amino acids 26–297) was generated using the Swiss‐Model server with a thermophilic fungal ADP/ATP Carrier crystal structure as the template (PDB ID: 6GCI) (Ruprecht et al, 2019). We also used AlphaFold2 generated cytoplasmic open state model for our study (Jumper et al, 2021). The SLC25A51 models quality estimation was performed using MolProbity, pLDDT scores and QMEANDisCo analysis (Williams et al, 2018; Studer et al, 2020; Jumper et al, 2021).
Setup used in molecular dynamics simulations
CharmmGUI webserver was used to build the starting coordinates of our system for MD simulations with a total dimension of 85 Å × 85 Å × 100 Å (Jo et al, 2007; Wu et al, 2014). The c‐state Swiss‐Model (apo–state or NAD+ docked), m‐state Swiss‐Model and apo‐c‐state AlphaFold2 SLC25A51 were embedded in lipid bilayer systems with phosphatidylcholine (POPC), phosphatidylethanolamine (POPE) and cardiolipin (TLCL2) in a ratio 2:3:2 as experimentally determined previously (Comte et al, 1976). We chose TLCL2 cardiolipin with a negative 2 charge that agrees with the recent studies on ionization properties of the cardiolipin molecules (Olofsson & Sparr, 2013; Sathappa & Alder, 2016). The protein and the lipid bilayer were solvated with TIP3P water molecules extending up to 22.5 Å on both sides of the membrane. The system also contains neutralizing K+ and Cl− ions to make the final salt concentration 150 mM. The NAD+ was docked on the final coordinate of SLC25A51 after 1 μs simulation of AlphaFold2 SLC25A51 model which was embedded in a lipid bilayer membrane of POPC lipids with all the other parameters same as the other models described above.
Molecular dynamics simulation
The simulations were performed with Gromacs (version 2022.1) using an all atom Charmm36m force field for protein, lipids, ions and ligands with periodic boundary conditions (Bekker et al, 1993; Abraham et al, 2015). We performed energy minimization of the system in 5,000 steps of steepest descent with position restraints on protein and NAD+ while lipids, water and ions were free to equilibrate. The system was equilibrated in a multi‐step procedure with decreasing positional and dihedral constraints with each step with following parameters: Two steps of 0.125 ns NVT simulation at 310.15 K, one step of 0.125 ns NPT simulation at 310.15 K and 1 bar, and three steps of 0.5 ns NPT simulation at 310.15 K and 1 bar. MD simulations were run for different lengths of time using leap‐frog algorithm as an integrator with 2 fs time step. Particle Mesh Ewald was used for non‐bonded long‐range interactions with a 12 Å cutoff and Leonard‐Jones potential cutoff was 12 Å. Temperature was maintained at 310.15 K using Nose‐Hoover thermostat with a 1 ps coupling parameters and pressure at 1 bar using Parrinello‐Rahman barostat with 5 ps coupling and 4.5 × 10−5 bar−1 compressibility.
Trajectory analysis
The analysis was done using in‐built tools available in the gromacs package. The coordinates were viewed, and structural graphics and videos were prepared using Pymol. The cutoff distance was 3.5 Å for hydrogen bonds and for salt bridges while 4 Å for distance between any atom of Nicotinamide ring of NAD+ and side chain of amino acid. The mean percentage occupancy (mpo) represents the percentage of simulation time where the distance between the atoms was under the cut‐off distance. Time evolution of distances and RMSD is shown as moving averages of 5 values on each side. The videos are made with every 10th ns frame of the trajectories.
APBS electrostatics
APBS electrostatics was performed using Pymol plugin. The molecule was prepared using pdb2pqr method with 0.5 grid spacing and the electrostatic potential was projected to Connolly Surface in the −2 to +2 range.
Cell culture
HeLa (ATCC: CCL‐2) and HEK293 (ATCC: CRL‐1573) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g/L glucose, 1 mM sodium pyruvate and 4 mM L‐glutamine supplemented with 10% fetal bovine serum, 1× penicillin–streptomycin and 12.5 mM HEPES buffer (pH 7.4). Cell lines are tested monthly for mycoplasma contamination.
