Abstract
We present single molecule studies demonstrating the capabilities of the FRET-PAINT method to detect secondary structures that would be challenging to detect with alternative methods, particularly single molecule FRET (smFRET). Instead of relying on the change in end-to-end separation as in smFRET, we use the change in accessibility to a small probe as the criterion for secondary structure formation and relative stability. As a model system, we study G-triplex formation by human telomeric repeat sequences in different structural contexts.
Graphical Abstract:
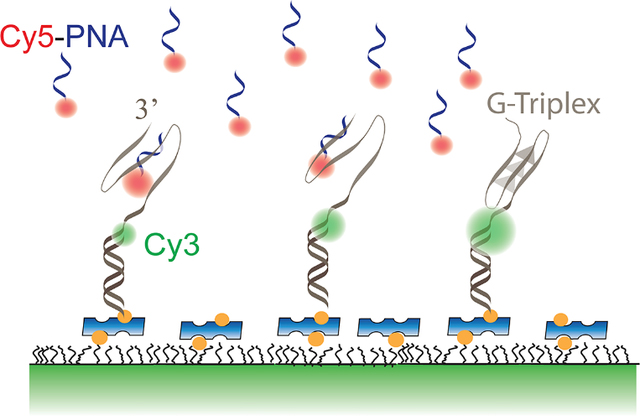
Accessibility of a nucleic acid to a complementary imager strand (Cy5-PNA) reduces when it forms a secondary structure (G-Triplex). This provides a way to detect structure formation regardless of how small the change in end-to-end separation is.
Single molecule Förster Resonance Energy Transfer (smFRET) is a potent tool for detecting structural changes in biomolecules 1. When the separation between the donor and acceptor fluorophores is optimized, it is possible to detect sub-nanometer variations in separation. However, such optimization may not always be feasible or practical depending on the construct, which makes it challenging to study the formation and stability of secondary structures that result in modest variations in donor-acceptor separation. Secondary structures that form by short oligonucleotides, which already have a small end-to-end separation before folding, or competing structures that have similar end-to-end separations are examples of cases that pose complications for smFRET. We propose FRET-PAINT (FRET-Point Accumulation In Nanoscale Topography) as an alternative method to overcome such complications.
FRET-PAINT combines the strengths of smFRET with those of DNA-PAINT and relies on detecting the binding of a short complementary probe (imaging strand) to a specific sequence 2,3. Typically, a donor fluorophore is placed on a DNA/RNA construct immobilized on the surface while the acceptor is on the imager strand. Not requiring a label on the strand of interest might be significant to minimally disturb the relevant structure(s) or enable studying long sequences that are challenging to label. This feature has enabled studying telomeric overhangs with physiologically relevant lengths (~150 nt), which could not be obtained commercially in labeled form 4. In the case of commonly used partial duplex DNA (pdDNA) constructs, binding of the imager strand to the single stranded overhang results in a FRET burst. The frequency and dwell time of these bursts can be optimized by modulating the imager strand and its concentration 5. Binding to different segments of the overhang results in bursts at different FRET levels. The frequency of such binding events represents the accessibility of different regions of the construct 4. If a secondary structure forms in the overhang or if a protein blocks the binding site of the imager strand, the binding frequency decreases. Therefore, the folding of a secondary structure or a conformational change between two structures can be detected if they result in different accessibility to the imager strand, even if end-to-end separation does not change. In this respect, FRET-PAINT provides an orthogonal method to smFRET in the study of secondary structure formation.
We have used these concepts to study the accessibility of telomeric overhangs of physiological lengths in the absence and presence of various co-factors 6. In this earlier investigation, we demonstrated the feasibility of resolving periodic patterns that depend on the length and folding topography in long telomeric overhangs using FRET-PAINT. In that case, the large number of folding states with potentially overlapping FRET distributions made the system too complicated to study using the variations in end-to-end separation. In the current investigation, we demonstrate the advantages of using FRET-PAINT over smFRET at detecting formation of secondary structures formed by short nucleic acids, which result in small or undetectable changes in end-to-end separation upon folding.
