Abstract
A sensitive lawn-based format has been developed to screen bead-tethered combinatorial chemical libraries for antimicrobial activity. This method has been validated with beads linked to penicillin V via a photocleavable chemical linker in several analyses including a spike-and-recover experiment. The lawn-based screen sensitivity was modified to detect antibacterial compounds of modest potency, and a demonstration experiment with a naive combinatorial library of over 46,000 individual triazines was evaluated for antibacterial activity. Numerous hits were identified, and both active and inactive compounds were resynthesized and confirmed in traditional broth assays. This demonstration experiment suggests that novel antimicrobial compounds can be easily identified from very large combinatorial libraries of small, nonpeptidic compounds.
Rapid emergence of drug-resistant bacterial pathogens has led to a demand for novel antimicrobial compounds (4, 5, 18, 22). A potentially rich source of new antimicrobial compounds may be found in combinatorial chemistry libraries. The chemical diversity generated by combinatorial methods offers microbiologists a potential of heretofore-unseen drug leads; however, the sheer number of compounds presents a practical problem in screening and identifying those of interest (10, 11). This work addresses the fundamental technology challenge of developing assays and formats which allow the discrete screening of large (>10,000-member) combinatorial libraries.
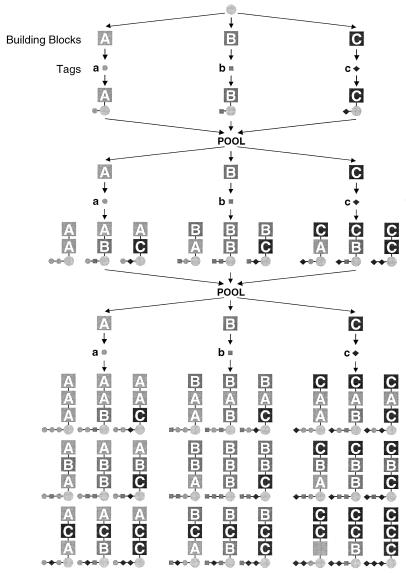
Extensive reviews on the preparation and utilization of combinatorial libraries in the drug discovery process have been published (7, 12, 17). Combinatorial libraries are often made by using split-and-pool synthesis, as first described by Furka et al. (9), and as exemplified in Fig. 1 for a 3 × 3 × 3 or 27-member library (for simplicity, a linear peptide library is shown). Beads are distributed into three reaction vessels, and an amino acid (A, B, or C) is coupled to the beads. The beads are then pooled and redistributed to the same three reaction vessels, where the same amino acids are coupled, resulting in a dipeptide. This results in a set of 2 × 3 peptides: AA, AB, AC, etc. The process is repeated once more to create a set of 27 peptides. A fundamental consequence of this approach is that while there can be millions of beads used in this synthesis, each bead carries only one of the 27 compounds that were synthesized.
FIG. 1.
Split-and-pool synthesis.
The compounds are tethered to the beads via cleavable linkers to allow release of the compound for assay. Frequently, photo-labile linkers (photolinkers) are used for this purpose (13). When the beads are exposed to high-intensity long-wave UV light, the linker is cleaved and the compound is released. The compound may then be assayed in a soluble form.
Due to the large number of compounds generated during a combinatorial synthesis, several approaches have been used to identify the structure of the compound carried on an individual bead (2). One solution, referred to as chemical encoding, is to utilize a collection of chemical identifier tags which can be detected more efficiently than the library compound which they represent. This process of chemical encoding includes the addition of chemical tags to the beads after each synthetic step, as illustrated in Fig. 1. For example, after the addition of amino acid A to a bead, tag “a” is also added to the same bead but is joined at a distinct chemical site on the bead. Each bead then carries a record of the synthesis of the compound also carried on that bead. By “reading” this tag, one can deduce the identity of the compound carried on the bead. Numerous tags and analytical methods for reading these tags have been developed (15, 16, 20, 21, 23–26).
Large encoded combinatorial libraries on beads present a major challenge to the assayer: how to screen thousands of compounds efficiently while maintaining separation of the beads. Jayawickreme et al. presented the first evidence that single-bead activity from peptides could be detected on acid-cleavable beads in a lawn format assay (14). The work presented here demonstrates how we developed and implemented a system to simultaneously screen thousands of compounds against bacterial cells in a lawn assay. In a primary demonstration, penicillin V (PenV), a known antibiotic, was coupled to beads via a photolinker to test the concept and develop the method. Ultimately, a 36 × 36 × 36 (46,656-member) library of triazines was screened by using a two-dimensional (2D) agar-based whole-cell format, showing that the lawn-based format can be applied to large naive libraries as a first step in the application of combinatorial chemistry to antimicrobial lead discovery.