Cloning
SLC25A51 (amino acids 29–297) and YFP were cloned in pMW7 bacterial expression vector using NEBuilder® HiFi DNA Assembly Master Mix. Q5 site‐directed mutagenesis kit from NEB was used to make mutations in pMW7‐SLC25A51 (amino acids 29–297) plasmid and the pCMV‐Flag‐HA‐SLC25A51 (amino acids 1–297)‐IRES‐puro plasmid (Luongo et al, 2020).
Generation of HEK293 SLC25A51 knockout lines
CRISPR‐Cas9‐engineered HEK293 cells lacking a functional gene for SLC25A51 were generated as described before (Ran et al, 2013). Briefly, hybridized oligonucleotides 5′‐ CACCGTAGTTGTCTTCTGCATCAA and 5′‐ AAACTTGATGCAGAAGACAACTAC were inserted into high‐fidelity BbsI‐digested plasmid pSpCas9(BB)‐2A‐Puro (PX459) V2.0 (Addgene #62988). A total of 800,000 HEK293 cells were seeded in a 6‐well plate and after 24 h were transfected using X‐tremeGENE 9. Twenty‐four hours post transfection, about 5% each of the transfected cells were distributed onto 10 cm dishes and incubated with puromycin (2 μg/ml) for 48 h. Five to seven days after puromycin withdrawal, cell clones were picked and expanded. Genome editing within the targeted genomic DNA sequence was confirmed after PCR amplification from isolated genomic DNA using primers 5′‐ TTGTGTGGCTGCTGTGCAGC and 5′‐ GAGCACTGTGAGTCGTTGCG followed by Sanger sequencing using the primer 5′‐ CTTTCGACAACAGCTGTATGG. We confirmed that the CRISPR‐plasmid did not integrate itself by immunoblot analyses to monitor Flag‐Cas9 expression and retention of puromycin sensitivity. The knockout of SLC25A51 was validated at protein level using Western blotting and at the genomic level by sequencing.
Generation of HEK293 SLC25A51 Knockout lines stably expressing NAD+ biosensor.
The lines were generated following a similar protocol used for HeLa sensor lines described previously (Luongo et al, 2020). We transduced HEK293 SLC25A51 knockout cell line with lenti virus encoding MitocpVenus or MitoSensor. Stable integration was selected with 10 μg/ml Blasticidin for 1 week. Cells were sorted for fluorescence and validated using sensor response to FlagSLC25A51 and FlagNdt1 overexpression.
Mitochondrial steady state NAD + measurements
The pCMV‐Flag‐HA‐SLC25A51 (amino acids 1–297)‐IRES‐puro plasmids were transiently transfected:
HeLa mitocpVenus and HeLa mitoSensor cells using polyethylenimine (PEI, 1 mg/ml) with a PEI:DNA ratio of 5:1 and relative NAD+ steady state measurements were taken 24 h post‐transfection using flow cytometry.
HEK293 SLC25A51 KO mitocpVenus and HEK293 SLC25A51 KO mitoSensor cells using Lipofectamine 2000 (Invitrogen, 11668019) and relative steady state measurements were taken 48 h post‐transfection using flow cytometry.
FlowJo was used to calculate the ratiometric measurement of the NAD+ biosensor and CpVenus control by dividing the fluorescence emission when excited at 488 nm with measurement when excited at 405 nm for each cell. The geometric mean of the ratiometric fluorescence measurement was taken for 10,000 cells for both NAD+ biosensor and CpVenus. This was used to calculate the relative fluorescence as follows:
Western blotting for overexpression studies
HeLa or HEK293 cells were lysed with 2× Laemmli sample buffer containing DTT. Lysates from 200,000 or 75,000 cells were resolved using NuPAGE™ 10% or 4–12% Bis‐Tris protein gels (Invitrogen) and were transferred to Bio‐Rad 0.45 μm nitrocellulose membrane. The nitrocellulose membrane was blocked with 5% BSA in pH 7.4 Tris‐buffered saline with 0.1% (v/v) Tween 20 (TBST). The antibodies anti‐Flag M2 (Sigma, F1804, 1:3,000), and anti‐HSP60 Monoclonal Antibody (Invitrogen, 4B9/89, 1:3,000) were used for immunoblotting at 4°C overnight and were prepared in TBST with 1% BSA. After 3 washes of 5 min each, anti‐Mouse IgG (H+L), Alexa Fluor™ Plus 800 (Invitrogen, A32730, 1:10,000) prepared in TBST with 1% BSA was hybridized for 1 h at room temperature. LiCOR Odyssey CLx was used for imaging after 3 washes with 1×TBST.
