Abstract
Biological age captures a person’s age-related risk of unfavorable outcomes using biophysiological information. Multivariate biological age measures include frailty scores and molecular biomarkers. These measures are often studied in isolation, but here we present a large-scale study comparing them. In 2 prospective cohorts (n = 3 222), we compared epigenetic (DNAm Horvath, DNAm Hannum, DNAm Lin, DNAm epiTOC, DNAm PhenoAge, DNAm DunedinPoAm, DNAm GrimAge, and DNAm Zhang) and metabolomic-based (MetaboAge and MetaboHealth) biomarkers in reflection of biological age, as represented by 5 frailty measures and overall mortality. Biomarkers trained on outcomes with biophysiological and/or mortality information outperformed age-trained biomarkers in frailty reflection and mortality prediction. DNAm GrimAge and MetaboHealth, trained on mortality, showed the strongest association with these outcomes. The associations of DNAm GrimAge and MetaboHealth with frailty and mortality were independent of each other and of the frailty score mimicking clinical geriatric assessment. Epigenetic, metabolomic, and clinical biological age markers seem to capture different aspects of aging. These findings suggest that mortality-trained molecular markers may provide novel phenotype reflecting biological age and strengthen current clinical geriatric health and well-being assessment.
Keywords: DNA methylation, Frailty, Mortality
Age is the most prominent risk indicator for common chronic diseases, frailty, and mortality (1–3). However, there are large interindividual differences in the biological aging process and rate of functional decline. Hence, standardized markers that reflect biological age and can provide aging rate phenotypes to be studied in depth are needed in aging research. Geriatricians use the comprehensive geriatric assessment (CGA) to identify the medical, social, and functional needs of older patients (4) and determine whether invasive treatments are suitable for older patients (5). Although considered the gold standard for treating frail patients (4), CGA is time- and resource-consuming, and primarily narrative based (4). To date, no consensus exists for a multivariate molecular biomarker to accurately capture the complexity of the aging process and serve as a biological age phenotype for aging research or an overall health indicator in the clinic.
Simultaneously, consensus is lacking on the operationalization of frailty in research practice, leading to the introduction of a variety of frailty measures with their own approaches (2,6–10). The frailty index (FI) assesses frailty as an accumulation of deficits over a wide range of health domains (11), while the frailty phenotype (FP), also known as Fried frailty (2), is a widely used measure that focuses on physical frailty. Recently, the FP has been translated into a continuous score called continuous frailty phenotype (CFP) (7). The Tilburg Frailty Indicator (TFI) is metric combining the physical, psychological, and social domains (8). Finally, the Multidimensional Prognostic Index (MPI) (9) quantifies the CGA, providing information on the medical, social, and functional status of participants.
Furthermore, several attempts have been made to capture the discrepancy between an individual’s chronological age and their age based on biological and clinical information, known as biological age, in a biomarker. In the past 2 decades, large-scale molecular data were used to develop several molecular markers of biological age based on, for example, telomeres, DNA methylation (DNAm), and metabolomics (12). Well-known are the DNAm or epigenetic aging clocks. These epigenetic aging clocks are biomarkers based on methylation values at a combination of specific CpG sites by which chronological age is best reflected. The first-generation epigenetic aging clocks, DNAm Horvath (13) and DNAm Hannum (14), DNAm Lin (15) were trained on chronological age and outperformed other aging biomarkers, such as telomere length, in the reflection and prediction of the aging process (12). To develop a mitotic-age biomarker that is correlated with chronological age, DNAm epiTOC was trained on chronological age using CpGs that map to Polycomb group target gene promoters, which are constitutively unmethylated in fetal tissue (16). Since physiological deficits resulting from and contributing to aging do not develop in a regular, clock-like manner, the second-generation epigenetic aging biomarkers trained CpG-models on outcomes that incorporate information on biopsychology or mortality, or both. DNAm DunedinPoAm (17) was trained on the Pace of Aging (18), a score based on 18 biomarkers measured 3 times between the ages of 26 and 38 years. DNAm PhenoAge was trained on a multi-system proxy for physiological dysregulation (19). DNAm GrimAge (20) and DNAm Zhang (21) were trained on mortality risk. More recently, metabolomics-based aging biomarkers were established using a nuclear magnetic resonance platform (22,23). These biomarkers of biological age were trained on either chronological age (MetaboAge (24)) or mortality (MetaboHealth (25)).
Previous studies have shown that second-generation aging biomarkers outperform the first-generation epigenetic aging biomarkers in reflecting frailty, physical health outcomes, cognitive and physical capacity, and prediction of overall mortality (26–28). However, the performances of the newly developed metabolomic biological age biomarkers have not been compared with either the first- or second-generation epigenetic aging biomarkers. Moreover, whether epigenetic and metabolomic aging biomarkers capture different aspects of the aging process is unknown. Lastly, it is unclear whether aging biomarkers have added value to the CGA and, thus, their possible clinical applicability.
The current study compares the reflection of biological age of the first and second-generation epigenetic and metabolomic aging biomarkers by determining their association with 5 different frailty scores and with mortality. These outcomes largely reflect the aging process.
Method
Study Cohorts
The current study is a nested cohort study of data from the second and third cohorts of the population-based Rotterdam Study (RS) (29) and the second generation of the Leiden Longevity Study (LLS) (30). In the RS (29)1 347 participants were grouped into 2 subcohorts based on the platform of their epigenetic data, 450K-data (n = 611) or EPIC-data (n = 736). From the LLS, 1 875 participants with metabolomic information were selected as the external validation cohort for our findings of whom in a subcohort of 591 participants additional information on DNAm was available. A more detailed description of both cohorts and inclusion criteria can be found in the Supplementary Methods.
DNA Methylation
Genome-wide DNAm data was obtained from whole blood. In 687 RS participants and the LLS, we analyzed the samples using Illumina Infinium Human Methylation 450 K (450K) array (31,32). In the other 737 RS samples, we used Illumina Infinium MethylationEPIC BeadChip v1 manifest B5 (EPIC) arrays (33). The quality control procedures are described in the Supplementary Methods.
Metabolomics
Metabolomic biomarkers from EDTA plasma were measured using high-throughput NMR metabolomics (Nightingale Health Ltd., Helsinki, Finland; biomarker quantification version 2016) (23). This technique quantifies over 200 metabolic measures, including routine lipids, lipoprotein subclass profiling with lipid concentrations within 14 subclasses, fatty acid composition, and various low-molecular-weight metabolites in molar concentration units (22,23).
Biomarkers of Biological Aging
We calculated DNAm Horvath (13), DNAm Hannum (14), DNAm Lin (15), DNAm epiTOC (16), DNAm PhenoAge (19), DNAm DunedinPoAm (17), DNAm GrimAge (20), and DNAm Zhang (21) using the coefficients, R and Python scripts, and packages provided by the researchers who developed these measures and the R methylclock package (34). To calculate epigenetic aging biomarkers, missing information for 3 339 CpG sites on the EPIC array and 2 831 sites in RS and 1 638 in LLS on the 450K-array was imputed with the mean value from the GOLD consortium, as previously described (35). Unfortunately, 2 out of the 10 CpG-sites needed to calculate DNAm Zhang were missing on both our EPIC arrays as in the GOLD consortium. For these 2 CpGs, we imputed the mean value from the 450K-subcohort, where information on all 10 CpGs was present. The metabolomic biomarkers were used to compose MetaboAge (24) and MetaboHealth (25). MetaboAge and MetaboHealth were calculated using MiMIR (36), the dedicated R-shiny package, on the raw metabolomic biomarkers (24,25)
Finally, we calculated the chronological age-independent part of the above-mentioned variables that we defined as the biomarkers of biological aging to use in all analyses in this study. We did so by taking the residual from the linear regression model of chronological age on the before-mentioned epigenetic and metabolomic variables. The presented biomarkers, thus, represent the chronological age-independent part of the biomarkers.
