Abstract
Many resistance genes deployed against pathogens in crops are intracellular nucleotide-binding (NB) leucine-rich repeat (LRR) receptors (NLRs). The ability to rationally engineer the specificity of NLRs will be crucial in the response to newly emerging crop diseases. Successful attempts to modify NLR recognition have been limited to untargeted approaches or depended on previously available structural information or knowledge of pathogen-effector targets. However, this information is not available for most NLR-effector pairs. Here, we demonstrate the precise prediction and subsequent transfer of residues involved in effector recognition between two closely related NLRs without their experimentally determined structure or detailed knowledge about their pathogen effector targets. By combining phylogenetics, allele diversity analysis, and structural modeling, we successfully predicted residues mediating interaction of Sr50 with its cognate effector AvrSr50 and transferred recognition specificity of Sr50 to the closely related NLR Sr33. We created synthetic versions of Sr33 that contain amino acids from Sr50, including Sr33syn, which gained the ability to recognize AvrSr50 with 12 amino-acid substitutions. Furthermore, we discovered that sites in the LRR domain needed to transfer recognition specificity to Sr33 also influence autoactivity in Sr50. Structural modeling suggests these residues interact with a part of the NB-ARC domain, which we named the NB-ARC latch, to possibly maintain the inactive state of the receptor. Our approach demonstrates rational modifications of NLRs, which could be useful to enhance existing elite crop germplasm.
Keywords: NLRs, plant immunity, rational engineering, wheat, wheat stem rust
Nucleotide-binding leucine-rich repeat (NB-LRR) receptors (NLRs) are intracellular immune receptors that recognize pathogen-derived molecules, called effectors, that are secreted into the plant cell by a pathogen with the goal of subverting plant immune responses. NLRs deploy different means of monitoring the cell by either directly binding to effectors or recognizing the action of an effector on a plant component. Plant components whose modification is recognized by NLRs are referred to as guardees or decoys. The recognition of an effector leads to the activation of downstream responses that often culminate in localized cell death, referred to as hypersensitive response (Dodds and Rathjen 2010). Although plant NLRs have been shown to evolve rapidly and recognize a wide range of structurally diverse effectors, robust approaches for rational modifications of NLRs and their recognition specificities remain limited (Tamborski and Krasileva 2020).
Most NLRs share a common, multi-domain structure including a variable N-terminal domain, a central NB-ARC domain, and a C-terminal LRR domain. There are three types of N-terminal domains that enable separating NLRs into three categories: coiled-coil (CC) domains in CNLs, Toll/interleukin-1 receptor domains in TNLs, and Resistance to Powdery Mildew 8 (RPW8)–like domains in RNLs (Shao et al. 2016). Furthermore, a subset of NLRs have one or more atypical plant protein domains integrated into their canonical NLR structure and are referred to as NLR-IDs (integrated domains) (Grund et al. 2019). NLR-IDs usually function in pairs and are often genetically linked in a head-to-head orientation and are thought to exemplify the specialization of NLRs into a sensor and signaler function (Feehan et al. 2020). The integrated domains act as effector binding platforms and have been shown to directly interact with effectors (Cesari et al. 2013; Maqbool et al. 2015).
Several studies have already successfully engineered recognition specificities of NLRs. In direct-binder NLRs, recognition was transferred from one NLR to another through chimeras (Ellis et al. 1999; Shen et al. 2003; Slootweg et al. 2017) and effector recognition was expanded through the means of random and targeted mutagenesis (Chapman et al. 2014; Giannakopoulou et al. 2015; Racman et al. 2001; Segretin et al. 2014). Also, the modification of a guardee has proven successful through the introduction of effector substrate sites (DeYoung et al. 2012; Kim et al. 2016; Pottinger et al. 2020). Several studies have provided additional insights into the interaction of effectors with IDs or related host targets that foretell a promising future for engineering NLRs with IDs (Maidment et al. 2021; Ortiz et al. 2017). Some previous work showcased major advances in engineering the IDs of NLR-IDs by broadening or changing their recognition specificity (Cesari et al. 2022; De la Concepcion et al. 2019; Liu et al. 2021). A very innovative approach to NLR engineering involved the integration of nanobodies into the canonical NLR domain structure to recognize epitope-tagged viral particles (Kourelis et al. 2021a). Despite such efforts in NLR-ID engineering, however, we have yet to make significant progress in engineering NLRs that bind to effectors through their LRR domain.
The recent cryo-electron microscopy (cryo-EM) structures of plant NLRs have greatly enhanced our understanding of activated immune receptor function (Förderer et al. 2022; Ma et al. 2020; Martin et al. 2020; Wang et al. 2019a, b; Zhao et al. 2022). The CNLHOPZ-ACTIVATED RESISTANCE 1 (ZAR1) and the kinase RESISTANCE-RELATED KINASE 1 (RKS1) guard the PBS1-LIKE 2 (PBL2) kinase and initiate immune signaling upon its modification (Lewis et al. 2010; Wang et al. 2015). ZAR1 and RKS1 exist in pre-formed complexes that assemble into pentamers upon effector-mediated modification of PBL2 (Wang et al. 2019a, b). The CC domains in this pentamer form a funnel-like structure shown to be a calcium-permeable channel (Bi et al. 2021; Wang et al. 2019a). Two groups independently defined the structures of the two direct-binder TNLs, RECOGNITION OF PERONOSPORA PARASITICA 1 (RPP1) and RECOGNITION OF XOPQ1 (ROQ1), and found that they assemble into tetramers upon activation through their cognate effectors (Martin et al. 2020; Ma et al. 2020). Two recent cryo-EM structures of Sr35 with its cognate effector AvrSr35 have demonstrated that the complex assembles into a pentameric resistosome structure (Förderer et al. 2022; Zhao et al. 2022). Some residues within the last eight LRR units of Sr35 mediate interaction with AvrSr35, and the authors were able to transfer residues of Sr35 onto orphan receptors to enable their recognition of AvrSr35 (Förderer et al. 2022). These studies confirmed the importance of intramolecular interactions to maintain the inactive state of NLRs and intermolecular interactions for resistosome assembly and immune signaling.
NLRs have provided a valuable source of disease resistance in many crop species. Wheat stem rust disease is caused by the fungal agent Puccinia graminis f. sp. tritici and has been a major threat to wheat production, especially since the emergence of the highly virulent P. graminis f. sp. tritici race Ug99 (Singh et al. 2011). A large number of resistance genes that protect wheat against Ug99 have been cloned and deployed in recent years (Arora et al. 2019; Chen et al. 2017; Saintenac et al. 2013; Steuernagel et al. 2016; Zhang et al. 2021). Among them are Sr50 and Sr33, two CNL resistance genes that are highly similar, with an amino acid identity of approximately 80% (Mago et al. 2015; Periyannan et al. 2013). They have been introgressed into wheat from Secale cereale (rye) and Aegilops tauschii, respectively, and are closely related homologs of the barley MILDEW RESISTANCE LOCUS A (MLA) or R GENE HOMOLOG 1) (RGH1) family (Halterman et al. 2001; Mago et al. 2015; Periyannan et al. 2013; Wei et al. 2002). This family has been associated with high allelic and functional diversification, exemplified by the more than 30 barley MLA alleles that confer resistance to powdery mildew (Seeholzer et al. 2010).
Sr50 has been shown to directly bind to its cognate effector AvrSr50 (Chen et al. 2017). The effector recognized by Sr33 remains unknown to date but is assumed to be distinct from AvrSr50, based on resistance to different P. graminis f. sp. tritici races (Mago et al. 2015). Sr50 and Sr33 are interesting candidates to investigate, as their autoactivity seems directly linked to the level of expression, as observed by Cesari et al. (2016) and this study, which suggests that they might be in a sensitive on-off state. Moreover, with 80% sequence identity the recognition specificities of these NLRs must be defined by a small number of amino acids in their LRR regions.
Here, we report the engineering of Sr33 to recognize the effector AvrSr50. We used a combination of phylogenetics, sequence diversity analysis with Shannon entropy, and structural modeling to predict the region within the Sr50 LRR that mediates binding to AvrSr50 and to identify individual residues involved in the interaction. We confirmed that the predicted region was sufficient to enable effector recognition through the generation of a chimeric NLR that we named Sr33/50. This chimeric NLR was able to recognize and initiate cell death to AvrSr50 in wheat protoplasts and Nicotiana benthamiana. We subsequently generated the synthetic NLRSr33syn, in which groups of residues within the Sr33 LRR were changed to the corresponding sites of Sr50. The exchange of 12 amino acids was sufficient for Sr33 to initiate cell death in wheat protoplasts and N. benthamiana in the presence of AvrSr50. We also observed that Sr50 could exhibit autoactivity in the wheat protoplast cell-death assay, whereas Sr33 did not. We therefore used structural modeling to identify potential sites that could mediate this phenotype. We identified two residues that appear to be in close contact with the NB-ARC domain, and mutation of these sites led to a stark reduction in autoactivity of Sr50. This study therefore demonstrates that the combination of sequence diversity analyses and structural modeling can be sufficient to identify and subsequently alter sites mediating effector recognition and autoactivity in direct binder NLRs.
Results
We have previously identified highly variable NLRs (hvNLRs) by analyzing natural diversity of pangenomes of two model species, Arabidopsis thaliana and Brachypodium distachyon (Prigozhin and Krasileva 2021). The highly variable amino acid residues within hvNLR orthologous groups together with structural modeling were predictive of the cognate effector-binding sites in A. thaliana (Prigozhin and Krasileva 2021). We decided to translate this knowledge to crop species and test if the same approach can accurately predict residues mediating NLR-effector interaction and subsequently guide rational engineering of NLRs. For this work, we chose hvNLRs Sr33 and Sr50 that have been independently introgressed into bread wheat from Aegilops tauschii and rye, respectively (Fig. 1A). We initially assessed sequence diversity of the Sr33 and Sr50 homologous groups with sequences obtained with BLAST search against the nonredundant database in The National Center for Biotechnology Information. The multiple sequence alignments (MSA) of each homologous group indicated that there was sufficient sequence diversity to predict binding residues in Sr50 (D. Prigozhin personal communication) (Krasileva et al. 2023). This approach proved to be an easily accessible way to obtain sequences for Shannon entropy calculation. However, as Sr33 and Sr50 displayed approximately 80% sequence identity, many collected homologs were shared by the two groups. It was, therefore, necessary to refine the initial analysis to curate Sr33 and Sr50 homologous groups with non-overlapping sets of sequences. We inferred the phylogenetic tree with MLA family members collected from Pooideae species, 14 wheat genomes (Supplementary Table S1), and selected reference NLRs (Kourelis et al. 2021b). Despite complex NLR evolution within Triticeae species, we were able to resolve the NLRs into four distinct phylogenetic subfamilies representing Sr50, Sr33, HvMLA1, and HcMLA1 based on similar phylogenetic distances (Fig. 1B).
Fig. 1.
