Summary
Background
Alcohol consumption is widespread with over half of the individuals over 18 years of age in the U.S. reporting alcohol use in the last 30 days. Moreover, 9 million Americans engaged in binge or chronic heavy drinking (CHD) in 2019. CHD negatively impacts pathogen clearance and tissue repair, including in the respiratory tract, thereby increasing susceptibility to infection. Although, it has been hypothesized that chronic alcohol consumption negatively impacts COVID-19 outcomes; the interplay between chronic alcohol use and SARS-CoV-2 infection outcomes has yet to be elucidated.
Methods
In this study we employed luminex, scRNA sequencing, and flow cytometry to investigate the impact of chronic alcohol consumption on SARS-CoV-2 anti-viral responses in bronchoalveolar lavage cell samples from humans with alcohol use disorder and rhesus macaques that engaged in chronic drinking.
Findings
Our data show that in both humans (n = 6) and macaques (n = 11), the induction of key antiviral cytokines and growth factors was decreased with chronic ethanol consumption. Moreover, in macaques fewer differentially expressed genes mapped to Gene Ontology terms associated with antiviral immunity following 6 month of ethanol consumption while TLR signaling pathways were upregulated.
Interpretation
These data are indicative of aberrant inflammation and reduced antiviral responses in the lung with chronic alcohol drinking.
Funding
This study was supported by NIH 1R01AA028735-04 (Messaoudi), U01AA013510-20 (Grant), R24AA019431-14 (Grant), R24AA019661 (Burnham), P-51OD011092 (ONPRC core grant support). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Keywords: Alcohol, SARS-CoV-2, Lung, Non-human primate, scRNA-seq, Inflammation
Research in context.
Evidence before this study
Alcohol use disorder (AUD) impacts 8% of all adults in the United States. Clinical and experimental studies have shown that AUD interferes with anti-microbial responses and tissue repair resulting in increased susceptibility to infection. Indeed, AUD has been implicated in increased severity of COVID-19; however, the exact mechanisms have yet to be elucidated.
Added value of this study
For this study, we utilized a rhesus macaque model of voluntary ethanol self-administration to uncover the impact of chronic alcohol consumption on the acute response to SARS-CoV-2 infection. This model accurately recapitulates human patterns of drinking and immunological consequences. We report that macrophages are a prominent immune cell target of SARS-CoV-2 infection in the lung and that chronic alcohol consumption led to a heightened inflammatory response, but a dampened interferon stimulated gene expression response.
Implications of all the available evidence
Collectively, these findings provide evidence that chronic alcohol consumption leads to a heightened inflammatory state at baseline and dysregulates acute response to SARS-CoV-2, skewing it towards aberrant inflammatory outcomes and away from anti-viral responses. Finally, the non-human primate model utilized in this study provides insight into pathophysiology of AUD and consequences on respiratory infection.
Introduction
Alcohol consumption is widespread in the United States, with 55% of the American population over 18 years of age reporting alcohol use within the last 30 days. Alarmingly, 25% and 6.3% of adults over 18 years of age are classified as binge or heavy drinkers, respectively (National Survey on Drug Use and Health 2020). Both binge and heavy drinking can contribute to an Alcohol Use Disorder (AUD) diagnosis, defined as the inability to control or cease alcohol use despite experiencing negative social, occupational, or health-related consequences.1 In 2019, 9 million men and 5.5 million women were diagnosed with AUD.2 Excessive alcohol consumption impairs lung,3 liver,4 pancreas,5 spleen,6 and heart7,8 function. Alcohol consumption is also associated with increased incidence of cancers.9, 10, 11 Moreover, excessive alcohol consumption leads to an increased rate12 and length of hospitalizations13 including admission to the intensive care unit.14 Individuals with AUD are also at increased risk of nosocomial infections following trauma15 and wound-related infections.16 Overall, excessive alcohol use in the United States is associated with a $249 billion loss to the American economy17 and $28 billion in healthcare-related costs.17,18 Moreover, it is also associated with more than 260 alcohol-related deaths per day, making AUD the third leading cause of preventable deaths in the United States.19
Data from clinical and experimental studies indicate that chronic excessive alcohol consumption disrupts cellular mechanisms responsible for pathogen clearance and tissue repair,20,21 leading to increased susceptibility to infection and delayed wound healing.22,23 Specifically, the incidence of K. pneumonia, S. pneumonia, Mycobacterium tuberculosis infection, hepatitis C virus, and respiratory syncytial virus (RSV) are increased with chronic heavy drinking (CHD).23, 24, 25 Animal model studies have recapitulated the increased severity of bacterial and viral infections with CHD further highlighting the immunological basis of these adverse outcomes.26,27
Underlying mechanisms driving poor infectious disease outcomes are the focus of several studies that have reported hyper-inflammatory responses following LPS stimulation by circulating and tissue-resident myeloid cells from humans with or animal models of CHD.3,28 In contrast, stimulation with pathogens decreased the production of key immune mediators.3,29,30 A dysregulation of phagocytic capacity within myeloid populations has also been observed in conjunction with CHD.31,32 Additionally, in-vitro ethanol treatment of monocytes resulted in lower expression levels of MHC-II molecules,33 suggesting that CHD can lead to reduced antigen presentation capacity. Similarly, excessive alcohol consumption interferes with the phagocytic capacity of alveolar macrophages (AM) and their ability to generate anti-viral and anti-bacterial responses.3,34,35 Alcohol exposure in rats dampens the expression of GM-CSF receptors required for the differentiation of monocytes into macrophages upon infiltrating the lungs.34,36 In addition to impairing immune responses, CHD decreases barrier function in the respiratory tract, increasing susceptibility to respiratory infections.37 Individuals with CHD are at increased risk of developing acute respiratory distress syndrome (ARDS),38 leading to organ failure, sepsis, or death.39
The shelter-in-place orders associated with the COVID-19 pandemic led to increased alcohol consumption in the United States.40 Given the impact of chronic ethanol consumption on the respiratory system, it has been hypothesized that increased ethanol consumption could be one of the drivers for COVID-19 severity.41 Indeed, recent studies have highlighted increased COVID-19 disease severity and need for respiratory support in patients with AUD.42,43 Additionally, susceptibility to COVID-19 positively correlated with alcohol intake when compared to nondrinkers.41 However, the interplay between chronic ethanol consumption and the anti-viral response to SARS-CoV-2 remains poorly understood. Therefore, in this study, bronchoalveolar lavage (BAL) cell samples obtained from rhesus macaques before and after 6-months of voluntary ethanol self-administration, as well as those obtained from individuals with and without AUD were infected ex vivo with SARS-CoV-2. Response to infection was monitored by measuring immune mediator release and transcriptional changes using single cell RNA sequencing (scRNA-Seq).
Methods
Sample collection
Non-human primate (NHP) studies were approved by the Oregon National Primate Research Center (ONPRC) Institutional Animal Care and Use Committee (IACUC), an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). This study leveraged an NHP model of voluntary ethanol self-administration which recapitulates human drinking behavior and has a high genetic and physiological homology to humans. In this model, rhesus macaques have concurrent access to both water and a 4% w/v ethanol solution for 22 h/day.44 Drinking phenotypes are defined by the pattern and volume of ethanol consumption per day (g/kg/day) and classified into four categorical levels (LD: low drinking (>0.5 g/kg/day), BD: binge drinking (>2 g/kg/day), HD: heavy drinking (>3 g/kg/day), VHD: very heavy drinking (>4 g/kg/day)) defined in Baker et al.45 Blood samples were collected every 5–7 days, 7 h into the 22 h/day drinking session to measure blood ethanol concentration (BEC) via headspace gas chromatography.
For these studies, BAL samples were collected from the following macaques: 5 male (2 low drinkers and 3 very heavy drinkers) and six female rhesus macaques (4 low drinkers, 1 heavy drinker, and 1 very heavy drinker) at baseline (before induction) and after 6 months of chronic consumption. Table 1 summarizes the cohort demographics for the rhesus macaques used in this study (Cohort 18 on matrr.com). The average age of the animals was 6.5 ± 0.38 years old, which translates to young adults in their ∼20's. None of the animals were vaccinated against or infected with SARS-CoV-2. All animals were fed standard chow diet consisting of the Fiber-balanced monkey diet (15% calories from fat, 27% from protein, and 59% from carbohydrates; no. 5052; Lab Diet, St. Louis, MO) This diet is supplemented with seasonal fresh fruit and produce once daily. Municipal water was available ad libitum. All animals were housed in the same room in accordance with standards established by the US (United States) Federal Animal Welfare Act and The Guide for the Care and Use of Laboratory Animals. Animals received enrichment and clincal care as needed through the Division of Comparative Medicine at the Oregon National Primate Research Center.