Assessment of Mortality
Based on a linkage with the mortality registry of the municipality and the digitally connected medical records of the GPs working in the study area, we gathered information on the vital status of the participants on a bimonthly basis (37). The information on the vital status of participants in Rotterdam was last updated on the 20th of October 2022.
The vital status of the participants in the LLS was updated in January 2021 through the Personal Records Database, which is managed by the Dutch governmental service for identity information (38).
Frailty Assessment
We used interviews, physical examinations, blood sampling, and general practitioners’ records to obtain information on the participants’ frailty. Using this information, we constructed the FP (2), CFP (7), FI (6), TFI (39), and MPI (9). A more detailed description of the construction of these 5 frailty measures and the literature-described cut-offs to classify participants as either frail or nonfrail can be found in the Supplementary Methods.
Assessment of Covariates
A questionnaire at baseline provided information on the sex and chronological age at blood sampling for all participants. We weighted and measured participants when they visited the research center; based on this information, BMI was calculated (kg/m2). We classified participants as smokers or nonsmokers based on the answer to the question: “are you currently smoking?.” We defined cell counts as the measured white blood count percentage of lymphocytes and monocytes, making the percentage of granulocytes a given. Socioeconomic status was defined based on the highest level of attained education following UNESCO classification (40).
Statistical Analysis
The biomarkers of biological aging were constructed per dataset by calculating the residual of a linear regression of chronological age on the epigenetic and metabolomics measures. Spearman’s rank correlation was used to assess the correlation between the biomarkers of biological aging and frailty scores, and a Yeo–Johnson transformation using the bestNormalize R-package (41) was applied to the frailty indices to increase homoscedasticity. We decided upon using Yeo–Johnson transformations as it can incorporate zeroes (41), which were informative in our frailty scoring system and thus should not be lost in power transformations. Linear regression models were used to determine the association between cross-sectional continuous Yeo–Johnson transformed Z-scored frailty and Z-scored biological aging biomarkers. Standardization was performed in the subcohorts separately for the subcohort analyses and in the combined information on all participants for the analyses involving the overall study population. Z-scores were used to improve comparability of effect sizes. Logistic regression analyses were used for the associations with frailty as binary outcome. The linear and logistic regression analyses were adjusted for chronological age at blood sampling, sex, cell counts, BMI, and visit and cohort within RS. The analyses in the entire RS study population were additionally adjusted for underlying subcohort and, thereby, methylation array and metabolomics batch used. Moreover, a sensitivity analysis was conducted, in which adjustments were made for smoking status and socioeconomic status. Additionally, we tested for effect modification of sex, and a sex-stratified sensitivity analysis was performed for all aforementioned analyses.
We used the R-package survival (42) to create Cox Proportional hazard regression models to determine the association between standardized aging predictors and overall mortality. We used chronological age at blood sampling as the starting point and chronological age at censoring as the endpoint of the analysis. We performed the analyses in 4 models in the RS and validated only the first 2 models in LLS as the MPI was not available in the LLS. The first model adjusted for sex, cell counts, BMI, and study-specific covariates. In the second model, we additionally adjusted for smoking status and socioeconomic status. Thirdly, we adjusted for the same covariates as in the first model, but additionally for the MPI. The MPI mimics the CGA, as used in the clinic, best. Adjusting for the MPI provides an opportunity to determine the value of aging predictors beyond ongoing practice. Fourthly, we additionally adjusted the third model for smoking status and socioeconomic status. Moreover, we performed sensitivity analyses to examine effect modification by including interaction terms with sex in the model. Additionally, we conducted survival analyses stratified by sex. We determined the association between frailty measures per standard deviation increase and overall mortality with a Cox Proportional Hazard model with again age at blood sampling and age at censoring as timescale. We adjusted the analyses for the first 2 abovementioned models.
To investigate the role of frailty in linking biomarkers and mortality, we performed a formal mediation analysis using the mediation R-package (43) with all-cause mortality as main outcome. We have fitted a parametric regression with Weibull distribution using the survival R-package (42) and a linear regression. To correct for multiple comparisons, we applied a Benjamini–Hochberg false discovery rate (FDR) correction (44). We performed all analyses in R version 4.1.3. Figure 1 was created with BioRender.com.
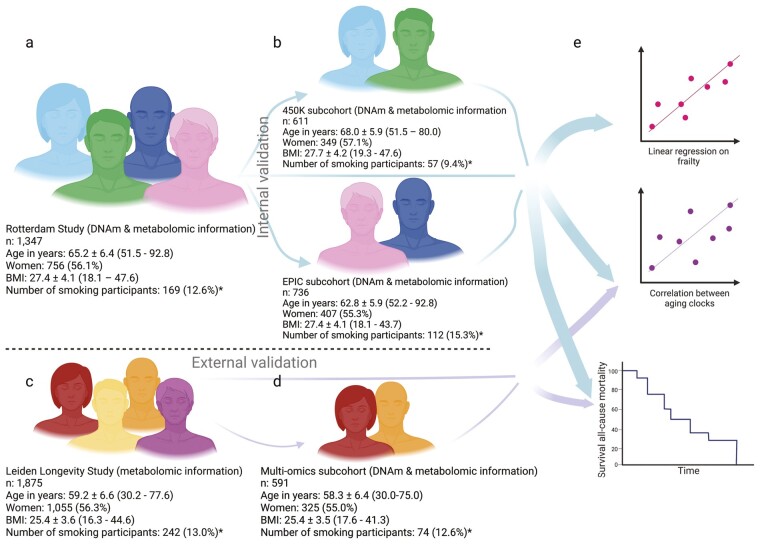
Figure 1.
Outline of the study and study population characteristics. *Smoking status was unknown for 5 Rotterdam Study participants (3 participants of the 450K-subcohort, 2 participants of the EPIC-subcohort) and for 17 LLS participants, of whom 4 belonged to the multiomics subcohort. (A) The Rotterdam Study overall study population with population characteristics; (B) the Rotterdam Study 2 subcohorts, 450K and EPIC, stratified by the DNA methylation array used and their population characteristics; (C) the external validation cohort, the Leiden Longevity Study with population characteristics; (D) the subcohort of the Leiden Longevity Study, where epigenetic information was available with population characteristics; and (E) we used both the overall Rotterdam Study population and its two subcohorts (i) to determine the correlations between each of the biological aging biomarkers, (ii) to perform a linear regression for the association between the biological aging biomarkers and frailty, and (iii) to determine the association between each of the biological aging biomarkers and all-cause mortality. We externally validated the correlations between the biological aging biomarkers and the association between the biological aging biomarkers and all-cause mortality in the Leiden Longevity Study and its subcohort. In A–D, BMI indicates body mass index; DNAm, DNA methylation; and n, size of the study population. Population characteristics in A–D are shown as a number for the population size; mean ± standard deviation (range) for age and BMI; and number (percentage) for the number of women and the number of participants currently smoking.
Results
We used 2 distinct cohorts for our analyses: the RS (29), a population-based study, and the LLS (30), a long-living family study (Figure 1, Supplementary Table 1). The RS was separated into 2 subcohorts, where the distinguishing factor was the DNAm array used, either 450K (n = 611) or EPIC (n = 736). For the LLS, the study population consisted of all participants with metabolomics information (n = 1 849) with a multiomics subcohort (n = 584) of offspring and their partners from families without a family history of longevity, thereby selecting a subcohort closest to the population at large (45). We calculated the biomarkers of biological aging as the age-independent part of the aging biomarkers and used this metric in all further analyses (Methods: Biomarkers of biological aging).