We used the G-Triplex (G3) structure formed by the human telomeric sequence as a model system 7. The human telomeric sequence consists of repeats of the GGGTTA sequence, which will be referred to as a G-Tract (GTr). G3 structures are proposed to fold in telomeric overhangs when only three G-Tracts are available at the ends or between consecutive folded G-quadruplex (G4) structures, which require four repeats 8,9. G3 is also detected as a folding intermediate along the G4 folding pathway 10–12. We investigated the variation in the binding frequency of an imager strand (Cy5-PNA: TAACCCTT-Cy5) to telomeric overhangs (via underlined nucleotides in Cy5-PNA) that contain 1–3 GGGTTA repeats (Figure 1A–B, constructs 1GTr, 2GTr, and 3GTr). Binding of Cy5-PNA to the overhang results in FRET bursts (Figure 1C). Histograms of these FRET bursts are illustrated in Figure 1D (after subtracting the donor-only peak where no binding event takes place) 13. As expected, the 1GTr, 2GTr, and 3GTr histograms contain one, two, or three peaks, which represent binding to the available G-Tracts. In case of 3GTr construct (bottom histogram in Figure 1D), the peak with highest FRET efficiency is due to binding of Cy5-PNA to the G-Tract closest to Cy3 (junction between double and single stranded DNA), while the peak with lowest FRET efficiency is due to binding of Cy5-PNA to G-Tract at the 3’-end. The binding of Cy5-PNA to the first or third G-Tract only differs by 12 nt single stranded DNA, which is very compact due to the small persistence length of ssDNA (~2 nm at physiological ionic conditions) 14. Nevertheless, the FRET distributions obtained via FRET-PAINT are detailed enough to show multiple peaks when two or three G-Tracts are available on the overhang. In contrast, the distributions obtained through a traditional smFRET assay, by placing the fluorophores across the overhang (within a similar FRET range), are largely featureless and differ minimally from each other for 1GTr, 2GTr, and 3GTr constructs in 150 mM K+ (Figure 1E). These distributions are also effectively identical to the corresponding distributions in 150 mM Li+ (Figure 1F).
Figure 1.
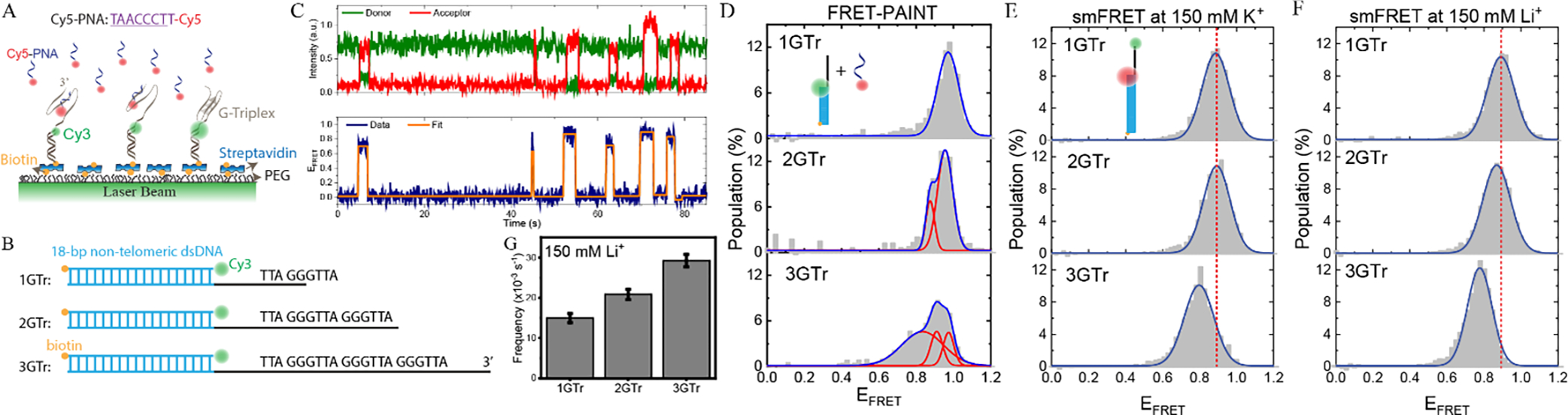
FRET-PAINT assay to study G-Triplex formation. (A) Schematic for FRET-PAINT assay where pdDNA constructs are immobilized on a surface and a Cy5-PNA probe (complementary to telomeric sequence) scans the surface-immobilized molecules for accessible sites. Folding of G3 reduces access of Cy5-PNA. (B) Schematic of pdDNA constructs containing 1, 2, or 3 G-Tracts in the overhang. (C) A single molecule time trace illustrating multiple binding events of Cy5-PNA. The FRET trace is fitted