Western blotting for SLC25A51 knockout validation
HEK293 SLC25A51 knockout cells were washed with ice‐cold PBS and lysed directly with 100 μl 2× Laemmli sample buffer containing DTT. 400,000 cells were electrophoresed on 10% Bis‐Tris gels (Invitrogen) and transferred to 0.45 μm nitrocellulose membrane. Membranes were blocked with 5% nonfat milk in TBST at RT for 1 h. The primary antibody was prepared in 1% nonfat milk in TBST and hybridized at 4°C overnight. The secondary antibody was prepared in 1% nonfat milk in TBST and hybridized at room temperature for 1 h. Antibodies used and their dilutions are as follows: anti‐SLC25A51 (MyBiosource, MBS1496255, 2 μg/ml), anti‐HSP60 Monoclonal Antibody (Invitrogen, 4B9/89, 1:3,000), anti‐rabbit IgG H&L IRDye 800CW (Abcam, ab216773, 1:10,000), and anti‐mouse IgG H&L Alexa Fluor 680 (Invitrogen, A10038, 1:10,000). Membranes were imaged using the LiCOR Odyssey CLx.
Immunofluorescence
Cells were transfected with pCMV‐Flag‐HA‐SLC25A51 (amino acids 1–297)‐IRES‐puro or the mutants and 24 h post‐transfection were seeded on coverslips. At 48 h post‐transfection, cells were washed and fixed in 500 μl using 4% PFA and 4% sucrose in PBS for 10 min at room temperature. Samples were washed three times with 1 ml of PBS followed by 10 min of blocking and permeabilization in solution containing: 5% Normal Goat Serum, 1% BSA, and 0.3% Triton‐X 100 in PBS. After blocking, 50 μl of primary antibodies anti‐flag D6W5B (Cell signaling, 14793) and anti‐CoxIV Mouse (abcam ab33985) at 1:500 dilution in blocking solution were added on top of the coverslip for 1 h at room temperature. The coverslips were washed three times with 200 μl PBS and 50 μl of secondary antibodies were added onto the coverslips for 1 h at room temperature. This was followed by three times wash with PBS. Coverslips were mounted on slides using 5 μl VECTASHIELD Vibrance antifade mounting medium (NC1601054) and were allowed to set for 1 h at room temperature before imaging. Images were taken using an Olympus IX83 confocal microscope with an 100× objective and were analyzed using ImageJ.
Flag immunoprecipitation of FlagSLC25A51 and variants
HEK293T cells were transfected using PEI with wildtype FlagSLC25A51 and the variant with R82Q, R174Q, and R270Q mutations. Cells were harvested 2 days post‐transfection, washed with ice‐cold PBS and lysed with the lysis buffer containing 1% DDM, 20 mM Tris pH 8.0, 200 mM NaCl, and 2× Pierce™ Protease Inhibitor (A32965) for 1 h at 4°C. The lysates were centrifuged at 13,000 g at 4°C for 15 min. The supernatant was collected and incubated with Anti‐DYKDDDDK affinity resin beads (Thermo Scientific Pierce, A36801) for 2 h at 4°C. After spinning down and removing the supernatant, the beads were washed three times using the lysis buffer. To elute the bound protein, 3× flag peptide (0.5 mg/ml) was added to the beads and incubated for 30 min at 4°C. The elution steps were repeated a total of three times. Finally, the three eluates were combined for the cardiolipin binding assay.