Correlation Between Biological Aging Biomarkers
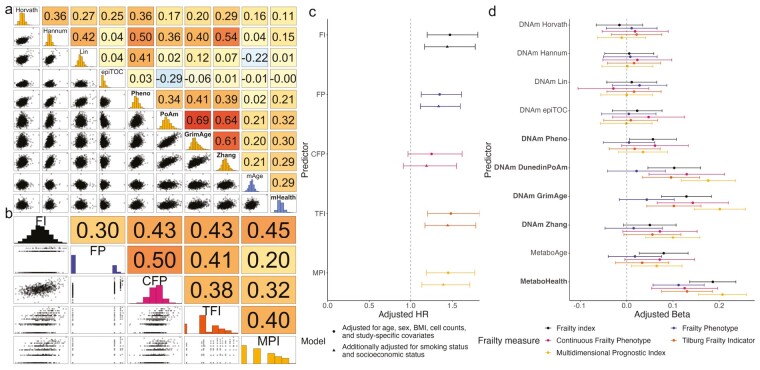
Spearman’s rank correlation coefficients between 8 epigenetic and 2 metabolomic aging biomarkers were low negative to moderate positive, ranging between −0.29 and 0.69 in the overall population (Figure 2A). The highest correlation was found between DNAm DunedinPoAm and DNAm GrimAge. The metabolomic aging biomarkers had the highest correlations with each other and with DNAm DunedinPoAm, DNAm GrimAge, and DNAm Zhang. The correlations between biomarkers trained on chronological age (clocks) from different molecular origins, ie, DNAm Horvath, DNAm Hannum, or DNAm Lin versus MetaboAge, were low negative to low positive, namely 0.04 between MetaboAge and DNAm Hannum, 0.16 between MetaboAge and DNAm Horvath and −0.22 between MetaboAge and DNAm Lin. Interestingly, these correlations were lower than the associations between MetaboAge and the pace of aging-trained DNAm DunedinPoAm (0.21) and those between MetaboAge and the mortality-trained DNAm GrimAge (0.20) and DNAm Zhang (0.21). The mortality-trained epigenetic aging biomarkers DNAm GrimAge and DNAm Zhang had a correlation with the metabolomic-based mortality biomarker of, respectively, 0.30 and 0.29. Comparable correlation patterns were observed across methylation arrays (within RS) and between RS and LLS cohorts (Supplementary Figure 1). Post hoc analysis revealed similar correlation patterns among participants who survived the study period and those who died, except for a decline in correlation between epigenetic clocks and MetaboAge observed in RS but not LLS (Supplementary Figure 2).
Figure 2.
Correlations between biological age measures and the association between biomarkers of biological aging and frailty. (A) Spearman’s correlation of the different biological aging biomarkers in 1 424 Rotterdam Study participants with the histograms of epigenetic aging biomarkers in yellow and metabolomic-based aging biomarkers in blue. Labels in bold indicate aging biomarkers trained on outcomes including phenotypic and/or mortality information; the regular font, an aging biomarker trained on chronological age. Biomarkers are arranged by omics layer, ordered from fully age-trained to fully mortality-trained. Values after r = represent Spearman’s rank coefficient; values after p = represent the p value; the background color is darker for higher correlations. epiTOC = DNAm epiTOC; GrimAge = DNAm GrimAge; Hannum = DNAm Hannum; Horvath = DNAm Horvath; Lin = DNAm Lin; mHealth = MetaboHealth; Pheno = DNAm PhenoAge; PoAM = DNAm DunedinPoAm; Zhan = DNAm Zhang; mAge = MetaboAge. (B) Spearman’s correlation between the different Yeo–Johnson-transformed frailty measures in the 746 Rotterdam Study participants with information on all 5 frailty measures. Values represent Spearman’s rank coefficient; the background color is darker for higher correlations. CFP = continuous frailty phenotype; FI = frailty index; FP = frailty phenotype; MPI = Multidimensional Prognostic Index; TFI = Tilburg Frailty Indicator. (C) Risk of all-cause mortality per standard deviation increase of the Yeo–Johnson transformed FI (n cases = 130/n = 1 330), FP (n cases = 132/n = 1 328), CFP (n cases = 69/n = 743), TFI (n cases = 129/n = 1 328), MPI (n cases = 132/n = 1 333) in the RS overall study population. The figure represents the adjusted hazard ratios and 95%-confidence intervals. (D) Associations of standardized biological aging biomarkers with standardized FI (n = 1 341), FP (n = 1 339), CFP (n = 748), TFI (n = 1 339), and MPI (n = 1 344) based on linear regression analyses in all participants for whom data on biological aging biomarkers and frailty were available in the overall Rotterdam Study dataset. Analyses were adjusted for age, sex, BMI, cell counts, subcohort, and Rotterdam Study cohort and visit. The figure represents the adjusted betas and 95%-confidence intervals. Biomarkers are arranged by omics layer, ordered from fully age-trained to fully mortality-trained. DNAm Zhang is missing information on 2 out of 10 CpGs in the EPIC-subcohort (736 of the 1 347 participants). BMI = body mass index.
Frailty
Frailty measures, like the biological aging biomarkers, were developed to capture the individual aging process (11,12,46); they represent measures of biological age. We assessed the interchangeability of various frailty measures developed based on different rationales by measuring 5 different frailty scores in our study population: FI (6), FP (2), CFP (7), TFI (8,39), and MPI (9) (Supplementary Texts 1 and 2). When elements from these frailty measures were lacking in our data set, we used proxies (Supplementary Text 2).
As shown in Figure 2B, the correlation between the different frailty measures ranged from 0.20 to 0.50. The highest correlation was observed between the 2 physical frailty measures, FP and CFP, and the lowest between FP and the MPI, the latter being the frailty measure directly derived from the CGA. All frailty measures were associated with an increased risk of overall mortality. We observed higher hazard ratios for broad frailty scores than for the physical frailty measures, yet this difference was not statistically significant (Figure 2C, Supplementary Table 2).
We concluded that the frailty measures could not be used interchangeably. Therefore, we examined 10 biological aging biomarkers (8 epigenetic and 2 metabolomic aging biomarkers) for their association with all 5 frailty measures. All biological aging biomarkers and the frailty scores were standardized to improve comparability. We observed that an increase in biomarkers trained on biophysiological or mortality information had consistently more prominent associations with frailty than we observed for the clocks, with MetaboAge among the clocks showing the most prominent association with frailty scores. The strongest associations were found for MetaboHealth (adjusted beta in the RS combined study population per standard deviation increase [B 0.20 95% confidence interval {CI}: 0.15; 0.25]) and DNAm GrimAge (B 0.21 [CI: 0.16; 0.26]) with the MPI (Figure 2D, Supplementary Figure 2, Supplementary Table 3). These associations exhibited higher magnitudes than the associations previously observed between frailty scores and all-cause mortality (Figure 2C, Supplementary Table 2). Furthermore, our observations revealed variability among frailty measures in the association with biological age biomarkers. The physical-oriented FP especially displayed a distinct association pattern from other frailty measures. FP showed a weaker association with epigenetic biomarkers trained on biophysiological measures associated with health and/or mortality and with MetaboAge. This drop was most prominent in the association with DNAm DunedinPoAm and DNAm GrimAge. Broad frailty measures were significantly associated with multiple biological aging biomarkers, with FI and MPI linked to 4 out of 10 biomarkers after adjustment for multiple testing, and TFI with 3. Surprisingly, constructing CFP using the same information as FP resulted in associations more similar to those of FI, TFI, and MPI, and we observed a significant association with 3 biological aging biomarkers.