by an automated bias-free algorithm (AutostepFinder 19) to determine the binding events. (D) FRET histograms formed by combining the several hundred binding events for each construct in 150 mM K+. The histograms for 1GTr, 2GTr, and 3GTr constructs are fitted to 1, 2, or 3 Gaussian peaks, respectively. The following are peak positions and number of molecules (N): xc=0.97±0.06 and N=124 for 1GTr, xc1=0.96±0.04 and xc2=0.88±0.02 and N=87 for 2GTr, and xc1=0.97±0.03, xc2=0.91±0.03, and xc3=0.83±0.11 and N=146 for 3GTr. The uncertainties in the peak positions are standard deviations due to Gaussian fitting. (E-F) Corresponding histograms obtained using a traditional smFRET assay where donor-acceptor fluorophores are placed across the overhang in (E) 150 mM K+ and (F) 150 mM Li+. There are minimal variations among the three overhangs, with the peak positions at xc=0.90±0.05 for 1GTr, xc=0.89±0.05 for 2GTr, and xc=0.80±0.06 for 3GTr in K+ and xc=0.89±0.09 for 1GTr, xc=0.87±0.09 for 2GTr, and xc=0.78±0.08 for 3GTr in Li+. Each histogram had more than 100 molecules. The uncertainties in the peak positions are standard deviations due to Gaussian fitting. The fitting errors are consistently ±0.01 or less for all constructs. (G) The Cy5-PNA binding frequencies for constructs containing 1–3 G-Tracts in 150 mM Li+, at which secondary structure formation is not expected. In agreement with expectations, the frequency increases as the number of binding sites increases. The number of binding events were 248, 306, and 402 for 1GTr, 2GTr, and 3GTr, respectively.
Furthermore, for very short oligonucleotides, the change in end-to-end separation upon folding of an unstructured DNA into a secondary structure (such as G3) could be small enough to pose a challenge for detection with smFRET 15. To illustrate, we provide comparative smFRET histograms for the 3GTr construct in K+ or Li+ (up to 500 mM concentration), which are essentially identical regardless of how effective the cations are in stabilizing the G3 structure (Supplementary Figure S1).
FRET-PAINT data also allows the analysis of the variation in the binding frequency (total number of binding events divided by total observation time) of the imager strand Cy5-PNA. In the absence of any secondary structures, the binding frequency should monotonically increase as more binding sites (G-Tracts) become available. This prediction is confirmed when the FRET-PAINT measurements are performed in 150 mM Li+, at which we do not expect any stable secondary structures (Figure 1G). The binding frequencies are (15.0±1.1)x10−3 s−1, (20.9±1.3)x10−3 s−1, (29.3±1.5)x10−3 s−1 for 1GTr, 2GTr, and 3GTr constructs, respectively.