Cardiolipin binding assay
Equivalent amounts of immunoprecipitated wildtype and mutant FlagSLC25A51 from HEK293T cells were incubated with 20 μl of cardiolipin‐conjugated beads or control beads (Echelon Biosciences, P‐BCLP) for 2 h at 4°C. Supernatant was removed and beads were washed three times in buffer containing 1% DDM, 20 mM Tris pH 8.0, 200 mM NaCl, and 2× Pierce™ Protease Inhibitor (A32965). 2× Laemmli sample buffer containing DTT was added to each sample of washed beads and analyzed by Western blotting using antibodies against anti‐Flag M2 (Sigma, F1804, 1:3,000).
E. coli uptake assay
The protocol was adopted from a previously described uptake assay for ADP/ATP carrier (Haferkamp et al, 2002). The pMW7 expression plasmids encoding YFP, SLC25A51 (amino acids 29–297) or its mutants were transformed into BL21 DE3 pLysS E. coli strain. The colonies were picked for each mutant and grown overnight. The overnight culture was diluted to 0.5 OD600 and induced for 1 h with 1 mM IPTG. Cells equivalent to an OD600 of 5 in a final reaction volume of 50 μl were aliquoted in tubes, spun down at 4,000 g for 5 min and resuspended in 25 μl of uptake buffer (120 mM KCl, 5 mM KH2PO4, 1 mM EGTA and 2 mM HEPES‐NaOH pH 7.4). The reaction was started by adding 25 μl of uptake buffer with the final reaction concentration of 100 μM of unlabeled NAD+ traced with 3.33 nM of 32P‐NAD+. The reaction was stopped after 1 h by diluting it 20‐fold with ice cold uptake buffer containing 200 μM NAD+. The reactions were spun down at 4,000 g for 5 min, the supernatant was discarded, and the cells were resuspended in 500 μl of uptake buffer (For uptake at different time points in Fig EV3E, this step was skipped). The samples were filtered through 0.22 μm mixed‐cellulose ester filter using a filtration assembly with attached vacuum pump. The filter was washed two times with 5 and 3 ml uptake buffer. The filters were placed on a supporting sheet, wrapped with clingwrap and exposed to BioRad phosphorimaging screen in an exposure cassette for 18 h. Typhoon FLA 9500 Imager was used to image and ImageJ was used to quantify the dots. The intensity of the YFP control dot was subtracted for all the samples to account for non‐specific binding or any non‐active transport.
E. coli membrane enrichment and Western blot
The cultures were grown overnight and induced as described. Equivalent cells from the induced cultures for YFP, SLC25A51 (amino acids 29–297) and the mutants were spun down at 4,000 g for 5 min and washed with 1×PBS. The pellets were resuspended in 1 ml of 10 mM Tris/150 mM NaCl containing 1× Pierce™ Protease Inhibitor (A32965). Sonication was performed at 50% amplitude with 15 s ON/15 s OFF cycles for a total ON time of 5 min. The sonicated samples were spun down at 12,000 g for 30 min at 4°C. Supernatant was ultracentrifuged to pellet the membrane fraction at 100,000 g for 45 min at 4°C. The membrane pellet was resuspended in 100 μl of 50 mM Tris/150 mM NaCl with 1× protease inhibitor. 2× Laemmli sample buffer containing DTT was added to each sample and the samples were resolved with NuPAGE™ 10% Bis‐Tris protein gels (Invitrogen), transferred to Bio‐Rad 0.45 μm nitrocellulose membrane and blocked with 5% BSA in TBST. Immunoblotting was done using the following antibodies at 4°C overnight: anti‐SLC25A51 (Prosci‐Inc, 55‐424, 1:500), anti‐TolC (Mavridou lab at UT Austin, 1:2,000) and anti‐DNAK E. coli (Enzo ADI‐SPA‐880‐D, 1:5,000). After 3 washes in 1× TBST, the anti‐Mouse IgG (H+L), Alexa Fluor™ Plus 800 (Invitrogen, A32730, 1:10,000) or anti‐Rabbit IgG (H+L), Alexa Fluor™ 680 (Invitrogen, A27042, 1:10,000) was hybridized for 1 h at room temperature. This was followed by 3 wash steps in 1× TBST and imaging using LiCOR Odyssey CLx.