The analyses were adjusted for age despite using chronological age-independent biological aging biomarkers to address the inherent correlation of frailty measures, for example, the FI, with chronological age. BMI was included as a covariate as both epigenetic- (47) and metabolomic-based (24) aging biomarkers are known to associate with a higher BMI and BMI information is included in all frailty scores (2,6–9) (Supplementary Texts 1 and 2). We performed a sensitivity analysis adjusting for smoking status and socioeconomic status to determine whether the inclusion of smoking-pack years in the construction of the DNAm GrimAge was driving the results or that factors associated with socioeconomic status influenced the performance of the biomarkers. The sensitivity analysis did not remarkably alter the results (Supplementary Table 3). We tested for effect modification of sex on the aging biomarkers by adding an interaction term between the two in the model. None of these contrasts were FDR-corrected significant. A sex-stratified analysis did not indicate sex differences in the association between biomarkers of biological age and frailty (Supplementary Table 4).
Subsequentially, we determined whether the associations with the frailty measures of the best-performing epigenetic and best-performing metabolomic aging biomarkers were independent of each other. Both DNAm GrimAge and MetaboHealth remained independently associated with frailty in a linear regression adjusted for the same covariates as the univariable analyses (Table 1, Supplementary Table 5). There were some small improvements in the explained variance of the models when both DNAm GrimAge and MetaboHealth were included, for example, the explained variance of the association with FI improved from 0.22 for DNAm GrimAge and 0.23 for MetaboHealth to 0.24 in the combined model (Supplementary Tables 3 and 5). Additionally, adjusting for smoking status did, again, not considerably change the results (Supplementary Table 5), and the results were not statistically significantly different between men and women (Supplementary Table 6). Furthermore, the same pattern appeared when categorizing participants as nonfrail and frail using the traditional cut-offs (Supplementary Text 1) of the frailty measures (Supplementary Table 7).
Table 1.
Results of Multivariable Regression Models Including Both DNAm GrimAge and MetaboHealth as Exposures and Frailty Measures as Outcome
| DNAm GrimAge | MetaboHealth | ||||
|---|---|---|---|---|---|
| Adjusted* Beta Per SD (CI) | pFDR | Adjusted* Beta Per SD (CI) | pFDR | ||
| FI | n = 1 339 | 0.08 (0.03; 0.14) | .02 | 0.17 (0.11; 0.22) | 7.35 × 10−9 |
| FP | n = 1 339 | 0.01 (-0.05; 0.07) | .79 | 0.11 (0.05; 0.17) | 5.51 × 10−4 |
| CFP | n = 748 | 0.11 (0.03; 0.19) | .02 | 0.10 (0.02; 0.17) | .02 |
| TFI | n = 1 339 | 0.07 (0.01; 0.13) | .06 | 0.11 (0.06; 0.17) | 3.38 × 10−4 |
| MPI | n = 1 344 | 0.15 (0.10; 0.21) | 6.06 × 10−6 | 0.17 (0.12; 0.22) | 8.08 × 10−9 |
Notes: CFP = continuous frailty phenotype; CI = confidence interval; FI = frailty index; FP = frailty phenotype; MPI = multidimensional prognostic index; n = number of participants for whom we have information on this frailty score; pFDR = p value after adjustment for multiple testing by the false discovery rate method; SD = standard deviation; TFI = Tilburg Frailty Indicator.
*Adjusted for chronological age at blood sampling, sex, body mass index, cell counts, subcohort, and visit and cohort within the Rotterdam Study.
Mortality
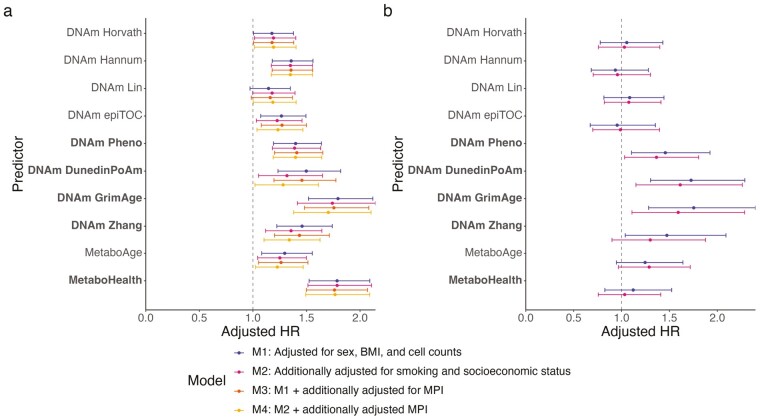
Beyond reflecting on an individual’s current state, biomarkers of biological age are believed to capture predictive information on the aging process (12). We, therefore, determined the association of biological aging biomarkers with mortality during 11 281 person-years of follow-up in the RS. The median follow-up time was 8.6 years. During follow-up, 132 participants died. A higher score on all biomarkers of biological age was associated with an increased risk of overall mortality in the combined study population. Moreover, DNAm GrimAge and MetaboHealth were consistently associated with increased mortality risk in both RS-subcohorts. The highest risk estimates for all-cause mortality were observed for DNAm GrimAge (adjusted hazard ratio in the RS combined study population per standard deviation increase [HR] 1.79 95%-CI [1.52;2.12]) and MetaboHealth (HR 1.79 [CI 1.52;2.09]; Figure 3A, Supplementary Figure 3, Supplementary Table 8). Yet, the observed hazard ratios were a bit more stable for DNAm GrimAge than for MetaboHealth. The Cox Proportional Hazard models were adjusted for the same covariates as used in the linear regression except for age, which was included in the timescale. A sensitivity analyses to determine whether smoking status and socioeconomic status influenced the risk entailed by the aging biomarkers, again, did not noteworthy shift the results (Supplementary Table 8).
Figure 3.
Aging predictors and their univariable risk of all-cause mortality per SD. Risk of all-cause mortality per standard deviation increase of the aging biomarkers in (A) the overall Rotterdam Study population (n = 1 336). DNAm Zhang is missing information on 2 out of 10 CpGs in the EPIC-subcohort (727 of the 1 336 participants); and (B) the subcohort of the Leiden Longevity Study with information on the epigenetic aging predictors (n = 584). CpGs = methylation sites; BMI = body mass index; HR = hazard ratio; MPI= multidimensional prognostic index; SD = standard deviation. Biomarkers are arranged by omics layer, ordered from fully age trained to fully mortality trained.
We then assessed whether the observed associations between the aging biomarkers and mortality were explained by frailty. For this, we used the MPI since it is the frailty score most closely related to the CGA. Earlier, we showed that the MPI itself is associated with an increased risk of overall mortality (Figure 2C, Supplementary Table 2). Nevertheless, the associations between the aging predictors and mortality were independent of and not notably changed by the MPI (Figure 3A, Supplementary Figure 3, Supplementary Table 8). These results remained, yet again, unchanged in a sensitivity analysis adjusting for smoking status and socioeconomic status (Figure 3A, Supplementary Figure 3, Supplementary Table 8). We evaluated the potential modification of sex on the impact of aging biomarkers by including an interaction term between these variables in our model. Nevertheless, none of the identified interaction terms achieved statistical significance after applying FDR correction. A sex-stratified analysis did not reveal any differences in the association between the biological age biomarkers and mortality based on sex (Supplementary Table 9). Lastly, to further assess the associations between frailty, biomarkers of biological age, and all-cause mortality, mediation analyses were conducted. Our results gave no indication of frailty mediating the association between biomarkers of biological age and mortality (Supplementary Table 10).
To determine whether the risk of mortality captured by the best-performing epigenetic aging predictor and metabolomic aging predictor were mutually independent, we performed a Cox proportional hazard analysis including both aging predictors and adjusted for sex, BMI, and cell count. When combining DNAm GrimAge and MetaboHealth in a model, both showed an independent risk of all-cause mortality, respectively, DNAm GrimAge (HR 1.56 [CI 1.31;1.85]) and MetaboHealth (HR 1.60 [CI 1.35;1.89]). The concordance increased slightly from 0.69 when only using DNAm GrimAge and 0.67 when only including MetaboHealth to 0.70 in the combined model. These results were robust among the RS overall study population and subcohorts (Supplementary Table 11). These results remained, again, similar when adjusting for the MPI and smoking status and socioeconomic status and no-sex differences were found (Table 2, Supplementary Tables 11 and 12).