Using this concept, we tested G-Triplex formation in 150 mM K+, at which we expect G3 formation for constructs that have three G-Tracts. To mimic various structural contexts that might occur in long telomeric overhangs, we placed the potential G3 structures in the vicinity of a double stranded DNA (dsDNA), G4, or free ends. Figure 2A shows the structural variations we tested: (i) dsDNA on one side and free-end on the other (nGTr constructs in Figure 1); (ii) dsDNA on one side and G4 on the other (nGTr-G4 constructs); (iii) G4 on one side and free-end on the other (G4-nGTr constructs); (iv) G4s on both sides (G4-nGTr-G4 constructs. In all these cases, we increased the number of G-Tracts from one to three (n=1–3) and monitored the change in the binding frequency of Cy5-PNA (Figure 2B). A large drop in binding frequency for the construct with three G-Tracts was interpreted as effective G-Triplex formation, while a modest drop was interpreted as evidence for G-Triplex destabilization due to neighboring structures. To avoid interference between G4 forming sequence and G3 forming sequence, we did not use the telomeric G4 forming sequence. Instead, we used the (GGGT)3GGG sequence, which breaks the symmetry in the sequence and is much more stable than the G3 or G4 formed by telomeric sequences 16.
Figure 2.
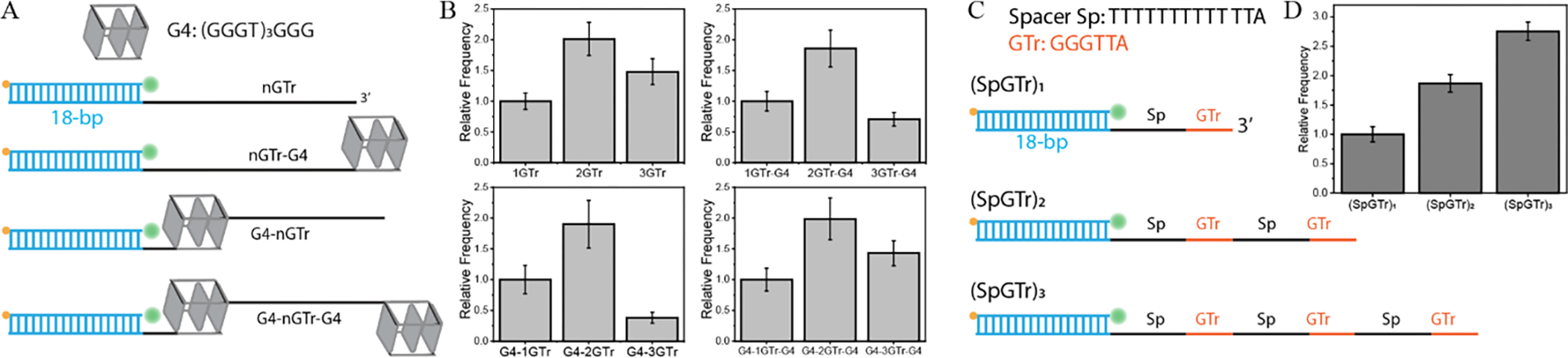
DNA constructs and frequency analysis in 150 mM K+. (A) G3 formation was tested in four different structural contexts where we had 1, 2, or 3 G-Tracts in each group (a total of 12 pdDNA constructs). (B) Binding frequency histograms. Binding frequencies for each group are analysed, and results are normalized with respect to the construct that contains a single G-Tract. In all cases, the frequency increases when the available G-Tracts are increased from one to two but decreases when they are increased from two to three, illustrating the impact of G3 formation on Cy5-PNA accessibility. Depending on neighbouring structures, G3 formation efficiency shows large variations, most efficient for G4–3GTr and least efficient for G4–3GTr-G4. The number of binding events were 356, 289, and 297 for 1GTr, 2GTr, and 3GTr, respectively; 410, 294, and 299 for 1GTr-G4, 2GTr-G4, and 3GTr-G4, respectively; 135, 299, and 154 for G4–1GTr, G4–2GTr, and G4–3GTr, respectively; 170, 355, and 224 for G4–1GTr-G4, G4–2GTr-G4, and G4–3GTr-G4, respectively. (C) Schematics of DNA constructs where spacer sequences between consecutive G-Tracts are increased from 3 nt (TTA) to 13 nt (TTTTTTTTTTTTA), which prohibits G3 formation. (D) Results of FRET-PAINT measurements illustrating a systematic increase in Cy5-PNA binding frequency as the number of G-Tracts increases. The number of binding events were 156, 148, and 430 for (SpGTr)1, (SpGTr)2, and (SpGTr)3, respectively.