Statistical analysis and figure graphics
The results are presented as mean ± SD in box and whisker graphical format. The data was analyzed, and figures were prepared using GraphPad Prism version 9, Microsoft Excel, and adobe illustrator. P‐values were determined using unpaired two‐sided t‐test (for two group comparison) and one‐way ANOVA (for 3 or more group comparisons) with post‐hoc Dunnett's test. P‐values under 0.05 were considered significant and *P < 0.05, **P < 0.01 and ***P < 0.001.
Author contributions
Shivansh Goyal: Conceptualization; data curation; software; formal analysis; investigation; writing – original draft; writing – review and editing. Akhilesh Paspureddi: Software; formal analysis. Mu‐Jie Lu: Investigation; writing – review and editing. Hsin‐Ru Chan: Investigation; writing – review and editing. Scott N Lyons: Investigation; writing – review and editing. Crystal N Wilson: Validation. Marc Niere: Resources; writing – review and editing. Mathias Ziegler: Resources; writing – review and editing. Xiaolu A Cambronne: Conceptualization; supervision; funding acquisition; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Source Data for Expanded View
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Acknowledgements
We thank Dr. Ron Elber and Dr. Liao Chen for valuable discussions. We also thank Dr. Despoina Mavridou for providing us with the TolC antibody and Dr. Gennaro Agrimi for the pMW7 plasmid. Supported by the Texas Advanced Computing Center, NIH DP2 GM126897, the Pew Charitable Trust, and CPRIT RP210079.
EMBO reports (2023) 24: e56596
Data availability
Full view images, all data points, and simulation models are available via the Texas Data Repository, Cambronne XA Lab Dataverse https://doi.org/10.18738/T8/TIHMRO.
References
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindah E (2015) GROMACS: high performance molecular simulations through multi‐level parallelism from laptops to supercomputers. SoftwareX 1–2: 19–25 [Google Scholar]
- Bekker H, Berendsen HJC, Dijkstra EJ, Achterop S, Vondrumen R, Vanderspoel D, Sijbers A, Keegstra H, Renardus MKR (1993) Gromacs: a parallel computer for molecular dynamics simulations. In Physics Computing 92, de Groot RA, Nadrchal J (eds), pp 252–256. Singapore: World Scientific; [Google Scholar]
- Cambronne XA, Stewart ML, Kim D, Jones‐Brunette AM, Morgan RK, Farrens DL, Cohen MS, Goodman RH (2016) Biosensor reveals multiple sources for mitochondrial NAD+ . Science 352: 1474–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimadamore‐Werthein C, Baron SJ, King MS, Springett R, Kunji ER (2023) Human mitochondrial ADP/ATP carrier SLC25A4 operates with a ping‐pong kinetic mechanism. EMBO Rep 24: e57127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comte J, Maǐsterrena B, Gautheron DC (1976) Lipid composition and protein profiles of outer and inner membranes from pig heart mitochondria. Comparison with microsomes. Biochim Biophys Acta 419: 271–284 [DOI] [PubMed] [Google Scholar]
- Crichton PG, Lee Y, Ruprecht JJ, Cerson E, Thangaratnarajah C, King MS, Kunji ERS (2015) Trends in thermostability provide information on the nature of substrate, inhibitor, and lipid interactions with mitochondrial carriers. J Biol Chem 290: 8206–8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehez F, Pebay‐Peyroula E, Chipot C (2008) Binding of ADP in the mitochondrial ADP/ATP carrier is driven by an electrostatic funnel. J Am Chem Soc 130: 12725–12733 [DOI] [PubMed] [Google Scholar]
- Dowhan W (1997) Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem 66: 199–232 [DOI] [PubMed] [Google Scholar]
