Summary
Misfolded glycoprotein recognition and endoplasmic reticulum (ER) retention are mediated by the ER glycoprotein folding quality control (ERQC) checkpoint enzyme, UDP-glucose glycoprotein glucosyltransferase (UGGT). UGGT modulation is a promising strategy for broad-spectrum antivirals, rescue-of-secretion therapy in rare disease caused by responsive mutations in glycoprotein genes, and many cancers, but to date no selective UGGT inhibitors are known. The small molecule 5-[(morpholin-4-yl)methyl]quinolin-8-ol (5M-8OH-Q) binds a CtUGGTGT24 “WY” conserved surface motif conserved across UGGTs but not present in other GT24 family glycosyltransferases. 5M-8OH-Q has a 47 μM binding affinity for CtUGGTGT24in vitro as measured by ligand-enhanced fluorescence. In cellula, 5M-8OH-Q inhibits both human UGGT isoforms at concentrations higher than 750 μM. 5M-8OH-Q binding to CtUGGTGT24 appears to be mutually exclusive to M5-9 glycan binding in an in vitro competition experiment. A medicinal program based on 5M-8OH-Q will yield the next generation of UGGT inhibitors.
Subject areas: Chemistry, Cell biology, Functional aspects of cell biology
Graphical abstract
Highlights
-
•
A ligand of UGGT was discovered by fragment-based lead discovery
-
•
The ligand’s Kd for a fungal UGGT was measured as 47 μM by ligand-enhanced fluorescence
-
•
The ligand inhibits both human UGGT1 and UGGT2 above 750 μM in cellula
-
•
The ligand and an M5-9 glycan mix compete for overlapping sites on UGGT
Chemistry; Cell biology; Functional aspects of cell biology
Introduction
In the endoplasmic reticulum (ER) of eukaryotic cells, the ER glycoprotein folding quality control (ERQC) system ensures ER retention of immature glycoproteins and assists their folding.1 Glycoprotein ERQC is central to glycoproteostasis, which in turn plays a major role in health and disease.2,3 Glycoprotein ERQC is reliant on detection of glycoprotein misfolding, affected by its checkpoint enzyme, UDP-glucose glycoprotein glucosyltransferase (UGGT). UGGT is capable of detecting non-native and slightly misfolded glycoproteins and re-glucosylates its clients to flag them for ER retention.4,5
While other components of ERQC have been studied as drug targets,6,7,8 cellular consequences of pharmacological modulation of UGGT have been relatively understudied—partly because of the risks associated with targeting core cell housekeeping machineries, and partly because there are no known UGGT selective inhibitors. UGGT is inhibited by its product uridine diphosphate (UDP)9 and squaryl derivatives of UDP10; by the non-hydrolyzable UDP-Glucose (UDP-Glc) cofactor analog UDP-2-deoxy-2-fluoro-D-glucose (U2F); and by synthetic analogs of the N-linked Man9GlcNAc2 glycan substrate,11,12 but obviously none of these molecules are UGGT specific. Selective and potent UGGT modulators would be important reagents for interrogating the cell biology of the secretory pathway, as well as having therapeutic potential in several areas of medical science (such as virology,13,14,15 metabolic and rare genetic disease,16,17,18 immunology,5 and cancer19,20,21), biotechnology, and agricultural science.22,23,24,25
We set out to search for ligands of UGGT by fragment-based lead discovery (FBLD) using X-ray crystallography, an approach which requires the growth of hundreds of well-diffracting crystals of the target.26,27,28,29 No crystal structures of mammalian UGGTs have been obtained so far, but atomic resolution structures of UGGTs from thermophilic fungi have been determined.30,31,32 None of the crystals of full-length UGGT we grew so far diffracted past 2.8 Å,30,32 but 1.35 and 1.4 Å crystal structures of the catalytic domain of Thermomyces dupontii UGGT (TdUGGTGT24), in complex with UDP and UDP-Glc, respectively, have been described.31 Although compounds binding the UGGT N-terminal folding-sensor domains of the enzyme would also be potential UGGT inhibitors, we decided to target the UGGT C-terminal catalytic domain (belonging to the GlycosylTransferase Family 24 (GT24) fold), given the high 70% similarity and 60% identity between human and fungal sequences in this portion of the enzyme.
Toward the FBLD of ligands of the UGGT C-terminal catalytic domain, we cloned in the pHLsec vector for secreted mammalian expression33 the catalytic domain of Chaetomium thermophilum UGGT (CtUGGTGT24), without its C-terminal ER-retrieval motif, and expressed, purified, and crystallized the protein.34 We then used those CtUGGTGT24 crystals for our FBLD effort, in which each crystal was soaked with a different chemical compound from a molecular fragment library.34 The study’s best hit was a 2.25 Å crystal structure of CtUGGTGT24 in complex with the fragment ligand 5-[(morpholin-4-yl)methyl]quinolin-8-ol, 5M-8OH-Q for short in what follows.
Here, we describe the 1.65 Å structure of a co-crystal of CtUGGTGT24 and 5M-8OH-Q (5M−8OH−QCtUGGTGT24), as well as the crystal structures of apo CtUGGTGT24 and CtUGGTGT24 in complex with the U2F cofactor analog (U2FCtUGGTGT24). We measure the 5M-8OH-Q affinity for CtUGGTGT24 and human UGGT1 in vitro and show that in human cells the molecule inhibits both human paralogs of UGGT, UGGT1, and UGGT2, at concentrations higher than 750 μM. We present an in silico model of the GlcNac2Man9 N-linked glycan in the catalytic site of UGGT, suggesting that the ligand interferes with N-glycan binding, therefore likely acting as a competitive inhibitor. This hypothesis is supported by a competition assay in vitro, in which the N-linked glycan displaces the inhibitor from its binding site in the UGGT catalytic domain. A medicinal chemistry program to generate more potent and selective UGGT inhibitors starting from 5M-8OH-Q is in progress.
Results
The active site of CtUGGTGT24 undergoes structural rearrangements upon binding the U2F cofactor analog
The crystal structures of CtUGGTGT24 in absence of the UDP-Glc cofactor and of the same protein in complex with the U2F cofactor analog (U2FCtUGGTGT24) were determined by X-ray crystallography. Tables S1 and S2 list the X-ray data collection statistics and structure refinement statistics, respectively. These structures constituted the basis for the FBLD effort that discovered 5M-8OH-Q as a CtUGGTGT24 ligand.34
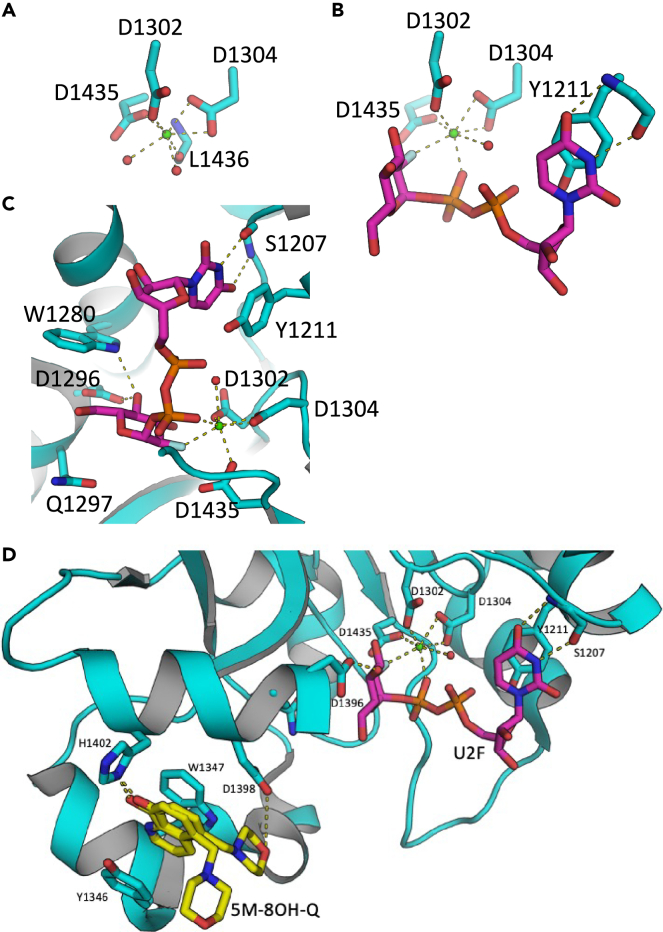
The CtUGGTGT24 active site undergoes structural changes binding the U2F cofactor analog. Half of the coordination sphere of the Ca2+ ion in the CtUGGTGT24 active site is common to both structures: the side chains of D1302 and D1304 (belonging to the UGGT conserved DAD motif35) and the side chain of the conserved D1435 always take up three invariant coordination sites around the Ca2+ ion (Figures 1A and 1B). In the 1.8 Å structure of apo CtUGGTGT24 (PDB ID 7ZKC), two water molecules occupy two of the three remaining coordination sites around the Ca2+ ion, with the main chain carbonyl oxygen of L1436 completing the ion’s octahedral coordination (Figure 1A). In the U2FCtUGGTGT24 structure (PDB ID 7ZLU) these two water molecules are replaced by an O atom from the β phosphate and by the F atom on the Glc ring of U2F (Figure 2A); the main chain of L1436 moves away from the Ca2+ ion, and a water molecule occupies its Ca2+ coordination site (Figures 1B, 1C, and 2A).
Figure 1.
CtUGGTGT24 crystal structures
(A–C) The active sites of CtUGGTGT24 and U2FCtUGGTGT24. Protein atoms in sticks representation; C cyan (but U2F C magenta, and 5M-8OH-Q C atoms yellow), O red, N blue, P orange, F light green. H-bonds and Ca2+-coordination bonds are in yellow dashed lines. The Ca2+ ion is a green sphere and its coordinating water molecules are red spheres. The side chains of residues D1302, D1304, and D1435 coordinate the Ca2+. (A) apo CtUGGTGT24 (PDB ID 7ZKC). The octahedral coordination sphere of the Ca2+ ion is completed by two water molecules and the main chain of L1436. (B) U2FCtUGGTGT24 (PDB ID 7ZLU). L1436 moves away from the Ca2+ ion, and two coordination sites are taken up by the U2F β phosphate and the F atom at position 2′ of the Glc ring. The uracyl O4 atom accepts an H-bond from the S1207 main chain NH. Only one Ca2+-coordinating water molecule remains. (C) the UGGT active site selects UDP-Glc over UDP-Gal36,37,38,39: in UDP-Glc the glucose O4′ atom forms hydrogen bonds to the side chains of conserved W1280 and D1396, but these interactions would be lost in UDP-Gal (because of the difference in stereochemistry between Glc and Gal in position 4). The side chain of Y1211 and the main chain of S1207 coordinate the uracyl ring.
(D) The U2FCtUGGTGT24 structure (PDB ID 7ZLU) overlaid with the 5M-8OH-Q ligand from the 5M−8OH−QCtUGGTGT24 structure (PDB ID 7ZLL), in the enzyme active site region. The CtUGGT clamp, the conserved motif, H1402, Y1211, and the main chain of S1207 are in stick representation. Only two of the many poses of the 5M-8OH-Q inhibitor are shown.
Figure 2.