MATERIALS AND METHODS
Growth of cultures.
Bacillus subtilis ATCC 6633 and Staphylococcus aureus ATCC 29523 were obtained from the American Type Culture Collection. Broth microdilution assays were run in accordance with National Committee for Clinical Laboratory standards with the modification that the concentration of bacteria was ∼105/ml (19). The MIC was determined as the lowest dilution of compound prohibiting visible growth of the organism.
PenV beads.
The hydroxymethyl-nitroveratryl photolinker (13) was converted to the benzyl bromide with dibromotriphenylphosphorane in dichloromethane and then treated with the cesium salt of PenV (Sigma) in 50% dimethyl sulfoxide–dimethyl formamide overnight. After the resin was washed, analysis by solid-phase magic-angle-spinning 1H nuclear magnetic resonance (8) indicated an 80% coupling yield. The linkage and integrity of PenV were consistent with high-performance liquid chromatography (HPLC) and mass spectroscopy (MS) analyses of photolysates of washed beads.
Photolysis of PenV and triazine beads.
For lawn-based assays, beads were distributed (see below) and subjected to variable periods of exposure to 365-nm light at an intensity of 10 mW/cm2. For analysis of the PenV beads, 0.5-mg samples of PenV-linked beads were suspended in methanol and exposed to 365-nm light at 10 mW/cm2 for 30 min. The methanol was subsequently removed from the beads for further analysis of released PenV.
Lawn-based assays.
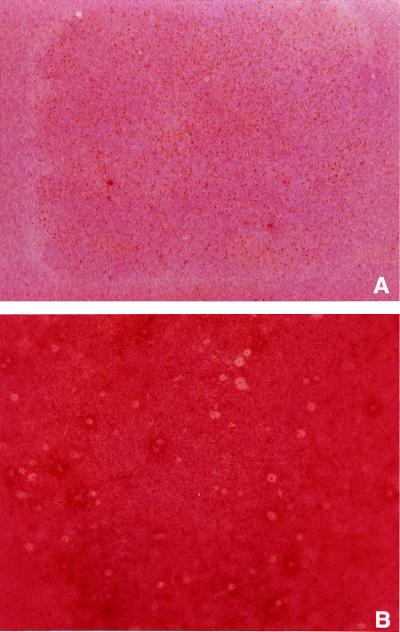
B. subtilis was selected for these studies based on its sensitivity to PenV. SeaPlaque GTG agarose (FMC Bioproducts) was used for all lawn-based assays. For the PenV assays, 30 mg of unmodified TentaGel-SH beads (130-μm diameter, 0.21 mmol/g; Rapp Polymere) was mixed with a given number of PenV beads (see below and Fig. 4) in 4 ml of 0.4% agarose in phosphate-buffered saline (PBS; Sigma) and plated onto Luria-Bertani (LB) agar (Difco Laboratories). Following 5 min of photolysis as described above, the beads were covered with 8 ml of 0.8% LB agarose mixed with ∼6 × 107 CFU of B. subtilis. The plates were incubated overnight at 30°C. To test the tiered release of PenV, the initial photolysis time was reduced to 2 min. Following overnight incubation, the entire growth inhibition zone (GIZ) was excised, placed on a polycarbonate filter (0.4-μm pore size; Millipore), and repeatedly washed with 37°C 6 N NaI (Bio 101), followed by PBS. The filter was placed on an LB agar plate. The beads were manually dispersed, suspended in PBS agarose, photolysed for an additional 3 min, and inoculated as before. For the more sensitive triazine library screening, Omni Trays (Nunc) with a base layer of 0.8% LB agarose were used. Library beads were manually spread on 105-μm-pore-size polyester mesh (Spectrum) and placed on a nitrocellulose membrane (Bio-Rad) previously spread with in 1.4 ml of 0.4% PBS agarose. Following 30 min of photolysis, the plates were covered with a 4-ml layer of 0.4% LB agarose containing ∼107 CFU of B. subtilis and incubated as before. To better visualize the GIZs, the plates were flooded with 0.1% triphenyl tetrazolium chloride (TTC) in LB broth (Difco Laboratories), which imparts a strong red color to redox-active cells. GIZs are then visualized as white zones, with a bead in the center, on a red lawn.