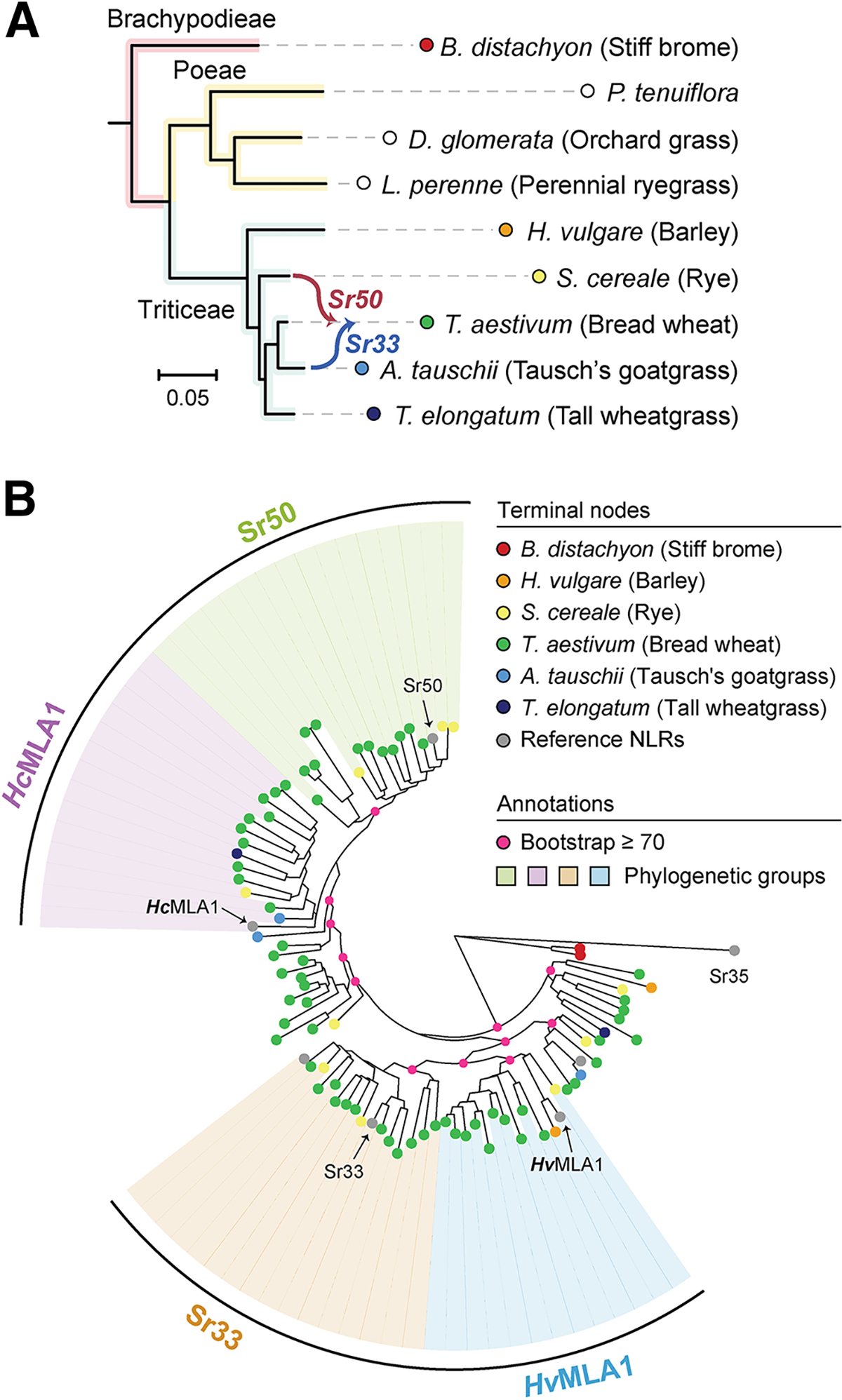
Expanded evolutionary analyses of Sr33- and Sr50-related MILDEW RESISTANCE LOCUS A sequences (MLAs). A, The species tree of Pooideae members. The source of Sr33 and Sr50 introgression is indicated, and branches are colored based on tribe membership. No MLA family members were identified from the Poeae species, and these species are indicated with empty circles. B, The phylogenetic tree of selected MLA family members. The bootstraps are provided only for nodes used to distinguish the four phylogenetic groups or higher nodes. The four phylogenetic groups were named based on the representative nucleotide-binding leucine-rich repeat receptors (NLRs) from RefPlantNLR placed within each group (Kourelis et al. 2021b): Sr33, Sr50, HcMLA1 from Hordeum chilense, and HvMLA1 from Hordeum vulgare. The phylogenetic groups were curated based on the bootstrapping values ≥98, except for the HcMLA1 family, which has a bootstrapping value of 63. The tree was rooted with the outgroup Sr35.
This analysis allowed us to calculate Shannon entropy for each phylogenetic group with at least 13 unique NLR sequences (Fig. 2A; Supplementary Table S2). The entropy plots of these four phylogenetic groups appear almost identical across the whole NLR, suggesting that the same positions of variable or invariable sites would be conserved across the MLA gene family. To incorporate the location of highly variable sites within the NLR structure, we visualized Shannon entropy measures of the individual residues within each LRR unit that form the beta sheets, which together compose the inner concave surface of the LRR (Fig. 2B). We noticed that LRRs 8 to 16 of the Sr50 group contained a cluster of sites with high Shannon entropy that we hypothesized to be a potential area effector-binding pocket (Fig. 2B). High Shannon entropy scores occur similarly towards the end of the LRRs in units 8 to 16 in all four phylogenetic groups. This suggests more specifically that the sites mediating effector recognition are in the same LRR region in the MLA family members, despite their ability to recognize distinct effectors. These results indicate that highly variable sites within the NLRs of this family could be where functional diversification of paralogs occurs.
Fig. 2.
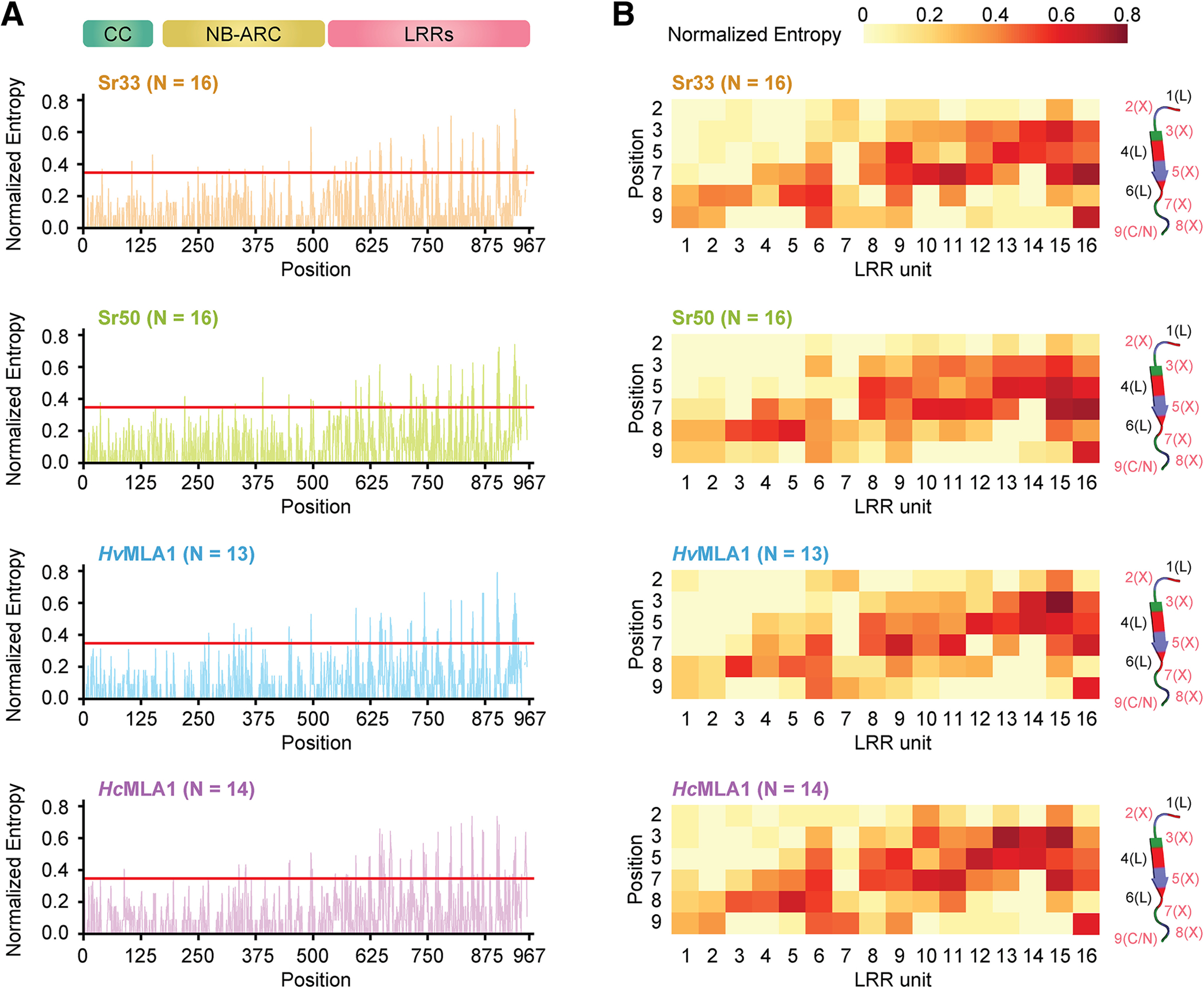
Shannon entropy scores of Sr33- and Sr50-related MILDEW RESISTANCE LOCUS A (MLAs) reveal variable hotspots in the nucleotide-binding (NB) leucine-rich repeat (LRR) receptor structure. A, The normalized Shannon entropy of four phylogenetic groups, ranging from 0 (no variability) to 1 (complete variability). The red line indicates the previously used cut-off (Prigozhin and Krasileva 2021) to determine highly variable residues (Shannon entropy = 1.5 or normalized Shannon entropy = 0.347). The normalized Shannon entropy was calculated with the indicated number of members (N) in each phylogenetic group. B, The distribution of the normalized Shannon entropy across the LRR domain for the four phylogenetic groups. The heat map provides a detailed view of the normalized entropy for each individual LRR repeat. Each LRR unit and each position within the LRR unit was defined according to the general motif LXXLXLXX(C/N), as annotated in the secondary structure.
To test which LRR region confers effector recognition, we created a chimeric protein consisting of Sr33 and LRRs 8 to 14 of Sr50, based on the presence of restriction sites in both genes, which we will refer to as Sr33/50 (Supplementary Fig. S1A; Supplementary Table S3). We utilized a wheat protoplast cell-death assay to test the recognition specificities of the synthetic NLRs that we generated, as previously described (Saur et al. 2019; Yoshida et al. 2009). In this assay, wheat protoplasts are transfected with luciferase and a combination of NLRs and effectors. A positive interaction between NLR and effector leads to cell death as exemplified by a lack of luciferase activity 2 days after transfection. Co-transfection of Sr50 with AvrSr50 led to cell death and strongly reduced luminescence, whereas co-transfection of Sr33 with AvrSr50 did not lead to a reduction as compared with NLRs alone (Supplementary Fig. S1B). Although Sr33/50 displayed an intermediate autoactive phenotype in heterologous N. benthamiana (Supplementary Fig. S2A; Supplementary Table S4), the wheat protoplast assay demonstrated that Sr33/50 was able to recognize AvrSr50 (Supplementary Fig. S1B). This confirmed that the transfer of LRRs 8 to 14 from Sr50 to Sr33 was sufficient to transfer effector specificity. We therefore decided to focus our engineering efforts on LRRs 8 to 14 and aimed to identify the individual amino acids in this region that are involved in the recognition of AvrSr50.
Among 184 amino acids within LRRs 8 to 14, 26 positions differ between Sr50 and Sr33 (Supplementary Fig. S1A; Supplementary Table S3). To define specific LRR residues that mediate interaction between Sr50 and AvrSr50, we selected amino acids that fulfilled most of the following criteria: high Shannon entropy signature, differing amino acid side chain property between Sr33 and Sr50, and exposure at the surface of the beta sheets that face the NB-ARC domain (Fig. 3A, B, and C; Supplementary Fig. S3). We synthesized several iterations of Sr33 carrying mutations in these predicted amino acid residues by transferring the corresponding sites of Sr50 and tested these synthetic NLRs in wheat protoplasts. Sr3310AA contained 10 amino acids transferred from Sr50; this was the minimum number of substitutions among the constructs we generated (Supplementary Table S4). Other synthetic Sr33 variants had one to three additional substitutions introduced into Sr3310AA (Supplementary Table S4).
Fig. 3.