Table 1.
Demographics of the NHP cohort utilized in this study.
| ID | Sex | Drinking phenotype | Mean daily ethanol intake (g/kg/day) | Average blood ethanol content (Avg mg%) |
|---|---|---|---|---|
| 10,331 | F | LD | 0.5 | 0 |
| 10,332 | F | LD | 1.23 | 3.15 |
| 10,334 | F | LD | 1.78 | 24.32 |
| 10,339 | F | LD | 1.74 | 30.42 |
| 10,340 | F | VHD | 3.53 | 40.24 |
| 10,333 | F | HD | 3.04 | 46.01 |
| 10,341 | M | LD | 1.94 | 32.64 |
| 10,335 | M | LD | 1.32 | 16.78 |
| 10,337 | M | VHD | 4.02 | 158.62 |
| 10,338 | M | VHD | 5.93 | 209.11 |
| 10,342 | M | VHD | 4.04 | 161.72 |
Human BAL samples were obtained, with IRB approval, from the University of Colorado School of Medicine's Colorado-Pulmonary Alcohol Research Collaborative (CoPARC) (https://medschool.cuanschutz.edu/coparc) where informed consent was acquired from all subjects. Three samples were collected from individuals with a history of only smoking and three samples were collected from individuals with a history of alcohol use disorder and smoking. Table 2 summarizes the self-reported demographics of the human participants involved in this study. Only samples from male subjects were available for this study. None of the individuals were vaccinated against SARS-CoV-2.
Table 2.
Demographic information from the human BAL samples utilized in this study.
| Smoking n = 3 | Smoking and alcohol constumption n = 3 | |
|---|---|---|
| Age (years) | 54.7 ± 6.1 | 58.0 ± 2.6 |
| Sex | Males: 3 Females: 0 |
Males: 3 Females: 0 |
| Race | ||
| African American | 1 | 1 |
| White | 1 | 1 |
| Hispanic | 1 | 1 |
The macaque samples were obtained under IACUC number IP0001263 (Oregon National Primate Research Center). Human Samples were obtained under IRB # COMIRB 12-0181 (University of Colorado).
Bronchoalveolar lavage (BAL) cell isolation
BAL samples were centrifuged at 2000 rpm for 10 min. Pellets were resuspended in 10% DMSO/FBS solution, stored in a Mr. Frosty Freezing container (Thermo Fisher Scientific, Waltham, MA) at −80 °C for 24 h, and then transferred to a cryogenic unit for long-term storage.
Flow cytometry
BAL cells were thawed and 1 × 106 were stained with a live dead stain, Ghost Dye Violet 510 (Tonbo Biosciences Cat# 13-0870), and the following antibody panel: CD20 (BioLegend/Thermo Fisher Scientific Cat# 302304/25-0209-42, RRID:AB_314252/RRID:AB_1834471), CD3 (BD Biosciences Cat# 556611, RRID:AB_396484), CD4 (BioLegend Cat# 317410, RRID:AB_571955), CD8a (Tonbo Biosciences Cat# 25-0088), CD16 (BioLegend Cat# 302032/302010, RRID:AB_2104003/RRID:AB_314210), CD14 (BioLegend Cat# 301822, RRID:AB_493747) HLA-DR (BioLegend Cat# 307618, RRID:AB_493586), CD169 (BioLegend Cat# 346014, RRID:AB_2750264), CD163 (BioLegend Cat# 333624, RRID:AB_2564201), and CD206 (BD Biosciences Cat# 555954, RRID:AB_396250) for 30 min at 4 °C, washed with FACS buffer (2% FBS, 2% 0.5 M EDTA in PBS), and acquired using an Attune NxT Flow Cytometer (ThermoFisher Scientific, Waltham, MA) and two different forward and side scatter values to capture small lymphocytes and innate immune cells respectively. Results were analyzed using FlowJo software (Ashland, OR). The remainder of the cells were subjected to SARS-CoV-2 infection.
SARS-CoV-2 propagation and infection
SARS-CoV-2 (strain USA-AZ1/2020) was obtained from BEI Resources (NR-52383) and grown on Vero E6 cells (ATCC, Manassas, Virginia). Vero E6 cells were cultured at 37 °C in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES (pH 7.3), and 10 μg/mL of puromycin antibiotic. The virus was passaged using a multiplicity of infection (MOI) of 0.01 and titrated by focus forming assay (FFA) on Vero E6 cells as described previously by Jureka et al.46
To quantify infectious viral titer, samples were serially diluted 10-fold and added to a monolayer of Vero E6 cells that were plated 24 h prior in a 96-well flat bottom plate. The plates were then incubated at 37 °C, 5% CO2 for 1 h to allow attachment and entry of the virus into the cells, then overlaid with 2% (w/v) methylcellulose in DMEM supplemented with 5% FBS overnight at 37 °C, 5% CO2. After incubation, the cells were fixed using 5% paraformaldehyde for 15 min, before being washed with 1X PBS. The plates were washed using FFA buffer 1X PBS, 0.05% Triton X-100, before adding 500 ng/mL anti-Spike (mAb 2165) in FFA staining buffer for overnight incubation at 4 °C. The cells were washed with FFA buffer, then placed in HRP-conjugated goat anti-human IgG secondary antibody (Sigma–Aldrich, St. Louis, MO) in FFA buffer at room temperature for 1 h. After incubation with secondary antibody, the cells were washed with FFA buffer before TrueBlue peroxidase substrate (KPL) was used to stain and develop the plates before counting the foci on an EliSpot analyzer (Advanced Imaging Devices GmbH, Straßberg, Germany).46 All virus experiments were completed in a Biosafety level 3 (ABSL-3) facility.
1 × 106 live BAL cells were infected with SARS-CoV-2 at an MOI of 1 or left untreated for 24 h at 37 °C, 5% CO2. An MOI of 1 was selected to achieve robust infection of the primary cells while avoiding excess cell death. The 24 h time point was select to maximize likelihood of virus uptake/entry while minimizing cell death given the fragile nature of primary BAL cells. At the end of the infection, cultures were spun down, and supernatants were stored at −80 °C for Luminex analysis while cells were immediately processed for single cell RNA sequencing.
Luminex
Immune mediators of BAL cell culture supernatant were measured before and after stimulation with SARS-CoV-2. NHP samples were analyzed via an R&D 36-plex NHP XL Cytokine Premixed Kit, which contained the following analytes: BDNF, CCL2, CCL5, CCL11, CCL20, CD40 Ligand, CXCL2, CXCL10, CXCL11, CXCL13, FGF basic, G-CSF, GM-CSF, Granzyme B, IFNα, IFNβ, IFNγ, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12 p70, IL-13, IL-15, IL-17, IL-21, PD-L1, PDGF-AA, PDGF-BB, TGFα, TNFα, VEGF (Bio-Techne, Minneapolis, MN). Human samples were analyzed via a customized human 29-plex kit containing the following analytes: TNFα, IL-6, PD-L1, PDGF-BB, S100B, IL-7, IFN-β, IL-10, CCL2, VEGF, CXCL13, IL-1RA, CCL3, CCL4, IL-4, IL-17, IL-2, IL-15, GM-CSF, IL-8, CXCL9, IFNγ, IL-12P70, IL-1β, CXCL11, CXCL10, IL-23, CCL11, and IL-18. The samples were analyzed on a MAGPIX instrument (Luminex, Austin, TX). A standard curve was established using the xPONENT™ software and a 7-parameter logistic curve.
Single cell RNA library preparation
After overnight stimulation, BAL cells were washed twice with 0.04% BSA in PBS, before labeling with cell multiplexing oligos (CMOs) (10X genomics, Pleasanton, CA) at 4 °C for 20 min. Live cells were counted in duplicate and then pooled at a concentration of 1.5 × 106 cells/mL in 1% BSA in PBS. The pools were stained with Sytox green (Invitrogen) and ∼200,000 live cells from each group were sorted using a WOLF cell sorter (Nanocellect Biomedical Inc., San Diego, CA). The sorted cells were resuspended and loaded into a 10x Genomics Chromium Controller at a concentration of 1600 cells/μL for a target recovery of 20,000 cells. The libraries were prepared using the v3.1 chemistry of the Chromium Single Cell 3′Feature Barcoding Library Kit (10x Genomics, Pleasanton, CA) according to the manufacturer's instructions.