Modeling of the GlcNAc2Man9 glycan bound to the CtUGGTGT24 domain
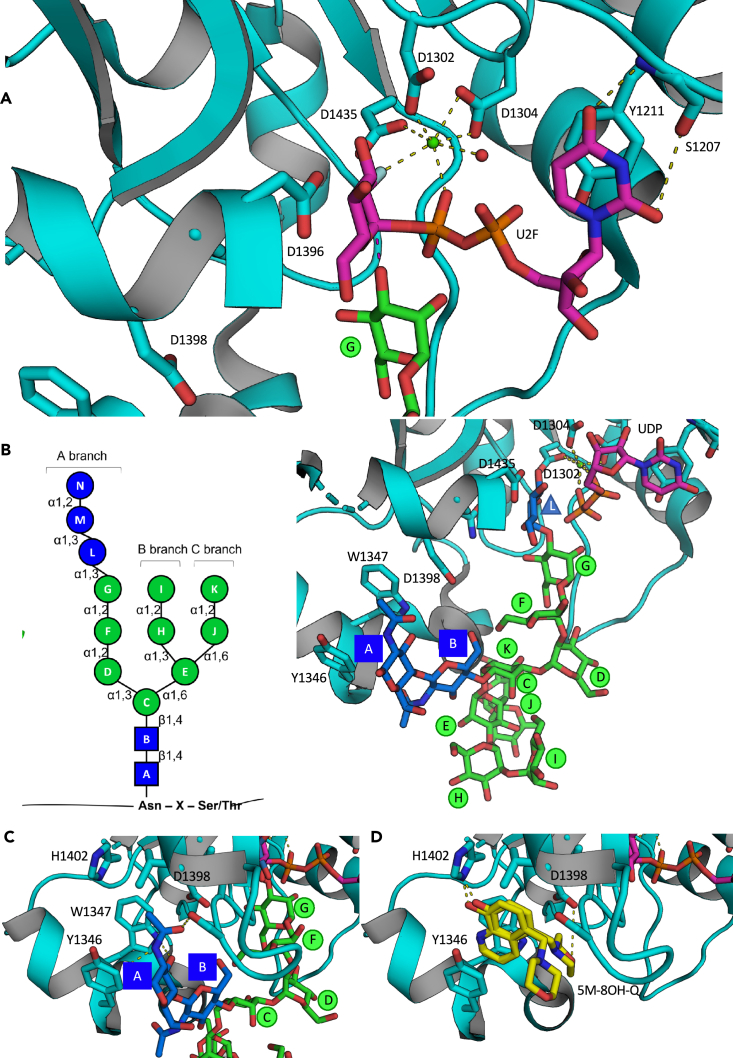
(A) Man ”G″ placement next to the UDP-Glc binding site, in an orientation suitable for the nucleophilic attack of its O3 oxygen to the glucose anomeric center (red dashed line), to yield the β(1–3) Glc-Man bond.
(B) GlcNAc2Man9Glc3 glycan nomenclature and final model of the GlcNAc2Man9Glc1 glycan docked onto the CtUGGTGT24 domain. Saccharide moieties are color-coded according to the scheme on the left hand side.40
(C and D) The docked GlcNAc2 moiety of the Man9GlcNAc2N-linked glycan and 8-OH-Q share a binding pocket.
In the CtUGGTGT24 binding site, U2F adopts a conformation equivalent to that of UDP-Glc described in Caputo et al.34 This conformation likely represents the initial stage of the cofactor binding process: the ribose ring points toward the solvent (Figures 1B and 1C and 2A). The uracyl ring O4 atom accepts a hydrogen bond from the main chain NH of S1207, and its N3 atom donates one hydrogen bond to the main chain O of the same residue (Figures 1B and 1C); the uracyl ring also forms a π-stacking interaction with the conserved CtUGGT Y1211, whose side chain rotates slightly when compared to the apo structure, to accommodate the ligand. The molecule’s pose suggests that the UGGT active site selects UDP-Glc over UDP-Gal36,37,38,39: in UDP-Glc the glucose O4′ atom forms hydrogen bonds to the side chains of conserved W1280 and D1396, but these interactions would be lost in UDP-Gal, because of the difference in stereochemistry between Glc and Gal in position 4 (Figure 1C).
UGGT binds 5M-8OH-Q via a conserved patch on the surface of its catalytic domain
To confirm the 5M-8OH-Q:CtUGGTGT24 binding pose observed in the FBLD soaked crystal,34 we grew a CtUGGTGT24:5M-8OH-Q co-crystal and obtained a 1.65 Å crystal structure (5M−8OH−QCtUGGTGT24, PDB ID 7ZLL). The structure confirms that the compound binds to a conserved patch on the surface of the CtUGGTGT24 domain, about 15 Å away from the UDP-Glc binding site (Figures 1D, S1A, and S1B). The morpholine ring is partially disordered in the crystal, but one of its ring placements is 4.2 Å from the conserved motif coordinating the Glc ring of UDP-Glc or U2F (Figures 1D and 2A); the ligand also causes a displacement of the side chain of CtUGGTGT24 1346Y.34 Through this displacement, the 8OH-quinoline ring inserts and is sandwiched between the aromatic side chains of the conserved residues —which we propose to call the “YW clamp”. The two aromatic side chains stabilize the quinoline ring forming an aromatic trimer41; the 8OH group of the quinoline also establishes a hydrogen bond to the side chain of 1402H (Figures 1D and S1B).
5M-8OH-Q and M9 glycan-binding sites overlap
To gain insight into how 5M-8OH-Q UGGT binding compares with UGGT substrate binding, we built an in silico model of the Man9GlcNAc2 glycan bound to CtUGGT using a combination of knowledge-based docking and molecular dynamics (see STAR Methods).
The surface of the UGGT catalytic domain on which the glycan docks according to our model is highly conserved across eukaryotic UGGT1s and UGGT2s.42 The A branch of the Man9GlcNAc2 glycan stretches toward the UGGT active site, while B and C branches point toward the solvent, fitting into shallower grooves, binding the protein with fewer interactions (Figure 2B). These observations are consistent with previous work showing that UGGT is able to glucosylate misfolded glycoproteins bearing GlcNAc2Man8 (Man ”I″ trimmed) and GlcNAc2Man7 (Man ”I″ and Man ”K″ trimmed) glycans (Figure 2B) albeit with lower efficiency than those bearing GlcNAc2Man9.43
Importantly, the model suggests how UGGT recognizes the first GlcNAc: the glycan’s first N-acetamide group faces directly into the hydrophobic cavity formed by residues Y1346, W1347, and L1392, its acetyl oxygen hydrogen-bonded to the L1392 backbone nitrogen, and the S1391 hydroxyl group (Figures 2C and 2D), in agreement with the published finding that the first GlcNAc is required for the Man9GlcNAc2 glycan to bind to UGGT.43,44
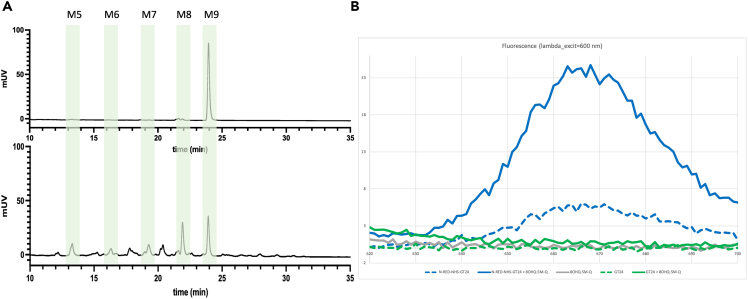
To test the hypothesis that 5M-8OH-Q and the N-linked glycan of a client glycoprotein compete for overlapping sites, we set up assays in vitro. Initially, the affinity of 5M-8OH-Q for full-length human UGGT1 (UGGT1) was measured by saturation transfer difference (STD) Nuclear Magnetic Resonance (NMR) spectroscopy, but no signal was measurable below 100 μM 5M-8OH-Q concentration, and a weak binding event with a 613 μM Kd was measured—the significance of which remains unclear (Figure S3A). For the remaining binding assays, we decided to exploit detection of fluorescence, from either of two kinds of fluorescently labeled molecules: 2-anthranylic acid-labeled N-linked glycans (2AA-glycans, Figure 3A) or N-NHS-RED-labeled CtUGGTGT24 protein (Figure 3B).
Figure 3.
Fluorescence from 2AA-labeled glycans and N-NHS-RED-labeled CtUGGTGT24
(A) HPLC elution profiles for the purification of 2AA-labeled glycans obtained from recombinantly expressed HIV gp120. Top panel, black trace: 2AA-labeled glycans purified from cells treated with 10 μM kifunensine (predominantly GlcNAc2Man9, i.e., 2AA-M9 glycan). Bottom panel: 2AA-labeled glycans purified from cells not treated with kifunensine: mostly 2AA-M9 glycan, but containing 2AA-M5, 2AA-M6, 2AA-M7 and 2AA-M8 glycans as well. We call this mixture 2AA-M5-9.
(B) Fluorescence spectra of 5M-8OH-Q, unlabelled CtUGGTGT24 and NT-RED-NHS-labeled CtUGGTGT24. λExcit = 600 nm. Solid and dashed lines refer to samples with or without 5M-8OH-Q, respectively. Gray: 5M-8OH-Q 2.5 mM; green dashed: unlabelled CtUGGTGT24 1.7 μM; green: unlabelled CtUGGTGT24 1.7 μM plus 5M-8OH-Q 2.5 mM; blue dashed: NT-RED-NHS-labeled CtUGGTGT24 1.7 μM; blue: NT-RED-NHS-labeled CtUGGTGT24 1.7 μM plus 5M-8OH-Q 2.5 mM.
Fluorescence from 2-anthranylic acid-labeled GlcNAc2Man9 glycan (2AA-M9) was used as the basis of detection only in one experiment, in which we followed its binding to the CtUGGTGT24 domain in vitro (Figure S4A) using fluorescence polarization anisotropy (FPA). The FPA-estimated dissociation constant for the binding of CtUGGTGT24 to the 2AA-M9 N-linked glycan is Kd = 117 32 μM. No measurement of the affinity of UGGT for an N-linked glycan has been published before, although a Michaelis Menten Km = 18 μM was reported for misfolded soybean agglutinin and bovine thyroglobulin in reglucosylation assays mediated by full-length rat UGGT.35
The remaining in vitro binding assays followed fluorescence from N-NHS-RED-labeled CtUGGTGT24 protein. This signal was preliminarly characterized by acquisition of fluorescence spectra in the 620–700 nm range, using λexcit = 600 nm ((Figure 3B)). Fluorescence spectra from solutions containing either 5M-8OH-Q or CtUGGTGT24 (with or without N-NHS-RED-labeling), or both, were measured. No fluorescence was detected from 5M-8OH-Q (gray fluorescence spectrum in Figure 3B), nor from unlabeled CtUGGTGT24 protein, with or without 5M-8OH-Q (green fluorescence spectra in Figure 3B). N-NHS-RED-labeled CtUGGTGT24 fluoresced at a low level (dashed blue spectrum in Figure 3B). Addition of 5M-8OH-Q to N-NHS-RED-labeled CtUGGTGT24 appeared to enhance its fluorescence 5-fold (solid blue spectrum in Figure 3B).45 Since no difference in fluorescence was observed from SDS/heat-denatured N-NHS-RED-labeled CtUGGTGT24 protein with or without 5M-8OH-Q (data not shown), it appears that the observed 5M-8OH-Q-induced enhancement of N-NHS-RED-labeled CtUGGTGT24 fluorescence depends on binding of 5M-8OH-Q to the labeled CtUGGTGT24 in its native structure/fold (ligand-enhanced fluorescence, LEF).46
Three in vitro experiments followed binding of ligands to N-NHS-RED-CtUGGTGT24, either by LEF or by microscale thermophoresis (MST). Those are as follows.
-
1.
Binding of N-NHS-RED-CtUGGTGT24 to 5M-8OH-Q was assayed by measuring LEF of a fixed amount of NHS-RED-CtUGGTGT24 along a dilution series of 5M-8OH-Q (Figure S3B). The equilibrium dissociation constant of the N-NHS-RED-CtUGGTGT24:5M-8OH-Q complex is estimated as Kd5M−8OH−Q = 47 0.7 μM.
