Abstract
Tobacco etch virus Protease (TEVp), a cysteine protease, is renowned for its remarkable specific proteolysis, making it an invaluable tool for removing fusion tags from recombinant proteins. However, TEV protease's inherent insolubility limits its broad application. Fusion constructs like an N-terminal MBP fusion, known for its improved solubility, have been employed for TEVp production to address this issue. In this study, we fused the TEVp with the N-terminal domain of the spider silk protein, specifically utilizing a charge-reversed mutant (D40K/K65D) of the N-terminal domain of major ampullate spidroin-1 protein from Euprosthenops australis, referred to as NT*. This fusion construct contains a TEVp cleavage site, enabling intracellular self-processing and the release of a His7-tagged protease. The significant increase in soluble protein expression allowed us to purify approximately 90–100 mg of TEVp from a 1-L E. coli culture, surpassing previous findings by a considerable margin. The enzyme remained stable and catalytically active even after several months of storage in a deep freezer (−80 °C).
Keywords: NT*, Fusion protein, TEV protease, Cleavage site, Purification, Enzymatic activity
Graphical abstract
Highlights
-
•
NT* fusion tag offers the benefit of high expression and solubility for TEV protease production.
-
•
Expresses soluble intracellular self-processed protease amenable to a one-step affinity purification method.
-
•
Impressive purification yield, ∼90–100 mg of TEV protease from a 1-L culture of E. coli.
-
•
A stable enzyme, active even after six months of storage (−80 °C).
1. Introduction
Several fusion tags are widely employed to boost the synthesis and purity of proteins (Yadav et al., 2016). The chances of the fusion tags affecting the protein's structure or function are typically very high. Consequently, complete and specific removal of the fusion tags is preferred after expression and purification. Although various chemical methods have been employed for this purpose (Arnau et al., 2006, LaVallie et al., 1994), they usually lack the high specificity of a natural proteolytic enzyme. Therefore, the favored approach for most biomolecular applications involves the removal of fusion tags using natural proteases, which provide enhanced specificity and typically achieve cleavage under mild conditions.
In the past two decades, numerous viral proteases have been identified and assessed (Huang et al., 2013; Nallamsetty et al., 2004; Zhang et al., 2009). Within this group, certain members display exceptional proteolytic specificity. They commonly possess a chymotrypsin-like fold characterized by an unconventional catalytic triad, wherein cysteine replaces serine, and they require the presence of glutamine at the P1 position of their substrate (Dougherty et al., 1989). The nuclear inclusion protease from the tobacco etch virus (TEV) is the most thoroughly researched enzyme in this category (Phan et al., 2002). Another notable member is the human rhinovirus 3C protease (HRV 3Cp) (Matthews et al., 1994). Unlike other prominent proteases such as Factor-Xa, Thrombin, and Enteropeptidase, there have been no reports of these viral proteases cleaving at non-canonical sites within engineered fusion proteins (Waugh, 2011).
The usage of fusion tags and site-specific proteases has played a crucial role in streamlining laboratory-scale protein production workflows (Nallamsetty and Waugh, 2007; Waugh, 2005). Aligned with this observation, it's not surprising that TEV protease has earned broad acclaim among protein engineering and structural biology enthusiasts, firmly cementing its status as an indispensable tool for generating native proteins. The progress in TEVp research has been intriguing (Cesaratto et al., 2016). Over the years, researchers began to optimize TEVp and develop variants with improved properties, such as enhanced solubility, stability, and activity at different temperatures and pH ranges (Cabrita et al., 2007; Kapust et al., 2001; Nam et al., 2020; Sanchez and Ting, 2020; Wong et al., 2017). These engineered variants expanded the utility of TEVp in diverse experimental conditions and facilitated its application to a broader range of substrates. Today, it is also widely used in other research areas, including cell biology, enzymology, and functional studies of proteins. The main challenge with recombinant TEVp synthesis has been protein insolubility and low yield. Several reports of high protease production showcased different fusion tags and mutants (Blommel and Fox, 2007; Fang et al., 2007; Kapust et al., 2001; Van Den Berg et al., 2006; Wu et al., 2009). We report here the NT* fusion of the TEVp that markedly improved the production of soluble active protease. NT* tag is a charge-reversed mutant (D40K/K65D) of the N-terminal domain of major ampullate spidroin-1 protein (MaSp) from Euprosthenops australis (Kronqvist et al., 2017). The protease is expressed as a fusion but self-processed intracellularly using an internal TEVp recognition site. A His7 tag is present at the N-terminus of the cleaved protease to facilitate purification. One liter of bacterial culture yielded around 100 mg or about 15 mg of protein per gram of wet pellet. The purification is straightforward with a single immobilized metal affinity step, and the protein remains stable, retaining both its secondary structure and activity even after six months of storage in a deep freezer. This article further explores enzymatic activity and protein stability, offering potential guidelines for the broader application of the protease.