Single cells RNA sequencing analysis
The resulting libraries were sequenced using a NovaSeq6000. The reads were aligned and quantified via the Cell Ranger Single-Cell Software Suite and STAR aligner (version 4.0, 10x Genomics) against the Mmul_8 rhesus macaque reference genome for the macaque samples and the human reference genome GRCh38 for the human samples. Downstream analysis was performed separately using Seurat (version 4.1.1). Briefly, samples were de-multiplexed based on their unique CMOs followed by the removal of ambient RNA (<200 feature counts) and dead cells (>5% mitochondrial gene expression).47 The samples were normalized and scaled based on cell cycle and mitochondrial gene expression using NormalizeData and ScaleData, respectively. Data sets were integrated using reciprocal PCA via Seurat's FindIntegrationAnchors. Once the anchor points were found, the samples were combined into a single data frame using IntegrateData. These integrated data were again scaled using ScaleData, and a PCA was generated to determine the dimensionality of the data set via RunPCA. UMAP generation and cell clustering was performed using RunUMAP (dims = 1:50), FindNeighbors (dims = 1:20), and FindClusters (resolution = 0.5). The identity of the resulting clusters was then determined using the canonical markers found using FindAllMarkers and a Log2 fold change cutoff of 0.4. Differential gene expression analysis was performed using the default settings of MAST in Seurat and comparing SARS-CoV-2 infected to non-infected cells at each timepoint. Differentially expressed genes (DEGs) were defined as those with FDR p ≤ 0.05 and Log2 fold change ≥0.25. When comparing infection to non-infection in human samples, the Log2 fold change cut-off was removed. Functional enrichment was performed through Metascape.48
Statistical analysis
As these studies leveraged established repositories, no power calculation was conducted prior to initiating these studies. Statistical analyses for non scRNA-Seq data sets were performed using the GraphPad Prism software (GraphPad Software Inc., La Jolla, CA). All data sets were tested for Gaussian distribution using a Shapiro–Wilk normality test (α = 0.05). Subsequent tests were performed non-parametrically if the data set failed normality. Statistical analyses were performed in a pairwise (paired T-test) fashion when comparing pre- and post-infection results from the same subjects/macaques, such as when comparing cluster frequencies of immune subsets between baseline and 6 months (6mo) since these were within subject comparisons. Unpaired T-tests were used for comparison between AUD and non-AUD subjects as this was a cross-sectional and not within subject comparison. We used an RM 1-way analysis of variance (ANOVA) with Holm-Šídák post hoc multiple comparisons (passed normality) or Friedman test with Dunn's post hoc multiple comparisons (failed normality) when comparing more than two groups, such as module scores between SARS-CoV-2 infected and non-infection samples at each timepoint/condition. In the event there were missing values, we utilized a Kruskal–Wallis 1-way ANOVA with Dunn's post hoc multiple comparisons. For two groups, T tests were carried out using a non-parametric Mann–Whitney test or Wilcoxon matched-pairs signed ranked test if failing normality or a parametric paired T-test if passing normality.
Role of funders
The Funders did not have any role in the study design, data collection, data analyses, interpretation, or writing of report.
Results
CHD skews the lung environment towards a heightened inflammatory state
To understand the impact of chronic ethanol consumption on the immunological landscape of the lung, BAL samples were collected from male and female rhesus macaques before and after 6 months of daily voluntary ethanol self-administration (Table 1) and subjected to flow cytometry (Fig. 1a, Fig. S1a and b). This analysis revealed that at baseline the relative abundance of CD8 and CD4 T cells was significantly higher in females compared to males (Fig. S1c). In contrast, the frequency of B cells was higher in males than females at baseline (Fig. S1d). Following 6 months of ethanol self-administration, the frequency of CD8 T cells was significantly reduced in females while that of natural killer (NK) cells was significantly reduced in males (Fig. S1c–e). Conversely, the relative abundance of alveolar macrophages (AM) was increased in both males and females (Fig. S1f). This increase was primarily mediated by an expansion of activated AM (CD163+), and a decrease in interstitial macrophages (IM) (Fig. S1f and g). The frequency of dendritic cells (DC) also significantly increased after 6 months of CHD in males, whereas the opposite trend was observed in females (Fig. S1h). The frequency of infiltrating monocytes was greater in females than males at baseline but decreased with drinking (Fig. S1i). Nevertheless, ethanol exposure led to an expansion of nonclassical (activated) monocytes and a decrease in classical monocytes in both males and females (Fig. S1i).
Fig. 1.
Response to SARS-CoV-2 infection is mitigated in rhesus macaques after chronic alcohol consumption. a) BALs were collected from rhesus macaques before and after 6 months of drinking to assess the impact of alcohol consumption on the acute innate immune response by lung-resident immune to SARS-CoV-2 infection. b) Uniform manifold approximation and projection (UMAP) representation of 151, 269 BAL immune cells with and without infection at baseline and after 6 months drinking. Sixteen unique clusters were determined. c) Bubble plot of key cluster marker genes. Percent of cells that express each transcript is represented by the size of the bubble while the average expression of those cells is denoted by color, ranging from blue (low) to red (high). d) Relative frequency of each cluster before and after infection at baseline and after 6 months of chronic drinking. Non-infected and infected samples within the same time point were compared in a pairwise fashion (Error bars: Mean and SEM, n = 5 M/6 F, [Wilcoxon matched-pairs signed ranked test] ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001). e) Scatter bar plots comparing the Log2 fold change in response to SARS-CoV-2 infection of select cytokine and chemokines within BAL cell culture supernatants at baseline and after 6 months of open access to alcohol in male and female subjects (n = 5 M/6 F, [Wilcoxon matched-pairs signed ranked test], ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001). f) Feature plot of SARS-CoV-2 expression within the UMAP. g) Bubble plot of SARS-CoV-2 expression within each cluster stratified by timepoint. Average expression of the SARS-CoV-2 transcript is represented by bubble size while the significance of baseline to 6 months comparison is denoted by color. These data were collected over 2–3 independent experiments.
We next investigated the impact of chronic ethanol consumption on the transcriptional landscape of immune cells using scRNA-Seq (Fig. 1a). A total of 16 unique clusters were identified, including 9 AM subsets (defined by markers MRC1, FABP4, MARCO), 1 infiltrating monocyte group (CD14), 1 conventional DC subset (MAMU-DRA, BATF3), CD4 and CD8 T cells (CD3D, CD4, GZMB), B cells (MS4A1, CD79B), proliferating cells (MKI67), and epithelial cells (EPCAM, CLDN4) populations (Fig. 1b and c). Each cluster was constituted of cells from all subjects, regardless of the time point of collection and SARS-CoV-2 infection (Fig. S2a). The 9 AM clusters were distinct based on several inflammatory and regulatory markers (e.g., FABP4, STAT1, CD9.1, MMP10, ISG20, IL1B, CD14, MARCO, LCN2, HYOU1, LYZ, MAMU-DRA) (Fig S2b).
In line with the data from flow cytometry, the relative abundance of T cells decreased while that of myeloid cells increased following 6 months of ethanol consumption (Fig. 1d). Ethanol consumption also resulted in transcriptional shifts in the immune landscape of the lung. Specifically, within myeloid populations (AM, infiltrating monocytes), scores of modules associated with migration were decreased while those associated with hypoxia (HIF1α signaling) and other inflammatory pathways (TLR, viral/bacterial responses) were increased (Fig. S2c). Similarly, genes differentially expressed (DEG) with alcohol consumption in myeloid subsets mapped to GO terms such as “response to oxygen levels” (HlF-1α), innate immune responses (CD14), and positive regulation of cell migration (IL-1α, IL-1β, MMP14, CD74, MIF) (Fig. S2d and e). Within CD4 and CD8 T cell subsets, scores of modules associated with Th2 and viral/bacterial response signaling pathways were increased with chronic ethanol consumption (Fig. S3a). DEGs in lymphoid populations map to GO terms such as cell killing (GZMB), lymphocyte-mediated immunity (IL-2Rβ, CD40LG), in addition to response to hypoxia (Fig. S3b and c). These data indicate that chronic ethanol consumption alters the frequency and transcriptional profile of BAL-resident immune cells.
CHD alters immune mediator responses to SARS-CoV-2 infection and cellular distribution
We next assessed the impact of ethanol consumption on immune mediator production in response to SARS-CoV-2 infection (Fig. 1a). The increase in the production of immune mediators over no infection condition was comparable between males and females (Table S1) so the data were combined. SARS-CoV-2 infection induced a robust response demonstrated by increased induction of several cytokines, chemokines, and growth factors in culture supernatants at both timepoints relative to non-infection conditions (Fig. 1e, Table S1). Chronic ethanol consumption resulted in an increase in the induction of growth factors BDNF and FGFβ and a modest increase in the induction of inflammatory cytokine TNFα. In contrast, induction of growth factors VEGFA and TGFα, inflammatory cytokines IFNβ and IL-15, T cell activation marker PD-L1, and chemokine CCL2 were significantly decreased after 6 months of drinking (Fig. 1e). These results demonstrate that chronic ethanol consumption alters the secrotome in response to viral infection, suggestive of a dysregulated SARS-CoV-2 responses.