Table 2.
The Multivariable Risk of All-Cause Mortality of DNAm GrimAge and MetaboHealth
| Model 1 | Model 2 | Model 3 | Model 4 | |||||
|---|---|---|---|---|---|---|---|---|
| n/N = 132/1 336 | n/N = 132/1 325 | n/N = 132/1 331 | n/N = 131/1 325 | |||||
| HR (CI) | pFDR | HR (CI) | pFDR | HR (CI) | pFDR | HR (CI) | pFDR | |
| DNAm GrimAge | 1.56 (1.31; 1.85) | 9.95 × 10−6 | 1.51 (1.22; 1.86) | 3.01 × 10−4 | 1.55 (1.30; 1.84) | 9.95 × 10−6 | 1.50 (1.21; 1.85) | 3.79 × 10−4 |
| MetaboHealth | 1.60 (1.35; 1.89) | 2.60 × 10−7 | 1.67 (1.40; 1.98) | 1.12 × 10−7 | 1.59 (1.34; 1.89) | 3.49 × 10−7 | 1.66 (1.39; 1.97) | 1.13 × 10−7 |
| Concordance | 0.70 | 0.71 | 0.71 | 0.71 | ||||
Notes: Model 1: Adjusted for sex, BMI, cell count, batch, and cohort within the Rotterdam Study. Model 2: Model 1 + additionally adjusted for smoking status and socioeconomic status. Model 3: Model 1 + additionally adjusted for the MPI. Model 4: Model 3 + additionally adjusted for smoking status and socioeconomic status. CI = confidence interval; DNAm = DNA methylation; HR = hazard ratio; MPI = multidimensional prognostic index; n = cases; N = persons at risk; pFDR = p value after false discovery rate adjustment for multiple testing.
In our external validation cohort, the LLS, 147 participants died during 23 977 person-years of follow-up (median 13.3 years), of whom 43 were part of the multiomics subcohort representing 7 553 person-years of follow-up (median 13.2 years). In the subcohort, where we could validate both the epigenetic and the metabolomic aging predictors, the results of DNAm PhenoAge, DNAm DunedinPoAm, DNAm GrimAge, DNAm Zhang, and MetaboAge were comparable to the results in the RS. By contrast, the associations between the other aging predictors, especially MetaboHealth, and mortality drops significantly (Figure 3B, Supplementary Table 8). In the overall population of the LLS, MetaboHealth outperformed MetaboAge in the prediction of mortality, yet the mortality risk of both aging predictors was smaller than in the RS overall population and more comparable to the EPIC-subcohort (Supplementary Figure 3, Supplementary Table 8).
Discussion
There is no consensus on which biomarkers that can be measured in human studies in standardized fashion reflect biological age. In the present study, we considered frailty measures and mortality as phenotypes representing biological age. We compared 5 frailty measures in a population-based cohort and 8 epigenetic and 2 metabolomic biomarkers of biological age in 2 distinct cohorts of mainly nonclinically frail older participants. Our most prominent findings were: (a) the rather weak to moderate correlations between DNAm and metabolomics biological aging biomarkers especially between biomarkers from different origins trained on chronological age; (b) the outperformance of the mortality- and biophysiological-trained biomarkers, especially MetaboHealth and DNAm GrimAge, in reflection of biological age, as represented by frailty and mortality, compared to clocks and frailty measures; (c) the mutually independent associations between the mortality-trained biomarkers of biological age, DNAm GrimAge, and MetaboHealth with frailty and mortality; and (d) the independence of the mortality association of the biomarkers of biological age from the MPI, the frailty measure directly derived from the CGA, smoking and socioeconomic status. These findings stress that the different molecular markers of biological age complement each other in estimating frailty and mortality risk and potentially complement standardized health assessment in the clinical setting.
Similar to previous reports (9,48–54), we found an association between higher frailty scores and an increased mortality risk for all frailty measures of interest. Despite the wide variety of frailty measures, most studies focused on the FI and FP (54–57). Our results are consistent with previous studies reporting a higher risk of overall mortality for the FI compared to FP (56,57). However, this difference was not statistically significant in our study, which may be due to a lack of power. Given (55) the varied findings regarding the association between biomarkers of biological age and different frailty measures in our study, we recommend carefully choosing the frailty measure best suited to the research question.
Our results show that epigenetic aging biomarkers trained on longitudinal data outperform epigenetic clocks. This is in line with earlier reports on epigenetic biomarkers. A number of studies determined the association between various epigenetic biomarkers and frailty measures, and other hallmarks of aging and showed that only biomarkers trained on mortality and biophysiological information were associated with FI (28,58), CFP (58), cognitive and physical capacity (26,27), hallmarks of aging (59), and all-cause mortality (26,60).
A limited number of previous studies compared frailty and aging biomarkers for their ability to predict mortality. A previous study reported that DNAm Horvath, DNAm Hannum, DNAm PhenoAge, DNAm GrimAge, and FI were all associated with mortality when separately analyzed. When these measures were combined in a model with 6 other biological age measures, the largest effect sizes were reported for FI and DNAm GrimAge (61).
One modestly sized study solely focused on DNAm Horvath in comparison to the FI, but did not find an association of DNAm Horvath with mortality (62). Our results did show an association between DNAm Horvath and all-cause mortality. This is probably due to the larger sample size of our study. However, the association between DNAm Horvath and mortality was also weaker in our study than the association with either mortality- or biophysiological-trained biomarkers of biological age. In our study, the longitudinal-based epigenetic and metabolomic biomarkers showed a profound association with mortality independent of the MPI. Our study, therefore, is in line with a previous study emphasizing the added value of using biomarkers in frailty assessment as they report higher discriminative ability when both the biomarker-based frailty and FI are included in mortality prediction (55).
The performance of the metabolomic-based aging biomarkers trained on chronological age and mortality has, to our knowledge, not been evaluated in other studies. In 2 RS cohorts, the outperformance of the mortality-trained markers in reflecting frailty and predicting mortality was evident, as was the case for the comparison of metabolomics aging predictors in the LLS overall study population. The lower performance of MetaboHealth in the LLS multiomics subcohort might be caused by the small sample size since in the original studies of this cohort (24,25) the metabolomic markers predicted adverse outcomes equally well in RS and LLS. Another possible explanation might be the difference in follow-up time between the 2 cohorts. Due to the small sample size, we cannot check the latter. In the RS, we observed somewhat unstable results for DNAm Horvath and MetaboAge, and to a lesser extent, MetaboHealth. This might have resulted from the usage of the age-independent part of the aging biomarkers. We used the age-independent part of our biomarkers by regressing out chronological age. This approach is more vulnerable to outliers when fewer participants are included in the study. In case of the epigenetic biomarkers, some CpGs were lacking on the EPIC-platform; in the case of DNAm Zhang, even 2 out of 10 were lacking, which may have led to differences between the subcohorts in RS. We also noted a decline in overall mortality risk identified by the frailty measure in the EPIC-subcohort compared to the 450K-subcohort. Demographic differences between the subcohorts may have played a role in the differences in performance of the aging biomarkers between the subcohorts, the 450K-subcohort was older and had a higher mortality incidence. Further analysis of the performance of these aging biomarkers in small studies is recommended to determine their applicability in studies with smaller sample sizes. The molecular markers of biological age could potentially be used in the clinical setting to improve health and resilience estimates and as response monitors in intervention studies. In both future applications, the performance in limited sample-sized groups and ultimately even in individual patients is crucial.