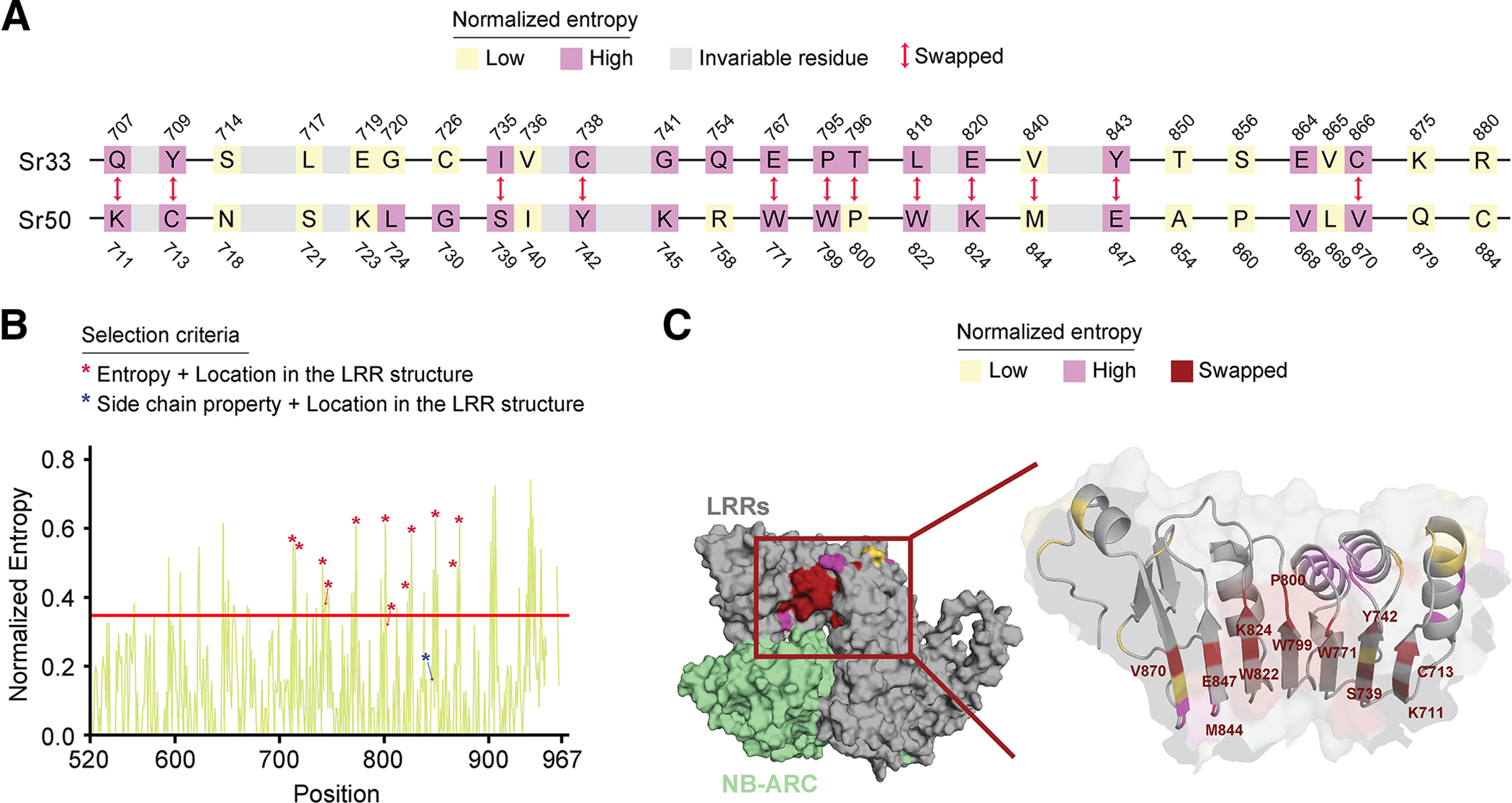
Diversity and structural modeling guided selection of amino acids mediating effector recognition. A, Amino-acid residues in Sr33 and Sr50 targeted for swapping. The amino acids depicted include all variable positions in leucine-rich repeats (LRRs) 8 to 14 between Sr33 and Sr50. Invariable amino acids within these LRRs are indicated as gray boxes if they exist between closely located variable residues; otherwise, the invariable positions were omitted. Low and high entropy positions were determined based on the normalized Shannon entropy cut-off of 0.347 and are indicated in different colors. The 12 amino-acid sequences used to generate Sr33syn are indicated as ‘swapped’. B, The normalized Shannon entropy of the Sr50 phylogenetic group in the LRR region. The asterisks indicate amino acids chosen for the generation of Sr33syn. The residues annotated with asterisks were in the inner concave surface of the LRR and were selected based on high entropy in the Sr33 or Sr50 phylogenetic groups (red asterisks) or the side chain property (blue asterisks). The blue asterisk indicates Sr50 M844. C, Variable amino acids mapped to the predicted Sr50 structure. The variable residues identified from LRRs 8 to 14 were mapped to the predicted structure. A portion of the Sr50 LRR (710 to 890) was zoomed in to highlight the variable residues. The swapped residues are colored in red, and other variable residues are colored based on the entropy of the Sr50 phylogenetic group.
Among the constructs we tested, the synthetic NLR Sr33syn carries 12 selected amino-acid polymorphisms (Fig. 3A) and initiated cell death in wheat protoplasts in an AvrSr50-dependent manner, while Sr33syn− did not (Fig. 4A). These two synthetic NLRs differ only in one amino-acid residue that seems to affect the ability to recognize AvrSr50. While Sr33syn carries a methionine in position 840 transferred from Sr50, Sr33syn− carries a valine residue (Fig. 3A; Supplementary Fig. S2E; Supplementary Table S4). Our observation is, therefore, that 12 amino-acid residues were the minimal amount needed to transfer recognition of AvrSr50 to Sr33 among the synthetic variants we tested (Supplementary Table S4). To substantiate this observation, we infiltrated these mutants in N. benthamiana with AvrSr50. It has been published that Sr50 and Sr33 can exhibit autoactivity when overexpressed in N. benthamiana but not when expressed under the weaker native Sr50 promoter (Cesari et al. 2016). We also only observed autoactivity in N. benthamiana when NLR expression was driven by the 35S cauliflower mosaic virus promoter (p35S) (Supplementary Fig. S2C) but not when we used the previously described weaker promoter RECOGNITION OF PERONOSPORA PARASITICA 13 promoter (pRPP13) (Rentel et al. 2008) (Fig. 4B; Supplementary Fig. S2A to D). We therefore decided to drive expression of NLRs with pRPP13, as it led to little or no autoactivity of Sr50 and Sr33 in our conditions, which allowed us to differentiate between autoactivity and effector recognition in N. benthamiana. We infiltrated our synthetic NLR Sr33syn to test if it can induce cell death in an effector-dependent manner in N. benthamiana. Sr33syn showed a clear cell-death response when co-infiltrated with AvrSr50, which confirms that the transfer of 12 amino acids is sufficient for gain of effector recognition (Fig. 4B). In contrast, the effector variant AvrSr50_QCMJC that evades recognition by Sr50 is also not recognized by Sr33syn, demonstrating that the gained effector recognition is AvrSr50-specific. These results demonstrate that we successfully predicted amino acids involved in the recognition of AvrSr50 through Sr50 and that effector recognition can be transferred to a closely related NLR.
Fig. 4.
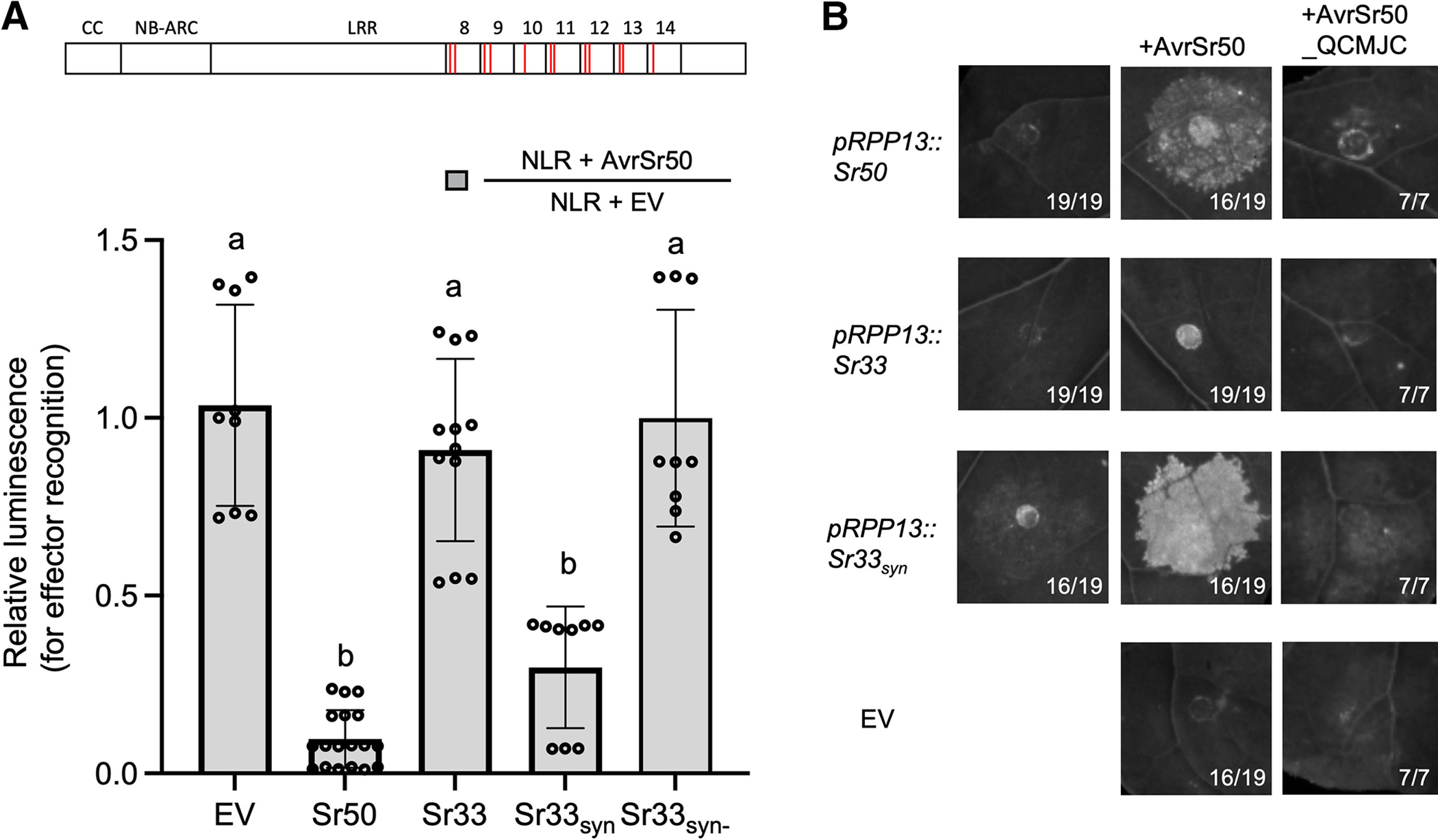
The transfer of 12 amino acids between closely related nucleotide-binding leucine-rich repeat (LRR) receptors (NLRs) is sufficient to transfer recognition specificity. A, Twelve amino-acid changes on Sr33 are sufficient to enable recognition of AvrSr50, as exemplified by the synthetic NLR Sr33syn. Sr33 is unable to induce cell death in wheat protoplasts in response to AvrSr50. In contrast to Sr33, Sr33syn with 12 amino-acid substitutions is able to recognize and execute cell death in an AvrSr50-dependent manner. The synthetic NLR Sr33syn−, which contains only 11 amino-acid substitutions does not recognize AvrSr50. Schematic above the graphs shows Sr33syn and all substitutions are marked as red lines in the LRRs. Numbers above the LRR signify which LRR repeat this region corresponds to. Luciferase activity was determined approximately 36 h after transfection, as a proxy for cell death. The relative luminescence reflects the changes driven by effector recognition, and the low relative luminescence level represents cell deaths by effector recognition. This value was obtained by normalizing the luminescence of each NLR construct (or empty vector [EV] as a control) in the AvrSr50 co-transfection by the luminescence of that NLR construct in the EV co-transfection. All values obtained in at least three independent experiments are indicated by dots, and error bars indicate standard deviation. The equality of group means was assessed by one-way analysis of variance and subsequent Tukey post-hoc tests. The samples assigned the same letter (a or b) do not show statistically significant differences (P ≥ 0.05), while those assigned different letters do (P < 0.05). B, Sr33syn induces cell death in response to AvrSr50 in Nicotiana benthamiana. None of the NLRs initiated cell death in response to the effector variant AvrSr50_QCMJC. Leaves of 4- to 5-week-old N. benthamiana plants were infiltrated with Agrobacterium tumefaciens carrying NLRs and effectors at an optical density at 600 nm of 0.3 and 0.4, respectively. UV images were taken on a BioRad Gel Imager 3 to 5 days after infiltration. Numbers on images correspond to the numbers of leaves that showed the depicted phenotype. Expression of NLRs was driven by the pRPP13 promoter, whereas effectors were driven by p35S.