-
2.
Binding of a mixture of 2AA-GlcNAc2Man5-9 glycans (2AA-M5-9) to N-NHS-RED-labeled CtUGGTGT24 was measured using MST (Figure S4B). The average affinity of N-NHS-RED-labeled CtUGGTGT24 for the 2AA-M5-9 N-linked glycan mixture is Kd2AA−M5-9 = 250 39 μM, weaker than the Kd2AA−M9 = 117 32 μM we measured by FPA between CtUGGTGT24 and the 2AA-M9 N-linked glycan (Figure S4). These values are consistent with the loss of protein affinity expected for N-linked glycan species with fewer than 9 mannose residues.
-
3.
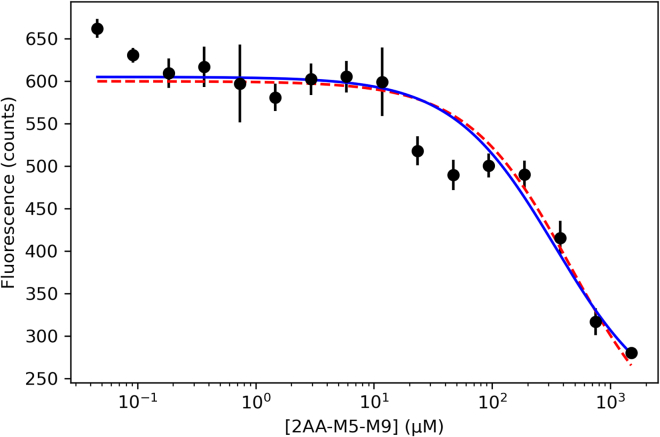
Binding of the 2AA-M5-9 mixture to CtUGGTGT24 in presence of 40 μM 5M-8OH-Q was assayed in an in vitro competition experiment. The changes of fluorescence of the 5M-8OH-Q:N-NHS-RED-labeled CtUGGTGT24 complex were followed along a 2AA-M5-9 dilution series (black data points in Figure 4). The same changes in fluorescence were then computed with a model in which two simultaneous equilibria are established, but no ternary complex can form; i.e., 5M-8OH-Q and 2AA-M5-9 N-linked glycan binding to N-NHS-RED-labeled CtUGGTGT24 are mutuallfy exclusive. The calculation used the two Kds measured in the experiments described earlier: Kd5M−8OH−Q = 47 0.7 μM and Kd2AA−M5-9 = 250 39 μM. The main qualitative trend of the 2AA-M5-9-induced displacement of 5M-8OH-Q from N-NHS-RED-labeled CtUGGTGT24 is well predicted by this model (red curve in Figure 4), suggesting that 5M-8OH-Q and the 2AA-M5-9 glycans compete for overlapping sites. A fit to the same data using a model with a single equilibrium gives an apparent dissociation constant of appKd2AA−M5-9 = 341 μM (blue dashed curve in Figure 4).
Figure 4.
5M-8OH-Q and the 2AA-M5-9 N-linked glycan mixture compete for N-NHS-RED-labeled CtUGGTGT24in vitro
Black filled circles: 2AA-M5-9 N-linked glycan dilution series from 1.5 mM to 45.8 nM, displacing 40 μM 5M-8OH-Q from 100 nM NT-RED-NHS-labeled CtUGGTGT24, as measured by LEF. λExcit = 650 nm λEmiss = 670 nm. Error bars are esds from four independent dilution series. Red dashed line: calculated fluorescence from NT-RED-NHS-labeled CtUGGTGT24 in the above conditions, using two mutually exclusive binding equilibria and the two measured Kd5M−8OH−Q = 47 μM and Kd2AA−M5-9 = 250 μM. Blue line: a fit to the data using a model with a single equilibrium gives appKd2AA−M5-9 = 341 μM.
5M-8OH-Q is a sub-millimolar inhibitor of human UGGTs in cellula
To ascertain if 5M-8OH-Q can be delivered to the ER and inhibit UGGT-mediated glucosylation in cellula, modified HEK293-6E cells were treated with the inhibitor, monoglucosylated glycoproteins isolated by affinity precipitation (with a glutathione S-transferase [GST]-calreticulin [GST-CRT] resin), and the eluate analyzed by immunoblotting.47,48 To ensure the CRT interaction resulted from UGGT glucosylation, and not from the initial glycan trimming that occurs during normal glycan maturation, CRISPR/Cas9 was used to knock out the alpha-1,3-glucosyltransferase 6 (ALG6) gene. ALG6 appends the first glucose to the Man9GlcNAc2 carbohydrate during the synthesis of the Glc3Man9GlcNAc2 N-linked glycan precursor at the ER membrane. Once the ALG-mediated synthesis of its precursor is complete, the Glc3Man9GlcNAc2 glycan is then appended to nascent glycoproteins by Oligosaccharyl Transferase (OST) and trimmed by glucosidases I and II to a monoglucosylated state, which in turn can bind to the ER lectin chaperones calnexin and calreticulin.49 Therefore, during glycan maturation in wild-type cells, CRT-affinity pull-downs would select two types of glycoproteins: either those with a glycan trimmed from Glc3 Man9GlcNAc2 to GlcMan9GlcNAc2 or those which underwent glucosylation of a Man9GlcNAc2 glycan by a UGGT.50 In our ALG6−/−cells, the CRT-affinity pull-down can only select monoglucosylated glycoproteins that were glucosylated by UGGT and not the ones produced by the ER glucosidases initial glycan trimming because in these cells the N-glycan precursors added to nascent glycoproteins initially lack the three glucoses.
In order to decide on the maximum assay concentration of 5M-8OH-Q, toxicity assays were carried out. In a trypan blue assay, toxic effects were observed around 1–2 mM 5M-8OH-Q and above in modified HEK293-6E cells: after 5 h of treatment with 1 or 2 mM 5M-8OH-Q the viability was about 75%–80% (Figure S5).
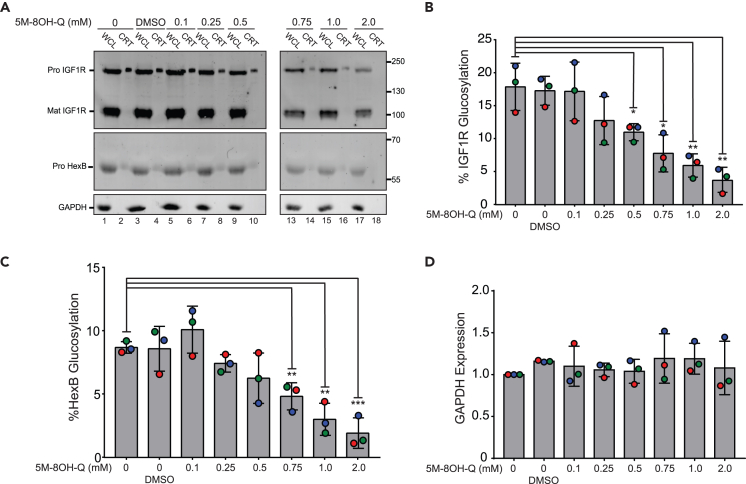
The ALG6−/− HEK293-6E cells were treated with increasing concentrations of 5M-8OH-Q, and—following incubation with the molecule—glucosylation of known UGGT substrate glycoproteins was analyzed by isolating monoglucosylated glycoproteins from the cell lysate. After GST-CRT precipitation, the eluate was probed for two known substrates of UGGT: the proprotein of human insulin like growth factor 1 receptor (IGF1R) (ProIGF1R, a UGGT1 substrate48) and the proprotein of hexosaminidase subunit beta (HexB) (ProHexB, a UGGT2 substrate48), and their glucosylation levels were quantified. The amount quantified in each GST-CRT pull-down was divided by the total amount found within the sample’s whole-cell lysate (WCL), resulting in the percent glucosylation at that dose of 5M-8OH-Q.48
Levels of monoglucosylated IGF1R and HexB in the ALG6−/− HEK293-6E cells decrease as the concentration of 5M-8OH-Q increases (Figure 5A, even-numbered lanes 2–18). In particular, a significant decrease in IGF1R and HexB glucosylation is observed at 500 and 750 μM 5M-8OH-Q, respectively. IGF1R and HexB glucosylation decreases from 17% to 4% and 9% to 2%, respectively, going from no treatment to 2 mM 5M-8OH-Q (Figures 5B–5D).
Figure 5.
5M-8OH-Q dose-dependent inhibition of UGGT in cellula
(A) ALG6−/− HEK293-6E cells were cultured and treated with increasing concentrations of 5M-8OH-Q. The “0 mM” group was treated with no drug or vehicle. The vehicle control group was incubated with DMSO. The lysate was split between a whole-cell lysate sample (20%, “WCL”) and a GST-CRT pull-down sample (60%, “CRT”), and resolved by 9% SDS-PAGE gel electrophoresis, before transferring the protein bands to a PVDF membrane. Imaged are immunoblots probed for IGF1R (whose proprotein HsProIGF1R is a UGGT1 substrate48), HexB (whose proprotein HsProHexB is a UGGT2 substrate48) and GAPDH (loading control). Each data point comes from three independent biological replicates.
(B and C) Quantification of HsProIGF1R and HsProHexB glucosylation over increasing amounts of 5M-8OH-Q from the experiments in A. Percent glucosylation was calculated by dividing the normalized CRT value by the normalized value from the WCL and multiplying by 100.
(D) Anti-GAPDH blot control. Protein samples were loaded to match the protein in the “0 mM” group for each condition. Error bars represent the standard deviation. Statistical significance levels: ∗: p 0.05; ∗∗: p 0.01; ∗∗∗: p 0.001.
Interestingly, the overall levels of IGF1R and HexB glycoproteins also seem to decrease with increasing levels of 5M-8OH-Q (WCL lanes in Figure 5A).
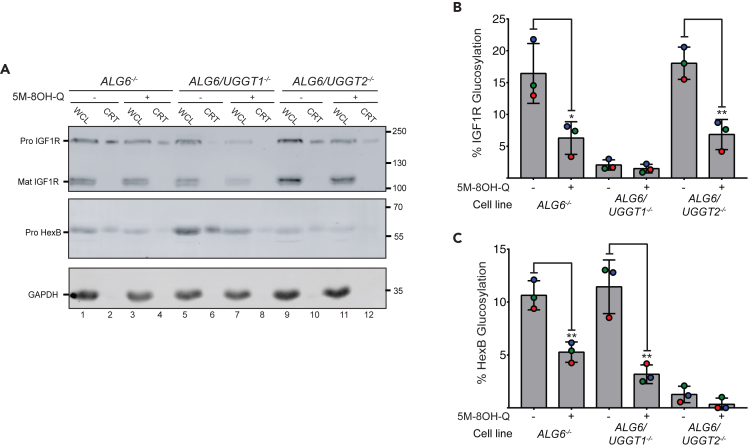
Next, we asked whether 5M-8OH-Q inhibits both human paralogs of UGGT (UGGT1 and UGGT2).48,51,52 ALG6/UGGT1−/− and ALG6/UGGT2−/− double knockout (KO) cells48 were exposed to 1 mM of the drug to measure glucosylation of IGF1R and HexB as described earlier (Figure 6A). As expected, glucosylation of IGF1R (a UGGT1 substrate48) is significantly inhibited in both the ALG6−/− and ALG6/UGGT2−/− cells, but not in the ALG6/UGGT1−/− cell line (Figure 6B). Similarly, glucosylation of the UGGT2 substrate HexB is inhibited in the ALG6−/− and ALG6/UGGT1−/− cells, but not in the ALG6/UGGT2−/− cell line (Figure 6C). The levels of inhibition within each of these UGGT KO cell lines agree well with the findings described earlier (Figure 5; Adams et al.48). In agreement to what is observed in Figure 5A, 5M-8OH-Q also decreases the levels of IGF1R and HexB in the WCL lanes (Figure 6A). Taken together these results suggest 5M-8OH-Q can reach the ER and inhibit both paralogs of UGGT.