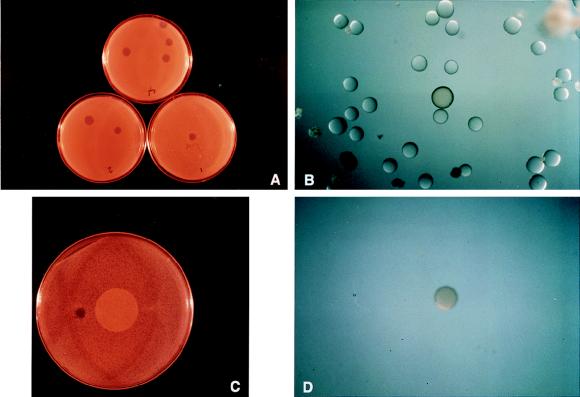
FIG. 4.
Validation with PenV linked to beads. (A) One, two, or four PenV beads (shown clockwise) were mixed with over 10,000 untreated beads, dispersed on LB agar petri dishes (premarked for the number of PenV beads added), and photolysed. The plates were covered with B. subtilis in LB agarose and incubated at 30°C for 16 h. (B) Magnification of a GIZ where one bead was combined with 10,000 TentaGel beads and then subjected to photolysis; note the presence of a brown bead. (C) A GIZ as shown in panel B was excised, placed on a filter, and extracted from the agarose (see Materials and Methods). The filter containing released beads was placed on an LB agar plate, and the beads were manually dispersed on the surface and treated as described for panel A. (D) Magnification of a GIZ showing an isolated brown PenV bead in the center of the clearing viewed postincubation.
Triazine library construction.
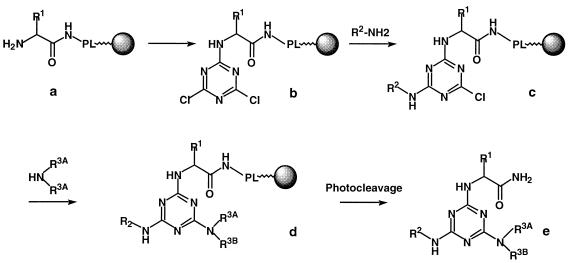
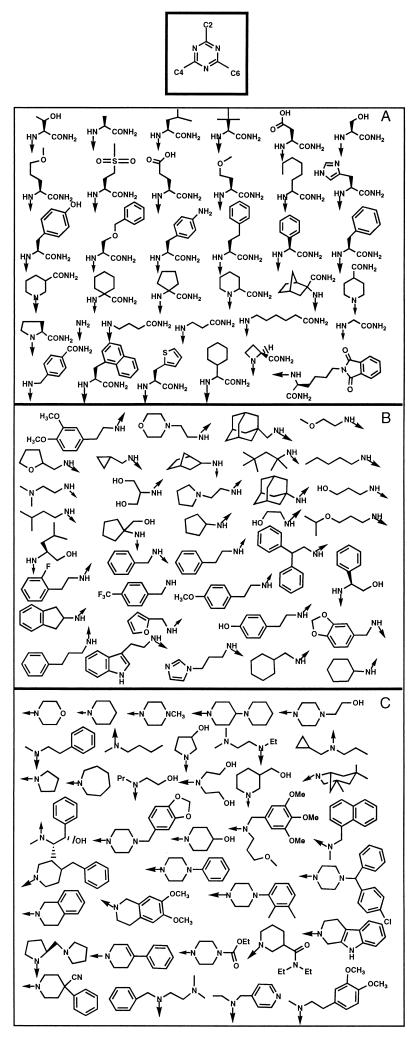
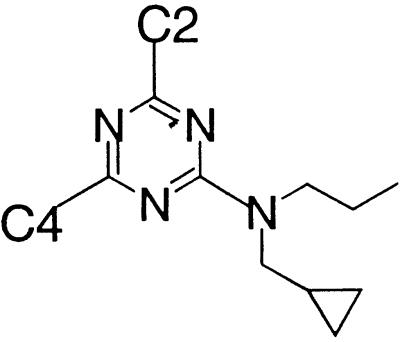
The triazine combinatorial library is based on a trichlorotriazine scaffold which is then modified with 36 chemical building blocks at each of three positions, resulting in a complete library size of 46,656 compounds. The outline of the synthetic route is shown in Fig. 2. The set of building blocks is shown in Fig. 3. The 9-fluorenylmethoxycarbonyl (Fmoc)-protected amino acid building blocks, with t-butyl-protected side chains, of Fig. 3A were attached to 130-μm-diameter TentaGel beads via amide bond formation to a photocleavable linker (13). After removal of the 9-fluorenylmethoxycarbonyl group, the amino group was then reacted with trichlorotriazine for attachment to the triazine scaffold. The tethered dichlorotriazine was then sequentially modified at the remaining two positions with amines (Fig. 3B and C) by using a split-and-pool strategy (9) (Fig. 1). Finally, side chain protecting groups were removed by treatment with 50% trifluoracetic acid-dichloromethane. The synthetic history of each bead was encoded by using secondary amine tagging, allowing for rapid structural analysis of the compound originally tethered to any given bead (23).