Similar to the Li+ data, the binding frequency is higher for all constructs that contain two G-Tracts compared to those that contain one G-Tract at 150 mM K+ (Figure 2B). To facilitate the comparison, all frequency data were normalized with respect to the respective construct that contains a single G-Tract. The binding frequency for 2GTr, 2GTr-G4, G4–2GTr, and G4–2GTr-G4 constructs were 2.01±0.27, 1.86±0.30, 1.90±0.39, and 1.99±0.34 fold higher than those of 1GTr, 1GTr-G4, G4–1GTr, and G4–1GTr-G4 constructs, respectively.
On the other hand, the Cy5-PNA binding frequency decreases for all constructs when the number of G-Tracts is increased from two to three in K+. We interpret this decrease in binding frequency as evidence for G3 formation, which prevents Cy5-PNA to access the overhang. In two of the cases, the G3 formation was prominent enough to reduce the binding frequency even below that of single G-Tract constructs. The binding frequencies for 3GTr, 3GTr-G4, G4–3GTr, and G4–3GTr-G4 constructs were 1.48±0.21, 0.71±0.11, 0.38±0.09, and 1.43±0.20 fold those of 1GTr, 1GTr-G4, G4–1GTr, and G4–1GTr-G4 constructs, respectively. One-way ANOVA test (Supplementary Table S2) showed that the observed differences in relative frequencies within each group are statistically significant (p=0.001). The non-normalized frequencies of the data in Figure 2B are given in Supplementary Table S3. In comparison, we observed an order of magnitude drop in the frequency between a construct that forms a G4 compared to that containing a single G-Tract 4, demonstrating the enhanced stability of G4 compared to G3.
To illustrate specificity of the decrease in frequency to G3 formation, we maintained identical ionic conditions (150 mM K+ and 2 mM Mg2+) but increased the length of spacers between the G-Tracts to 13 nt (TTTTTTTTTTTTA in Figure 2C, which adds 10T to the former TTA spacers). The complete sequences of these constructs are given in Supplementary Table S1. These spacers will need to be incorporated as loops within the G3 structure; however, long loops have high entropic cost and destabilize the G3. In this case, we observed a systematic increase in frequency with the number of G-Tracts (Figure 2D), demonstrating the lack of secondary structure formation. The associated FRET histograms formed by binding of Cy5-PNA to the constructs in Figure 2C are given in Supplementary Figure S2.
The decrease in imager probe binding frequency upon secondary structure formation is the primary premise of using FRET-PAINT to detect secondary structure formation, regardless of whether the end-to-end separation changes during the folding process. Figure 2B shows that G3 formation showed significant variation depending on neighboring structures. Interestingly, the most effective G3 formation is observed when the three G-Tracts are adjoined by a folded G4 on their 5′-side while their 3′-side is free (G4–3GTr construct). In the context of telomeric overhangs, this is the case when three G-Tracts are otherwise exposed at the 3′-end of the overhang (while a folded G4 is located on their 5′-side). In this case, the G-Triplex formation could serve as a protective mechanism against exonuclease activity or telomerase access when G4 formation is not possible. On the other hand, G3 formation is less effective when such repeats are between a G4 and dsDNA (3GTr-G4 construct), as would be the case for the junction region between double and single stranded telomere, or when they are located between two folded G4s (G4–3GTr-G4 construct), as would be the case at intermediate regions of the overhang. The junction region has been demonstrated to be effectively protected by the shelterin complex 6, while unfolded sequences in the intermediate regions of the overhang could be protected by POT1-TPP1 17,18.