Figure 6.
5M-8OH-Q inhibits both UGGT1 and UGGT2 in cellula
(A) ALG6−/−, ALG6/UGGT1−/− and ALG6/UGGT2−/− HEK293-6E cells were cultured and either not treated or treated with 1 mM 5M-8OH-Q to determine if the drug inhibits one or both of UGGT1 and UGGT2. After the cells were incubated with the inhibitor, they were lysed and split between a whole-cell lysate sample (20%, “WCL”) and a GST-CRT pull-down sample (60%, “CRT”), and resolved by 9% SDS-PAGE gel electrophoresis, before transferring the protein bands to a PVDF membrane. Imaged are immunoblots probed for IGF1R (UGGT1 substrate48) and HexB (UGGT2 substrate48). Glucosylation of human ProIGF1R and human ProHexB was observed in ALG6−/−, ALG6/UGGT2−/−, and ALG6−/−, ALG6/UGGT1−/− cell lines, respectively. Each data point represents three independent biological replicates. GAPDH was used as a loading control.
(B and C) Quantification of human ProIGF1R and human ProHexB glucosylation from (A) Percent glucosylation was calculated by dividing the normalized value from the CRT lane by the normalized WCL. The resulting value was multiplied by 100 to obtain percent glucosylation. Error bars represent the standard deviation. Statistical significance levels: ∗: p 0.05; ∗∗: p 0.01; ∗∗∗: p 0.001.
Discussion
Since its discovery in 1989,53 UGGT retains a central role in the standard model of glycoprotein ERQC.4 As such, and considering the importance of glycoprotein folding to health and disease,3 UGGT is a potential target for drugs to treat a variety of conditions.16,20,54 As of today, the only known UGGT inhibitors are its product, UDP,55 and some of its squaryl derivatives10; the UDP-Glc analog U2F; and synthetic analogs of its substrate (the N-linked Man9GlcNAc2 glycan).11,12 None of these molecules are good scaffolds for selective drug design, given that all eukaryotic genomes encode a plethora of proteins carrying a UDP-, a UDP-Glc-, or a glycan-binding site. Until the molecular mechanisms underpinning misfold recognition are elucidated, and the portions of UGGT involved in this process are discovered,32 the catalytic domain remains the most promising target for novel classes of compounds that inhibit UGGT-mediated glucosylation of misfolded glycoproteins in the ER.
We grew crystals of CtUGGTGT24 in order to hunt for novel ligands by FBLD and discovered 5M-8OH-Q as a CtUGGTGT24 ligand.34 The molecule was originally synthesized as a component for soluble aluminum complex dyes56 or fluorescent Zinc sensors.57 In the medical field, 8-hydroxyquinoline derivatives can be used as insecticides, antibacterial, fungicidal, neuroprotective, and anti-HIV agents.58,59 The 5M-8OH-Q Kd for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) main viral protease was estimated as 28.6 10−6 M by a recent in silico study.60
The 5M−8OH−QCtUGGTGT24 crystal structure shows that 5M-8OH-Q binds a conserved pocket on the surface of the protein, not far from the UDP-Glc binding site (Figures 1 and 2). In vitro, 5M-8OH-Q binds to CtUGGTGT24 with 47 μM Kd (Figure S3B). 5M-8OH-Q and M5-9 glycan binding appear to be mutually exclusive in an in vitro competition assay (Figure 4). These observations are consistent with the in silico model of the Man9GlcNAc2 glycan bound to the catalytic domain of CtUGGT which shows the 5M-8OH-Q binding site partially overlapping with the putative Man9GlcNAc2 glycan-binding site.
Our experiments in human cells show a concentration-dependent decrease in glucosylation of the HsProIGF1R and HsProHexB UGGT substrates upon treatment of HEK293-6E cells with 5M-8OH-Q (Figure 5), indicating that the molecule inhibits ER lumenal UGGTs. Both UGGT isoforms are inhibited (Figure 6), a result that agrees with the sequence and structure conservation of the 5M-8OH-Q binding site in the catalytic domain of the two proteins.42
Besides HsUGGT1 and HsUGGT2, the human genome encodes 10 more genes containing a GlycosylTransferase-A (GT-A) or a GlycosylTransferase-B (GT-B) domain. From sequence alignment, it appears that the YW clamp providing the 5M-8OH-Q binding platform is specific to UGGTs (GT24 family61; Figure S2). It is therefore unlikely that 5M-8OH-Q binds other GT-A and GT-B domains in human proteins in the same way it binds UGGTs.
Rather, 8OH-quinolines can chelate a great number of cations, including Cu2+, Bi2+, Mn2+, Mg2+, Fe3+, Al3+, Zn2+, and Ni3+,62 and are known to bind to a dozen mammalian metalloproteins (see Table S3), including human demethylases, 2-oxoglutarate/iron-dependent oxygenases, and α-ketoglutarate-dependent RNA demethylases.63,64,65 Metalloproteins66,67,68,69,70,71,72 are therefore more likely candidates for any 5M-8OH-Q off-target effects.
In summary, 5M-8OH-Q provides a useful starting point for the synthesis of UGGT modulators for the treatment of diseases caused by “responsive mutants”, as persistent UGGT-mediated glucosylation may prevent trafficking of slightly misfolded, but otherwise functional, glycoproteins to their correct cellular locations.16 UGGT inhibition may one day also find application as an anti-cancer strategy, as some UGGT substrate glycoproteins48 are selectively up-regulated in cancer cells.20 Replication of pathogenic enveloped viruses whose envelope glycoproteins fold under UGGT control may be impaired by UGGT inhibitors.54 Modulation of UGGT activity would also affect adaptive immune responses triggered by antigenic peptides.5 The strong conservation of UGGT sequence/function across eukaryotes3 broadens the potential impact of such molecules to many fields: examples are plants as in vivo models to study secretion73,74,75; stress-resistant genetically modified crops22; or expression systems for recombinant glycoproteins.76
Limitations of the study
The low potency of 5M-8OH-Q in cells could be either related to low efficiency in crossing the plasma and ER membranes, or to low-specificity/off-target binding. The latter would be hardly surprising, given that the molecule was discovered as a UGGT binding fragment during an FBLD effort34 and it has not been chemically modified to improve its potency and selectivity yet. As it is, 5M-8OH-Q is toxic in cellula at concentrations higher than 1 mM (Figure S5) and a dose-dependent reduction of the levels of the two UGGT substrates assayed (HsProIGF1R and HsProHexB) was observed in ALG6−/− HEK293-6E cells (“WCL” lanes in Figures 5 and 6). At present, it is unclear if these side effects are due to 5M-8OH-Q directly interacting with other cellular targets, or to indirect effects of UGGT inhibition on UGGT glycoprotein clients’ folding and levels: 5M-8OH-Q treatment, as well as inhibiting UGGT-mediated reglucosylation of HsProIGF1R and HsProHexB, may cause a decrease in their levels because both client glycoproteins fold under UGGT control.
A medicinal chemistry program that will yield the next generation of 5M-8OH-Q derivatives of improved potency and selectivity is in progress. In silico screening, chemical synthesis, and in vitro assays will be used to modify the M6-8OH-Q molecule. Chemical modifications are being introduced to the quinoline scaffold, the 5-morpholino-residue, or the 8-hydroxy group. Together with derivatives incorporating polar/non-polar residues on the remaining positions of the scaffold, these daughter molecules will generate structure-activity-relationship data toward drug-like compounds with improved UGGT inhibitory potency and selectivity.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-IGF1R | Cell Signaling | Cat#9750;RRID:AB_10950969 |
| Anti-HexB | Abcam | Cat#ab140649;RRID:AB_3065101 |
| Anti-GAPDH | Millipore Sigma | Cat#MAB374;RRID:AB_2107445 |
| Bacterial and virus strains | ||
| E.coli DH5-α | New England Bioscience | Cat# C2987I |
| Chemicals, peptides, and recombinant proteins | ||
| AgeI-HF | New England Biolabs | Cat# R3552S |
| KpnI-HF | New England Biolabs | Cat# R3142S |
| CutSmart Buffer | New England Biolabs | Cat# B7204 actually replaced by Cat# B6004S |
| QIAquick gel extraction kit | QIAGEN | Cat# 28706 |
| In-Fusion Cloning | TakaraBio Ltd | Cat# 638947 |
| Kifunensine | Cayman Chemical | Cat# 109944-15-2 |
| Anthranilic Acid | Sigma-Aldrich | Cat# A89855 |
| MORPHEUS Crystallisation Screen | Molecular Dimensions | Cat# MD1–47 |
| HEPES | Sigma-Aldrich | Cat# H3375 |
| Imidazole | Honeywell Fluka | Cat# 56750 |
| Deposited data | ||
| Python code | This paper | https://doi.org/10.5281/zenodo.8305097 |
| CtUGGTGT24 | This paper | PDB ID 7ZKC |
| U2FCtUGGTGT24 | This paper | PDB ID 7ZLU |
| 5M−8OH−QCtUGGTGT24 | This paper | PDB ID 7ZLL |
| Experimental models: Cell lines | ||
| HEK FreeStyleTM 293F cells | ThermoFisher Scientific | Cat# R79007 |
| HEK293-EBNA1-6E ALG6-/- | Adams et al.48 | |
| HEK293-EBNA1-6E ALG6/UGGT1-/- | Adams et al.48 | |
| HEK293-EBNA1-6E ALG6/UGGT2-/- | Adams et al.48 | |
| HEK293-EBNA1-6E ALG6/UGGT1/2-/- | Adams et al.48 | |
| Oligonucleotides | ||
| OPPF UGGT1 Fwd gcgtagctgaaaccggc GACTCAAAAGCCATTACAACCTCTCT |
Eurofins Scientific | NA |
| OPPF UGGT1 Rev gtgatggtgatgttt TTTCTGAGGACCTTCTCGGCTTGG |
Eurofins Scientific | NA |
| Recombinant DNA | ||
| UGGT1-pUC57 | Genscript | NA |
| pOPINTTGneo:hUGGT1 plasmid | This paper | NA |
| Software and algorithms | ||
| autoPROC | Vonrhein et al.77 | Version 1.0.5 |
| Coot | Emsley et al.78 | Version 0.9 |
| BUSTER | Blanc et al.79 | Version 2.10.3 |
| GLYCAM-web | Singh et al.,80 | Version 1.0 |
| AutoDock-Bias | Arcon et al.,81 | Version 1.0 |
| AutoDock4 | Morris et al.,82 | Version 4.0 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Pietro Roversi (pietro.roversi@cnr.it).
Materials availability
The pOPINTTGneo:hUGGT1 plasmid generated in this study is available for distribution. This study did not generate any other unique reagents. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Pietro Roversi (pietro.roversi@cnr.it).
Experimental model and study participant details
E. coli strains for protein production
DH5α chemically competent E. coli was used to make the pHLsec:CtUGGT, pHLsec:CtUGGTGT24 and pOPINTTGneo:hUGGT1 plasmids.