- Duncan AL, Ruprecht JJ, Kunji ERS, Robinson AJ (2018) Cardiolipin dynamics and binding to conserved residues in the mitochondrial ADP/ATP carrier. Biochim Biophys Acta Biomembr 1860: 1035–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eller JM, Stewart ML, Slepian AJ, Markwardt S, Wiedrick J, Cohen MS, Goodman RH, Cambronne XA (2019) Flow cytometry analysis of free intracellular NAD+ using a targeted biosensor. Curr Protoc Cytom 88: 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eller J, Goyal S, Cambronne XA (2023) Improved yield for the enzymatic synthesis of radiolabeled nicotinamide adenine dinucleotide. ACS Bio Med Chem Au 3: 46–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ (2016) Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 115: 905–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi E, Agrimi G, Goldmann U, Fiume G, Lindinger S, Sedlyarov V, Srndic I, Gürtl B, Agerer B, Kartnig F et al (2020) Epistasis‐driven identification of SLC25A51 as a regulator of human mitochondrial NAD import. Nat Commun 11: 6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferkamp I, Hackstein JHP, Voncken FGJ, Schmit G, Tjaden J (2002) Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur J Biochem 269: 3172–3181 [DOI] [PubMed] [Google Scholar]
- Heidkämper D, Müller V, Nelson DR, Klingenberg M (1996) Probing the role of positive residues in the ADP/ATP carrier from yeast. The effect of six arginine mutations on transport and the four ATP versus ADP exchange modes. Biochemistry 35: 16144–16152 [DOI] [PubMed] [Google Scholar]
- Hung YP, Albeck JG, Tantama M, Yellen G (2011) Imaging cytosolic NADH‐NAD+ redox state with a genetically encoded fluorescent biosensor. Cell Metab 14: 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Im W (2007) Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PloS One 2: e880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Iyer VG, Im W (2008) CHARMM‐GUI: a web‐based graphical user interface for CHARMM. J Comput Chem 29: 1859–1865 [DOI] [PubMed] [Google Scholar]
- Jones SA, Gogoi P, Ruprecht JJ, King MS, Lee Y, Zögg T, Pardon E, Chand D, Steimle S, Copeman DM et al (2023) Structural basis of purine nucleotide inhibition of human uncoupling protein 1. Sci Adv 9: eadh4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Chen L (2023) Structural basis for the binding of DNP and purine nucleotides onto UCP1. Nature 620: 226–231 [DOI] [PubMed] [Google Scholar]
- King MS, Kerr M, Crichton PG, Springett R, Kunji ERS (2016) Formation of a cytoplasmic salt bridge network in the matrix state is a fundamental step in the transport mechanism of the mitochondrial ADP/ATP carrier. Biochim Biophys Acta Bioenerg 1857: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kory N, de Bos J u, van der Rijt S, Jankovic N, Güra M, Arp N, Pena IA, Prakash G, Chan SH, Kunchok T et al (2020) MCART1/SLC25A51 is required for mitochondrial NAD transport. Sci Adv 6: eabe5310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer EM, Ravaud S, Dehez F, Frelet‐Barrand A, Pebay‐Peyroula E, Chipot C (2009) High‐chloride concentrations abolish the binding of adenine nucleotides in the mitochondrial ADP/ATP carrier family. Biophys J 97: L25–L27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunji ERS, Robinson AJ (2006) The conserved substrate binding site of mitochondrial carriers. Biochim Biophys Acta Bioenerg 1757: 1237–1248 [DOI] [PubMed] [Google Scholar]
- Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y et al (2016) CHARMM‐GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput 12: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luongo TS, Eller JM, Lu MJ, Niere M, Raith F, Perry C, Bornstein MR, Oliphint P, Wang L, McReynolds MR et al (2020) SLC25A51 is a mammalian mitochondrial NAD+ transporter. Nature 588: 174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majd H, King MS, Palmer SM, Smith AC, Elbourne LDH, Paulsen IT, Sharples D, Henderson PJF, Kunji ERS (2018) Screening of candidate substrates and coupling ions of transporters by thermostability shift assays. Elife 7: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Yao S, Yi Q, Xu ZM, Cang X (2021) Function‐related asymmetry of the specific cardiolipin binding sites on the mitochondrial ADP/ATP carrier. Biochim Biophys Acta Biomembr 1863: 183466 [DOI] [PubMed] [Google Scholar]