Conclusions
As illustrated by this model system, FRET-PAINT provides detailed information regarding secondary structure formation which would otherwise be challenging to attain using alternative methods, such as smFRET. As FRET-PAINT relies on variations in the binding frequency of a short imaging strand, rather than variations in end-to-end separation, it provides an orthogonal means to smFRET and has advantages for studying structures whose folding does not result in a significant change in end-to-end separation or for distinguishing between structures that might have similar sizes but different accessibilities. In addition, FRET-PAINT provides additional flexibility in construct design compared to smFRET as it requires placing only one fluorophore on the nucleic acid construct while the other fluorophore is placed on the imager strand. The implications of our data for most likely sites for G-Triplex formation in long telomeric overhangs, depending on neighboring structures, are also discussed.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (1R15GM123443 and 1R15GM146180).
Footnotes
Conflicts of interest
There are no conflicts to declare.
Electronic Supplementary Information (ESI) available: Materials and Methods, Sequences of DNA constructs. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Maleki P, Budhathoki JB, Roy WA and Balci H, Molecular Genetics and Genomics, 2017, 292, 483–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jungmann R, Steinhauer C, Scheible M, Kuzyk A, Tinnefeld P and Simmel FC, Nano Lett, 2010, 10, 4756–4761. [DOI] [PubMed] [Google Scholar]
- 3.Auer A, Strauss MT, Schlichthaerle T and Jungmann R, Nano Lett, 2017, 17, 6428–6434. [DOI] [PubMed] [Google Scholar]
- 4.Shiekh S, Mustafa G, Kodikara SG, Hoque ME, Yokie E, Portman JJ and Balci H, Proc Natl Acad Sci U S A, DOI: 10.1073/pnas.2202317119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J, Park S and Hohng S, Mol Brain, DOI: 10.1186/s13041-018-0414-3. [DOI] [Google Scholar]
- 6.Shiekh S, Jack A, Saurabh A, Mustafa G, Kodikara SG, Gyawali P, Hoque ME, Pressé S, Yildiz A and Balci H, Nucleic Acids Res, 2022, 50, 12885–12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koirala D, Mashimo T, Sannohe Y, Yu Z, Mao H and Sugiyama H, Chem Commun (Camb), 2012, 48, 2006–2008. [DOI] [PubMed] [Google Scholar]
- 8.Abraham Punnoose J, Ma Y, Hoque ME, Cui Y, Sasaki S, Guo AH, Nagasawa K and Mao H, Biochemistry, 2018, 57, 6946–6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abraham Punnoose J, Cui Y, Koirala D, Yangyuoru PM, Ghimire C, Shrestha P and Mao H, J Am Chem Soc, 2014, 136, 18062–18069. [DOI] [PubMed] [Google Scholar]
- 10.Stadlbauer P, Trantírek L, Cheatham TE, Koča J and Šponer J, Biochimie, 2014, 105, 22–35. [DOI] [PubMed] [Google Scholar]
- 11.Hou XM, Ben Fu Y, Wu WQ, Wang L, Teng FY, Xie P, Wang PY and Xi XG, Nucleic Acids Res, 2017, 45, 11401–11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Hou XM, Wang PY, Xi XG and Li M, J Am Chem Soc, 2013, 135, 6423–6426. [DOI] [PubMed] [Google Scholar]
- 13.Mustafa G, Shiekh S, Gc K, Abeysirigunawardena S and Balci H, Nucleic Acids Res, 2021, 49, 3371–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy MC, Rasnik I, Cheng W, Lohman TM and Ha T, Biophys J, 2004, 86, 2530–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aznauryan M, Søndergaard S, Noer SL, Schiøtt B and Birkedal V, Nucleic Acids Res, 2016, 44, 11024–11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magbanua E, Zivkovic T, Hansen B, Beschorner N, Meyer C, Lorenzen I, Grotzinger J, Hauber J, Torda AE, Mayer G, Rose-John S and Hahn U, RNA Biol, 2013, 10, 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor DJ, Podell ER, Taatjes DJ and Cech TR, J Mol Biol, 2011, 410, 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang H, Kreig A, Calvert J, Lormand J, Kwon Y, Daley JM, Sung P, Opresko PL and Myong S, Structure, 2014, 22, 842–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loeff L, Kerssemakers JWJ, Joo C and Dekker C, Patterns, DOI: 10.1016/j.patter.2021.100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