In accordance with the published observation that overexpression of Sr50 and Sr33 can lead to cell death (Cesari et al. 2016), we noticed that Sr50 often exhibited autoactivity in our protoplast cell-death assay, in which expression is driven by a strong, constitutive promoter (pZmUBQ) (Supplementary Fig. S4). In contrast, this autoactive phenotype was much weaker for Sr33 (Fig. 5A). Since the two NLRs are highly similar, we reasoned that we could reduce the Sr50 autoactive phenotype by changing certain amino acids to the corresponding sites in Sr33. Given that many synthetic Sr33 variants we generated exhibited autoactivity (Supplementary Fig. S4A; Supplementary Table S4), we hypothesize that the residues governing autoactivity are present among the 13 substituted residues that we tested. Together with our data from Sr50, Sr33, and Sr33syn, we selected two residues, M844 and V868, to be mutated in Sr50 that we hypothesized to affect the autoactivity phenotype in wheat protoplasts. We tested the autoactivity of the Sr50 mutants in the wheat protoplast cell-death assay and observed that two single mutations reduced Sr50 autoactivity while preserving effector recognition (Fig. 5A), whereas the double mutant Sr50_M848V_V868E displayed autoactivity similar to that of wild-type Sr50. This result suggests that the level of autoactivity in Sr50 may be modulated through a complex interplay of residues but can still be altered to control the autoactive phenotype of Sr50. We also tested whether effector recognition is retained and remains specific in N. benthamiana. We infiltrated Sr50 and the single and double mutants with both AvrSr50 and AvrSr50_QCMJC (Fig. 5C;Supplementary Fig. S5). All Sr50 variants induce cell death in an AvrSr50-dependent manner, while no cell death was observed with the AvrSr50_QCMJC effector variant (Fig. 5C). Notably, Sr50_M848V_V868E that displayed similar autoactive phenotype as Sr50 in the wheat protoplast cell death assay did not induce the cell death in N. benthamiana in the absence of AvrSr50. We therefore show that closely related NLRs can also provide information about activation mechanisms and be utilized to reduce autoactivity without affecting effector recognition specificity.
Fig. 5.
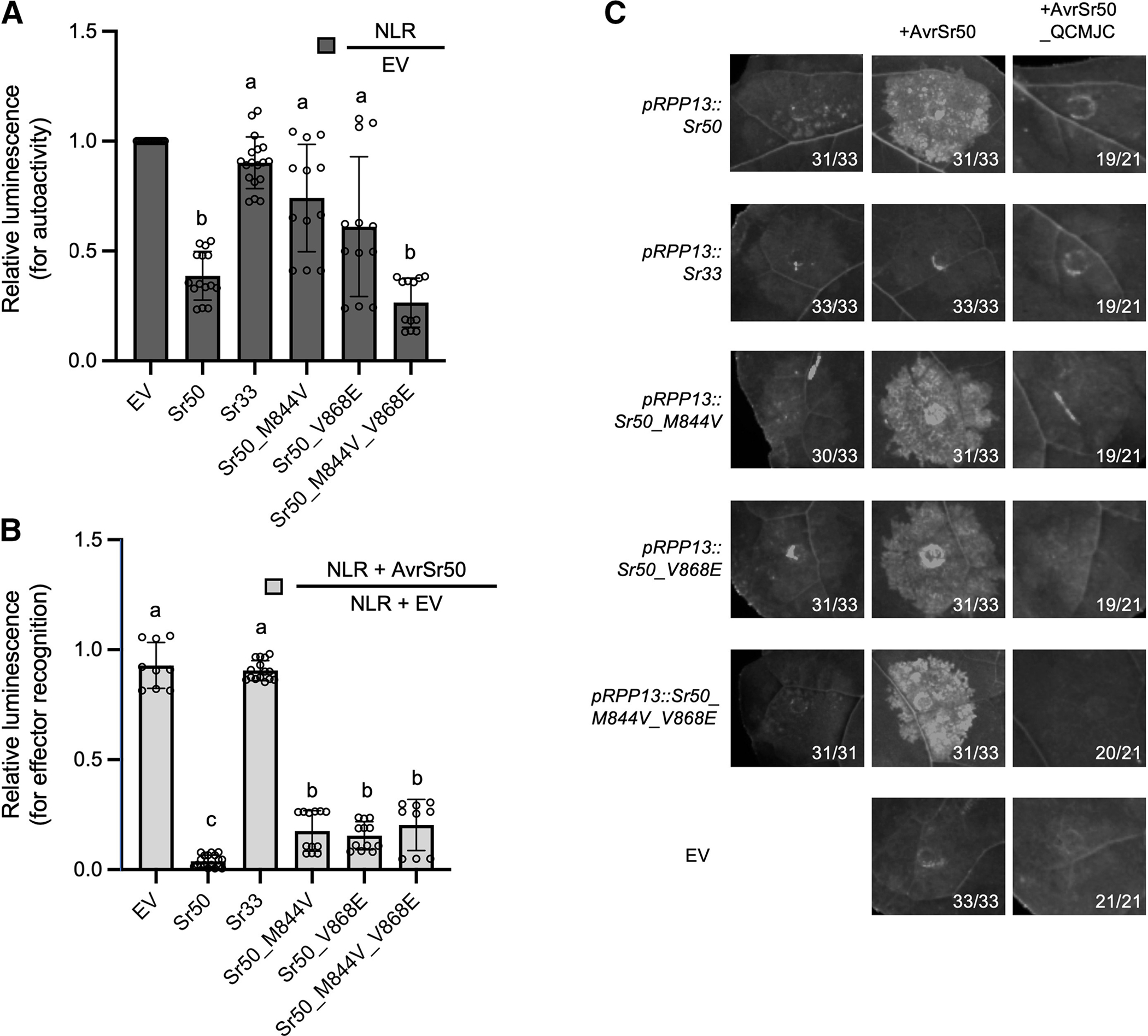
Sr50 autoactivity is decreased through mutations in its leucine-rich repeats (LRRs). A, Single mutants of Sr50 show decreased autoactivity in wheat protoplasts compared with wild-type Sr50, while the double mutant shows wild type–like autoactivity. We introduced single and double amino-acid changes into the nucleotide-binding LRR receptor (NLR) Sr50 to decrease its autoactivity. The sites were changed to the corresponding amino acids in that position within Sr33, as this NLR does not exhibit the same autoactivity phenotype in wheat protoplasts. Luciferase activity was determined approximately 36 h after transfection as a proxy for cell death. The relative luminescence reflects changes driven by autoactivity, and the low relative luminescence level indicates cell deaths by strong autoactivity. This value was obtained by normalizing the luminescence of each NLR construct by the luminescence obtained from the empty vector (EV) transfection. All values obtained in at least three independent experiments are indicated by dots and error bars indicate standard deviation. The equality of group means was assessed by one-way analysis of variance (ANOVA) and subsequent Tukey post-hoc tests. The samples assigned the same letter (a or b) do not show statistically significant differences (P ≥ 0.05), while those assigned different letters do (P < 0.05). B, Single mutants and double mutants of Sr50 induce cell death in response to AvrSr50. Luciferase activity was determined approximately 36 h after transfection as a proxy for cell death. The relative luminescence reflects the changes driven by effector recognition, and the low relative luminescence level represents cell deaths by effector recognition. This value was obtained by normalizing the luminescence of each NLR construct (or EV as a control) in the AvrSr50 co-transfection by the luminescence of that NLR construct in the EV co-transfection. All values obtained in at least three independent experiments are indicated by dots, and error bars indicate standard deviation. The equality of group means was assessed by one-way ANOVA and subsequent Tukey post-hoc tests. The samples assigned the same letter (a, b, or c) do not show statistically significant differences (P ≥ 0.05), while those assigned different letters do (P < 0.05). C, Single and double mutants of Sr50 induce cell death in Nicotiana benthamiana in response to AvrSr50. None of the NLRs initiated cell death in response to the effector variant AvrSr50_QCMJC. Leaves of 4- to 5-week-old N. benthamiana plants were infiltrated with Agrobacterium tumefaciens carrying NLRs and effectors at an optical density at 600 nm = 0.3 and 0.4, respectively. UV images were taken on a BioRad Gel Imager 3 to 5 days after infiltration. Numbers on images correspond to the numbers of leaves that showed the depicted phenotype. Expression of NLRs was driven by the pRPP13 promoter, whereas effectors were driven by p35S.
To understand why certain amino-acid sites had such a drastic effect on Sr50 autoactivity, we modeled the structure of Sr50 with AlphaFold2. The predicted Sr50 structure, despite its high accuracy, displayed discrepancy on the NB-ARC domain position when superposed against the activated Sr35 structure (Protein Data Bank [PDB]: 7XC2) (Förderer et al. 2022). This suggested that the predicted Sr50 structure likely represents the inactive state. The model showed that LRR residues M844 and V868 we identified to affect autoactivity in Sr50 and effector recognition inSr33syn are in proximity to the NB-ARC domain (Fig. 6A).The NB-ARC residues in contact appeared as a short flexible stretch of random coils with potential to be dislodged. We named these structural features that define this possible inter-domain interaction the NB-ARC/LRR latch. We then hypothesized that the two LRR residues are important in maintaining the inactive state of the NLR through the interaction with the NB-ARC domain. We further investigated whether the sites in the NB-ARC latch correspond to any highly conserved major or minor NB-ARC motifs and found that they did not (Fig. 6B; Supplementary Table S5). For the MLA family members, sequence conservation of the NB-ARC latch was not as high as that of the major motifs (Fig. 6B); however, a glycine residue (G496) was nearly perfectly conserved (Fig. 6C). We postulated that this glycine is important to make a loop in the protein tertiary structure and give rise to the structural characteristic of the NB-ARC latch. Indeed, we found that the NB-ARC latch consistently appeared to be in contact with the LRR domain in the cryo-EM structures of two distantly related CNLs, ZAR1 (PDB: 6J5T) (Wang et al. 2019a) and Sr35 (PDB: 7XC2) (Förderer et al. 2022), with a conserved glycine residue.
Fig. 6.