- Mavridou V, King MS, Tavoulari S, Ruprecht JJ, Palmer SM, Kunji ERS (2022) Substrate binding in the mitochondrial ADP/ATP carrier is a step‐wise process guiding the structural changes in the transport cycle. Nat Commun 131: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia EM, Hatch GM (2015) Mitochondrial phospholipids: role in mitochondrial function. J Bioenerg Biomembr 482: 99–112 [DOI] [PubMed] [Google Scholar]
- Mifsud J, Ravaud S, Krammer EM, Chipot C, Kunji ERS, Pebay‐Peyroula E, Dehez F (2013) The substrate specificity of the human ADP/ATP carrier AAC1. Mol Membr Biol 30: 160–168 [DOI] [PubMed] [Google Scholar]
- Miniero DV, Monné M, Di Noia MA, Palmieri L, Palmieri F (2022) Evidence for non‐essential salt bridges in the M‐gates of mitochondrial carrier proteins. Int J Mol Sci 23: 5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monné M, Palmieri F, Kunji ERS (2013) The substrate specificity of mitochondrial carriers: mutagenesis revisited. Mol Membr Biol 30: 149–159 [DOI] [PubMed] [Google Scholar]
- Morris GM, Ruth H, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Software news and updates AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Felix CM, Swanson JM (1998) Highly conserved charge‐pair networks in the mitochondrial carrier family. J Mol Biol 277: 285–308 [DOI] [PubMed] [Google Scholar]
- Nury H, Dahout‐Gonzalez C, Trézéguet V, Lauquin G, Brandolin G, Pebay‐Peyroula E (2005) Structural basis for lipid‐mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett 579: 6031–6036 [DOI] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M, Varlamov A, Vaskin Y, Efremov I, German Grehov OG, Kandrov D, Rasputin K, Syabro M et al (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28: 1166–1167 [DOI] [PubMed] [Google Scholar]
- Olofsson G, Sparr E (2013) Ionization constants pKa of cardiolipin. PloS One 8: e73040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Müller S, Wollscheid B (2014) Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30: 884–886 [DOI] [PubMed] [Google Scholar]
- Palmieri F (2013) The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Aspects Med 34: 465–484 [DOI] [PubMed] [Google Scholar]
- Palmieri F, Rieder B, Ventrella A, Blanco E, Do PT, Nunes‐Nesi A, Trauth AU, Fiermonte G, Tjaden J, Agrimi G et al (2009) Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J Biol Chem 284: 31249–31259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R (2007) COBALT: constraint‐based alignment tool for multiple protein sequences. Bioinformatics 23: 1073–1079 [DOI] [PubMed] [Google Scholar]
- Pebay‐Peyroula E, Dahout‐Gonzalez C, Kahn R, Trézéguet V, Lauquin GJM, Brandolin G (2003) Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426: 39–44 [DOI] [PubMed] [Google Scholar]
- Pierri CL, Palmieri F, De Grassi A (2014) Single‐nucleotide evolution quantifies the importance of each site along the structure of mitochondrial carriers. Cell Mol Life Sci 71: 349–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR‐Cas9 system. Nat Protoc 8: 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravaud S, Bidon‐Chanal A, Blesneac I, MacHillot P, Juillan‐Binard C, Dehez F, Chipot C, Pebay‐Peyroula E (2012) Impaired transport of nucleotides in a mitochondrial carrier explains severe human genetic diseases. ACS Chem Biol 7: 1164–1169 [DOI] [PubMed] [Google Scholar]
- Robinson AJ, Kunji ERS (2006) Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc Natl Acad Sci USA 103: 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson AJ, Overy C, Kunji ERS (2008) The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc Natl Acad Sci USA 105: 17766–17771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht JJ, Kunji ERS (2021) Structural mechanism of transport of mitochondrial carriers. Annu Rev Biochem 90: 535–558 [DOI] [PubMed] [Google Scholar]