Method details
UGGT1 cloning, protein expression and purification
The C-terminally His-tagged construct encoding human UGGT1 residues 43-1551 was PCR-amplified from the commercially sourced vector UGGT1-pUC57 (GenScript) with primers: OPPF_UGGT1_Fwd: gcgtagctgaaaccggcGACTCAAAAGCCATTACAACCTCTCT OPPF_UGGT1_Rev: gtgatggtgatgtttTTTCTGAGGACCTTCTCGGCTTGG. These primers were designed to surround the insert with an N-terminal AgeI restriction site and a C-terminal KpnI site (after the C-terminal 6xHis tag and the stop codon). The amplified DNA was run on a 0.8% agarose gel and the correctly-sized fragment excised and purified using the QIAquick Gel Extraction Kit (QIAgen). The pOPINTTGneo plasmid was linearised with 20 units of both AgeI-HF and KpnI-HF restriction enzymes, incubated with 1x CutSmart Buffer (New England BioLabs) and 500ng of pHLSec DNA and digested at 37°C overnight. Both the linearised pOPINTTGneo and the UGGT1 insert DNA were run on a 0.8% agarose gel and the correctly-sized fragments excised and purified using the QIAquick Gel Extraction Kit (QIAgen). DNA ligation of the linearised pOPINTTGneo vector and the human UGGT1 insert was achieved by In-fusion™ligation-independent cloning (Takara Ltd.)
Transfection of HEK293F cells with the pOPINTTGneo-hUGGT1 plasmid and expression of the recombinant human UGGT1 protein were carried out a protocol equivalent to the one described for expression of CtUGGT,42 using the FreeStyle 293 Expression System (Thermo Fisher Scientific) and following the manufacturer’s protocol.
Immobilised Metal Affinity Chromatography (IMAC): after 5 days, the cells’ supernatant was applied onto a Ni-affinity column equilibrated with PBS binding buffer. The protein was eluted with a 20 Column Volumes linear gradient elution at a flow rate of 1 ml/min increasing from 0% to 100% elution buffer (PBS plus 500 mM Imidazole).
Size Exclusion Chromatography (SEC): the IMAC step eluate was pooled and concentrated to 0.5 mL using a 100kDa spin concentrator. The sample was then loaded on a 0.5 mL loop and applied to a 10/300 Sephadex 200 column running at 1 mL/min. The SEC buffer was 20 mM MES pH 6.5, 50 mM NaCl, 1 mM , 1 mM UDP. The latter buffer was arrived at by Differential Scanning Fluorimetry (DSF): the stability of UGGT1 is greatly increased through the addition of , with an increase in melting temperature Tm of 3.0°C and addition of UDP, with an increase in Tm of 1.1 °C. The DSF experiment also showed a clear preference for lower salt concentrations and a slightly more acidic pH.
CtUGGTGT24 cloning, protein expression and purification
CtUGGTGT24 was cloned, expressed and purified as described in.34
Crystal growth
Crystals were grown at 18°C in sitting drops by the vapour diffusion method, set up with a Mosquito liquid handling robot (TTP Labtech). Crystallisation drops had an initial volume of 200 nL. The volume ratio of protein to precipitant was either 1:1 or 2:1.
CtUGGTGT24 crystallisation
A crystal of CtUGGTGT24 grew in one week in a 1:1 mixture of CtUGGTGT24 at 6 mg/mL and Morpheus screen condition 1-1 composed of 0.06 M Divalents, 0.1 M Buffer System 1 pH 6.5, 30% v/v Precipitant Mix 1.83,84
CtUGGTGT24:U2F co-crystallisation
U2F was synthesised as described in.85 A crystal of CtUGGTGT24:U2F grew in one week in a 1:1 mixture of CtUGGTGT24 at 12 mg/mL, 2 mM CaCl2, 1.25 mM U2F and Morpheus screen condition 2-17 composed of 0.12 M Monosaccharides, 0.1 M Buffer System 2 pH 7.5, 30% v/v Precipitant Mix 1.83,84
FBLD of UGGT ligands
Details of the study are available in.34
5M-8OH-QCtUGGTGT24 co-crystallisation
A crystal of 5M-8OH-QCtUGGTGT24 grew in one week in a 1:1 mixture of CtUGGTGT24 at 6.5 mg/mL, 10 mM 5M-8OH-Q in DMSO and Morpheus screen condition 1-1 composed of 0.06M Divalents, 0.1 M Buffer System 1 pH 6.5, 30% v/v Precipitant Mix 1.83,84
X-ray data collection, processing, and model refinement
X-ray data collection beamlines and data collection parameters are listed in Table S1. Data processing was carried out in autoPROC.77 The model refinement and ligand fitting were carried out with BUSTER79,86 and Coot.78,87 Refinement statistics are listed in Table S2.
In silico modeling of the CtUGGTGT24:Man9GlcNAc2 complex
Due to the limitations of conventional docking methods in dealing with oligosaccharides larger than five units,88 we used a hierarchical approach that combined biased docking and Molecular Dynamics (MD) in order to build a model of the Man9GlcNAc2 glycan (M9) bound to CtUGGTGT24.
As a rule, carbohydrate ligands bind to proteins in a conformation close to one of the gas-phase energy minima. The latter mainly depend on the values of the dihedral angles of each glycosidic bond.89 Although each of these can only assume a few possible conformations, the M9 glycan has 70 torsional degrees of freedom overall (including OH and CH3 groups, glycosidic linkages, etc.). This number is such that docking algorithms cannot handle full torsional optimisation.90
We therefore generated nine initial Man9GlcNAc2 conformations using the GLYCAM-web server at https://glycam.org/lib/load/hmlib/.80 Each of these structures was then optimized using MD in explicit solvent,91 thus broadening the M9 conformational space spanned by the structures.
The results were clustered using only the poses of furanose rings with a 1.4 A of tolerance92 and 250 representative Man9GlcNAc2 conformations were selected and underwent the analysis described here below:
-
1.
we first aligned the acceptor Man residue of the Man9GlcNAc2 N-linked glycan (i.e. the terminal Man residue of its A-branch, Man ”G” (Figure 2B) such that its C1 atom pointed towards the O3 atom of the UDP-Glc molecule in our U2FCtUGGTGT24 structure (see also the structure of TdUGGTGT24 in complex with UDP-Glc, PDB ID 5H18,31). This assumes that this Man ”G” residue docks in the active site such that upon Glc transfer, a β(1-3) linkage will form;
-
2.
then, using that Man ”G” residue orientation as a constraint, we performed multiple docking simulations of the Man9GlcNAc2 ligand, using the AutoDock-Bias protocol (81, modified as described in88), and keeping all torsional degrees of freedom fixed;
-
3.
the results were clustered and the three best ranking poses selected for further refinement using MD simulations. Starting from each complex, Molecular Dynamics was used to relax the Man9GlcNAc2 structure onto the CtUGGTGT24 domain, using the protocol described in93;
-
4.
since the final pose for each of the three best MD refinements was almost identical (RMSD 2 Å), we performed a final single-point energy calculation with AutoDock482 to select the best complex.
Estimation of 5M-8OH-Q: human UGGT1 by STD NMR in vitro
For each 5M-8OH-Q concentration, a 1 μM solution of human UGGT1 was incubated with 5M-8OH-Q in PBS prepared in D2 O. Briefly, 100 μL of a human UGGT1 stock solution at 2 μM and 100 μL of the dilution series of molecule at twice the desired final concentration were mixed and left to equilibrate for at least one hour. High and low concentrations were measured alternately to remove any time effects. As a further control the first sample was remeasured after the last one to confirm that the STD had not changed. The signal/noise was not high enough at 5M-8OH-Q concentrations below 100 μM. No STD was observed with the maximum tested dose (2 mM 5M-8OH-Q) in absence of human UGGT1.
Measurements of N-NHS-RED-CtUGGTGT24 by LEF in vitro
Fluorescence spectra were measured in a quartz cuvette on a Cary Eclipse fluorescence spectrophotometer. λexcit=600 nm, λemiss=620-700(5) nm. 5M-8OH-Q fluorescence: 1 μL of 5M-8OH-Q 250 mM in DMSO was added to 99 μL of a buffer 100 mM NaCl and 20 mM HEPES pH 7.4. CtUGGTGT24 fluorescence: 27.6 μL of a 6.15 μM solution of CtUGGTGT24 were diluted to 1.7 μM with the addition of 71.4 μL of the same buffer. After the spectrum was measured, 1 μL of 5M-8OH-Q 250 mM in DMSO was added and the spectrum measured again. N-NHS-RED-CtUGGTGT24 fluorescence: a spectrum was first measured from 99 μL of a 1.7 μM solution of N-NHS-RED-CtUGGTGT24; a second spectrum was measured after addition of 1 μL of 5M-8OH-Q 250 mM in DMSO.
Purification of the 2AA-M9 and 2AA-M5-9 N-glycans
N-glycans were cleaved from HIV gp120 protein expressed in HEK293F cells in the presence of 5 μM kifunensine,94 labelled with 2-anthranylic acid (2AA) and purified by HPLC following the protocol in.42 A 2AA calibration curve was obtained by measuring 2AA fluorescence on a BMG Labtech ClarioSTAR spec, with λexcit=320(15) nm, λemiss=420(20) nm, for a dilution curve of 2AA in a Greiner 384 wells plate between 730 μM to 273 nM. Using this calibration curve, the concentration of the purified 2AA-Man9GlcNAc2 glycan was estimated as 2 mM and the one of the 2AA-Man5-9GlcNAc2 glycan was estimated as 3 mM.
Estimation of 2AA-Man5-9:CtUGGTGT24 Kd by MST and LEF in vitro
Measurements were carried out in quartz capillaries on a NanoTemper Monolith X. Initial fluorescence and thermophoresis were measured with λExcit=650 nm, λEmiss=670 nm. Each of three independent 16-point dilution series of 2AA-Man5-9GlcNAc2 glycan from 1.5 mM to 45.8 nM was mixed with NT-RED-NHS-labelled CtUGGTGT24 100 nM and a buffer containing NaCl 100 mM, HEPES 20 mM pH 7.4 and 0.05% Tween.
The 2AA-M5-9 glycan : CtUGGTGT24 binding was characterised by microscale thermophoresis (MST). The data were fitted with one equilibrium model using the instrument’s data analysis software.
The same measurements were repeated with samples made 40 μM 5M-8OH-Q and the binding characterised by LEF (the enhanced NT-RED-NHS-labelled CtUGGTGT24 fluorescence once 5M-8OH-Q binds to the labelled protein precludes the use of MST to follow glycan binding in presence of 5M-8OH-Q). The data were analysed by custom-written Python code. A single equilibrium model was used to obtain an apparent dissociation constant, by solving a system of 4x16=64 equations in 3x16+3=51 unknowns. For the ith data point in the 16-points 2AA-M5-9 dilution series, the four equations read:
| (Equation 1) |
where P=NT-RED-NHS-labelled CtUGGTGT24 and L=2AA-M5-9.
The 51 variables are the 16x3 values of and for i=1 to 16, plus a, b and appKd2AA-M5-9. The solution gave appKd2AA-M5-9=341 μM ; a= 605.1 counts; b= 205.6 counts. These values were used in the last equation of the system (1) to compute the fluorescence in the desired interval of [2AA-M5-9]tot (blue curve in Figure 4).
The calculated fluorescence curve expected by the two simultaneous and competing equilibria was computed by first solving one system of 5 equations in 5 unknowns for each i-th data point in the dilution series, i=1 to 16:
| (Equation 2) |
Once the values of [P]i, [5M-8OH-Q]i, [P:5M-8OH-Q]i, [2AA-M5-9]i and [P:2AA-M5-9]i were obtained for each of the 16 values of [2AA-M5-9]tot, i, a least-squares fit was carried out to obtain the coefficients A and B from a fit to the experimental data, using the 16 equations in the dilution series, i=1,16:
| (Equation 3) |
The solution gave A = 1,621 counts/μM and B = 11,105 counts/μM. Using these values, the calculated fluorescence curve was computed using Equation 3 for the values of [2AA-M5-9]tot in the desired interval (red dashed line in Figure 4).