We then leveraged scRNA Seq to investigate the impact of chronic ethanol consumption on immune subset distribution and SARS-CoV-2 detection. A pairwise comparison of the relative abundance of clusters after infection showed a significantly reduced frequency of the AM3, AM5, and DC clusters and a notable decrease in the AM7, Mono, CD4, and CD8 clusters at baseline (Fig. 1d). After 6 months of ethanol consumption, significant decrease in frequency was evident in AM1 and CD4 clusters, with a notable decrease in the Mono cluster and an increase in the Epithelial cluster was observed in response to SARS-CoV-2 infection (Fig. 1d). Also, this analysis revealed that myeloid clusters contained most of the SARS-CoV-2 transcript, with the AM7 cluster exhibiting the greatest expression (Fig. 1f and g). Interestingly, the average expression of SARS-CoV-2 transcripts was significantly decreased across all AM clusters, except AM4 and AM5, after 6 months of ethanol consumption (Fig. 1g). In contrast, levels of viral transcripts were increased in the CD4 and CD8 T cell subset after 6 months of chronic ethanol consumption (Fig. 1g).
Chronic ethanol consumption alters transcriptional response to SARS-CoV-2
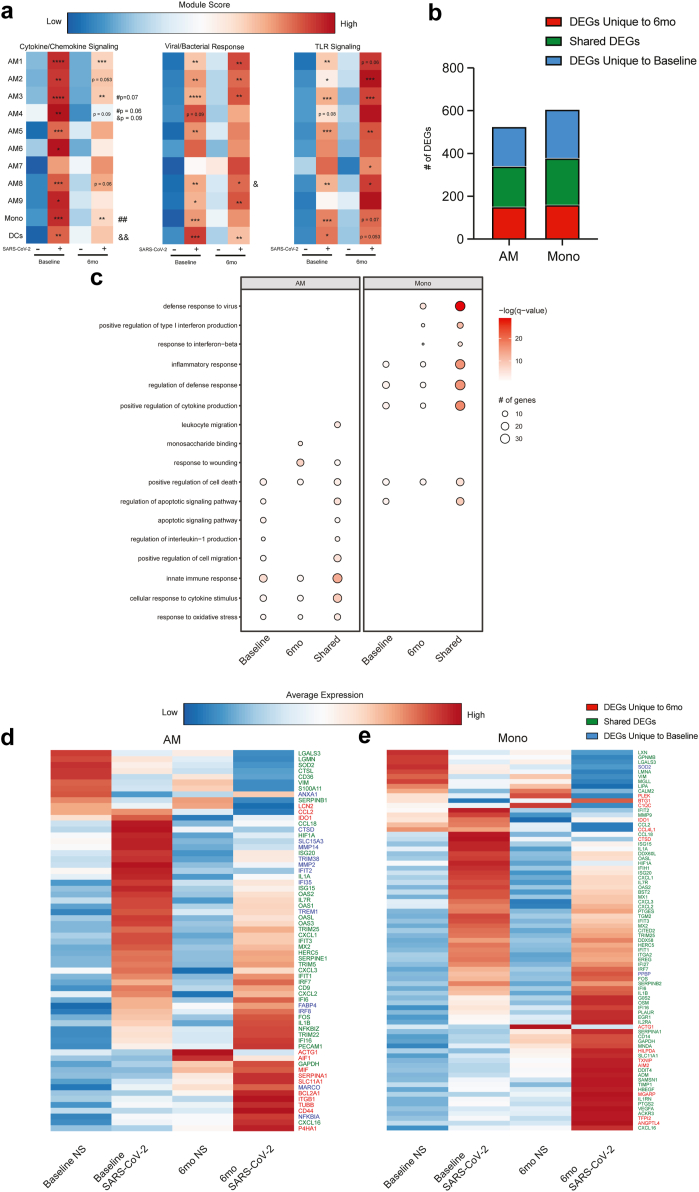
Because SARS-CoV-2 was predominately detected in myeloid cells, we focused our analysis on these clusters. To interrogate the impact of ethanol consumption on the anti-SARS-CoV-2 response, we measured module scores associated with cytokine/chemokine signaling, viral/bacterial response, and toll-like receptor (TLR) signaling (Fig. 2a, Table S2). All three module scores increased after SARS-CoV-2 infection at both time points within several myeloid clusters (Fig. 2a). After infection, there was a significant increase in module scores of cytokine/chemokine signaling and anti-viral/bacterial responses within more myeloid clusters at baseline, whereas scores associated with TLR signaling were increased within a greater number of clusters after ethanol consumption (Fig. 2a). Furthermore, module scores for cytokine/chemokine signaling were significantly lower in AM3, AM4, and Mono at 6mo when compared to baseline after infection. These observations are in line with the decreased induction of IL-15, VEGF, IFNβ and increased induction of TNFa observed in the Luminex analysis (Fig. 1e). All three module scores increased in DCs after infection in the samples obtained at baseline whereas a significant increase in the expression of genes only within the anti-viral/bacterial module were noted after 6 months of ethanol consumption (Fig. 2a).
Fig. 2.
CHD predominately affects myeloid cells response to SARS-CoV-2. a) Heatmap comparing module scores for cytokine/chemokine signaling, viral/bacterial responses, and TLR signaling between samples based on infection status and timepoint. The average module score for each cluster ranges from low (blue) to high (red). Significance displayed is relative to non-infected conditions within timepoints (n = 5 M/6 F, [Wilcoxon matched-pairs signed ranked test], ∗p ≤ 0.05, ∗∗ ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001). # Denotes significance when comparing infected conditions and & denotes significance when comparing non infected conditions (#/& p ≤ 0.05, && ≤ 0.01). b) Stacked bar graph showing the number of unique and shared DEGs for myeloid clusters. c) Bubble plot of significant and notable gene ontology (GO) terms from functional enrichment of myeloid clusters. d) Heatmap of the average expression of select DEGs in AM clusters across conditions and timepoint. The average expression for each transcript ranges from low (blue) to high (red). e) Heatmap of the average expression of select DEGs in mono clusters across conditions and timepoint. The average expression for each transcript ranges from low (blue) to high (red). These data were collected over 2–3 independent experiments.
We next performed differential gene expression analysis comparing infected to noninfected cells within each time point. For this analysis, the AM clusters were combined into one cluster. SARS-CoV-2 infection was associated with a robust transcriptional response at baseline and after 6mo of chronic drinking in both AM and Mono clusters (Fig. 2b). While a significant overlap between was noted between the two time points, many DEGs unique to baseline or post-ethanol consumption conditions were also identified (Fig. 2b). DEGs unique to baseline enriched to processes associated with innate immunity and apoptosis (e.g. “regulation of IL1 production” and “apoptotic signaling pathway”) (Fig. 2c). Some of the notable unique DEG include cathepsin D (CTSD), metalloproteases (MMP2 and MMP4) as well viral restriction factors (TRIM36, IFI35) and regulatory molecules (TREM1) (Fig. 2d). On the other hand, DEGs unique to post-ethanol consumption enriched to gene ontology (GO) terms “monosaccharide binding” and “response to wounding” including MIF, SERPINA1, LCN2, CCL2, and IDO1 (Fig. 2c and d). Within the infiltrating monocytes, a greater number of genes enriched to GO terms “defense response to virus” and “positive regulation of type I IFN” (Fig. 2c). DEG unique to the 6mo time point include TXNIP, IDO1, CC4L1, and C1Q in line with heightened TLR signaling pathways (Fig. 2e).
Although small numbers of SARS-CoV-2 transcripts were detected within T cells (Fig. 1f), transcriptional responses to infection were noted (Fig. S3d). DEGs that play a role in type I and II interferon and anti-viral pathways were more evident at baseline. Those detected only after chronic drinking enriched primarily to apoptotic pathways (Fig. S3d). Overall, these findings indicate that ethanol consumption skews the transcriptional profiles of alveolar cells towards a hyperinflammatory state while dampening antiviral responses.