DNAm GrimAge, DNAm Zhang, and MetaboHealth were developed using prospective mortality data. MetaboHealth and DNAm Zhang were trained directly, while the DNAm GrimAge model used DNAm surrogates of plasma proteins and smoking-pack years developed in elastic net Cox regression on overall mortality. DNAm PhenoAge was trained on phenotypic age, a predictor of mortality consisting of 9 biomarkers and chronological age. Phenotypic age had a correlation with chronological age of 0.94 in NHANES IV (19). DNAm DunedinPoAm was trained on the longitudinally measured page of aging score, DNAm epiTOC on age-associated hyper- and hypomethylation, and DNAm Horvath, DNAm Hannum, and MetaboAge were trained directly on chronological age. MetaboHealth, DNAm GrimAge, and DNAm DunedinPoAm, along with DNAm Zhang in the 450K-subcohort, where all CpGs needed were present, outperformed other aging biomarkers not solely in mortality prediction but also in reflecting frailty, according to our study. Therefore, we believe that our study provides further support for the benefits of training on longitudinal information regardless of the omics layer used. Our findings align with the theory that fast-agers die sooner and consequently contribute less to the construction of biomarkers based on age, while biomarkers containing longitudinal information, such as mortality, suffer less from this selection bias (63). This finding could have important implications for the development of future biomarkers. Specifically, it suggests that aging predictors using longitudinal outcomes seem better equipped to capture the physiological heterogeneity that increases with aging. However, the performance of biological aging biomarkers on short-term outcomes still needs to be evaluated.
The low correlation of the epigenetic and metabolomic aging biomarkers in combination with the independent association of DNAm GrimAge and MetaboHealth with frailty and mortality suggests that metabolomic and epigenetic aging biomarkers capture different aspects of the aging process. However, there was only a slight increase in the explained variance when using both DNAm GrimAge and MetaboHealth. Furthermore, since the associations of these aging biomarkers were also independent of the MPI, there is an indication that these aspects are not captured in the CGA. This could imply that using these aging biomarkers would strengthen clinical geriatric risk assessment. The fact that we observed no indication of frailty mediating the association between aging biomarkers and all-cause mortality strengthens the hypothesis that different methods of determining biological age capture different parts of the aging process. Yet, as we had information on aging biomarkers and frailty around the same time, we possibly have underestimated the effect of mediation of the association. Therefore, further research into the different aspects of aging captured by the different aging biomarkers, frailty, and their applicability in the clinic and research would be advisable.
The main strengths of this study are the relatively large study population for which we had information on both DNAm and metabolomics, internal validation as well as external validation, and only a small loss to follow-up in the mortality data. Furthermore, with 5 different frailty measures, we had data on a wide variety of aspects of aging and were able to give insight into the distinct features of frailty measures. Besides, having information on both frailty and mortality gave us the opportunity to determine the associations of biological aging biomarkers with mortality adjusted for the MPI and, thereby, obtain an indication of the performance of molecular markers of biological aging beyond ongoing clinical practice.
However, there are some limitations to the current study. Firstly, participants needed to be fit enough to visit the research centers to provide blood samples and participate in several assessments for the frailty examinations. This requirement led to a selection bias towards healthy individuals. Secondly, RS and LLS were included in study selecting metabolites included in MetaboHealth, together accounting for 13.7% of the 44 168 study participants. The inclusion of these cohorts in the creation of MetaboHealth may have resulted in an overestimation of its association with frailty and mortality. Thirdly, we did not have all the original measurements on which the frailty measures are usually based. When a specific measurement was not present, we used proxies (Supplementary Text 2). We chose the proxies carefully with the help of a geriatrician; however, this could have had some impact on the estimations. Lastly, our study population consisted of White individuals aged 30 to 98 years; thus, validation of our project in other study populations and other (middle-aged) aspects of biological age is needed to assess the robustness of our results.
To our knowledge, this is the first study comparing the performance of both epigenetic and metabolomic-based aging biomarkers in reflecting frailty and mortality risk as measures of biological age. Furthermore, this is the first study to include information on 5 different frailty measures as well as information on molecular biomarkers of biological age. We showed that epigenetic and metabolomic-based aging biomarkers trained on longitudinal information, especially DNAm GrimAge, and MetaboHealth reflected these biological age measures better than aging predictors trained on age or phenotypic age. The associations of DNAm GrimAge and MetaboHealth with frailty and mortality are independent of each other, suggesting that they capture information on different aspects of aging and may both be studied as novel phenotypes in research aimed at finding determinants of biological ageing. For the age and health categories we have studied, it is also relevant that the associations of the biological age markers with mortality are partly independent of the MPI, a proxy for the standardized geriatric health assessment CGA as used in the clinic. These findings suggest that DNAm GrimAge and MetaboHealth could be valuable to complement the current health, well-being, and risk assessments in clinical practice. Therefore, further research into the potential integration of these biomarkers of biological aging in a clinical setting is warranted as well as increasing the informativity of these markers on the level of the individual patient.
Supplementary Material
Supplementary data are available at The Journals of Gerontology, Series A: Biological Sciences and Medical Sciences online.
Acknowledgments
The contribution of all participants in the Rotterdam Study and the Leiden Longevity Study is gratefully acknowledged; this research would not have been possible without them.
Contributor Information
Lieke M Kuiper, Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands; Center for Nutrition, Prevention and Health Services, Bilthoven, The Netherlands.
Harmke A Polinder-Bos, Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands.
Daniele Bizzarri, Molecular Epidemiology, Department of Biomedical Data Sciences, Leiden University Medical Centre, Leiden, The Netherlands; Pattern Recognition and Bioinformatics, Delft University of Technology, Delft, The Netherlands.
Dina Vojinovic, Molecular Epidemiology, Department of Biomedical Data Sciences, Leiden University Medical Centre, Leiden, The Netherlands; Department of Epidemiology, Erasmus MC, Rotterdam, The Netherlands.
Costanza L Vallerga, Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands.
Marian Beekman, Molecular Epidemiology, Department of Biomedical Data Sciences, Leiden University Medical Centre, Leiden, The Netherlands.
Martijn E T Dollé, Center for Health Protection, National Institute for Public Health and the Environment (RIVM), Bilthoven, The Netherlands.
Mohsen Ghanbari, Department of Epidemiology, Erasmus MC, Rotterdam, The Netherlands.
Trudy Voortman, Department of Epidemiology, Erasmus MC, Rotterdam, The Netherlands; Division of Human Nutrition and Health, Wageningen University & Research, Wageningen, The Netherlands.
Marcel J T Reinders, Molecular Epidemiology, Department of Biomedical Data Sciences, Leiden University Medical Centre, Leiden, The Netherlands; Pattern Recognition and Bioinformatics, Delft University of Technology, Delft, The Netherlands.
W M Monique Verschuren, Center for Nutrition, Prevention and Health Services, Bilthoven, The Netherlands; Julius Center for Health Sciences and Primary Care Utrecht, University Medical Center Utrecht, Utrecht University, Utrecht, The Netherlands.
P Eline Slagboom, Molecular Epidemiology, Department of Biomedical Data Sciences, Leiden University Medical Centre, Leiden, The Netherlands; Max Planck Institute for the Biology of Ageing, Cologne, Germany.
Erik B van den Akker, Molecular Epidemiology, Department of Biomedical Data Sciences, Leiden University Medical Centre, Leiden, The Netherlands; Pattern Recognition and Bioinformatics, Delft University of Technology, Delft, The Netherlands.
Joyce B J van Meurs, Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands; Department of Orthopaedics and Sports Medicine, Erasmus MC, Rotterdam, The Netherlands.