FIG. 2.
The combinatorial synthetic route for triazines from amino acids and free amines. The synthesis begins as an amino acid (with side chain R1) is coupled through its carboxyl terminus to a photolinker (PL) on a bead (a). The core scaffold, trichlorotriazine is then coupled to the free amino group of the amino acid to yield structure b. The second center of diversity derives from the primary amine R2, which substitutes one (and only one, under the conditions used) of the Cl groups, resulting in structure c. This is repeated with secondary amine R3AR3BNH to yield structure d. Upon photocleavage, the free triazine (structure e) is released into solution.
FIG. 3.
Components of the 36 × 36 × 36 triazine library. (A) Amino acid building blocks, C-2; (B) amine building blocks, C-4; (C) amine building blocks, C-6.
Bead selection and isolation from lawn-based triazine library assays.
Beads located in the center of GIZs (white zones on red lawns) were selected for decoding. In addition, a number of beads not associated with GIZs were selected as inactive beads. All beads were manually isolated from the assay plates by using capillary tubes and transferred to microtubes for further analysis.
Chemical decoding and structure determination.
All of the isolated beads recovered from lawn-based assays were rephotolysed in methanol, and the solution was subjected to MS analysis. Following removal of the methanol, the beads were recovered and treated appropriately to determine the chemical encoding, and hence, the synthetic history of the bead (23). MS analysis was used as a confirmatory method of compound identification.
Triazine compound synthesis.
“Active” triazines (Table 1) were prepared by solution-phase synthesis. The chlorine atoms on the trichlorotriazine scaffold were sequentially displaced with one equivalent of primary amine (structures as in C-4) (see Fig. 4) at 0°C, followed by reaction with 0.5 N NH3 in p-dioxane at 40°C and then with N-propylcyclopropanemethylamine at 80°C. The “inactive” compounds (see above and Table 1) were resynthesized in bulk by solid-phase chemistry similar to the previously described methods for triazine construction. The photolysates were concentrated and purified by HPLC. All compounds were characterized by 1H nuclear magnetic resonance and MS analyses, and HPLC indicated a purity of >95%.
TABLE 1.
Antibacterial activity of purified compounds identified from two-dimensional screeninga
| Active beads
|
Inactive beads
|
||||||
|---|---|---|---|---|---|---|---|
| Compound structure
|
MIC (μg/ml) for:
|
Compound structure
|
MIC (μg/ml) for:
|
||||
| C-2 | C-4 | B. subtilis | S. aureus | C-2 | C-4 | B. subtilis | S. aureus |
 |
 |
||||||
Compounds identified by decoding were synthesized in solid-phase reactions and purified as described in the text. The compounds were tested in a microdilution assay for antibacterial activity (see Materials and Methods).
RESULTS
To validate a lawn-based antibacterial assay, antibiotic-tethered beads were prepared. PenV was selected for biological activity and structural compatibility with photolinker-coupling chemistry (Materials and Methods). In comparison to untreated PenV solutions, PenV stock solutions prepared in PBS and subjected to photolysis conditions retained full activity. The results indicated that photolysis conditions do not alter PenV potency (data not shown). Examination of the PenV bead photolysates indicated an approximate loading efficiency of 200 pmol of PenV per bead. In addition, the photolysate of an aliquot of 100 PenV beads in PBS was tested and activity was confirmed in a MIC assay with B. subtilis. The PenV beads were first tested for antibacterial activity in a 2D agar format by dispensing from one to five beads, in molten agarose, onto a standard LB medium plate. The plate was exposed to 365-nm light for 10 to 30 min and then overlaid with a final layer of agarose inoculated with B. subtilis. Following incubation, GIZs were visualized as clear areas around the photolysed beads. PenV beads not subjected to photolysis did not generate GIZs, indicating that no spontaneous hydrolysis of the linkage between the antibiotic and the bead had occurred during the course of the assay (data not shown).