Dysregulation of anti-SARS-CoV-2 response by AUD in humans
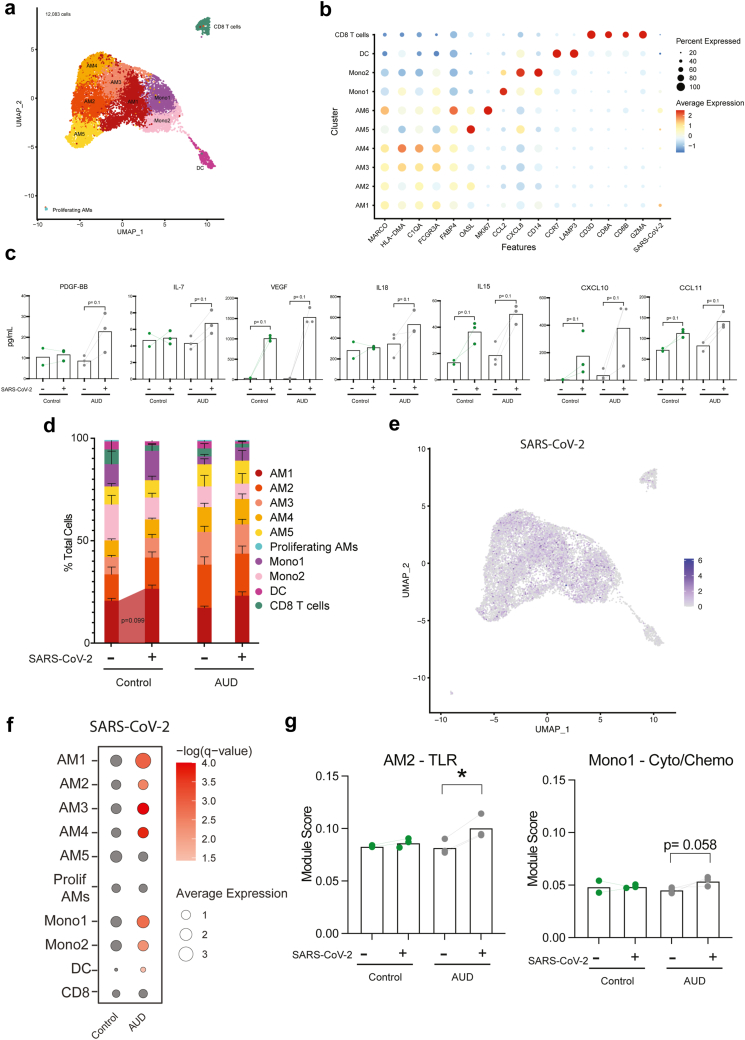
To place the observations obtained from the NHP model after 6 months of drinking in the context of long-term chronic heavy drinking in humans, BAL samples were obtained from individuals with AUD and smoking or smoking only (n = 3/group) (Table 2). Flow cytometry revealed a significant increase in the B cell population within the control group compared to the AUD group (Table 3). Transcriptional responses to ethanol consumption were assessed by scRNA-Seq. Using canonical gene markers, we identified 6 AM clusters (MARCO, HLA-DMA, C1QA, GCGR3A, FABP4, OASL) with one of the clusters being a Proliferating AM cluster (MKI67), 2 infiltrating monocyte clusters (CCL2, CXCL8, CD14), 1 DC cluster (CCR7, LAMP5), and 1 CD8 T cell cluster (CD3D, CD8A, CD8B, GZMA) (Fig. 3a and b). The clusters were distributed among all samples and timepoints (Fig. S4a). The 5 non-proliferating AM clusters were found to be distinct based on unique expression patterns of CXCL15, IL1B, FOS, CD14, SLC3A2, ARK1C3, FN1, FBP1, LIPA, C1QB, CTSD, HLA-DQB1, CXCL8, ATF3, OASL (Fig. S4b).
Table 3.
Frequencies of immune cell populations determined by flow cytometery.
| Cell type | Control (%) |
AUD (%) |
Derived | ||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | ||
| AM | 61.7 | 74.1 | 80.9 | 72.6 | 86.1 | 53.6 | % of Live Cells |
| AM 163- | 90.5 | 90.8 | 95.3 | 85.7 | 89 | 90.2 | % of Live AM |
| AM 163+ | 6.55 | 6.91 | 3.02 | 12.3 | 9.1 | 5.67 | % of Live AM |
| IM | 5.87 | 5.21 | 5.03 | 7.76 | 4.38 | 12.6 | % of Live Cells |
| DC | 1.88 | 2.26 | 0.37 | 3.15 | 0.6 | 1.33 | % of Live Cells |
| Monocytes | 1.18 | 3.09 | 0.79 | 3.74 | 0.51 | 1.71 | % of Live Cells |
| Classical Monocytes | 97.6 | 98.3 | 97 | 98.9 | 95.4 | 94.5 | % of Live Monocytes |
| Non-classical Monocytes | 2.41 | 1.67 | 2.95 | 1.09 | 4.6 | 5.54 | % of Live Monocytes |
| CD4+ | 15.4 | 18.4 | 5.99 | 17.1 | 30.9 | 8.96 | % of Live Cells |
| CD8+ | 27.3 | 17.2 | 5.55 | 27.6 | 4.8 | 9.47 | % of Live Cells |
| NK | 1.48 | 2.52 | 2.69 | 2.4 | 1.67 | 3.18 | % of Live Cells |
| B Cellsa | 0.74 | 1.01 | 0.67 | 0.15 | 0.38 | 0.46 | % of Live Cells |
Significant difference in cell populations when comparing the control group to the AUD group.
Fig. 3.
Alcohol consumption in humans dysregulates innate immune responses to SARS-CoV-2. a) UMAP visualization of 12,083 cells from individuals who smoke only (control) and individuals with AUD and smoking (AUD) prior to and after SARS-CoV-2 stimulation. b) Bubble plot of canonical marker genes used to identify cell clusters. Percent of cells that express each transcript is represented by the size of the bubble while the average expression of those cells is denoted by color, ranging from blue (low) to red (high). c) Bar plot of immune mediator production stratified by condition and timepoint (EtOH non-infected, Control SARS, and EtOH SARS, Control non-infected n = 3 for each, [Mann–Whitney test], trending p ≤ 0.1). d) Cluster frequency stratified by condition and timepoint (Error bars: Mean and SEM, n = 3 Control non-infected and n = 3 Control SARS [Paired-T test], trending p ≤ 0.1). e) Average expression of SARS-CoV-2 in the UMAP and compared between control and AUD groups. f) Bubbleplot of SARS-CoV-2 expression within each cluster stratified by timepoint. Average expression of the SARS-CoV-2 transcript is represented by bubble size while the significance of expression when comparing baseline to 6 months is denoted by color. g) Comparison of module scores of genes mapped to TLR signaling, cytokine/chemokine signaling, and viral/bacterial response, stratified by stimulation condition and collection timepoint (EtOH non-infected, Control SARS, and EtOH SARS, Control non-infected n = 3 for each, [Kruskal–Wallis 1-way ANOVA], ∗p ≤ 0.05, trending p ≤ 0.1). These data were collected over 2–3 independent experiments.
Similar to NHP, ethanol consumption resulted in transcriptional alteration of immune cell populations within the BAL. There was an increase in scores of modules associated with migration, oxidative stress, hypoxia, and signaling pathways (TLR, cytokine/chemokine, NFKB, HIF-1α) within myeloid cell populations (AM and monocytes) (Fig. S4c). Furthermore, DEGs in myeloid populations after ethanol consumption mapped to GO terms such as inflammatory response (CXCL1, CXCL3, FN1, LYZ) and positive regulation of cytokine production (CD36, CD84, IL-1α) (Fig. S4d and e). DEGs such as GNLY, CCL4, and GZMB were detected within lymphoid subsets (CD8) (Fig. S4e).
SARS-CoV-2 infection of BAL cells from non-AUD individuals was associated with modest increases in the secretion of VEGF, IL-15, CXCL10, and CCL11. AUD was associated with a modest increase in a larger number of immune mediators (PDGF-BB, IL-7, VEGF, IL-18, IL-15, CXCL10, CCL11) (Fig. 3c, Table S1). Infection with SARS-CoV-2 did not lead to significant alterations in cluster frequency other than a modest increase in AM1 in the control group (Fig. 3d).
As described for the macaque samples, SARS-CoV-2 transcripts were primarily detected in AM and infiltrating monocyte populations (Fig. 3e and f). In all AM clusters, except for AM5 and proliferating AMs, the expression of SARS-CoV-2 transcripts was higher in the AUD group compared to controls with viral transcripts more abundant in Mono1, Mono2, and DC (Fig. 3e and f). Module scores associated with TLR and cytokine/chemokine signaling were increased in the SARS-CoV-2 infected AUD samples within AM2 and Mono1 clusters respectively (Fig. 3g). Overall, DEGs in the AUD group played critical roles in anti-viral, inflammatory, and oxidative stress responses in both monocytes or CD8 T cells (Fig. S5a). Interestingly, several genes had different expression patterns with infection in AUD samples including ISG (ISG20, IFI6), cytokines (IL6, TNF), and chemokines (CXCL2, CXCL3, CCL22, CXCL10) in the monocytes and DCs clusters (Fig. S5b).