Estimation of 5M-8OH-Q:CtUGGTGT24 Kd by LEF in vitro
Measurements were carried out in quartz capillaries on a NanoTemper Monolith X. Fluorescence was measured with λExcit=650 nm at λEmiss=670 nm. Each of three independent 16-point dilution series of 5M-8OH-Q from 2.5 mM to 76.3 nM was mixed with NT-RED-NHS-labelled CtUGGTGT24 100 nM and a buffer containing NaCl 100 mM, HEPES 20 mM pH 7.4 and 0.05% Tween. The data were fitted by solving the following system of 4 equations:
| (Equation 4) |
in the four unknowns [P], [L], [PL] and Kd, depending on the two parameters a (the maximum observed fluorescence, when all the labelled protein is saturated with inhibitor) and b (the minimum observed fluorescence, when all the labelled protein is free).
The first three equations give the fraction of free protein fP as a function of the total concentrations of ligand and protein:
| (Equation 5) |
and the fourth equation of the system (4) is re-written as:
| (Equation 6) |
The fit to the data was effected by least-squares estimation of the a and b parameters.
Estimation of 2AA-M9:CtUGGTGT24 by FPA in vitro
Four dilutions series of CtUGGTGT24 to in 120 mM NaCl, 20 mM HEPES pH 7.2 (from 247 to 2.47 μM) in a Greiner 384 wells plate were mixed with 2.5 μL of a 2μM solution of 2AA-Man9GlcNAc2 glycan in water, and protein buffer added to a total volume of 25 μL. The final concentration of 2AA-Man9GlcNAc2 glycan was 200 nM.
The anisotropy of the 2AA-fluorescence polarisation was measured on a BMG Labtech ClarioSTAR spectrophotometer, with λexcit=360(15) nm, λemiss=490(20) nm, and the dichroic mirror set to 410 nm. Both instrument gain coefficients were set to 1,000. The curve was fitted with a single equilibrium constant, and a parameter for minimum value of the anisotropy (the maximum value of the anisotropy was set to 110 mA and kept fixed).
In cellula UGGT-mediated glucosylation assays
The in cellula UGGT-mediated glucosylation assays were carried out47,48 in presence of increasing amounts of 5M-8OH-Q.
Briefly, HEK293-6E cells were plated and grown for 24 hr before replacing with fresh media containing the drug, from a stock solution of 250 mM in 100% DMSO, diluted to the desired/tested concentration (no more than 1% final DMSO in the media).
After a 5 hr incubation time, the media was collected and the adhered cells were removed from the plate with lysis buffer. The media fraction was gently spun down (250 g for 5 min) to collect the dissociated cells and combined with the cells scraped off the plate. The combined samples were then shaken for 10 min at 4°C before being spun at 14,000 g for 10 min at 4°C prior to analyzing the soluble fraction.47,48
Fifty μL bed volume of glutathione beads was added to each sample and incubated for 1 hr at 4°C under gentle rotation to remove non-specific protein binding to the resin. The samples were then spun at 1,000 g for 5 min at 4°C to pellet the beads and the supernatant was collected. 20% of the supernatant was used for WCL and 60% was added to the GST-CRT conjugated glutathione beads,47,48 and incubated for 16 hr at 4°C under gentle rotation. The beads were collected by centrifuging at 1,000 g for 5 min at 4°C. The supernatant was aspirated and beads were washed twice with lysis buffer without protease inhibitors.
Beads were treated with reducing sample buffer (30 mM Tris-HCl pH 6.8, 9% SDS, 15% glycerol, 0.05% bromophenol blue). WCLs were trichloroacetic acid (TCA) precipitated by adding TCA to cell lysate to a final concentration of 10%. Cell lysate was then briefly rotated and allowed to incubate on ice for 15 min before centrifugation at 17,000 g for 10 min at 4°C. Supernatants were aspirated and washed twice with cold acetone and centrifuged at 17,000 g for 10 min at 4°C. Supernatants were aspirated and the remaining precipitant was allowed to dry for 5 min at room temperature and briefly at 65°C. Precipitated protein was resuspended in sample buffer. Samples were resolved on a 9% reducing SDS-PAGE and imaged by immunoblotting.
Viability assay for treated HEK293-6E cells
The viability of cells after drug treatment was determined using a LUNA II™ Automated Cell Counter. Briefly, untreated and treated cells were incubated with the drug 5 hr. After incubation cells were collected and washed twice with PBS and resuspended in 1 mL of culture media. Cells were mixed with trypan blue (50:50 mix) and viability was measured.
Quantification and statistical analysis
The percentage glucosylation was calculated by dividing the normalized amount of protein detected in the GST-calreticulin lane by the normalized total amount of protein in the WCL. This value was then divided by the amount of protein found in the WCL multiplied by 5 to account for the dilution factor and then multiplied by 100. The resulting value yielded the percent reglucosylation in each cell type.
The band intensities were determined using ImageJ v1.53i for pixel quantification. All statistics, biological replicates, and significance information are reported in the figure legends. Prism v8 was used for all quantifications and the error bars were calculated using the standard deviation of three independent biological replicates. Statistical significance was determined by using an unpaired t test with a confidence interval of 95%.
Acknowledgments
We thank the members of N.Z.’s laboratory for assistance in the lab. Edward Lowe, Patrick Collins, Alice Douangamath, Jose Brandao-Neto, and the staff at beamline I04-1, at the Diamond Light Source, Harwell, England, UK, assisted with crystal growing, soaking, fishing, and X-ray data collection. Jo Nettleship at the Oxford Protein Production Facility assisted in the cloning of UGGT1. Christina Redfield assisted with the NMR measurements. The work was funded by the Glycobiology Endowment and by a University of Oxford Confidence in Concept Scheme, grant reference MRC–MC_PC_16056 (to N.Z.). P.R. was the recipient of an LISCB Wellcome Trust ISSF award, grant ref. 204801/Z/16/Z and a Wellcome Trust Seed Award in Science, grant ref. 214090/Z/18/Z. J.C.H. and A.T.C. were funded by Wellcome Trust 4-Year Studentships 106272/Z/14/Z and 097300/Z/11/Z, respectively. This work was also supported by the US Foundation for the National Institutes of Health (GM086874 to D.N.H.) and a Chemistry-Biology Interface program training grant (T32 GM139789 to K.P.G). Michela Bollati kindly gave us access to her TECAN Infinite 200 COMPlex fluorimeter. Vincenzo Pisapia pointed us to a few references describing MST experiments. R.I. was the recipient of Sardinian Regional Government Erasmus and PhD scholarships. F.B. was funded by Project IR00011– EBRAINS-Italy - - PNRR Investment thread 3.1, Action 3.1.1 - Area ESFRI H&F, financed by the European Union – NextGeneration EU (CUP B51E22000150006). N.Z. is a Fellow of Merton College, Oxford.
Author contributions
P.R., N.Z., and D.N.H. conceived and funded the study. P.R., J.C.H., S.V., and R.I. cloned, expressed, and purified UGGT1. K.H. and S.G.W. synthesized U2F. P.R., J.D.L.C., R.I., A.L., A.T.C., M.H., and S.V. expressed and purified CtUGGTGT24. P.R., J.D.L.C., R.I., M.H., A.V.C., A.T.C., and A.L.K. determined and refined the crystal structures. Y.B. contributed to structure refinement. J.L.K. carried out the NMR in vitro binding assays. P.R., K.P.G., D.N.H., I.Z., M.D.B., A.L., A.S., J.D.L.C., and E.B. carried out inhibitor assays. J.I.B.C., C.P.M., and Ma.Ma. carried out the in silico docking. S.V. and J.B. purified the glycan samples. P.R., J.B., A.V.C., and S.V. carried out the fluorescence polarization anisotropy binding assays. P.R., F.B, Ma. Mi., and M.d.R. carried out the fluorescence and microscale thermophoresis binding assays and analyzed the binding data. All authors contributed to the writing of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: September 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107919.
Contributor Information
Carlos P. Modenutti, Email: cpmode@gmail.com.
Daniel N. Hebert, Email: dhebert@biochem.umass.edu.
Nicole Zitzmann, Email: nicole.zitzmann@bioch.ox.ac.uk.
Pietro Roversi, Email: pietro.roversi@cnr.it.
Supplemental information
Data and code availability
-
•
Crystal structure coordinates and structure factor files (mmCIF format) were deposited and are publicly accessible in the protein databank (PDB) as PDB IDs 7ZKC (CtUGGTGT24), 7ZLU (U2FCtUGGTGT24) and 7ZLL (5M-8OH-QCtUGGTGT24). Accession numbers are also listed in the key resources table.