- Ruprecht JJ, Hellawell AM, Harding M, Crichton PG, McCoy AJ, Kunji ERS (2014) Structures of yeast mitochondrial ADP/ATP carriers support a domain‐based alternating‐access transport mechanism. Proc Natl Acad Sci USA 111: E426–E434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht JJ, King MS, Zögg T, Aleksandrova AA, Pardon E, Crichton PG, Steyaert J, Kunji ERS (2019) The molecular mechanism of transport by the mitochondrial ADP/ATP carrier. Cell 176: 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathappa M, Alder NN (2016) The ionization properties of cardiolipin and its variants in model bilayers. Biochim Biophys Acta Biomembr 1858: 1362–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo N, Kandasamy S, Ogunbona OB, Baile MG, Lu Y, Claypool SM (2020) Cardiolipin, conformation, and respiratory complex‐dependent oligomerization of the major mitochondrial ADP/ATP carrier in yeast. Sci Adv 6: 780–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo N, Chinthapalli DK, Baile MG, Golla VK, Saha B, Ogunbona OB, Saba JA, Munteanu T, Valdez Y, Whited K et al (2023) Conserved cardiolipin‐mitochondrial ADP/ATP carrier interactions assume distinct structural and functional roles that are clinically relevant. bioRxiv 10.1101/2023.05.05.539595 [PREPRINT] [DOI]
- Springett R, King MS, Crichton PG, Kunji ERS (2017) Modelling the free energy profile of the mitochondrial ADP/ATP carrier. Biochim Biophys Acta Bioenerg 1858: 906–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer G, Rempfer C, Waterhouse AM, Gumienny R, Haas J, Schwede T (2020) QMEANDisCo‐distance constraints applied on model quality estimation. Bioinformatics 36: 1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todisco S, Agrimi G, Castegna A, Palmieri F (2006) Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae . J Biol Chem 281: 1524–1531 [DOI] [PubMed] [Google Scholar]
- Tomasello G, Armenia I, Molla G (2020) The Protein Imager: a full‐featured online molecular viewer interface with server‐side HQ‐rendering capabilities. Bioinformatics 36: 2909–2911 [DOI] [PubMed] [Google Scholar]
- Wang Y, Tajkhorshid E (2008) Electrostatic funneling of substrate in mitochondrial inner membrane carriers. Proc Natl Acad Sci USA 105: 9598–9603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, De Beer TAP, Rempfer C, Bordoli L et al (2018) SWISS‐MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46: W296–W303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB et al (2018) MolProbity: more and better reference data for improved all‐atom structure validation. Protein Sci 27: 293–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson DH, Lund P, Krebs HA (1967) The redox state of free nicotinamide‐adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem J 103: 514–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu EL, Cheng X, Jo S, Rui H, Song KC, Dávila‐Contreras EM, Qi Y, Lee J, Monje‐Galvan V, Venable RM et al (2014) CHARMM‐GUI membrane builder toward realistic biological membrane simulations. J Comput Chem 35: 1997–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Q, Li Q, Yao S, Chen Y, Guan MX, Cang X (2019) Molecular dynamics simulations on apo ADP/ATP carrier shed new lights on the featured motif of the mitochondrial carriers. Mitochondrion 47: 94–102 [DOI] [PubMed] [Google Scholar]
- Yi Q, Yao S, Ma B, Cang X (2022) The effects of cardiolipin on the structural dynamics of the mitochondrial ADP/ATP carrier in its cytosol‐open state. J Lipid Res 63: 100227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH (2002) Regulation of corepressor function by nuclear NADH. Science 295: 1895–1897 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Jin J, Hu Q, Zhou HM, Yi J, Yu Z, Xu L, Wang X, Yang Y, Loscalzo J (2011) Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metab 14: 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler M, Monné M, Nikiforov A, Agrimi G, Heiland I, Palmieri F (2021) Welcome to the family: identification of the NAD+ transporter of animal mitochondria as member of the solute carrier family SLC25. Biomolecules 11: 880 [DOI] [PMC free article] [PubMed] [Google Scholar]