Discussion
Chronic heavy drinking has been associated with increased incidence and severity of respiratory infections.49, 50, 51 Shelter-in-place orders during the COVID-19 pandemic have led to increased alcohol consumption in the United States.40 Data from several recent studies showed increased COVID-19 susceptibility and disease severity that correlated with ethanol consumption.41, 42, 43 However, the molecular mechanisms underlying dysregulated responses to SARS-CoV-2 remain poorly understood. In this study, we investigated the cellular and transcriptional impact of chronic alcohol consumption on responses to SARS-CoV-2 infection using BAL samples from female and male rhesus macaques that self-administered ethanol for six months6,52, 53, 54, 55 and human male subjects with AUD.
In macaques, most immune cell populations decreased in relative frequency following six months of ethanol consumption, with the exception of AM. The increase in the relative frequency of AM was more pronounced in male macaques, which is in accordance with data from rodent models.56 Moreover, ethanol consumption led to a shift toward inflammatory AM and non-classical monocytes in line with heightened inflammation.3,28,52,57 We also report that chronic ethanol consumption skews the immune landscape towards inflammatory responses, as indicated by increased expression of genes within the HIF1α as well as TLR signaling pathways.
SARS-CoV-2 transcripts were primarily detected within myeloid subsets in BAL. These observations align with earlier studies that have reported the detection of SARS-CoV-2 viral RNA or transcripts in BAL58, 59, 60 as well as scRNA-seq analysis of whole lung tissue that reported SARS-CoV-2 viral reads within macrophage, endothelial cells, DC, and monocyte populations.60 Studies have also shown that AM can express ACE261,62 and TMPRSS,62 both required for SARS-CoV-2 infection. However, it remains unclear whether monocytes/macrophages can support SARS-CoV-2 replication or whether the presence of the SARS-CoV-2 transcripts is simply a consequence of phagocytosis of dying infected cells or viral particles.63
In line with the detection of SARS-CoV-2 transcripts, the most significant transcriptional differences after infection were detected within myeloid subsets. Chronic ethanol consumption led to greater induction of TLR signaling pathways in line with previous studies that showed chronic inflammation increases oxidative stress and activates TLR signaling cascades.64,65 In contrast, after 6 months of chronic drinking, the induction of genes important for anti-viral responses was reduced across multiple subsets compared to baseline. Reduced ISG expression could be a result of reduced production of IFNβ. These data are similar to those from previous studies using the same rhesus macaque model of self-administration, which reported heightened pro-inflammatory responses and dampened expression of ISG in AM subsets in response to RSV infection.3 Decreased production of immune mediators involved in tissue repair mechanisms, such as VEGF, suggests a decreased tissue repair capacity.66 Reduced antiviral responses and tissue repair capacity may explain the exacerbated SARS-CoV-2 disease severity documented among AUD patients.42
Although T cells harbored very few SARS-CoV-2 transcripts, the expression of genes important for chemotaxis, anti-viral responses, cytokine signaling, and apoptosis were altered. These transcriptional responses are likely secondary to the inflammatory responses generated by the myeloid cells after CHD and align with previous reports of impaired T cell function after alcohol exposure.67, 68, 69 The lack of induction of genes mapping to leukocyte migration could be explained by the decrease in T-cell chemoattractant IL-15 following CHD.70
Myeloid cells were also the predominant cell type to harbor SARS-CoV-2 transcripts in BAL samples from humans with AUD. However, inflammatory mediators were produced at greater levels in samples from individuals with AUD compared to the control smoking group only. This differs from the data obtained with NHP control BAL samples which were obtained before beginning of ethanol self-administration, suggesting that smoking may abrogate inflammatory responses. Indeed, previous work has shown that tobacco smoking reduces both gene expression and production of proinflammatory cytokines (TNFα, IL-1β, and IL-16) in alveolar macrophages stimulated with TLR2 and TLR4 agonists.71 Additional studies utilizing electrochemiluminescence techniques to measure immune mediator production indicated that smoking with AUD resulted in decreased AM immune mediator responses to bacterial stimuli.72
This study is not without its limitations. Our work utilizes the SARS-CoV-2 USA-AZ1/2020 strain which has significantly mutated since these experiments were conducted. Future studies should examine the acute innate immune response to variants of concern, notably Delta and currently circulating Omicron. In addition, our study utilized samples from young adult rhesus macaques and humans. It is well established that increased age is associated with increased disease severity. Therefore, repeating these experiments with BAL samples from aged individuals would also be a critical future step. We were not able to assess sex as a biological variable for the human BAL samples due to limitations of the available samples. Moreover, all human BAL samples were obtained from individuals who smoked since smoking is often an associated comorbidity with AUD. Vaccination and prior infection can modulate subsequent responses to pathogens so interrogating these variables is also a crucial next step. Furthermore, the impact of CHD on the adaptive branch (T and B cells) is understudied in general, especially at mucosal sites such as the lung. Last but not least, it is now well established that the lung is not a sterile site but rather home to a unique microbial community. CHD may lead to dysbiosis of this community, which in turn can impact immune fitness at this site. Future studies should address how prior immunity, lymphocyte function and the microbiome are impacted by CHD and how those changes alter responses to SARS-CoV-2.
In summary, we show that CHD dysregulates acute innate immune response to SARS-CoV-2 in the lung. The innate branch of the immune system is a key modulator of COVID-19 severity and clinical outcomes.73 CHD reprograms innate immune cells, skewing their ability to respond to pathogens appropriately and efficiently by decreasing antiviral cytokines and transcriptionally altering the viral recognition and response pathways. Such reprogramming can induce aberrant inflammation and reduced antimicrobial responses.54 Our study supports this conclusion with the demonstration that impaired response to pathogens involves CHD-induced transcriptional changes that then increase both susceptibility and severity of SARS-CoV-2 infection.
Contributors
S.A.L., K.A.G., and I.M. conceived and designed the experiments. S.A.L., C.H., N.N, M.D. and B.D., performed the experiments. S.A.L., I.C., and M.B. analyzed the data. M.B., I.C., and I.M. wrote the paper. All authors have read and approved the final draft of the manuscript. S.A.L., I.C., B.D., M.B., and I.M. have accessed and verified the data.
Data sharing statement
The datasets utilized in this article are available on NCBI's Sequence Read Archive under project accession numbers PRJNA930878 (NHP) and PRJNA930381 (Human). Supplemental tables can be found on the online repository Mendeley Data under https://doi.org/10.17632/f44h3v2fn8.3 (Table S1) and https://doi.org/10.17632/729m9vxf4v.2 (Table S2).
Declaration of interests
There are no competing interests to report.
Acknowledgements
We would like to thank Dr. Delphine Malherbe for critical reading of the manuscript, all the members of Dr. Kathy Grant for expert animal care, the Division of Comparative Medicine at the Oregon National Primate Center for sample collection, and Dr. Ellen Burnham's team for providing human BAL samples.
We would also like to acknowledge the funding sources supporting this study: NIH 1R01AA028735-04 (Messaoudi), U01AA013510-20 (Grant), R24AA019431-14 (Grant), R24AA019661 (Burnham), P-51OD011092 (ONPRC core grant support). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104812.