-
•
All original code has been deposited at Zenodo (https://doi.org/10.5281/zenodo.8305097) and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Vincenz-Donnelly L., Hipp M.S. The endoplasmic reticulum: A hub of protein quality control in health and disease. Free Radic. Biol. Med. 2017;108:383–393. doi: 10.1016/j.freeradbiomed.2017.03.031. [DOI] [PubMed] [Google Scholar]
- 2.Hebert D.N., Molinari M. In and out of the ER: Protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 3.Hebert D.N., Lamriben L., Powers E.T., Kelly J.W. The intrinsic and extrinsic effects of N-linked glycans on glycoproteostasis. Nat. Chem. Biol. 2014;10:902–910. doi: 10.1038/nchembio.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammond C., Braakman I., Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc. Natl. Acad. Sci. USA. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sagert L., Winter C., Ruppert I., Zehetmaier M., Thomas C., Tampé R. The er folding sensor uggt1 acts on tapbpr-chaperoned peptide-free mhc i. Elife. 2023;12 doi: 10.7554/eLife.85432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caputo A.T., Alonzi D.S., Marti L., Reca I.-B., Kiappes J.L., Struwe W.B., Cross A., Basu S., Lowe E.D., Darlot B., et al. Structures of mammalian ER α-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals. Proc. Natl. Acad. Sci. USA. 2016;113:E4630–E4638. doi: 10.1073/pnas.1604463113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warfield K.L., Plummer E.M., Sayce A.C., Alonzi D.S., Tang W., Tyrrell B.E., Hill M.L., Caputo A.T., Killingbeck S.S., Beatty P.R., et al. Antiviral research; 2016. Inhibition of Endoplasmic Reticulum Glucosidases Is Required for in Vitro and in Vivo Dengue Antiviral Activity by the Iminosugar UV-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y., Huang X., Zheng Z., Yang X., Ba Y., Lian J. Role and mechanism of chaperones calreticulin and ERP57 in restoring trafficking to mutant HERG-A561V protein. Int. J. Mol. Med. 2021;48 doi: 10.3892/ijmm.2021.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trombetta E.S., Helenius A. Glycoprotein reglucosylation and nucleotide sugar utilization in the secretory pathway: identification of a nucleoside diphosphatase in the endoplasmic reticulum. EMBO J. 1999;18:3282–3292. doi: 10.1093/emboj/18.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abe J., Takeda Y., Kikuma T., Kizuka Y., Kajiura H., Kajihara Y., Ito Y. Squaryl group-modified udp analogs as inhibitors of the endoplasmic reticulum-resident folding sensor enzyme uggt. Chem. Commun. 2023;59:2803–2806. doi: 10.1039/D2CC06634C. [DOI] [PubMed] [Google Scholar]
- 11.Totani K., Ihara Y., Tsujimoto T., Matsuo I., Ito Y. The recognition motif of the glycoprotein-folding sensor enzyme udp-glc:glycoprotein glucosyltransferase. Biochemistry. 2009;48:2933–2940. doi: 10.1021/bi8020586. [DOI] [PubMed] [Google Scholar]
- 12.Kudo T., Hirano M., Ishihara T., Shimura S., Totani K. Glycopeptide probes for understanding peptide specificity of the folding sensor enzyme uggt. Bioorg. Med. Chem. Lett. 2014;24:5563–5567. doi: 10.1016/j.bmcl.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 13.Saeed M., Suzuki R., Watanabe N., Masaki T., Tomonaga M., Muhammad A., Kato T., Matsuura Y., Watanabe H., Wakita T., Suzuki T. Role of the endoplasmic reticulum-associated degradation (ERAD) pathway in degradation of hepatitis c virus envelope proteins and production of virus particles. J. Biol. Chem. 2011;286:37264–37273. doi: 10.1074/jbc.m111.259085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J., Yin H., Yin P., Jian X., Song S., Luan J., Zhang L. SR-BI Interactome Analysis Reveals a Proviral Role for UGGT1 in Hepatitis C Virus Entry. Front. Microbiol. 2019;10:2043. doi: 10.3389/fmicb.2019.02043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruan J., Rothan H.A., Zhong Y., Yan W., Henderson M.J., Chen F., Fang S. A small molecule inhibitor of ER-to-cytosol protein dislocation exhibits anti-dengue and anti-zika virus activity. Sci. Rep. 2019;9:10901. doi: 10.1038/s41598-019-47532-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amara J.F., Cheng S.H., Smith A.E. Intracellular protein trafficking defects in human disease. Trends Cell Biol. 1992;2:145–149. doi: 10.1016/0962-8924(92)90101-r. [DOI] [PubMed] [Google Scholar]
- 17.Parodi A.J., Caramelo J.J., D’Alessio C. Handbook of Glycosyltransferases and Related Genes. Springer Japan; 2014. UDP-Glucose: Glycoprotein Glucosyltransferase 1,2 (UGGT1,2) pp. 15–30. [Google Scholar]
- 18.Kuribara T., Imagawa A., Hirano M., Ito Y., Totani K. Metabolic syndrome perturbs deglucosylation and reglucosylation in the glycoprotein folding cycle. FEBS Lett. 2020;594:1759–1769. doi: 10.1002/1873-3468.13780. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Ye Y. Proteostasis regulation at the endoplasmic reticulum: a new perturbation site for targeted cancer therapy. Cell Res. 2011;21:867–883. doi: 10.1038/cr.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tax G., Lia A., Santino A., Roversi P. Modulation of ERQC and ERAD: A Broad-Spectrum Spanner in the Works of Cancer Cells? JAMA Oncol. 2019;2019 doi: 10.1155/2019/8384913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahon M., Samali A., Chevet E. Springer International Publishing; 2021. Maintenance of Endoplasmic Reticulum Protein Homeostasis in Cancer: Friend or Foe; pp. 197–214. ISBN 978-3-030-67696-42021. [DOI] [PubMed] [Google Scholar]
- 22.Liu J.-X., Srivastava R., Che P., Howell S.H. Salt stress responses in arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007;51:897–909. doi: 10.1111/j.1365-313x.2007.03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valente M.A.S., Faria J.A.Q.A., Soares-Ramos J.R.L., Reis P.A.B., Pinheiro G.L., Piovesan N.D., Morais A.T., Menezes C.C., Cano M.A.O., Fietto L.G., et al. The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. J. Exp. Bot. 2009;60:533–546. doi: 10.1093/jxb/ern296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia X.-Y., Xu C.-Y., Jing R.-L., Li R.-Z., Mao X.-G., Wang J.-P., Chang X.-P. Molecular cloning and characterization of wheat calreticulin (CRT) gene involved in drought-stressed responses. J. Exp. Bot. 2008;59:739–751. doi: 10.1093/jxb/erm369. [DOI] [PubMed] [Google Scholar]
- 25.Sekiya M., Maruko-Otake A., Hearn S., Sakakibara Y., Fujisaki N., Suzuki E., Ando K., Iijima K.M. EDEM function in ERAD protects against chronic ER proteinopathy and age-related physiological decline in drosophila. Dev. Cell. 2017;41:652–664.e5. doi: 10.1016/j.devcel.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciulli A., Williams G., Smith A.G., Blundell T.L., Abell C. Probing hot spots at protein-ligand binding sites: a fragment-based approach using biophysical methods. J. Med. Chem. 2006;49:4992–5000. doi: 10.1021/jm060490r. [DOI] [PubMed] [Google Scholar]
- 27.Murray C.W., Blundell T.L. Structural biology in fragment-based drug design. Curr. Opin. Struct. Biol. 2010;20:497–507. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Chen X., Qin S., Chen S., Li J., Li L., Wang Z., Wang Q., Lin J., Yang C., Shui W. A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline. Sci. Rep. 2015;5:8361. doi: 10.1038/srep08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radoux C.J., Olsson T.S.G., Pitt W.R., Groom C.R., Blundell T.L. Identifying Interactions that Determine Fragment Binding at Protein Hotspots. J. Med. Chem. 2016;59:4314–4325. doi: 10.1021/acs.jmedchem.5b01980. [DOI] [PubMed] [Google Scholar]
- 30.Roversi P., Marti L., Caputo A.T., Alonzi D.S., Hill J.C., Dent K.C., Kumar A., Levasseur M.D., Lia A., Waksman T., et al. Interdomain conformational flexibility underpins the activity of uggt, the eukaryotic glycoprotein secretion checkpoint. Proc. Natl. Acad. Sci. USA. 2017;114:8544–8549. doi: 10.1073/pnas.1703682114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satoh T., Song C., Zhu T., Toshimori T., Murata K., Hayashi Y., Kamikubo H., Uchihashi T., Kato K. Visualisation of a flexible modular structure of the ER folding-sensor enzyme UGGT. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-12283-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modenutti C.P., Blanco Capurro J.I., Ibba R., Alonzi D.S., Song M.N., Vasiljević S., Kumar A., Chandran A.V., Tax G., Marti L., et al. Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of udp-glucose: Glycoprotein glucosyltransferase. Structure. 2021;29:357–370.e9. doi: 10.1016/j.str.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 34.Caputo A.T., Ibba R., Le Cornu J.D., Darlot B., Hensen M., Lipp C.B., Marcianò G., Vasiljević S., Zitzmann N., Roversi P. Crystal polymorphism in fragment-based lead discovery of ligands of the catalytic domain of UGGT, the glycoprotein folding quality control checkpoint. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.960248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tessier D.C., Dignard D., Zapun A., Radominska-Pandya A., Parodi A.J., Bergeron J.J., Thomas D.Y. Cloning and characterization of mammalian UDP-glucose glycoprotein: glucosyltransferase and the development of a specific substrate for this enzyme. Glycobiology. 2000;10:403–412. doi: 10.1093/glycob/10.4.403. [DOI] [PubMed] [Google Scholar]
- 36.Sprong H., Degroote S., Nilsson T., Kawakita M., Ishida N., van der Sluijs P., van Meer G. Association of the Golgi UDP-galactose transporter with UDP-galactose:ceramide galactosyltransferase allows UDP-galactose import in the endoplasmic reticulum. Mol. Biol. Cell. 2003;14:3482–3493. doi: 10.1091/mbc.E03-03-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maszczak-Seneczko D., Olczak T., Olczak M. Subcellular localization of UDP-GlcNAc, UDP-Gal and SLC35B4 transporters. Acta Biochim. Pol. 2011;58:413–419. [PubMed] [Google Scholar]
- 38.Nakajima K., Kizuka Y., Yamaguchi Y., Hirabayashi Y., Takahashi K., Yuzawa Y., Taniguchi N. Identification and characterization of UDP-mannose in human cell lines and mouse organs: Differential distribution across brain regions and organs. Biochem. Biophys. Res. Commun. 2018;495:401–407. doi: 10.1016/j.bbrc.2017.10.173. [DOI] [PubMed] [Google Scholar]
- 39.Miyagawa A., Totani K., Matsuo I., Ito Y. Promiscuous activity of er glucosidase ii discovered through donor specificity analysis of uggt. Biochem. Biophys. Res. Commun. 2010;403:322–328. doi: 10.1016/j.bbrc.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 40.McNicholas S., Agirre J. Glycoblocks: a schematic three-dimensional representation for glycans and their interactions. Acta Crystallogr. D Struct. Biol. 2017;73:187–194. doi: 10.1107/S2059798316013553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanzarotti E., Defelipe L.A., Marti M.A., Turjanski A.G. Aromatic clusters in protein-protein and protein-drug complexes. J. Cheminf. 2020;12:30. doi: 10.1186/s13321-020-00437-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roversi P., Marti L., Caputo A.T., Alonzi D.S., Hill J.C., Dent K.C., Kumar A., Levasseur M.D., Lia A., Waksman T., et al. Interdomain conformational flexibility underpins the activity of uggt, the eukaryotic glycoprotein secretion checkpoint. Proc. Natl. Acad. Sci. USA. 2017;114:8544–8549. doi: 10.1073/pnas.1703682114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sousa M.C., Ferrero-Garcia M.A., Parodi A.J. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 1992;31:97–105. doi: 10.1021/bi00116a015. [DOI] [PubMed] [Google Scholar]
- 44.Sousa M.C., Parodi A.J. The interaction of the UDP-GLC:glycoprotein glucosyltransferase with the acceptor glycoprotein. Cell. Mol. Biol. 1996;42:609–616. [PubMed] [Google Scholar]
- 45.Lee E., Shim S.-H., Cho M. Fluorescence enhancement of a ligand-activated fluorescent protein induced by collective noncovalent interactions. Chem. Sci. 2018;9:8325–8336. doi: 10.1039/C8SC03558J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartoschik, T., Zoephel, A., Rumpel, K., Ciulli, A., and Heffern, C. MSTMicroScale thermophoresis (MST)and TRICTemperature related intensity change (TRIC) Technology to Reliably Study PROTACProteolysis-targeting chimeras (PROTACs) Binary and Ternary Binding in Drug Development ( 115–133). New York, NY: Springer US. ISBN 978-1-0716-1665-9 (2021):( 115–133). URL: https://doi.org/10.1007/978-1-0716-1665-9_6. doi:10.1007/978-1-0716-1665-9_6