As a statistical test of the assay method and reagents, either one, two, or four PenV beads were added to pools of approximately 10,000 unmodified 130-μm-diameter beads. The PenV-bead-spiked pools were then resuspended in molten agarose, plated, photolysed, and inoculated as described above. Following overnight incubation, the plates displayed one, two, and four GIZs, respectively (Fig. 4A).
To demonstrate that GIZs are generated by single PenV beads, and that beads can successfully be recovered from the agarose, an experiment was designed to demonstrate the purification of one active bead from a pool of >10,000 unmodified 130-μm-diameter beads. As in the statistical test, one PenV bead was combined with >10,000 unmodified 130-μm-diameter beads in molten agarose and then photolysed briefly prior to overlay with the B. subtilis culture. One small GIZ was apparent following overnight incubation. Microscopic examination of the GIZ revealed the presence of approximately 30 colorless beads and 1 amber-colored bead (Fig. 4B). Since TentaGel beads acquire an amber color upon photolinker coupling and unmodified beads remain colorless, the presence of the PenV bead within the GIZ could be tentatively confirmed by virtue of its amber color. An agarose plug containing the beads in the GIZ was excised and immobilized over a vacuum filtration device, and the agarose was dissolved with NaI. The rinsed filter was transferred to a new LB agar plate, and the recovered beads were widely dispersed in molten agarose. A 10-min photolysis was performed, and the plate was inoculated as described previously. Overnight incubation of this plate revealed a single GIZ (Fig. 4C). Microscopic examination of the GIZ revealed the presence of the single amber bead in the very center of the GIZ (Fig. 4D), confirming the isolation of one PenV-containing bead from the original pool of >10,000 beads.
While the PenV model demonstrates the purification of one highly potent antibacterial bead from a population of essentially inactive beads, we were concerned that isolation of less potent compounds from naive chemical libraries in this format would require greater sensitivity. The method was therefore modified by applying the beads to a polyester mesh support and, following photolysis, overlaying them with a very thin layer of inoculated agarose. The sensitivity was further enhanced by staining the mature bacterial culture with the application of TTC, a redox indicator dye. TTC treatment stains viable cells a deep red color. GIZs are visualized as white zones around a bead in the midst of a red lawn. With the modified method, GIZs appear from PenV beads photolysed for only 30 s, a process previously requiring minutes of exposure (data not shown).
To demonstrate the efficacy of the method to identify novel antimicrobial compounds, a large naive combinatorial library was tested by using the modified method. An available library of triazines was chosen for its size and composition, as there was no evidence in the literature to suggest that triazines could function as antimicrobial agents (therefore the triazine library could be considered a “naive” library with respect to antimicrobial screening). The complete triazine library of 46,656 compounds (Fig. 3) was chosen to test the concept of identifying active compounds among thousands of inactive compounds. For purposes of this study, the 36 pools of beads generated in the final round of synthesis were preserved as individual samples, meaning that each pool of 1,296 compounds has one building block in common (Fig. 3). Antibacterial screening was performed on aliquots of the individual pools. For each of the 36 pools, more than one pool equivalent (1,400 to 1,900 beads; approximately 5 to 7 mg of beads) was screened for antibacterial activity by using the more sensitive format described above. GIZs, as shown in Fig. 5, appeared as white zones in contrast to the red lawn and are clearly associated with individual beads. As might be expected from the synthesis strategy and their common chemical signature, individual pools presented unique antibacterial qualities, as measured by the number and relative intensities of GIZs observed in each screen. For example, pool 25 revealed no strong GIZs (not shown) in contrast to pool 16, which had a few small well-defined GIZs (Fig. 5A), and pool 10, which generated numerous GIZs of various intensities (Fig. 5B).
FIG. 5.
Lawn-based screen of pools 16 and 10 from the triazine library versus B. subtilis. Beads from the library pool were separated and placed on the surface of LB agarose in a rectangular assay plate. The beads were photolysed and overlaid with B. subtilis. After a 16-h incubation at 30°C, the plates were flooded with TTC to visualize the GIZs (see Materials and Methods).