Appendix A. Supplementary data
Fig. S1
References
- 1.Alcoholism NIoAAa Understanding alcohol use disorder 2020. https://www.niaaa.nih.gov/publications/brochures-and-fact-sheets/understanding-alcohol-use-disorder Available from:
- 2.SAMHSA CfBHSaQ Table 5.4A – alcohol use disorder in past year among persons aged 12 or older, by age group and demographic characteristics: numbers in thousands, 2018 and 2019. 2019. https://www.samhsa.gov/data/sites/default/files/reports/rpt29394/NSDUHDetailedTabs2019/NSDUHDetTabsSect5pe2019.htm#tab5-4a Available from:
- 3.Lewis S.A., Doratt B.M., Sureshchandra S., et al. Ethanol consumption induces nonspecific inflammation and functional defects in alveolar macrophages. Am J Respir Cell Mol Biol. 2022;67(1):112–124. doi: 10.1165/rcmb.2021-0346OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruha R., Dvorak K., Petrtyl J. Alcoholic liver disease. World J Hepatol. 2012;4(3):81–90. doi: 10.4254/wjh.v4.i3.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Detlefsen S., Sipos B., Feyerabend B., Klöppel G. Fibrogenesis in alcoholic chronic pancreatitis: the role of tissue necrosis, macrophages, myofibroblasts and cytokines. Mod Pathol. 2006;19(8):1019–1026. doi: 10.1038/modpathol.3800613. [DOI] [PubMed] [Google Scholar]
- 6.Sureshchandra S., Stull C., Ligh B.J.K., Nguyen S.B., Grant K.A., Messaoudi I. Chronic heavy drinking drives distinct transcriptional and epigenetic changes in splenic macrophages. eBioMedicine. 2019;43:594–606. doi: 10.1016/j.ebiom.2019.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Keefe J.H., Bhatti S.K., Bajwa A., DiNicolantonio J.J., Lavie C.J. Alcohol and cardiovascular health: the dose makes the poison…or the remedy. Mayo Clin Proc. 2014;89(3):382–393. doi: 10.1016/j.mayocp.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Mukamal K.J., Rimm E.B. Alcohol's effects on the risk for coronary heart disease. Alcohol Res Health. 2001;25(4):255–261. [PMC free article] [PubMed] [Google Scholar]
- 9.Rumgay H., Shield K., Charvat H., et al. Global burden of cancer in 2020 attributable to alcohol consumption: a population-based study. Lancet Oncol. 2021;22(8):1071–1080. doi: 10.1016/S1470-2045(21)00279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freedman N.D., Schatzkin A., Leitzmann M.F., Hollenbeck A.R., Abnet C.C. Alcohol and head and neck cancer risk in a prospective study. Br J Cancer. 2007;96(9):1469–1474. doi: 10.1038/sj.bjc.6603713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen N.E., Beral V., Casabonne D., et al. Moderate alcohol intake and cancer incidence in women. J Natl Cancer Inst. 2009;101(5):296–305. doi: 10.1093/jnci/djn514. [DOI] [PubMed] [Google Scholar]
- 12.Smothers B.A., Yahr H.T. Alcohol use disorder and illicit Drug use in admissions to general hospitals in the United States. Am J Addict. 2005;14(3):256–267. doi: 10.1080/10550490590949433. [DOI] [PubMed] [Google Scholar]
- 13.de Wit M., Zilberberg M.D., Boehmler J.M., Bearman G.M., Edmond M.B. Outcomes of patients with alcohol use disorders experiencing healthcare-associated infections. Alcohol Clin Exp Res. 2011;35(7):1368–1373. doi: 10.1111/j.1530-0277.2011.01475.x. [DOI] [PubMed] [Google Scholar]
- 14.Delgado-Rodríguez M., Gómez-Ortega A., Mariscal-Ortiz M., Palma-Pérez S., Sillero-Arenas M. Alcohol drinking as a predictor of intensive care and hospital mortality in general surgery: a prospective study. Addiction. 2003;98(5):611–616. doi: 10.1046/j.1360-0443.2003.00353.x. [DOI] [PubMed] [Google Scholar]
- 15.Trevejo-Nunez G., Kolls J.K., de Wit M. Alcohol use as a risk factor in infections and healing: a clinician's perspective. Alcohol Res. 2015;37(2):177–184. [PMC free article] [PubMed] [Google Scholar]
- 16.Gentilello L.M., Cobean R.A., Walker A.P., Moore E.E., Wertz M.J., Dellinger E.P. Acute ethanol intoxication increases the risk of infection following penetrating abdominal trauma. J Trauma. 1993;34(5):669–674. doi: 10.1097/00005373-199305000-00009. discussion 74-5. [DOI] [PubMed] [Google Scholar]
- 17.Prevention CfDCa The cost of excessive alcohol use. https://www.cdc.gov/alcohol/onlinemedia/infographics/cost-excessive-alcohol-use.html Available from:
- 18.Sacks J.J., Gonzales K.R., Bouchery E.E., Tomedi L.E., Brewer R.D. 2010 National and state costs of excessive alcohol consumption. Am J Prev Med. 2015;49(5):e73–e79. doi: 10.1016/j.amepre.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 19.Prevention CfDCa Deaths from excessive alcohol use in the United States. 2022. https://www.cdc.gov/alcohol/features/excessive-alcohol-deaths.html Available from:
- 20.Kershaw C.D., Guidot D.M. Alcoholic lung disease. Alcohol Res Health. 2008;31(1):66–75. [PMC free article] [PubMed] [Google Scholar]
- 21.Jung M.K., Callaci J.J., Lauing K.L., et al. Alcohol exposure and mechanisms of tissue injury and repair. Alcohol Clin Exp Res. 2011;35(3):392–399. doi: 10.1111/j.1530-0277.2010.01356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malherbe D.C., Messaoudi I. Transcriptional and epigenetic regulation of monocyte and macrophage dysfunction by chronic alcohol consumption. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.911951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szabo G., Saha B. Alcohol's effect on host defense. Alcohol Res. 2015;37(2):159–170. [PMC free article] [PubMed] [Google Scholar]
- 24.Jerrells T.R., Pavlik J.A., DeVasure J., et al. Association of chronic alcohol consumption and increased susceptibility to and pathogenic effects of pulmonary infection with respiratory syncytial virus in mice. Alcohol. 2007;41(5):357–369. doi: 10.1016/j.alcohol.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simet S.M., Sisson J.H. Alcohol's effects on lung health and immunity. Alcohol Res. 2015;37(2):199–208. [PMC free article] [PubMed] [Google Scholar]
- 26.Meadows G.G., Wallendal M., Kosugi A., Wunderlich J., Singer D.S. Ethanol induces marked changes in lymphocyte populations and natural killer cell activity in mice. Alcohol Clin Exp Res. 1992;16(3):474–479. doi: 10.1111/j.1530-0277.1992.tb01403.x. [DOI] [PubMed] [Google Scholar]
- 27.Osterndorff-Kahanek E., Ponomarev I., Blednov Y.A., Harris R.A. Gene expression in brain and liver produced by three different regimens of alcohol consumption in mice: comparison with immune activation. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0059870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burnham E.L., Kovacs E.J., Davis C.S. Pulmonary cytokine composition differs in the setting of alcohol use disorders and cigarette smoking. Am J Physiol Lung Cell Mol Physiol. 2013;304(12):L873–L882. doi: 10.1152/ajplung.00385.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolls J.K., Xie J., Lei D., Greenberg S., Summer W.R., Nelson S. Differential effects of in vivo ethanol on LPS-induced TNF and nitric oxide production in the lung. Am J Physiol Lung Cell Mol Physiol. 1995;268(6):L991–L998. doi: 10.1152/ajplung.1995.268.6.L991. [DOI] [PubMed] [Google Scholar]
- 30.Mandrekar P., Dolganiuc A., Bellerose G., et al. Acute alcohol inhibits the induction of nuclear regulatory factor kappaB activation through CD14/toll-like receptor 4, interleukin-1, and tumor necrosis factor receptors: a common mechanism independent of inhibitory kappaBalpha degradation? Alcohol Clin Exp Res. 2002;26(11):1609–1614. doi: 10.1097/01.ALC.0000036926.46632.57. [DOI] [PubMed] [Google Scholar]
- 31.Karavitis J., Kovacs E.J. Macrophage phagocytosis: effects of environmental pollutants, alcohol, cigarette smoke, and other external factors. J Leukoc Biol. 2011;90(6):1065–1078. doi: 10.1189/jlb.0311114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gandhi J.A., Ekhar V.V., Asplund M.B., et al. Alcohol enhances Acinetobacter baumannii-associated pneumonia and systemic dissemination by impairing neutrophil antimicrobial activity in a murine model of infection. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0095707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szabo G., Mandrekar P., Catalano D. Inhibition of superantigen-induced T cell proliferation and monocyte IL-1 beta, TNF-alpha, and IL-6 production by acute ethanol treatment. J Leukoc Biol. 1995;58(3):342–350. doi: 10.1002/jlb.58.3.342. [DOI] [PubMed] [Google Scholar]
- 34.Joshi P.C., Applewhite L., Ritzenthaler J.D., et al. Chronic ethanol ingestion in rats decreases granulocyte-macrophage colony-stimulating factor receptor expression and downstream signaling in the alveolar macrophage. J Immunol. 2005;175(10):6837–6845. doi: 10.4049/jimmunol.175.10.6837. [DOI] [PubMed] [Google Scholar]