Funding
The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for the Health Research and Development (ZonMW); the Research Institute for Disease in the Elderly (RIDE); the Ministry of Education, Culture, and Science; the Ministry of Health, Welfare, and Sports; the European Commission; and the Municipality of Rotterdam. The Leiden Longevity Study is supported by the European Union’s Seventh Framework Programme (FP7/2007–2011) under grant agreement number 259679. This study was financially supported by the Innovation-Oriented Research Program on Genomics (SenterNovem IGE05007), the Centre for Medical Systems Biology, and the Netherlands Consortium for Healthy Ageing (grant 050-060-810), all in the framework of the Netherlands Genomics Initiative, Netherlands Organization for Scientific Research (NWO), by BBMRI-NL, a Research Infrastructure financed by the Dutch government (NWO 184.021.007 and 184.033.111). The current study was supported by VOILA (ZonMW 457001001) and Medical Delta (scientific program METABODELTA: Metabolomics for clinical advances in the Medical Delta). E.B.vdA. is funded by a personal grant from the Dutch Research Council (NWO; VENI: 09150161810095). M.E.T.D. is funded by the Ministry of Health, Welfare, and Sport & The National Institute for Public Health of the Netherlands (S/010003).
Conflict of Interest
None.
Author Contributions
L.K., H.P., M.B., M.D., T.V., M.R., E.S., M.V., E.vdA., and J.vM. conceptualized the study. L.K., D.B., D.V., MR, E.vdA., and J.vM. designed the methodology. L.K., D.B., D.V., and C.V. were involved in the formal analysis. M.B., M.G., T.V., P.S,. and J.vM. curated the data. D.B., D.V., M.B., E.S., and E.vdA. performed the validation study. D.B., M.R., and E.vdA. developed the software used. L.K. and D.B. visualized the analyses. L.K. wrote the first draft of the manuscript. All authors provided valuable feedback for the revision of the manuscript. M.D., M.R., M.V., E.S., E.vdA., and J.vM. supervised the project. M.B., M.R., M.V., E.S., and J.vM. acquired funding for the study.
References
- 1. Rando TA, Chang HY. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 2012;148(1–2):46–57. doi: 10.1016/j.cell.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fried LP, Tangen CM, Walston J, et al. Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci. 2001;56(3):M146–M157. doi: 10.1093/gerona/56.3.M146 [DOI] [PubMed] [Google Scholar]
- 3. Niccoli T, Partridge L. Ageing as a risk factor for disease. Curr Biol. 2012;22(17):R741–R752. doi: 10.1016/j.cub.2012.07.024 [DOI] [PubMed] [Google Scholar]
- 4. Parker SG, Mccue P, Phelps K, et al. What is Comprehensive Geriatric Assessment (CGA)? An umbrella review. Age Ageing. 2018;47(1):149–155. doi: 10.1093/ageing/afx166 [DOI] [PubMed] [Google Scholar]
- 5. Sourdet S, Brechemier D, Steinmeyer Z, Gerard S, Balardy L. Impact of the comprehensive geriatric assessment on treatment decision in geriatric oncology. BMC Cancer. 2020;20(1):384. doi: 10.1186/s12885-020-06878-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schoufour JD, Erler NS, Kiefte-de Jong JC, et al. Design of a frailty index among community living middle-aged and older people: the Rotterdam study. Maturitas. 2017;97:14–20. doi: 10.1016/j.maturitas.2016.12.002 [DOI] [PubMed] [Google Scholar]
- 7. Wu C, Geldhof GJ, Xue QL, Kim DH, Newman AB, Odden MC. Development, construct validity, and predictive validity of a continuous frailty scale: results from 2 large US cohorts. Am J Epidemiol. 2018;187(8):1752–1762. doi: 10.1093/aje/kwy041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gobbens RJJ, Boersma P, Uchmanowicz I, Santiago LM. The Tilburg frailty indicator (TFI): new evidence for its validity. Clin Interv Aging. 2020;15:265–274. doi: 10.2147/CIA.S243233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pilotto A, Ferrucci L, Franceschi M, et al. Development and validation of a multidimensional prognostic index for one-year mortality from comprehensive geriatric assessment in hospitalized older patients. Rejuvenation Res. 2008;11(1):151–161. doi: 10.1089/rej.2007.0569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dent E, Kowal P, Hoogendijk EO. Frailty measurement in research and clinical practice: a review. Eur J Intern Med. 2016;31:3–10. doi: 10.1016/j.ejim.2016.03.007 [DOI] [PubMed] [Google Scholar]
- 11. Mitnitski AB, Mogilner AJ, Rockwood K. Accumulation of deficits as a proxy measure of aging. ScientificWorldJournal. 2001;1:323–336. doi: 10.1100/tsw.2001.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jylhävä J, Pedersen NL, Hägg S. Biological Age Predictors. EBioMedicine. 2017;21:29–36. doi: 10.1016/j.ebiom.2017.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(3156):R115. doi: 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin Q, Weidner CI, Costa IG, et al. DNA methylation levels at individual age-associated CpG sites can be indicative for life expectancy. Aging (Milano). 2016;8(2):394–401. doi: 10.18632/aging.100908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang Z, Wong A, Kuh D, et al. Correlation of an epigenetic mitotic clock with cancer risk. Genome Biol. 2016;17(1):205. doi: 10.1186/s13059-016-1064-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Belsky DW, Caspi A, Arseneault L, et al. Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. Elife. 2020;9:1–56. doi: 10.7554/eLife.54870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Belsky DW, Caspi A, Houts R, et al. Quantification of biological aging in young adults. Proc Natl Acad Sci U S A. 2015;112(30):E4104–E4110. doi: 10.1073/pnas.1506264112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10(4):573–591. doi: 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY). 2019;11(2):303–327. doi: 10.18632/aging.101684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y, Wilson R, Heiss J, et al. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat Commun. 2017;8:14617. doi: 10.1038/ncomms14617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Soininen P, Kangas AJ, Würtz P, Suna T, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet. 2015;8(1):192–206. doi: 10.1161/CIRCGENETICS.114.000216 [DOI] [PubMed] [Google Scholar]
- 23. Würtz P, Kangas AJ, Soininen P, Lawlor DA, Davey Smith G, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: a primer on -Omic technologies. Am J Epidemiol. 2017;186(9):1084–1096. doi: 10.1093/aje/kwx016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van Den Akker EB, Trompet S, Barkey Wolf JJH, et al. Metabolic age based on the BBMRI-NL 1H-NMR metabolomics repository as biomarker of age-related disease. Circ Genom Precis Med. 2020;13:541–547. doi: 10.1161/CIRCGEN.119.002610 [DOI] [PubMed] [Google Scholar]
- 25. Deelen J, Kettunen J, Fischer K, et al. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat Commun. 2019;10(1):1–8. doi: 10.1038/s41467-019-11311-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCrory C, Fiorito G, Hernandez B, et al. GrimAge outperforms other epigenetic clocks in the prediction of age-related clinical phenotypes and all-cause mortality. J Gerontol A Biol Sci Med Sci. 2021;76(5):741–749. doi: 10.1093/gerona/glaa286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maddock J, Castillo-Fernandez J, Wong A, et al. DNA methylation age and physical and cognitive aging. J Gerontol A Biol Sci Med Sci. 2020;75(3):504–511. doi: 10.1093/gerona/glz246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Verschoor CP, Lin DTS, Kobor MS, et al. Epigenetic age is associated with baseline and 3-year change in frailty in the Canadian Longitudinal Study on Aging. Clin Epigenetics. 2021;13(1):163. doi: 10.1186/s13148-021-01150-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ikram MA, Brusselle G, Ghanbari M, et al. Objectives, Design and Main Findings until 2020 from the Rotterdam Study. Vol. 35. Springer Netherlands; 2020. doi: 10.1007/s10654-020-00640-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schoenmaker M, de Craen AJM, de Meijer PHEM, et al. Evidence of genetic enrichment for exceptional survival using a family approach: the Leiden Longevity Study. Eur J Hum Genet. 2006;14(1):79–84. doi: 10.1038/sj.ejhg.5201508 [DOI] [PubMed] [Google Scholar]