To provide examples of the triazine library for further characterization, pool 10 was selected for further study. The large number of GIZs suggested that the building block common to all members of pool 10 (N-propyl-N-cyclopropylmethyl amine) (see Table 1) was likely to confer antimicrobial activity to a variety of decorated triazines. Furthermore, in contrast to the PenV pilot experiment, the small size of the zones meant that single beads could be readily isolated. The primary screening plate, containing an aliquot of approximately 1,900 beads of pool 10, was examined under the microscope. GIZs containing single beads were identified, and the beads (active beads) were removed from the agarose with a fine capillary tube. In addition, beads clearly not associated with GIZs (inactive beads) were selected to determine the structure of compounds with no detectable activity in the lawn-based screen. All isolated beads were submitted for analysis and determination of compound structure as described in Materials and Methods.
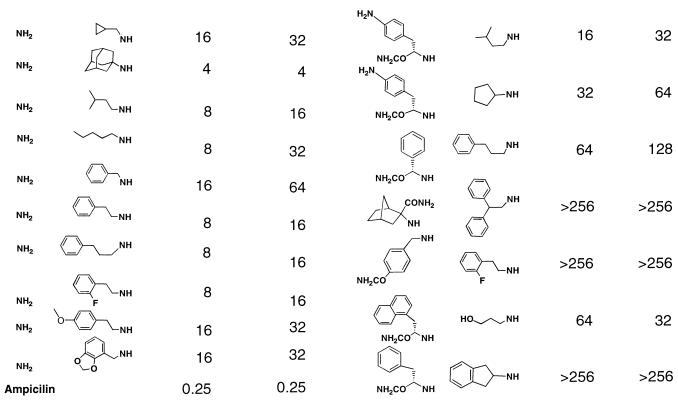
The deduced structures of compounds associated with active and inactive beads from the screening of pool 10 are shown in Table 1. The most striking feature of the data is the conservation of one of the 36 building blocks (the NH2 group at C-2) in all of the structures deduced from active beads. None of the compounds decoded from inactive beads possess an NH2 group, further strengthening this correlation. To assess their antimicrobial activity, all 16 compounds were resynthesized, purified, and tested in a standard broth microdilution assay against B. subtilis. The MICs presented in Table 1 show that the compounds identified within GIZs (active compounds) have measurable activity against B. subtilis from 4 to 32 μg/ml. That several of the compounds have activities of less than 10 μg/ml demonstrates the successful identification of active compounds from one selected pool of a 46,656-member library.
Although the values obtained from the purified active compounds would not be considered extraordinarily potent, the purpose of primary screening is to isolate new compounds that can be used as the basis of iterative synthesis and standard broth assay to increase potency. Examination of compounds from the inactive beads show a range of activities from 16 to >256 μg/ml, a potency range much reduced from that of the active compounds. The overlap of MICs between the set of active compounds and the set of inactive compounds is neither surprising nor particularly detrimental in the final evaluation. A large-scale primary screening method is expected to be qualitative rather than quantitative in nature, and several sources of variability, including compound loading per bead and variation in compound diffusion, will blur the threshold between active and inactive compounds (see Discussion). The more compelling issues are that (i) active compounds of moderate potency are identified by this method and (ii) as previously discussed, altering assay conditions allows one to change the sensitivity of the assay. The latter, in turn, changes the threshold zone between active and inactive compounds, making it possible to adjust the stringency of a screen for a given target strain, compound library, and desired potency. However, it should be noted that due to the previously mentioned variability, some active compounds may be missed.
DISCUSSION
We have presented a sensitive lawn-based assay for evaluating the antimicrobial activity of large combinatorial chemistry libraries. The method was initially developed and validated by using TentaGel bead-linked PenV and then further optimized to identify less potent compounds in large naive combinatorial libraries. A 46,656-member combinatorial library, not specifically designed to include antimicrobial compounds, was screened by this method, and selected hits were further characterized.
When tested against B. subtilis in the standard broth microdilution assay, the active compounds have MICs that fall within the range of 4 and 16 μg/ml, including six compounds with MICs of less than 10 μg/ml. All 16 compounds were also tested for their activity against S. aureus, a less sensitive organism. The contrast in MICs between the active and inactive compounds from pool 10 is maintained with this more clinically relevant organism, consistent with results generated in the lawn-based screen.