- 35.Staitieh B.S., Egea E.E., Fan X., Amah A., Guidot D.M. Chronic alcohol ingestion impairs rat alveolar macrophage phagocytosis via disruption of RAGE signaling. Am J Med Sci. 2018;355(5):497–505. doi: 10.1016/j.amjms.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guilliams M., De Kleer I., Henri S., et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210(10):1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simet S.M., Wyatt T.A., Devasure J., Yanov D., Allen-Gipson D., Sisson J.H. Alcohol increases the permeability of airway epithelial tight junctions in beas-2B and NHBE cells. Alcohol Clin Exp Res. 2012;36(3):432–442. doi: 10.1111/j.1530-0277.2011.01640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Y., Yeligar S.M., Brown L.A.S. Chronic-alcohol-Abuse-induced oxidative stress in the development of acute respiratory distress syndrome. Sci World J. 2012;2012:1–9. doi: 10.1100/2012/740308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modrykamien A.M., Gupta P. The acute respiratory distress syndrome. Proc (Bayl Univ Med Cent) 2015;28(2):163–171. doi: 10.1080/08998280.2015.11929219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Administration SAaMHS 2020 national Survey of Drug use and health (NSDUH) releases. https://www.samhsa.gov/data/release/2020-national-survey-drug-use-and-health-nsduh-releases Available from:
- 41.Dai X.-j., Tan L., Ren L., Shao Y., Tao W., Wang Y. COVID-19 risk appears to vary across different alcoholic beverages. Front Nutr. 2022;8 doi: 10.3389/fnut.2021.772700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhalla S., Sharma B., Smith D., et al. Investigating unhealthy alcohol use as an independent risk factor for increased COVID-19 disease severity: observational cross-sectional study. JMIR Public Health Surveill. 2021;7(11) doi: 10.2196/33022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei B., Liu Y., Li H., Peng Y., Luo Z. Impact of alcohol consumption on coronavirus disease 2019 severity: a systematic review and meta-analysis. J Med Virol. 2023;95(2) doi: 10.1002/jmv.28547. [DOI] [PubMed] [Google Scholar]
- 44.Grant K.A., Leng X., Green H.L., Szeliga K.T., Rogers L.S., Gonzales S.W. Drinking typography established by scheduled induction predicts chronic heavy drinking in a monkey model of ethanol self-administration. Alcohol Clin Exp Res. 2008;32(10):1824–1838. doi: 10.1111/j.1530-0277.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker E.J., Farro J., Gonzales S., Helms C., Grant K.A. Chronic alcohol self-administration in monkeys shows long-term quantity/frequency categorical stability. Alcohol Clin Exp Res. 2014;38(11):2835–2843. doi: 10.1111/acer.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jureka A.S., Silvas J.A., Basler C.F. Propagation, inactivation, and safety testing of SARS-CoV-2. Viruses. 2020;12(6):622. doi: 10.3390/v12060622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hao Y., Hao S., Andersen-Nissen E., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y., Zhou B., Pache L., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey K.L., Sayles H., Campbell J., et al. COVID-19 patients with documented alcohol use disorder or alcohol-related complications are more likely to be hospitalized and have higher all-cause mortality. Alcohol Clin Exp Res. 2022;46(6):1023–1035. doi: 10.1111/acer.14838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mehta A.J., Yeligar S.M., Elon L., Brown L.A., Guidot D.M. Alcoholism causes alveolar macrophage zinc deficiency and immune dysfunction. Am J Respir Crit Care Med. 2013;188(6):716–723. doi: 10.1164/rccm.201301-0061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeligar S.M., Chen M.M., Kovacs E.J., Sisson J.H., Burnham E.L., Brown L.A. Alcohol and lung injury and immunity. Alcohol. 2016;55:51–59. doi: 10.1016/j.alcohol.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lewis S.A., Sureshchandra S., Doratt B., et al. Transcriptional, epigenetic, and functional reprogramming of monocytes from non-human primates following chronic alcohol drinking. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.724015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sureshchandra S., Raus A., Jankeel A., et al. Dose-dependent effects of chronic alcohol drinking on peripheral immune responses. Sci Rep. 2019;9(1):7847. doi: 10.1038/s41598-019-44302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barr T., Girke T., Sureshchandra S., Nguyen C., Grant K., Messaoudi I. Alcohol consumption modulates host defense in rhesus macaques by altering gene expression in circulating leukocytes. J Immunol. 2016;196(1):182–195. doi: 10.4049/jimmunol.1501527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barr T., Lewis S.A., Sureshchandra S., Doratt B., Grant K.A., Messaoudi I. Chronic ethanol consumption alters lamina propria leukocyte response to stimulation in a region-dependent manner. Faseb J. 2019;33(6):7767–7777. doi: 10.1096/fj.201802780R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spitzer J.A. Gender differences in nitric oxide production by alveolar macrophages in ethanol plus lipopolysaccharide-treated rats. Nitric Oxide. 1997;1(1):31–38. doi: 10.1006/niox.1996.0101. [DOI] [PubMed] [Google Scholar]
- 57.O'Halloran E.B., Curtis B.J., Afshar M., Chen M.M., Kovacs E.J., Burnham E.L. Alveolar macrophage inflammatory mediator expression is elevated in the setting of alcohol use disorders. Alcohol. 2016;50:43–50. doi: 10.1016/j.alcohol.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiong Y., Liu Y., Cao L., et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microb Infect. 2020;9(1):761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Speranza E., Purushotham J.N., Port J.R., et al. Age-related differences in immune dynamics during SARS-CoV-2 infection in rhesus macaques. Life Sci Alliance. 2022;5(4) doi: 10.26508/lsa.202101314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Speranza E., Williamson B.N., Feldmann F., et al. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. Sci Transl Med. 2021;13(578) doi: 10.1126/scitranslmed.abe8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang C., Xie J., Zhao L., et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. eBioMedicine. 2020;57 doi: 10.1016/j.ebiom.2020.102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bertram S., Heurich A., Lavender H., et al. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stegelmeier A.A., van Vloten J.P., Mould R.C., et al. Myeloid cells during viral infections and inflammation. Viruses. 2019;11(2):168. doi: 10.3390/v11020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.El-Zayat S.R., Sibaii H., Mannaa F.A. Toll-like receptors activation, signaling, and targeting: an overview. Bull Natl Res Cent. 2019;43:187. [Google Scholar]
- 65.Hussain T., Tan B., Yin Y., Blachier F., Tossou M.C., Rahu N. Oxidative stress and inflammation: what polyphenols can do for us? Oxid Med Cell Longev. 2016;2016 doi: 10.1155/2016/7432797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eming S.A., Krieg T. Molecular mechanisms of VEGF-A action during tissue repair. J Invest Dermatol Symp Proc. 2006;11(1):79–86. doi: 10.1038/sj.jidsymp.5650016. [DOI] [PubMed] [Google Scholar]
- 67.Kapasi A.A., Patel G., Goenka A., et al. Ethanol promotes T cell apoptosis through the mitochondrial pathway. Immunology. 2003;108(3):313–320. doi: 10.1046/j.1365-2567.2003.01584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hemann E.A., McGill J.L., Legge K.L. Chronic ethanol exposure selectively inhibits the influenza-specific CD8 T cell response during influenza a virus infection. Alcohol Clin Exp Res. 2014;38(9):2403–2413. doi: 10.1111/acer.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pasala S., Barr T., Messaoudi I. Impact of alcohol abuse on the adaptive immune system. Alcohol Res. 2015;37(2):185–197. [PMC free article] [PubMed] [Google Scholar]
- 70.Wilkinson P.C., Liew F.Y. Chemoattraction of human blood T lymphocytes by interleukin-15. J Exp Med. 1995;181(3):1255–1259. doi: 10.1084/jem.181.3.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen H., Cowan M.J., Hasday J.D., Vogel S.N., Medvedev A.E. Tobacco smoking inhibits expression of proinflammatory cytokines and activation of IL-1R-associated kinase, p38, and NF-kappaB in alveolar macrophages stimulated with TLR2 and TLR4 agonists. J Immunol. 2007;179(9):6097–6106. doi: 10.4049/jimmunol.179.9.6097. [DOI] [PubMed] [Google Scholar]
- 72.Gaydos J., McNally A., Guo R., Vandivier R.W., Simonian P.L., Burnham E.L. Alcohol abuse and smoking alter inflammatory mediator production by pulmonary and systemic immune cells. Am J Physiol Lung Cell Mol Physiol. 2016;310(6):L507–L518. doi: 10.1152/ajplung.00242.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diamond M.S., Kanneganti T.-D. Innate immunity: the first line of defense against SARS-CoV-2. Nat Immunol. 2022;23(2):165–176. doi: 10.1038/s41590-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1