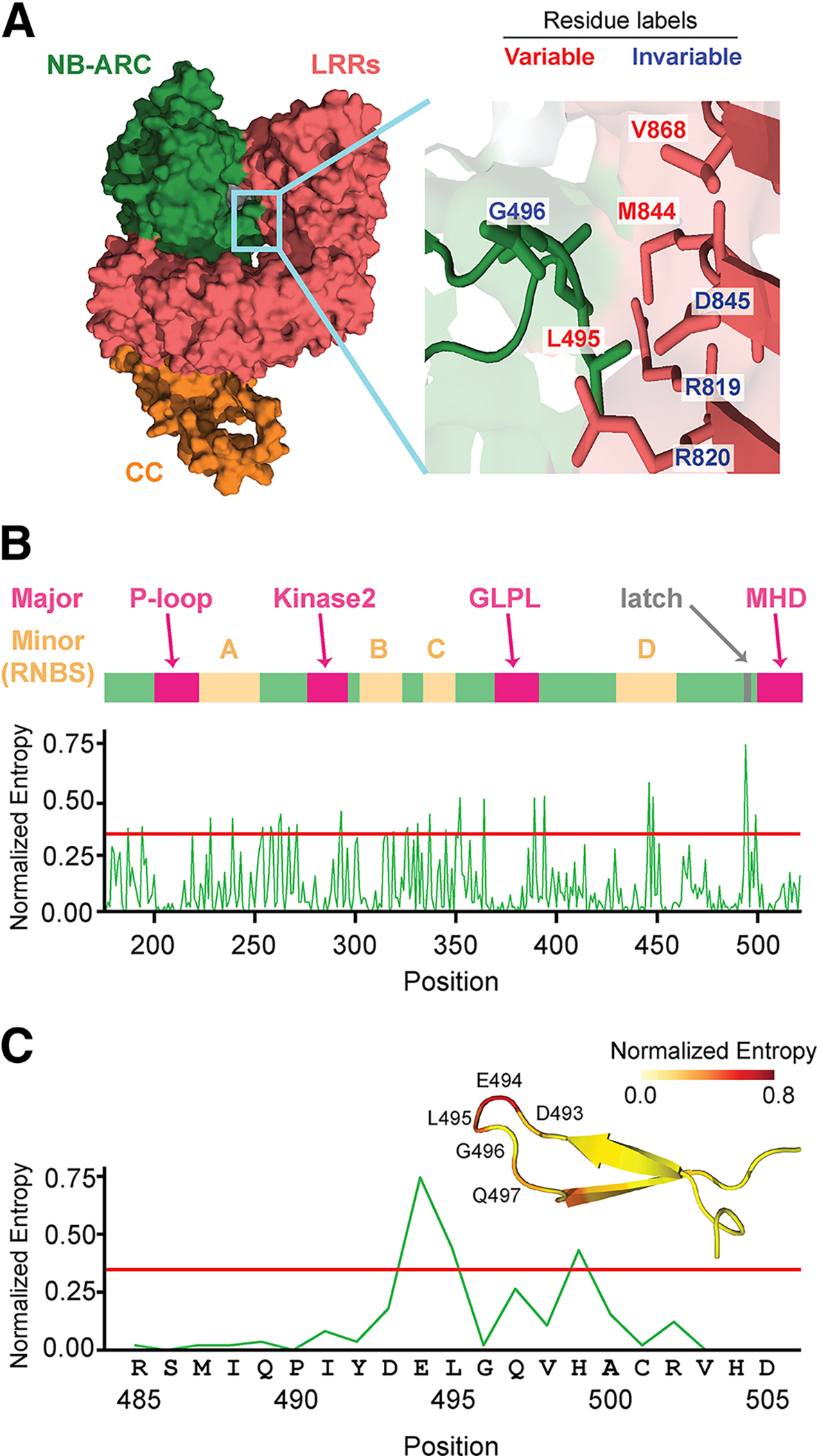
Amino acids in Sr50 leucine-rich repeats (LRRs) that govern autoactivity are in proximity to the nucleotide-binding (NB)-ARC domain. A, The predicted Sr50 structure and insert to the NB-ARC latch. The structure was predicted with AlphaFold2. The coiled-coil (CC), NB-ARC, and leucine-rich repeat (LRRs) domains are shown in orange, green and red, respectively. The light blue box enlarges the contact point between NB-ARC and LRR domains and the involved amino acids; we refer to this interaction as the NB-ARC/LRR latch. Some of the residues in this NB-ARC/LRR latch are depicted. If a homologous position of the displayed residue has the same amino acid in Sr33, the residue is indicated as ‘invariable’. Otherwise, it is indicated as ‘variable’. B, The sequence variation across the NB-ARC domain of all MILDEW RESISTANCE LOCUS A (MLA) family members. The normalized entropy was calculated with all MLA family members used in the study to examine the family-wise sequence variation of the NB-ARC domain. Major and minor motifs within the NB-ARC are shown in relation to the positions. The gray arrow indicates amino acid involved in NB-ARC/LRR latch interaction. C, The sequence variations around the NB-ARC latch. We zoomed in on positions 485 to 505 in B to highlight the sequence variations around the NB-ARC latch. The sequences of Sr50 in the given positions are indicated. The corresponding predicted structure of Sr50 is displayed with mapped Shannon entropy values and annotations on the loop region.
The residues in close spatial proximity may co-evolve to maintain their inter-residual interaction (de Juan et al. 2013). Since a single amino-acid substitution in the LRR domain could result in such a large change of the autoactive phenotype, we were curious how physically close the NB-ARC/LRR latch is and whether residues in the latch would co-evolve. The residues are typically considered to be in contact if their distance between Cβ falls within 0.8 nm (Graña et al. 2005). Upon calculation of the pair-wise residual distances in the predicted Sr50 structure, we found that two residues in the NB-ARC latch, E494 and L495, were in even closer contact with terminal LRR residues, R819, R820, M844, and D845 (<0.6 nm) (Figs. 6A and 7A). We collected 21,518 putative CNLs from 330 Viridiplantae species and tested co-evolution across the residues in the latch with direct coupling analysis (DCA) (Zerihun et al. 2020) and Gremlin (Ovchinnikov et al. 2014). However, while residues in contact typically displayed a signature of co-evolution, the NB-ARC/LRRs latch lacked any signal (Fig. 7B). The only inter-domain co-evolution between the NB-ARC and LRR domains was detected at the N-terminal LRR domain, possibly as this region needs to extend from the NB-ARC domain while maintaining optimal inter-domain interaction. Together, our functional and structural analyses highlight that a single LRR region, in which residues are in close physical proximity to the NB-ARC domain, can affect autoactivity of Sr50 and effector recognition of Sr33syn.
Fig. 7.
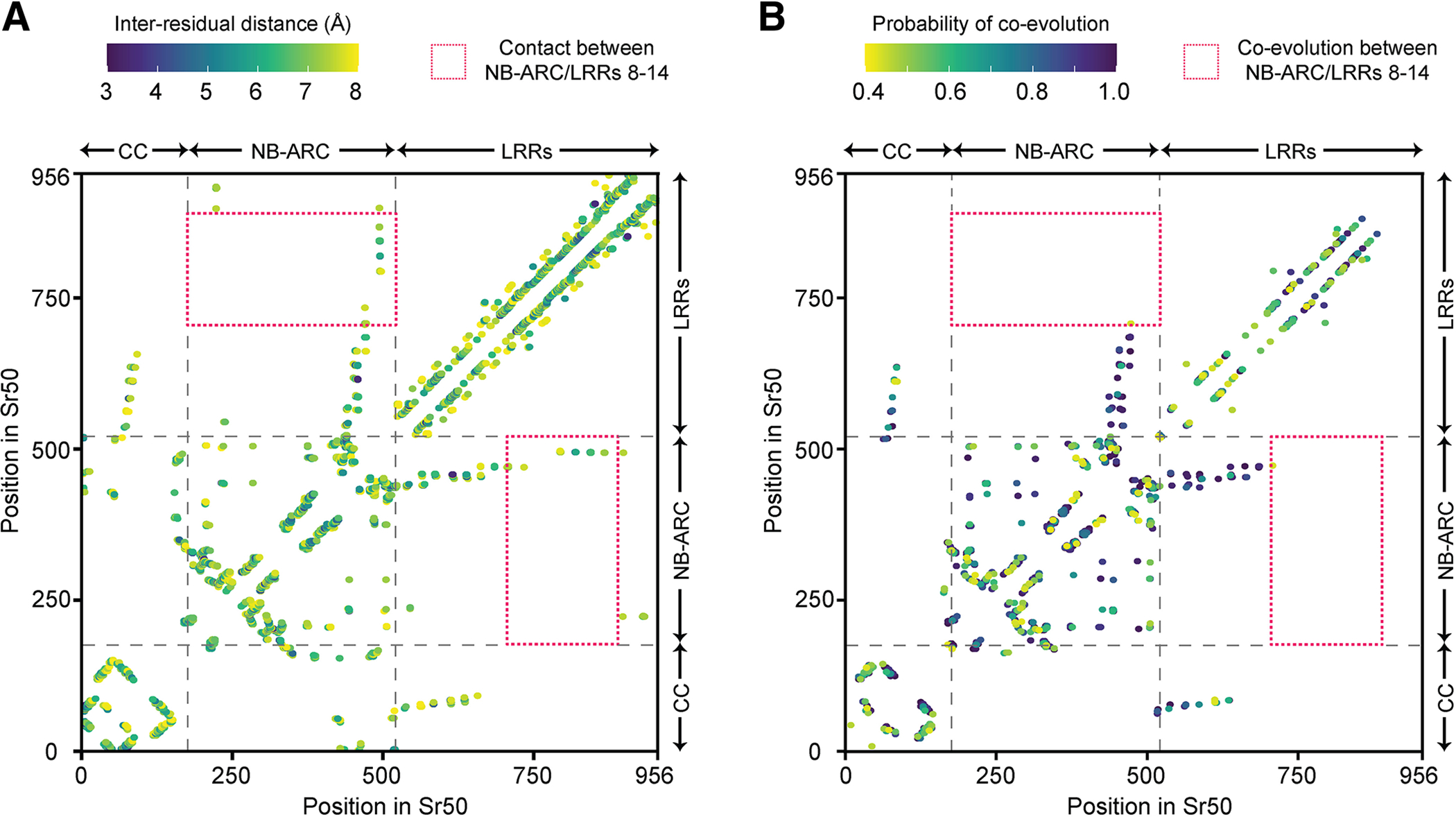
The signature of inter-residual co-evolution captured by Gremlin shows no co-evolution between nucleotide-binding (NB)-ARC and terminal leucine-rich repeats (LRRs). A, The inter-residual distances in the predicted Sr50 structure. When Cβ (Cα for glycine) atoms of two residues are within 0.8 nm, the residues are considered to be in contact. Only the long-range contact, which requires the pair of residues to be separated by at least 24 amino acids, or inter-domain contact are indicated. Multi-domain architecture (coiled coil [CC], NB-ARC, and LRRs) is provided along the axis. The red box highlights the residues of the NB-ARC domain and LRRs 8 to 14 in contact. B, The inter-residual co-evolution of NB-LRRs (NLRs) mapped to Sr50. The filtered multiple sequence alignment for Sr50 collected during structure prediction with AlphaFold2 was used to infer inter-residual co-evolution with Gremlin. The data visualized here is provided in Supplementary Table S6. Only the pairs forming long-range or inter-domain contact are indicated.
Discussion
Many deployed resistance genes are NLRs, which makes them excellent targets for plant health improvement. A bottleneck in the rational design of new NLR alleles has been the lack of understanding of effector recognition and subsequent receptor activation. Though it has been known for a long time that the LRR domains play a crucial role in effector specificity (Ellis et al. 1999; Krasileva et al. 2010; Shen et al. 2003), difficulties in structural biology have limited our ability to close in on the specific sites mediating effector recognition. To successfully generate new effector recognition specificities, we need to decipher sites within NLRs that mediate effector recognition and those required to maintain stability and the inactive state. Recent advances both in structural biology (Förderer et al. 2022; Martin et al. 2020; Wang et al. 2019a, b; Zhao et al. 2022) and NLR engineering (Cesari et al. 2022; De la Concepcion et al. 2019; Pottinger et al. 2020) have exemplified success in the engineering of NLR proteins.
Availability of genomic data from many different species and populations enables the analysis of NLRs with sufficient evolutionary depth. This made it possible for us to predict and engineer effector recognition specificity on NLRs from wheat and its relatives. It was previously shown that sequence diversity data and Shannon entropy can be used to predict where within the NLR effector recognition may occur (Prigozhin and Krasileva 2021). Here, we combined Shannon entropy analysis with structural modeling to predict potential effector recognition sites and guide rational engineering to transfer effector recognition specificity between closely related homologs. We used a structural model of Sr50 to map amino-acid polymorphisms between Sr50 and Sr33 on the exposed beta sheets of the LRR domain. This enabled us to use additional criteria to select residues for engineering as opposed to relying only on Shannon entropy scores. This approach greatly enhanced our ability to successfully engineer Sr33, as exemplified by the identification of Sr33 V840. Its corresponding site M844 in Sr50 has a low Shannon entropy score but was required for the transfer of effector recognition in Sr33syn. We therefore show that the application of several analyses can be complementary and help successfully engineer NLR recognition specificities, even in the absence of experimentally determined structural data.
The identification of 12 amino acids of Sr50 required for Sr33’s gain of recognition specificity for AvrSr50 may have additional importance. AvrSr50 escapes recognition with a single amino-acid substitution (Q121K) (Ortiz et al. 2022). The substitution of polar glutamine to positively charged lysine may disfavor the interaction with the LRR domain, and one or some of the 12 amino acids may be involved in this interaction. Future work may recover recognition specificity of Sr50 against the escape mutant by mutagenizing these highly variable LRRs.