- 47.Pearse B.R., Gabriel L., Wang N., Hebert D.N. A cell-based reglucosylation assay demonstrates the role of GT1 in the quality control of a maturing glycoprotein. J. Cell Biol. 2008;181:309–320. doi: 10.1083/jcb.200712068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams B.M., Canniff N.P., Guay K.P., Larsen I.S.B., Hebert D.N. Vol. 9. eLife; 2020. (Quantitative Glycoproteomics Reveals Substrate Selectivity of the ER Protein Quality Control Sensors UGGT1 and UGGT2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aebi M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta. 2013;1833:2430–2437. doi: 10.1016/j.bbamcr.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Adams B.M., Canniff N.P., Guay K.P., Hebert D.N. The Role of Endoplasmic Reticulum Chaperones in Protein Folding and Quality Control. Prog. Mol. Subcell. Biol. 2021;59:27–50. doi: 10.1007/978-3-030-67696-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnold S.M., Fessler L.I., Fessler J.H., Kaufman R.J. Two homologues encoding human UDP-glucose:glycoprotein glucosyltransferase differ in mRNA expression and enzymatic activity. Biochemistry. 2000;39:2149–2163. doi: 10.1021/bi9916473. [DOI] [PubMed] [Google Scholar]
- 52.Blanco-Herrera F., Moreno A.A., Tapia R., Reyes F., Araya M., D’Alessio C., Parodi A., Orellana A. The UDP-glucose: glycoprotein glucosyltransferase (UGGT), a key enzyme in ER quality control, plays a significant role in plant growth as well as biotic and abiotic stress in Arabidopsis thaliana. BMC Plant Biol. 2015;15:127. doi: 10.1186/s12870-015-0525-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trombetta S.E., Bosch M., Parodi A.J. Glucosylation of glycoproteins by mammalian, plant, fungal, and trypanosomatid protozoa microsomal membranes. Biochemistry. 1989;28:8108–8116. doi: 10.1021/bi00446a022. [DOI] [PubMed] [Google Scholar]
- 54.Dalziel M., Crispin M., Scanlan C.N., Zitzmann N., Dwek R.A. Emerging principles for the therapeutic exploitation of glycosylation. Science. 2014;343:1235681. doi: 10.1126/science.1235681. [DOI] [PubMed] [Google Scholar]
- 55.Trombetta E.S., Helenius A. Glycoprotein reglucosylation and nucleotide sugar utilization in the secretory pathway: identification of a nucleoside diphosphatase in the endoplasmic reticulum. EMBO J. 1999;18:3282–3292. doi: 10.1093/emboj/18.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mishra A., Periasamy N., Patankar M.P., Narasimhan K. Synthesis and characterisation of soluble aluminium complex dyes based on 5-substituted-8-hydroxyquinoline derivatives for oled applications. Dyes Pigments. 2005;66:89–97. doi: 10.1016/j.dyepig.2004.09.004. [DOI] [Google Scholar]
- 57.Wang F., Peng R., Sha Y. Selective dendritic fluorescent sensors for Zn(II) Molecules. 2008;13:922–930. doi: 10.3390/molecules13040922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumar S., Shah P., Tripathi S.K., Khan S.I., Singh I.P. Synthesis and In Vitro Evaluation of Hydrazonomethyl-Quinolin-8-ol and Pyrazol-3-yl-Quinolin-8-ol Derivatives for Antimicrobial and Antimalarial Potential. Med. Chem. 2022;18:949–969. doi: 10.2174/1573406418666220303144929. http://www.eurekaselect.com/article/121293 [DOI] [PubMed] [Google Scholar]
- 59.Al Busafi S.N., Suliman F., Al-Alawi Z.R. 2014. 8-hydroxyquinoline and its Derivatives: Synthesis and Applications.https://www.rroij.com/open-access/8hydroxyquinoline-and-its-derivatives-synthesis-and-applications-.php?aid=33914 [Google Scholar]
- 60.Yañez O., Osorio M.I., Uriarte E., Areche C., Tiznado W., Pérez-Donoso J.M., García-Beltrán O., González-Nilo F. In Silico Study of Coumarins and Quinolines Derivatives as Potent Inhibitors of SARS-CoV-2 Main Protease. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.595097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Albesa-Jové D., Cifuente J.O., Trastoy B., Guerin M.E. In: Chemical and Synthetic Biology Approaches To Understand Cellular Functions - Part A vol. 621 of Methods in Enzymology. Shukla A.K., editor. Academic Press; 2019. Chapter fifteen - quick-soaking of crystals reveals unprecedented insights into the catalytic mechanism of glycosyltransferases; pp. 261–279. [DOI] [PubMed] [Google Scholar]
- 62.Shoji E., Miyatake K., Hlil A.R., Hay A.S., Maindron T., Jousseaume V., Dodelet J.P., Tao Y., D’iorio M. Immiscible polymers in double spin-coated electroluminescent devices containing phenyl-substituted tris(8-hydroxyquinoline)aluminum derivatives soluble in a host polymer. J. Polym. Sci. A. Polym. Chem. 2003;41:3006–3016. [Google Scholar]
- 63.King O.N.F., Li X.S., Sakurai M., Kawamura A., Rose N.R., Ng S.S., Quinn A.M., Rai G., Mott B.T., Beswick P., et al. Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hopkinson R.J., Tumber A., Yapp C., Chowdhury R., Aik W., Che K.H., Li X.S., Kristensen J.B.L., King O.N.F., Chan M.C., et al. 5-carboxy-8-hydroxyquinoline is a broad spectrum 2-oxoglutarate oxygenase inhibitor which causes iron translocation. Chem. Sci. 2013;4:3110–3117. doi: 10.1039/C3SC51122G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aik W., Demetriades M., Hamdan M.K.K., Bagg E.A.L., Yeoh K.K., Lejeune C., Zhang Z., McDonough M.A., Schofield C.J. Structural basis for inhibition of the fat mass and obesity associated protein (fto) J. Med. Chem. 2013;56:3680–3688. doi: 10.1021/jm400193d. [DOI] [PubMed] [Google Scholar]
- 66.Moon H., Han S., Park H., Choe J. Crystal structures of human fih-1 in complex with quinol family inhibitors. Mol. Cell. 2010;29:471–474. doi: 10.1007/s10059-010-0058-3. [DOI] [PubMed] [Google Scholar]
- 67.Esposito C., Wiedmer L., Caflisch A. In silico identification of jmjd3 demethylase inhibitors. J. Chem. Inf. Model. 2018;58:2151–2163. doi: 10.1021/acs.jcim.8b00539. [DOI] [PubMed] [Google Scholar]
- 68.Xing J., Zhang R., Jiang X., Hu T., Wang X., Qiao G., Wang J., Yang F., Luo X., Chen K., et al. Rational design of 5-((1h-imidazol-1-yl)methyl)quinolin-8-ol derivatives as novel bromodomain-containing protein 4 inhibitors. Eur. J. Med. Chem. 2019;163:281–294. doi: 10.1016/j.ejmech.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 69.Chen W., Zhang H., Chen Z., Jiang H., Liao L., Fan S., Xing J., Xie Y., Chen S., Ding H., et al. Development and evaluation of a novel series of nitroxoline-derived bet inhibitors with antitumor activity in renal cell carcinoma. Oncogenesis. 2018;7:83. doi: 10.1038/s41389-018-0093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qin J., Xie P., Ventocilla C., Zhou G., Vultur A., Chen Q., Liu Q., Herlyn M., Winkler J., Marmorstein R. Identification of a novel family of brafv600e inhibitors. J. Med. Chem. 2012;55:5220–5230. doi: 10.1021/jm3004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buchler I., Akuma D., Au V., Carr G., de León P., DePasquale M., Ernst G., Huang Y., Kimos M., Kolobova A., et al. Optimization of 8-hydroxyquinolines as inhibitors of catechol o-methyltransferase. J. Med. Chem. 2018;61:9647–9665. doi: 10.1021/acs.jmedchem.8b01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McLean L.R., Zhang Y., Li H., Li Z., Lukasczyk U., Choi Y.-M., Han Z., Prisco J., Fordham J., Tsay J.T., et al. Discovery of covalent inhibitors for mif tautomerase via cocrystal structures with phantom hits from virtual screening. Bioorg. Med. Chem. Lett. 2009;19:6717–6720. doi: 10.1016/j.bmcl.2009.09.106. [DOI] [PubMed] [Google Scholar]
- 73.Vergara D., de Domenico S., Maffia M., Piro G., Di Sansebastiano G.P. Transgenic plants as low-cost platform for chemotherapeutic drugs screening. Int. J. Mol. Sci. 2015;16:2174–2186. doi: 10.3390/ijms16012174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marti L., Lia A., Reca I.-B., Roversi P., Santino A., Zitzmann N. In planta preliminary screening of er glycoprotein folding quality control (erqc) modulators. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19072135. https://www.mdpi.com/1422-0067/19/7/2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lia A., Gallo A., Marti L., Roversi P., Santino A. Efr-mediated innate immune response in arabidopsis thaliana is a useful tool for identification of novel erqc modulators. Genes. 2018;10:15. doi: 10.3390/genes10010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ko K., Ahn M.-H., Song M., Choo Y.-K., Kim H.S., Ko K., Joung H. Glyco-engineering of biotherapeutic proteins in plants. Mol. Cell. 2008;25:494–503. [PubMed] [Google Scholar]
- 77.Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta crystallographica. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S.M., Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 80.Singh A., Montgomery D., Xue X., Foley B.L., Woods R.J. GAG Builder: a web-tool for modeling 3D structures of glycosaminoglycans. Glycobiology. 2019;29:515–518. doi: 10.1093/glycob/cwz027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arcon J.P., Modenutti C.P., Avendaño D., Lopez E.D., Defelipe L.A., Ambrosio F.A., Turjanski A.G., Forli S., Marti M.A. AutoDock Bias: improving binding mode prediction and virtual screening using known protein–ligand interactions. Bioinformatics. 2019;35:3836–3838. doi: 10.1093/bioinformatics/btz152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Autodock4 and autodocktools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gorrec F. The MORPHEUS protein crystallization screen. J. Appl. Crystallogr. 2009;42:1035–1042. doi: 10.1107/S0021889809042022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gorrec F. The MORPHEUS II protein crystallization screen. Acta Crystallogr. F Struct. Biol. Commun. 2015;71:831–837. doi: 10.1107/S2053230X1500967X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morgan J.L.W., McNamara J.T., Fischer M., Rich J., Chen H.-M., Withers S.G., Zimmer J. Observing cellulose biosynthesis and membrane translocation in crystallo. Nature. 2016;531:329–334. doi: 10.1038/nature16966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O.S., Vonrhein C., TO W. BUSTER 2.10.3. 2017. BUSTER 2.10.3. [Google Scholar]
- 87.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 88.Modenutti C., Gauto D., Radusky L., Blanco J., Turjanski A., Hajos S., Marti M. Using crystallographic water properties for the analysis and prediction of lectin–carbohydrate complex structures. Glycobiology. 2015;25:181–196. doi: 10.1093/glycob/cwu102. [DOI] [PubMed] [Google Scholar]
- 89.Nivedha A.K., Makeneni S., Foley B.L., Tessier M.B., Woods R.J. Importance of ligand conformational energies in carbohydrate docking: Sorting the wheat from the chaff. J. Comput. Chem. 2014;35:526–539. doi: 10.1002/jcc.23517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Woods R.J. Predicting the structures of glycans, glycoproteins, and their complexes. Chem. Rev. 2018;118:8005–8024. doi: 10.1021/acs.chemrev.8b00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kirschner K.N., Yongye A.B., Tschampel S.M., González-Outeiriño J., Daniels C.R., Foley B.L., Woods R.J. Glycam06: A generalizable biomolecular force field. carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Blanco Capurro J.I., Di Paola M., Gamarra M.D., Martí M.A., Modenutti C.P. An efficient use of X-ray information, homology modeling, molecular dynamics and knowledge-based docking techniques to predict protein–monosaccharide complexes. Glycobiology. 2019;29:124–136. doi: 10.1093/glycob/cwy102. [DOI] [PubMed] [Google Scholar]
- 93.Makeneni S., Thieker D.F., Woods R.J. Applying pose clustering and md simulations to eliminate false positives in molecular docking. J. Chem. Inf. Model. 2018;58:605–614. doi: 10.1021/acs.jcim.7b00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dunlop D.C., Bonomelli C., Mansab F., Vasiljević S., Doores K.J., Wormald M.R., Palma A.S., Feizi T., Harvey D.J., Dwek R.A., et al. Polysaccharide mimicry of the epitope of the broadly neutralizing anti-HIV antibody, 2G12, induces enhanced antibody responses to self oligomannose glycans. Glycobiology. 2010;20:812–823. doi: 10.1093/glycob/cwq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Crystal structure coordinates and structure factor files (mmCIF format) were deposited and are publicly accessible in the protein databank (PDB) as PDB IDs 7ZKC (CtUGGTGT24), 7ZLU (U2FCtUGGTGT24) and 7ZLL (5M-8OH-QCtUGGTGT24). Accession numbers are also listed in the key resources table.
-
•
All original code has been deposited at Zenodo (https://doi.org/10.5281/zenodo.8305097) and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.