That the purified, resynthesized compounds have antibacterial activity validates the identification of active beads from GIZs. It should be noted that there was no rigorous correlation between the GIZ size and the MIC of the purified compound. Factors influencing the appearance of a GIZ around a bead include the relative solubility, and consequent partitioning, of a compound in agarose versus the bead matrix. Given the hydrophobic nature of many of the combinatorial building blocks included in this library, it is conceivable that a proportion of the compounds within the library simply do not diffuse away from the polymer matrix of the 130-μm-diameter beads. Inability to detect antimicrobial compounds with this property in the lawn-based screen may, in part, explain the presence of modest activity in some of the inactive compounds (1). This phenomenon has recently been observed with teicoplanin (28), an antibiotic of demonstrated potency, and argues for cautious interpretation in the initial screening of compounds followed by careful evaluation of selected active compounds, and their analogs, in a broth microdilution assay. Not every compound with an activity of <10 μg/ml may be detected in this format; however, the antimicrobial potency structure activity relationship is intended to be developed through subsequent rounds of synthesis and screening of chemical analogs. Active structures deduced from screening in this lawn-based format must then be considered a subset of all potentially active structures in a given library. Further synthesis of chemical analogs and evaluation of those compounds is intended to fully develop the antimicrobial structure activity relationship of a given scaffold. Thus, the lawn-based format satisfies the first technology challenge in that thousands of discrete combinatorial compounds may be tested simultaneously.
The lawn-based screening method was established to examine large combinatorial libraries of small molecules to find leads that can be further optimized. This strategy offers several advantages over current approaches to the discovery of novel antimicrobial compounds. Whole-cell assays eliminate the restriction of testing a known panel of target proteins. In addition, this method can be adapted to any species that can be cultured on an agar-based medium, including gram-negative bacteria, yeast (data not shown), and mammalian cells (27). Finally, hits from combinatorial libraries may be rapidly expanded to hundreds of second-generation analogs by using the powerful technique of automated parallel synthesis (3, 6). These analogs can then be tested to determine the spectrum of microbial action, cytotoxicity, and, by tests against specific enzymes, the probable mechanism of action. With systematic approaches, a new lead family can be generated and validated in a matter of days, rather than months. Practiced in series with traditional assay methods, the 2D agar format can be a useful tool to explore the diversity afforded by large, bead-based combinatorial chemistry in the discovery of novel antimicrobial compounds.
REFERENCES
- 1.Acar J F, Goldstein F W. Disk susceptibility test. In: Lorian V, editor. Antibiotics in laboratory medicine. Baltimore, Md: The Williams & Wilkins Co.; 1996. pp. 1–51. [Google Scholar]
- 2.Brenner S, Lerner R A. Encoded combinatorial chemistry. Proc Natl Acad Sci USA. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cargill J F, Maiefski R R. Automated combinatorial chemistry on solid phase. Lab Rob Autom. 1996;8:139–148. [Google Scholar]
- 4.Davies J. Inactivation of antibiotics and the dissemination of resistance genes. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 5.Desnottes J-F. New targets and strategies for the development of antibacterial agents. Trends Biotechnol. 1996;14:134–140. doi: 10.1016/0167-7799(96)10015-9. [DOI] [PubMed] [Google Scholar]
- 6.DeWitt S H, Schroeder M C, Stankovic C J, Strode J E, Czarnik A W. DIVERSOMER™ technology: solid phase synthesis, automation, and integration for the generation of chemical diversity. Drug Dev Res. 1994;33:116–124. [Google Scholar]
- 7.Ellman J A. Design, synthesis, and evaluation of small-molecule libraries. Acc Chem Res. 1996;29:132–143. [Google Scholar]
- 8.Fitch W L, Detre G, Holmes C P, Shoolery J N, Keifer P A. High-resolution 1H NMR in solid-phase organic synthesis. J Org Chem. 1994;59:7955–7956. [Google Scholar]