The ‘on’ and ‘off’ states of NLRs are tightly controlled, and recent cryo-EM structures of an indirect binder NLR, A. thaliana ZAR1, confirmed what has previously been inferred through domain interaction and swap data, i.e., the inactive and ADP-bound state was monomeric and had multiple intramolecular contact points between the LRR and NB-ARC domains (Wang et al. 2019a, b). Moreover, previous studies showed that the correct interactions of NB-ARC and LRR are required to avoid autoactivity of the NLR (Qi et al. 2012; Rairdan and Moffett 2006; Slootweg et al. 2013) and even for effector responsiveness (Ma et al. 2018). The differing autoactivity phenotypes of Sr33, Sr50, and our Sr33syn variants allowed us to investigate this, as the differences among all three proteins were confined to their LRR domains. The data from this study supports the notion that interactions between NB-ARC and LRR are crucial in maintaining the inactive state of the direct binder NLR Sr50. Interestingly, while single mutations of residues in the NB-ARC/LRR latch were able to reduce the Sr50 autoactive phenotype, introducing both of these mutations at the same time reverted this stabilizing effect. This result exemplifies that the NB-ARC/LRR interactions and how they affect autoactivity are still poorly understood. Further elucidation of this interaction and the factors that control NLR activation will be necessary for future NLR designs. A recent paper suggested that the interaction between the NB-ARC and LRR domains serves the purpose of preventing NLR oligomerization (Zhao et al. 2021). It would be interesting to see if residues in the NB-ARC/LRR latch also play a role in NLR oligomerization or whether the interaction is merely to prevent access to the sites mediating nucleotide exchange and oligomerization.
Through our introduction of amino acid changes in Sr50 and structural modeling, we identified a direct interaction point of NB-ARC and LRR that we termed the NB-ARC/LRR latch. This interaction most likely results in a conformation of the NLR that hinders the exchange of ADP to ATP in the NB-ARC domain or oligomerization into resistosomes. Strikingly, our data suggests that control of Sr50 autoactivity and Sr33syn effector specificity can be mediated by the same residues. We hypothesize that the effector competes with and disrupts the interaction of the NB-ARC/LRR latch leading to the activation of the NLR. In fact, the Cryo-EM structures of the Sr35 resistosome suggest that AvrSr35 contacts the NB-ARC latch. The location of the NB-ARC latch slightly differs between the Sr35 structure predicted by AlphaFold2 and the activated Sr35 structure, possibly suggesting that the NB-ARC latch may be repositioned during the interaction with the effector. This may be a part of the steric clash, in which the effector dislodges the NB-ARC domain, which subsequently activates the NLR (Förderer et al. 2022). Our data suggests that the interaction of the NB-ARC/LRR latch can also be disrupted through the exchange of amino-acid residues in the LRR domain. We therefore hypothesize that the disruption of the NB-ARC/LRR latch is a crucial step in NLR activation and can be caused by interaction with an effector or unstable intramolecular interactions. In this scenario, we would not expect to observe strong co-evolution between the residues in the NB-ARC latch and the LRR domain, as locking the conformation of the latch through optimal inter-residue interaction would disfavor NLR function. Overall, we suggest that the strength of the NB-ARC/LRR latch interaction is an important criterion to consider when undertaking NLR engineering approaches.
It has been proposed that NLRs exist in fluidity of active and inactive states and effector recognition stabilizes the active state, thereby shifting the equilibrium in its favor—this is referred to as the equilibrium-based switch model (Bernoux et al. 2016). We propose that the activation of hvNLRs, such as Sr50, follows the equilibrium-based switch model (Bernoux et al. 2016), based on our observations of expression-dependent autoactivity and our ability to change this phenotype with a few amino-acid substitutions. According to this model, there is an equal pool of activated and inactive receptors in each cell in the absence of a trigger. Recognition of the effector stabilizes the activated conformation and leads to the activation of downstream signaling pathways. This implies that the cell can cope with a certain number of activated receptors, and overexpression would lead to an excess of activated receptors. This could push the resulting cellular changes over the threshold required to initiate the cell death response, for instance, through increased calcium influx caused by the insertion of CC domains of activated, oligomerized receptors into the membrane. Collectively, our data, which showed that Sr50 exhibits varying degrees of autoactivity depending on its expression level (Fig. 5; Supplementary Fig. S2), suggests to us that this NLR follows the equilibrium switch model of activation. It will, therefore, be crucial to understand the limitations of deploying NLRs that follow this model of activation and ensure the use of appropriate promoters in the generation of new resistant plants with hvNLRs. We hypothesize that having multiple factors that can drastically change autoactivity phenotypes such as expression levels and intramolecular interaction strengths is what allows for the rapid generation of diversity in hvNLRs.
It is tempting to hypothesize that the amino-acid sites we identified in the latch would evolve together to maintain the rate at which the NLR can be activated. However, neither of the analyses we performed detected any co-evolutionary signature. Rather, they suggested that co-evolution between NB-ARC and terminal LRRs is lacking. Since there are multiple residues involved in the NB-ARC/LRR latch, it is conceivable that any co-evolutionary signature would be too diluted among all involved residues to be detected or that the activation mechanism of the NLR is too complicated to be summarized. The fluidity of the Sr50 activation state is another possible reason for the lack of measurable co-evolution, as the on and off states of this NLR are affected by many different factors. Moreover, the co-evolutionary analyses that are currently at our disposal require rather large datasets with an evolutionarily broad range of NLRs. If the co-evolution of the NB-ARC/LRR latch is specific to a subset of NLRs and multiple modes of interactions exist for these inter-domains, the datasets may obscure any co-evolutionary signatures. Elucidating the co-evolution of the NB-ARC/LRR latch will require better understanding of how NLRs are cycled between active and inactive states (there is no experimental structure of an inactive direct-binder NLR), how many different modes there may be, and which NLRs follow each mode of activation.
This study shows a major advance towards the successful engineering of new effector recognition specificities in direct-binder NLRs. We demonstrate that effector recognition sites predicted through diversity analysis can be successfully used to engineer or transfer recognition specificities. Our study also demonstrates that, for the successful engineering of an immune receptor, we also need to identify sites that are important for maintaining the inactive state of the NLR. Another study recently demonstrated similar engineering success of several closely related receptors. By using the cryo-EM structure of Sr35, authors were able to engineer orphan NLRs in wheat and barley, first by creating chimeric NLRs and subsequently transferring as few as eight to ten amino acids (Förderer et al. 2022). The transferred residues were similarly restricted to the concave LRR surface of the terminal LRRs. We calculated Shannon entropy for the residues transferred from Sr35 and found that almost all residues are highly variable (six of eight for TaSH1 and nine of ten for HvSH11). These findings therefore support the findings in this study.
The results of this study and Sr35 orphan NLR engineering demonstrate that this approach of transferring a small number of amino acids is applicable to several NLRs. This may open the potential to use existing NLRs that are available for deployment and edit their homologs in elite crop germplasms. The region required for the transfer of effector recognition involved as few as 12 amino-acid substitutions within 450 base pairs. If this region were to be introduced into a closely related homolog through homologous recombination, the engineered plants would follow the regulatory exemptions described in the SECURE rule from the United States Department of Agriculture, given this variation is already present in the gene pool of the edited plant (Hoffman 2021). The ability to transfer effector recognition between close homologs also gives us the ability to avoid interspecies transfer of genes, which is controversial for some opponents of transgenic plants. The information provided in this study may, therefore, help to drive forward the generation of resistant elite crop germplasm.
Materials and Methods
Plant materials and growth conditions.
N. benthamiana plants were grown in a Conviron growth chamber with 16 h of light at 24°C. Triticum aestivum cv. Fielder seeds were sterilized in a 10% bleach solution for 20 min, and then, they were placed on water and filter paper in a plastic cup and the cup was sealed by placing another cup on top and adhering them together with Millipore tape. The cups were kept on the west-facing windowsill for 7 to 14 days, depending on the season.
Cloning and vectors.
All vectors cloned for this study were cloned following the Golden Gate cloning technique (Weber et al. 2011; Werner et al. 2012) by utilizing the vectors deposited as MoClo Plant Parts (vector number 1000000047) and Tool kits (vector number 1000000044) at Addgene. For expression in wheat protoplasts, all genes were driven by the pZmUBQ promoter (pICSL12009) and the nopaline synthase terminator (tNos) (pICH41421), whereas, for expression in N. benthamiana, the pRPP13 promoter (Rentel et al. 2008) and tNos terminator as well as p35S (pICH51277) and t35S (pICH41414) were used. The following genes were domesticated for Golden Gate by removing internal restriction sites for BsaI and BpiI and were cloned into the corresponding level 0 acceptor, namely, Sr33, Sr50, AvrSr50, AvrSr50_QCMJC, and pRPP13. All coding sequences were cloned both with and without a stop codon. Mutants of Sr33 were created by synthesizing LRRs 8 to 13 with 10 amino acid substitutions (Sr3310AA) and ligating it together with PCR fragments of the start and end of Sr33. Additional mutations that eventually spanned to LRR 14 were added using site-directed mutagenesis PCR, by designing two overlapping primers with the desired mutation in the middle, amplifying the vector by Phusion PCR, and digesting the template with DpnI. Mutants of Sr50 were created by site-directed mutagenesis PCR. All vectors and primers used in this study are described in Supplementary Tables S7 and S8.
Wheat protoplast isolation, transfection, and cell-death assay.
Wheat protoplast isolation and transfection was adapted from the two previous studies (Saur et al. 2019; Yoshida et al. 2009). The epidermis of 7- to 14-day-old seedlings was peeled off by making a shallow cut in the adaxial epidermis with a sharp razor blade, folding the leaf over at the cutting edge, and pulling back the leaf tip with the abaxial epidermis. The peeled leaf sections were placed next to each other on masking tape with the abaxial side facing upwards and were placed into enzyme solution facing down. Washing and transfection were performed as previously described. Protoplasts were counted with a hemocytometer and the number was adjusted to 300,000 protoplasts/ml. All plasmids were transfected at a concentration of 15 μg and an empty vector was added to ensure all protoplasts were transfected with the same final DNA concentration. After transfection, protoplasts were incubated in a bovine serum albumin–coated 12-well plate at room temperature in the dark for 48 h. Subsequently, protoplasts were collected by centrifugation at 100 × g for 3 min. Supernatant was discarded and protoplasts were resuspended in 2× cell culture lysis reagent (Promega) and were incubated on ice for 20 min in the dark. The lysed cell contents were centrifuged at 1,000 × g for 3 min and were divided into a white 96-well plate (Greiner Bio-One) in triplicate. An equal amount of luciferase assay substrate (Promega) was added and luminescence was measured in a Tecan Infinite F Plex plate reader (20 min, settle time 100 ms, no attenuation).
Statistical analyses for wheat protoplast assays.
We followed Saur et al. (2019) to perform statistical analysis for the wheat protoplast assays. To exemplify autoactivity, the measured luminescence of each NLR construct was normalized by the luminescence detected from the empty vector (EV) transfection. This relative luminescence reflects the cell deaths modulated by NLR autoactivity. To exemplify effector recognition, the luminescence obtained for each NLR construct (or EV as a control) in the AvrSr50 co-transfection was normalized by the luminescence acquired for that NLR in the EV co-transfection. This relative luminescence reflects the cell deaths driven by effector recognition. The equality of group means between the relative luminescences of tested constructs was assessed by one-way analysis of variance and subsequent Tukey post-hoc tests. Groups with statistically significant differences were identified (P < 0.05).
Agrobacterium-mediated transformation of N.benthamiana.
Agrobacterium tumefaciens GV3101:pMP90 transformed with the corresponding construct were grown in Luria-Bertani medium (supplemented with the following antibiotics [per milliliter]: 50 mg of rifampicin, 25 mg of gentamycin, and 100 mg of carbenicillin) at 28°C. The cultures were centrifuged at 3,000 × g for 15 min and the pellet was resuspended in infiltration medium (10 mM MES, pH 5.6, 10 mM MgCl2). Optical density was adjusted to a final optical density at 600 nm = 0.3. If several constructs were co-infiltrated, the suspensions of individual constructs were mixed in a 1:1 ratio, while ensuring that all constructs reached a final density of 0.3. Leaves of 4- to 5-week-old N. benthamiana plants were inoculated with the suspension, using a blunt syringe. Pictures of leaves were taken 2 to 3 days after inoculation.
Manual curation of NLRs.