- 31. Sandoval J, Heyn HA, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. doi: 10.4161/epi.6.6.16196 [DOI] [PubMed] [Google Scholar]
- 32. Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 33. Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):208. doi: 10.1186/s13059-016-1066-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pelegí-Sisó D, de Prado P, Ronkainen J, Bustamante M, González JR. Methylclock: a bioconductor package to estimate DNA methylation age. Bioinformatics. 2021;37(12):1759–1760. doi: 10.1093/bioinformatics/btaa825 [DOI] [PubMed] [Google Scholar]
- 35. Mukherjee S, Stamatis D, Bertsch J, et al. Genomes OnLine Database (GOLD) v.8: overview and updates. Nucleic Acids Res. 2021;49(D1):D723–D733. doi: 10.1093/nar/gkaa983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bizzarri D, Reinders MJT, Beekman M, Slagboom PE, van den Akker EB. MiMIR: R-shiny application to infer risk factors and endpoints from Nightingale Health’s 1H-NMR metabolomics data. Bioinformatics. 2022;38(15):3847–3849. doi: 10.1093/bioinformatics/btac388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leening MJG, Kavousi M, Heeringa J, et al. Methods of data collection and definitions of cardiac outcomes in the Rotterdam Study. Eur J Epidemiol. 2012;27(3):173–185. doi: 10.1007/s10654-012-9668-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Government of the Netherlands. Personal Records Database (BRP). Accessed January 20, 2021. https://www.government.nl/topics/personal-data/personal-records-database-brp
- 39. Gobbens RJJ, van Assen MALM, Luijkx KG, Wijnen-Sponselee MT, Schols JMGA. The Tilburg frailty indicator: psychometric properties. J Am Med Dir Assoc. 2010;11(5):344–355. doi: 10.1016/j.jamda.2009.11.003 [DOI] [PubMed] [Google Scholar]
- 40. Rietveld CA, Medland SE, Derringer J, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science (1979). 2013;340(6139):1467–1471. doi: 10.1126/science.1235488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yeo I, Johnson RA. A new family of power transformations to improve normality or symmetry. Biometrika. 2000;87(4)954–959. doi: 10.1093/biomet/87.4.954 [DOI] [Google Scholar]
- 42. Therneau TM. Survival: A Package for Survival Analysis in R. R package version 238. 2021. [Google Scholar]
- 43. Tingley D, Yamamoto T, Hirose K, Keele L, Imai K. mediation: R package for causal mediation analysis. J Stat Softw. 2014;59(5):1–38. doi: 10.18637/jss.v059.i0526917999 [DOI] [Google Scholar]
- 44. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 45. Houwing-Duistermaat JJ, Callegaro A, Beekman M, Westendorp RG, Slagboom PE, van Houwelingen JC. Weighted statistics for aggregation and linkage analysis of human longevity in selected families: the Leiden Longevity Study. Stat Med. 2009;28(1):140–151. doi: 10.1002/sim.3421 [DOI] [PubMed] [Google Scholar]
- 46. Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19(6):371–384. doi: 10.1038/s41576-018-0004-3 [DOI] [PubMed] [Google Scholar]
- 47. Lundgren S, Kuitunen S, Pietiläinen KH, et al. BMI is positively associated with accelerated epigenetic aging in twin pairs discordant for body mass index. J Intern Med. 2022;292(4):627–640. doi: 10.1111/joim.13528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kane RL, Shamliyan T, Talley K, Pacala J. The association between geriatric syndromes and survival. J Am Geriatr Soc. 2012;60(5):896–904. doi: 10.1111/j.1532-5415.2012.03942.x [DOI] [PubMed] [Google Scholar]
- 49. Chang SF, Lin PL. Frail phenotype and mortality prediction: a systematic review and meta-analysis of prospective cohort studies. Int J Nurs Stud. 2015;52(8):1362–1374. doi: 10.1016/j.ijnurstu.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 50. Clegg A, Young J, Iliffe S, Rikkert MO, Rockwood K. Frailty in elderly people. The Lancet. 2013;381:752–762. doi: 10.1016/S0140-6736(12)62167-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gobbens RJJ, van Assen MALM, Augustijn H, Goumans M, van der Ploeg T. Prediction of mortality by the Tilburg Frailty Indicator (TFI). J Am Med Dir Assoc. 2021;22(3):607.e1–607.e6. doi: 10.1016/j.jamda.2020.07.033 [DOI] [PubMed] [Google Scholar]
- 52. Pilotto A, Rengo F, Marchionni N, et al. Comparing the prognostic accuracy for all-cause mortality of frailty instruments: a multicentre 1-year follow-up in hospitalized older patients. PLoS One. 2012;7(1):e29090. doi: 10.1371/journal.pone.0029090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pilotto A, Panza F, Ferrucci L. A multidimensional prognostic index in common conditions leading to death in older patients. Arch Intern Med. 2012;172(7):594. doi: 10.1001/archinternmed.2011.1891 [DOI] [PubMed] [Google Scholar]
- 54. Ravindrarajah R, Lee DM, Pye SR, et al. The ability of three different models of frailty to predict all-cause mortality: results from the European Male Aging Study (EMAS). Arch Gerontol Geriatr. 2013;57(3):360–368. doi: 10.1016/j.archger.2013.06.010 [DOI] [PubMed] [Google Scholar]
- 55. Mitnitski A, Collerton J, Martin-Ruiz C, et al. Age-related frailty and its association with biological markers of ageing. BMC Med. 2015;13(1). doi: 10.1186/s12916-015-0400-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shamliyan T, Talley KMC, Ramakrishnan R, Kane RL. Association of frailty with survival: a systematic literature review. Ageing Res Rev. 2013;12(2):719–736. doi: 10.1016/j.arr.2012.03.001 [DOI] [PubMed] [Google Scholar]
- 57. Rockwood K, Andrew M, Mitnitski A. A comparison of two approaches to measuring frailty in elderly people. J Gerontol. 2007;62A(7):738–743. doi: 10.1093/gerona/62.7.738 [DOI] [PubMed] [Google Scholar]
- 58. Seligman BJ, Berry SD, Lipsitz LA, Travison TG, Kiel DP. Epigenetic age acceleration and change in frailty in MOBILIZE Boston. J Gerontol A Biol Sci Med Sci. 2022;77(9):1760–1765. doi: 10.1093/gerona/glac019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Crimmins EM, Thyagarajan B, Levine ME, Weir DR, Faul J. Associations of age, sex, race/ethnicity, and education with 13 epigenetic clocks in a nationally representative U.S. sample: the Health and Retirement Study. J Gerontol A Biol Sci Med Sci. 2021;76(6):1117–1123. doi: 10.1093/gerona/glab016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li X, Zhang Y, Gào X, Holleczek B, Schöttker B, Brenner H. Comparative validation of three DNA methylation algorithms of ageing and a frailty index in relation to mortality: results from the ESTHER cohort study. EBioMedicine. 2021;74:103686. doi: 10.1016/j.ebiom.2021.103686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li X, Ploner A, Wang Y, et al. Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. Harper DM, Franco E, Moskalev A, eds. Elife. 2020;9:e51507. doi: 10.7554/eLife.51507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim S, Myers L, Wyckoff J, Cherry KE, Jazwinski SM. The frailty index outperforms DNA methylation age and its derivatives as an indicator of biological age. Geroscience. 2017;39(1):83–92. doi: 10.1007/s11357-017-9960-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nelson PG, Promislow DEL, Masel J. Biomarkers for aging identified in cross-sectional studies tend to be non-causative. J Gerontol A Biol Sci Med Sci. 2020;75(3):466–472. doi: 10.1093/gerona/glz174 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.