- 9.Furka A, Sebestyen F, Asgedom M, Dibo G. General method for rapid synthesis of multicomponent peptide mixtures. Int J Pept Protein Res. 1991;37:487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 10.Gallop M A, Barrett R W, Dower W J, Fodor S P A, Gordon E M. Applications of combinatorial technologies to drug discovery. 1. Background and peptide combinatorial libraries. J Med Chem. 1994;37:1233–1251. doi: 10.1021/jm00035a001. [DOI] [PubMed] [Google Scholar]
- 11.Gordon E M, Barrett R W, Dower W J, Fodor S P A, Gallop M A. Applications of combinatorial technologies to drug discovery. 2. Combinatorial organic synthesis, library screening strategies, and future directions. J Med Chem. 1994;37:1385–1401. doi: 10.1021/jm00036a001. [DOI] [PubMed] [Google Scholar]
- 12.Gordon E M, Gallop M A, Campbell D, Holmes C, Bermak J, Look G, Murphy M, Needels M, Jacobs J, Sugarman J, Chinn J, Fritsch B R, Jones D. Combinatorial organic synthesis: applications to drug discovery. Eur J Med Chem. 1995;30S:S337–S348. [Google Scholar]
- 13.Holmes C P, Jones D G. Reagents for combinatorial organic synthesis: development of a new o-nitrophenyl photolabile linker for solid phase synthesis. J Org Chem. 1995;60:2318–2319. [Google Scholar]
- 14.Jayawickreme C K, Graminski G F, Quillan J M, Lerner M R. Creation and functional screening of a multi-use peptide library. Proc Natl Acad Sci USA. 1994;91:1614–1618. doi: 10.1073/pnas.91.5.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerr J M, Banville S C, Zuckermann R N. Encoded combinatorial peptide libraries containing non-natural amino acids. J Am Chem Soc. 1993;115:2529–2531. [Google Scholar]
- 16.Krchnak V, Weichsel A S, Cabel D, Lebl M. Linear presentation of variable side-chain spacing in a highly diverse combinatorial library. Pept Res. 1995;8:198–205. [PubMed] [Google Scholar]
- 17.Lam K S, Lebl M, Krchnak V. The “one-bead-one-compound” combinatorial library method. Chem Rev. 1997;97:411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- 18.Mitsuhashi S. Drug resistance in bacteria: history, genetics and biochemistry. J Int Med Res. 1993;21:1–14. doi: 10.1177/030006059302100101. [DOI] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A3. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 20.Needels M C, Jones D G, Tate E H, Heinkel G L, Kochersperger L M, Dower W J, Barrett R W, Gallop M A. Generation and screening of an oligonucleotide-encoded synthetic peptide library. Proc Natl Acad Sci USA. 1993;90:10700–10704. doi: 10.1073/pnas.90.22.10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nester H P, Bartlett P A, Still W C. A general method for molecular tagging of encoded combinatorial chemistry libraries. J Org Chem. 1994;59:4723–4744. [Google Scholar]
- 22.Neu H C. The crisis in antibiotic resistance. Science. 1992;257:1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 23.Ni Z-J, Maclean D, Holmes C P, Murphy M M, Ruhland B, Jacobs J W, Gordon E M, Gallop M A. Versatile approach to encoding combinatorial organic syntheses using chemically robust secondary amine tags. J Med Chem. 1996;39:1601–1608. doi: 10.1021/jm960043j. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen J, Brenner S, Janda K D. Scientific methods for the implementation of encoded combinatorial chemistry. J Am Chem Soc. 1993;115:9812–9813. [Google Scholar]
- 25.Nikolaiev V, Stierandova A, Krchnak V, Seligmann B, Lam K S, Salmon S E, Lebl M. Peptide-encoding for structure determination of nonsequence-able polymers within libraries synthesized and tested on solid-phase supports. Pept Res. 1993;6:161–170. [PubMed] [Google Scholar]
- 26.Ohlmeyer M H, Swanson R N, Dillard L W, Reader J C, Asouline G, Kobayashi R, Wigler M, Still W C. Complex synthetic chemical libraries indexed with molecular tags. Proc Natl Acad Sci USA. 1993;90:10922–10926. doi: 10.1073/pnas.90.23.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salmon S E, Liu-Stevens R H, Zhao Y, Lebl M, Krchnak V, Wertman K, Sepetov N, Lam K S. High-volume cellular screening for anticancer agents with combinatorial chemical libraries: a new methodology. Mol Divers. 1996;2:57–63. doi: 10.1007/BF01718701. [DOI] [PubMed] [Google Scholar]
- 28.Sizemore C F, Seneci P, Kocis P, Wertman K F, Islam K. Combinatorial chemistry and natural products: determination of the biological activity of on-bead, double cleavable teicoplanin aglycone (TD) Protein Pept Lett. 1996;3:253–260. [Google Scholar]