To ensure the sequence diversity needed to calculate entropy, we manually curated a subset of NLRs from 14 wheat genomes (Walkowiak et al. 2020) (Supplementary Table S1). From the wheat reference genome, we first identified the NLRs that belong to the MLA family, guided by the previous study (Steuernagel et al. 2020). Their NB-ARC domain was searched with TBLASTN (e-value 1E-10) (Camacho et al. 2009) against the 14 wheat genomes. We identified matches that showed 85% or more sequence identity against the queries, and we extracted the putative NLR loci with 10,000–base pair flanking regions on both ends. We used MAKER v3.01.04 (Cantarel et al. 2008) to obtain gene models in these regions, relying on two ab initio predictors, Augustus v3.4.0 (–species wheat) (Stanke et al. 2006) and GeneMark v4.68_lic (Brůna et al. 2020), as well as the coding sequence and protein sequences of the wheat reference genome annotation. The gene models were loaded into Apollo genome browser v2.0.6 (Dunn et al. 2019) and were manually curated. The resulting annotation set included many redundant NLRs from the 14 wheat genomes, the sequences of which were nearly identical. The redundant sequences can inflate their weighting and copy numbers in entropy calculations and the phylogenetic analyses, which can mislead evolutionary conclusions. We therefore reduced the redundancy of the wheat NLRs, based on their sequence identity (95%), with CD-hit (-c 0.95) v4.8.1 (Li and Godzik 2006).
Phylogenetic analyses.
To infer a species tree of the selected Pooideae members, we first identified complete benchmarking universal single-copy ortholog (BUSCO) genes present in all nine species with BUSCO v5.2.2 (Seppey et al. 2019) and the poales_odb10 database and, then, randomly selected 500 (Supplementary Table S1). Each set of orthologous protein sequences was aligned with MAFFT v7.487 (–maxiterate 1000 –globalpair) (Katoh and Standley 2013). All MSAs were concatenated and trimmed with TrimAl v1.4.rev15 (-gt 0.3) (Capella-Gutiérrez et al. 2009) and were used to infer a phylogenetic tree with FastTree v2.1.10 (Price et al. 2010).
To infer the MLA family tree, we collected the MLA family members from the selected Pooideae species annotation sets and the filtered wheat NLR annotation sets and reference NLRs from RefPlantNLR (Kourelis et al. 2021b). The full-length NLR sequences were aligned with MAFFT, and all columns in which both Sr33 and Sr50 contained gaps were removed. The resulting MSA was used to infer a phylogenetic tree with FastTree (-slow). Sr35 was used as an outgroup to root the tree.
Protein structure prediction and structural analysis.
We used AlphaFold v2.0.0 to predict the structure of Sr50 (Evans et al. 2021; Jumper et al. 2021). The homologs were collected from the full databases, and homologous templates downloadable by July 20th, 2020, were included in the template database. Models 1, 3, 4, and 5 as well as ptm model 2 were used for structural inference. The best model was selected as a final structure based on the pLDDT scores. We measured the inter-residual distances in the predicted structure by calculating the Euclidean distances between the Cβ atoms (Cα for glycine) with Biopython v1.79(Cocketal.2009). The structure was visualized through PyMOL v2.5.
Co-evolutionary analyses.
To collect CNLs for co-evolutionary analyses, we extracted the protein sequences from the Viridiplantae species from the uniref100 database (2022–0222). The gene models containing the NB-ARC domain (PF00931) were identified as putative NLRs with hmmsearch 3.1b2 (–dome 1e-4-E 1e-4) and were annotated with InterProScan v5.52–86.0 (–appl Pfam-33.1) (Eddy 2011; Jones et al. 2014). Any sequences with Pfam entries other than the NB-ARC domain, the Rx N-terminal domain (PF18052), and LRRs (PF00560, PF07725, and PF13855) were excluded. The NB-ARC domain of the remaining NLRs were aligned with MAFFT (–auto –globalpair), were trimmed with TrimAl (-gt 0.3), and were used to infer a phylogenetic tree with FastTree. We identified any remaining TNL and RNL clades and removed the clade members.
We searched this CNL database with Sr50 as a query with jackhmmer (–domE 1e-10 -E 1e-10 -N 5). The MSA was trimmed with respect to Sr50, and homologs without ≥75%coverage were removed. We used this filtered MSA for DCA (Zerihun et al. 2020) and Gremlin (Ovchinnikov et al. 2014). The pseudolikelihood maximization algorithm of the pydca package was run for DCA (–max_iterations 1000 –apc). The filtered MSA was also submitted to the online Gremlin server (HHblits iterations = 0, coverage = 50, remove gaps = 50).
Shannon entropy calculation.
We computed the normalized Shannon entropy as follows:
where is the probability of observing an amino acid out of the 20 in the given position of the input MSA. Gaps were removed before the probabilities were computed. However, if the position of the MSA was composed of 50% or more gap characters, we did not calculate the entropy for that position. The normalized Shannon entropy ranges from 0 (no variability) to 1 (maximum variability). The normalized entropy score of 0.347 was used to define highly variable residues, following previous studies (Jumper et al. 2021; Prigozhin and Krasileva 2021).
Supplementary Material
Acknowledgments
We thank I. Saur and S. Wilson for helpful practical advice on the protoplast cell death assay. We thank D. Prigozhin for initial Shannon entropy analyses and effector binding site prediction on Sr50 and helpful comments on the manuscript. We are grateful for many personal discussions with E. Baggs about this project and thoughtful suggestions on the bioinformatics analyses. We are grateful to all members of the Krasileva lab for thoughtful comments and discussions of the presented material.
Funding:
The project has been funded by the Gordon and Betty Moore Foundation, the Foundation for Food and Agriculture Research, and 2Blades. K. Seong was supported by a University of California Berkeley Graduate Fellowship.
Footnotes
Competing interests: K. V. Krasileva, B. J. Staskawicz, and F. Liu have previously filed a patent application related to this work (patent publication number: WO/2021/113569; publication date: October 6, 2021; international application number: PCT/US2020/063203; international filing date: April 12, 2020).
e-Xtra: Supplementary material is available online.
Data availability.
All data and scripts used in this study are available in Github (https://github.com/krasileva-group/Sr33-Sr50_analysis).
Literature Cited
- Arora S, Steuernagel B, Gaurav K, Chandramohan S, Long Y, Matny O, Johnson R, Enk J, Periyannan S, Singh N, Asyraf Md Hatta M, Athiyannan N, Cheema J, Yu G, Kangara N, Ghosh S, Szabo LJ, Poland J, Bariana H, Jones JDG, Bentley AR, Ayliffe M, Olson E, Xu SS, Steffenson BJ, Lagudah E, and Wulff BBH 2019. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 37:139–143. [DOI] [PubMed] [Google Scholar]
- Bernoux M, Burdett H, Williams SJ, Zhang X, Chen C, Newell K, Lawrence GJ, Kobe B, Ellis JG, Anderson PA, and Dodds PN 2016. Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. Plant Cell 28:146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G, Su M, Li N, Liang Y, Dang S, Xu J, Hu M, Wang J, Zou M, Deng Y, Li Q, Huang S, Li J, Chai J, He K, Chen Y-H, and Zhou J-M 2021. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184:3528–3541.e12. [DOI] [PubMed] [Google Scholar]
- Brůna T, Lomsadze A, and Borodovsky M 2020. GeneMark-EP+: Eukaryotic gene prediction with self-training in the space of genes and proteins. NAR Genomics and Bioinformatics 2:lqaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, and Madden TL 2009. BLAST+: Architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, and Yandell M 2008. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18:188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, and Gabaldón T 2009. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S, Moore J, Chen C, Webb D, Periyannan S, Mago R, Bernoux M, Lagudah ES, and Dodds PN 2016. Cytosolic activation of cell death and stem rust resistance by cereal MLA-family CC-NLR proteins. Proc. Natl. Acad. Sci. U.S.A. 113:10204–10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, Morel J-B, Fournier E, Tharreau D, Terauchi R, and Kroj T 2013. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25:1463–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S, Xi Y, Declerck N, Chalvon V, Mammri L, Pugnière M, Henriquet C, deGuillen K, Chochois V, Padilla A, and Kroj T 2022. New recognition specificity in a plant immune receptor by molecular engineering of its integrated domain. Nat. Commun. 13:1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S, Stevens LJ, Boevink PC, Engelhardt S, Alexander CJ, Harrower B, Champouret N, McGeachy K, Van Weymers PSM, Chen X, Birch PRJ, and Hein I 2014. Detection of the virulent form of AVR3a from Phytophthora infestans following artificial evolution of potato resistance gene R3a. PLoS One 9:e110158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Upadhyaya NM, Ortiz D, Sperschneider J, Li F, Bouton C, Breen S, Dong C, Xu B, Zhang X, Mago R, Newell K, Xia X, Bernoux M, Taylor JM, Steffenson B, Jin Y, Zhang P, Kanyuka K, Figueroa M, Ellis JG, Park RF, and Dodds PN 2017. Loss of AvrSr50 by somatic exchange in stem rust leads to virulence for Sr50 resistance in wheat. Science 358:1607–1610. [DOI] [PubMed] [Google Scholar]
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, and de Hoon MJL 2009. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan D, Pazos F, and Valencia A 2013. Emerging methods in protein co-evolution. Nat. Rev. Genet. 14:249–261. [DOI] [PubMed] [Google Scholar]
- De la Concepcion JC, Franceschetti M, MacLean D, Terauchi R, Kamoun S, and Banfield MJ 2019. Protein engineering expands the effector recognition profile of a rice NLR immune receptor. Elife 8:e47713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Qi D, Kim S-H, Burke TP, and Innes RW 2012. Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell. Microbiol. 14:1071–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, and Rathjen JP 2010. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11:539. [DOI] [PubMed] [Google Scholar]
- Dunn NA, Unni DR, Diesh C, Munoz-Torres M, Harris NL, Yao E, Rasche H, Holmes IH, Elsik CG, and Lewis SE 2019. Apollo: Democratizing genome annotation. PLoS Comput. Biol. 15:e1006790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 70:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Lawrence GJ, Luck JE, and Dodds PN 1999. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J, Ronneberger O, Bodenstein S, Zielinski M, Bridgland A, Potapenko A, Cowie A, Tunyasuvunakool K, Jain R, Clancy E, Kohli P, Jumper J, and Hassabis D 2021. Protein complex prediction with AlphaFold-Multimer. bioRxiv 463034. [Google Scholar]
- Feehan JM, Castel B, Bentham AR, and Jones JD 2020. Plant NLRs get by with a little help from their friends. Curr. Opin. Plant Biol. 56:99–108. [DOI] [PubMed] [Google Scholar]
- Förderer A, Li E, Lawson AW, Deng Y-N, Sun Y, Logemann E, Zhang X, Wen J, Han Z, Chang J, Chen Y, Schulze-Lefert P, and Chai J 2022. A wheat resistosome defines common principles of immune receptor channels. Nature 610:532–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakopoulou A, Steele JFC, Segretin ME, Bozkurt TO, Zhou J, Robatzek S, Banfield MJ, Pais M, and Kamoun S 2015. Tomato I2 immune receptor can be engineered to confer partial resistance to the oomycete Phytophthora infestans in addition to the fungus Fusarium oxysporum. Mol. Plant. Microbe. Interact. 28:1316–1329. [DOI] [PubMed] [Google Scholar]
- Graña O, Baker D, MacCallum RM, Meiler J, Punta M, Rost B, Tress ML, and Valencia A 2005. CASP6 assessment of contact prediction. Proteins 61:214–224. [DOI] [PubMed] [Google Scholar]
- Grund E, Tremousaygue D, and Deslandes L 2019. Plant NLRs with integrated domains: Unity makes strength. Plant Physiol. 179:1227–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halterman D, Zhou F, Wei F, Wise RP, and Schulze-Lefert P 2001. The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 25:335–348. [DOI] [PubMed] [Google Scholar]
- Hoffman NE 2021. Revisions to USDA biotechnology regulations: The SECURE rule. Proc. Natl. Acad. Sci. U.S.A. 118:e2004841118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, and Hunter S 2014. InterProScan5: Genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, and Hassabis D 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, and Standley DM 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Qi D, Ashfield T, Helm M, and Innes RW 2016. Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 351:684–687. [DOI] [PubMed] [Google Scholar]
- Kourelis J, Marchal C, and Kamoun S 2021a. NLR immune receptor-nanobody fusions confer plant disease resistance. bioRxiv 2021.10.24.465418. [DOI] [PubMed] [Google Scholar]
- Kourelis J, Sakai T, Adachi H, and Kamoun S 2021b. RefPlantNLR is a comprehensive collection of experimentally validated plant disease resistance proteins from the NLR family. PLoS Biol. 19:e3001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva KV, Dahlbeck D, and Staskawicz BJ 2010. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22:2444–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva KV, Prigozhin DM, Staskawicz BJ, and Liu F January 2023. U. S. Patent 17781229.
- Lewis JD, Wu R, Guttman DS, and Desveaux D 2010. Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 6:e1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, and Godzik A 2006. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang X, Yuan G, Wang D, Zheng Y, Ma M, Guo L, Bhadauria V, Peng Y-L, and Liu J 2021. A designer rice NLR immune receptor confers resistance to the rice blast fungus carrying noncorresponding avirulence effectors. Proc. Natl. Acad. Sci. U.S.A. 118:e2110751118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Lapin D, Liu L, Sun Y, Song W, Zhang X, Logemann E, Yu D, Wang J, Jirschitzka J, Han Z, Schulze-Lefert P, Parker JE, and Chai J 2020. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370:abe3069. [DOI] [PubMed] [Google Scholar]
- Ma Y, Guo H, Hu L, Martinez PP, Moschou PN, Cevik V, Ding P, Duxbury Z, Sarris PF, and Jones JDG 2018. Distinct modes of derepression of an Arabidopsis immune receptor complex by two different bacterial effectors. Proc. Natl. Acad. Sci. U.S.A. 115:10218–10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mago R, Zhang P, Vautrin S, Šimková H, Bansal U, Luo M-C, Rouse M, Karaoglu H, Periyannan S, Kolmer J, Jin Y, Ayliffe MA, Bariana H, Park RF, McIntosh R, Doležel J, Bergès H, Spielmeyer W, Lagudah ES, Ellis JG, and Dodds PN 2015. The wheat Sr50 gene reveals rich diversity at a cereal disease resistance locus. Nat. Plants. 1:15186. [DOI] [PubMed] [Google Scholar]
- Maidment JHR, Franceschetti M, Maqbool A, Saitoh H, Jantasuriyarat C, Kamoun S, Terauchi R, and Banfield MJ 2021. Multiple variants of the fungal effector AVR-Pik bind the HMA domain of the rice protein OsHIPP19, providing a foundation to engineer plant defense. J. Biol. Chem. 296:100371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqbool A, Saitoh H, Franceschetti M, Stevenson CEM, Uemura A, Kanzaki H, Kamoun S, Terauchi R, and Banfield MJ 2015. Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. Elife 4:e08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Qi T, Zhang H, Liu F, King M, Toth C, Nogales E, and Staskawicz BJ 2020. Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science 370:abd9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz D, Chen J, Outram MA, Saur IML, Upadhyaya NM, Mago R, Ericsson DJ, Cesari S, Chen C, Williams SJ, and Dodds PN 2022. The stem rust effector protein AvrSr50 escapes Sr50 recognition by a substitution in a single surface-exposed residue. New Phytol. 234: 592–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz D, de Guillen K, Cesari S, Chalvon V, Gracy J, Padilla A, and Kroj T 2017. Recognition of the Magnaporthe oryzae effector AVR-Pia by the decoy domain of the rice NLR immune receptor RGA5. Plant Cell 29:156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov S, Kamisetty H, and Baker D 2014. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife 3:e02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periyannan S, Moore J, Ayliffe M, Bansal U, Wang X, Huang L, Deal K, Luo M, Kong X, Bariana H, Mago R, McIntosh R, Dodds P, Dvorak J, and Lagudah E 2013. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341:786–788. [DOI] [PubMed] [Google Scholar]
- Pottinger SE, Bak A, Margets A, Helm M, Tang L, Casteel C, and Innes RW 2020. Optimizing the PBS1 decoy system to confer resistance to potyvirus infection in Arabidopsis and soybean. Mol. Plant. Microbe. Interact. 33:932–944. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, and Arkin AP 2010. Fast Tree 2—Approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhin DM, and Krasileva KV 2021. Analysis of intraspecies diversity reveals a subset of highly variable plant immune receptors and predicts their binding sites. Plant Cell 33:998–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, DeYoung BJ, and Innes RW 2012. Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol. 158:1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racman DS, Mcgeachy K, Reavy B, Strukelj B, Zel J, and Barker H 2001. Strong resistance to potato tuber necrotic ringspot disease in potato induced by transformation with coat protein gene sequences from an NTN isolate of potato virus Y. Ann. Appl. Biol. 139:269–275. [Google Scholar]
- Rairdan GJ, and Moffett P 2006. Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18:2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel MC, Leonelli L, Dahlbeck D, Zhao B, and Staskawicz BJ 2008. Recognition of the Hyaloperonospora parasitica effector ATR13 triggers resistance against oomycete, bacterial, and viral pathogens. Proc. Natl. Acad. Sci. U.S.A. 105:1091–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintenac C, Zhang W, Salcedo A, Rouse MN, Trick HN, Akhunov E, and Dubcovsky J 2013. Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 341:783–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saur IML, Bauer S, Lu X, and Schulze-Lefert P 2019. A cell death assay in barley and wheat protoplasts for identification and validation of matching pathogen AVR effector and plant NLR immune receptors. Plant Methods. 15:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeholzer S, Tsuchimatsu T, Jordan T, Bieri S, Pajonk S, Yang W, Jahoor A, Shimizu KK, Keller B, and Schulze-Lefert P 2010. Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol. Plant. Microbe. Interact. 23: 497–509. [DOI] [PubMed] [Google Scholar]
- Segretin ME, Pais M, Franceschetti M, Chaparro-Garcia A, Bos JIB, Banfield MJ, and Kamoun S 2014. Single amino acid mutations in the potato immune receptor R3a expand response to Phytophthora effectors. Mol. Plant. Microbe. Interact. 27:624–637. [DOI] [PubMed] [Google Scholar]
- Seppey M, Manni M, and Zdobnov EM 2019. BUSCO: Assessing genome assembly and annotation completeness. Methods Mol. Biol. 1962:227–245. [DOI] [PubMed] [Google Scholar]
- Shao Z-Q, Xue J-Y, Wu P, Zhang Y-M, Wu Y, Hang Y-Y, Wang B, and Chen J-Q 2016. Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol.170:2095–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q-H, Zhou F, Bieri S, Haizel T, Shirasu K, and Schulze-Lefert P 2003. Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell 15:732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, Njau P, Herrera-Foessel S, Singh PK, Singh S, and Govindan V 2011. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu. Rev. Phytopathol. 49:465–481. [DOI] [PubMed] [Google Scholar]
- Slootweg E, Koropacka K, Roosien J, Dees R, Overmars H, Lankhorst RK, van Schaik C, Pomp R, Bouwman L, Helder J, Schots A, Bakker J, Smant G, and Goverse A 2017. Sequence exchange between homologous NB-LRR genes converts virus resistance into nematode resistance, and vice versa. Plant Physiol. 175:498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slootweg EJ, Spiridon LN, Roosien J, Butterbach P, Pomp R, Westerhof L, Wilbers R, Bakker E, Bakker J, Petrescu A-J, Smant G, and Goverse A 2013. Structural determinants at the interface of the ARC2 and leucine-rich repeat domains control the activation of the plant immune receptors Rx1 and Gpa2. Plant Physiol. 162:1510–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, and Morgenstern B 2006. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 34:W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuernagel B, Periyannan SK, Hernández-Pinzón I, Witek K, Rouse MN, Yu G, Hatta A, Ayliffe M, Bariana H, Jones JDG, Lagudah ES, and Wulff BBH 2016. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34:652–655. [DOI] [PubMed] [Google Scholar]
- Steuernagel B, Witek K, Krattinger SG, Ramirez-Gonzalez RH, Schoonbeek H-J, Yu G, Baggs E, Witek AI, Yadav I, Krasileva KV, Jones JDG, Uauy C, Keller B, Ridout CJ, and Wulff BBH 2020. The NLR-Annotator tool enables annotation of the intracellular immune receptor repertoire. Plant Physiol. 183:468–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamborski J, and Krasileva KV 2020. Evolution of plant NLRs: From natural history to precise modifications. Annu. Rev. Plant Biol. 71: 355–378. [DOI] [PubMed] [Google Scholar]
- Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J, Ramirez-Gonzalez RH, Kolodziej MC, Delorean E, Thambugala D, Klymiuk V, Byrns B, Gundlach H, Bandi V, Siri JN, Nilsen K, Aquino C, Himmelbach A, Copetti D, Ban T, Venturini L, Bevan M, Clavijo B, Koo D-H, Ens J, Wiebe K, N’Diaye A, Fritz AK, Gutwin C, Fiebig A, Fosker C, Fu BX, Accinelli GG, Gardner KA, Fradgley N, Gutierrez-Gonzalez J, Halstead-Nussloch G, Hatakeyama M, Koh CS, Deek J, Costamagna AC, Fobert P, Heavens D, Kanamori H, Kawaura K, Kobayashi F, Krasileva K, Kuo T, McKenzie N, Murata K, Nabeka Y, Paape T, Padmarasu S, Percival-Alwyn L, Kagale S, Scholz U, Sese J, Juliana P, Singh R, Shimizu-Inatsugi R, Swarbreck D, Cockram J, Budak H, Tameshige T, Tanaka T, Tsuji H, Wright J, Wu J, Steuernagel B, Small I, Cloutier S, Keeble-Gagnère G, Muehlbauer G, Tibbets J, Nasuda S, Melonek J, Hucl PJ, Sharpe AG, Clark M, Legg E, Bharti A, Langridge P, Hall A, Uauy C, Mascher M, Krattinger SG, Handa H, Shimizu KK, Distelfeld A, Chalmers K, Keller B, Mayer KFX, Poland J, Stein N, McCartney CA, Spannagl M, Wicker T, and Pozniak CJ 2020. Multiple wheat genomes reveal global variation in modern breeding. Nature 588:277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Roux B, Feng F, Guy E, Li L, Li N, Zhang X, Lautier M, Jardinaud M-F, Chabannes M, Arlat M, Chen S, He C, Noël LD, and Zhou J-M 2015. The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18:285–295. [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang H-W, Zhou J-M, and Chai J 2019a. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364:aav5870. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang J, Hu M, Wu S, Qi J, Wang G, Han Z, Qi Y, Gao N, Wang H-W, Zhou J-M, and Chai J 2019b. Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364: aav5868. [DOI] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, and Marillonnet S 2011. A modular cloning system for standardized assembly of multigene constructs. PLoS One 6:e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Wing RA, and Wise RP 2002. Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14:1903–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Engler C, Weber E, Gruetzner R, and Marillonnet S 2012. Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng. Bugs 3:38–43. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Saitoh H, Fujisawa S, Kanzaki H, Matsumura H, Yoshida K, Tosa Y, Chuma I, Takano Y, Win J, Kamoun S, and Terauchi R 2009. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21: 1573–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerihun MB, Pucci F, Peter EK, and Schug A 2020. pydca v1.0: A comprehensive software for direct coupling analysis of RNA and protein sequences. Bioinformatics 36:2264–2265. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hewitt TC, Boshoff WHP, Dundas I, Upadhyaya N, Li J, Patpour M, Chandramohan S, Pretorius ZA, Hovmøller M, Schnippenkoetter W, Park RF, Mago R, Periyannan S, Bhatt D, Hoxha S, Chakraborty S, Luo M, Dodds P, Steuernagel B, Wulff BBH, Ayliffe M, McIntosh RA, Zhang P, and Lagudah ES 2021. A recombined Sr26 and Sr61 disease resistance gene stack in wheat encodes unrelated NLR genes. Nat. Commun. 12:3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Chen Z, Wu Q, Cai Y, Zhang Y, Zhao R, Yan J, Qian X, Li J, Zhu M, Hong L, Xing J, Khan NU, Ji Y, Wu P, Huang C, Ding XS, Zhang H, and Tao X 2021. The Sw-5b NLR nucleotide-binding domain plays a role in oligomerization, and its self-association is important for activation of cell death signaling. J. Exp. Bot. 72: 6581–6595. [DOI] [PubMed] [Google Scholar]
- Zhao Y-B, Liu M-X, Chen T-T, Ma X, Li Z-K, Zheng Z, Zheng S-R, Chen L, Li Y-Z, Tang L-R, Chen Q, Wang P, and Ouyang S 2022. Pathogen effector AvrSr35 triggers Sr35 resistosome assembly via a direct recognition mechanism. Sci. Adv. 8:abq5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and scripts used in this study are available in Github (https://github.com/krasileva-group/Sr33-Sr50_analysis).