2. Materials and methods
2.1. Materials
The prepacked chromatography columns were purchased from Cytiva, USA. The ultrafiltration discs and tube concentrators were obtained from Merck Millipore, USA. Gateway cloning enzymes were obtained from Invitrogen Thermo Fisher Scientific, USA. For in vitro mutagenesis reactions, the QuikChange Lightning Site-Directed Mutagenesis Kit was employed (Agilent Technologies, USA). All restriction enzymes were obtained from New England Biolabs, USA. All other chemicals were molecular biology grade or above and purchased from Sigma-Aldrich, USA, and HiMedia Laboratories Private Limited, India. Pierce BCA Protein Assay Kit was purchased from Thermo Fisher Scientific. The Ubc9 expression vector pDN2405 was a gift from David Waugh.
2.2. Construct design
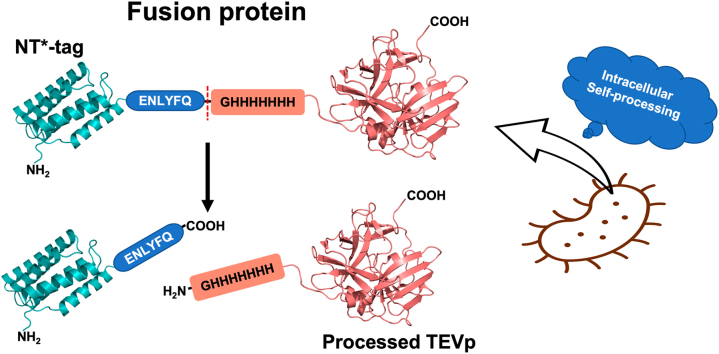
The construct design protocol was detailed in the supplementary portion of our prior article (Raran-Kurussi et al., 2022). Briefly, the pET-28b plasmid (Novagen, Merck Millipore) carrying the NT* open reading frame was converted to a gateway destination vector, pKRM025, using the Gateway Vector Conversion System (Invitrogen Thermo Fisher Scientific, USA). After that, the entry clone (pBHU07) carrying the TEVp coding sequence amplified from pDZ2087 (Addgene Plasmid #92414), and pKRM025 were recombined (LR reaction) to create the expression vector, pBHU10, which was later modified to pSRK51 by in vitro mutagenesis to obtain the desired TEVp expression clone. This mutagenesis step removed the His-tag from the NT* tag, while the His-tag at the N-terminus of TEVp remained unaltered. We designed the fusion construct with a TEVp recognition sequence (ENLYFQ/G) between the NT* tag and the TEVp. Hence, after the intracellular processing of the expressed protein, the TEVp had a short Gly-His7 sequence at the N-terminus, allowing self-processed TEVp to be captured by an immobilized metal affinity column. The NT* tag appeared in column effluent because it lacked an affinity tag.
2.3. Protein expression and purification
Plasmid encoding the NT*-TEV protease (pSRK51) was transformed into E.coli Rosetta2(DE3) cells for protein expression. Cells were grown to mid-log phase at 37 °C in LB broth containing 35 μg/mL Kanamycin, 30 μg/mL chloramphenicol, and 0.4% glucose. The overproduction of the fusion protein was induced with 1 mM of isopropyl b-D thiogalactopyranoside (IPTG) overnight at 22 °C. The cells were pelleted by centrifugation and stored at −80 °C till use. The purification of His7-TEV protease was carried out as described previously (Raran-Kurussi et al., 2017) with minor modifications. We coupled two 5-mL HisTrap FF columns (Cytiva, USA) in tandem to increase binding capacity during the immobilized metal affinity chromatography (IMAC) run. Following the affinity step, the eluted fractions containing TEV protease were supplemented with 5 mM dithiothreitol (DTT) and 2 mM ethylenediaminetetraacetic acid (EDTA) and then concentrated using an Amicon YM10 membrane (Merck Millipore). The concentrated protein was then applied onto an S-200 Hiload 16/600 superdex 200 pg column pre-equilibrated with buffer A (25 mM Na2HPO4, 100 mM NaCl, pH 7.5). The peak fractions were analyzed by SDS-PAGE, and the molecular weight was confirmed by electrospray ionization mass spectroscopy.
The Ubc9 expression and purification were carried out as described previously without any modifications (Hewitt et al., 2016).
2.4. Circular Dichroism (CD) analysis
The CD experiments were recorded using a JASCO J-1500 Spectrophotometer (Jasco, Japan) equipped with a Peltier temperature controller. The far-UV CD scans (190–260 nm) at 25 °C were collected for TEVp at 5 μM concentration in a 0.1 cm path length quartz cuvette. Protein samples were diluted in 10 mM sodium phosphate buffer, pH 7.2. Three scans with a scan speed of 50 nm/min were performed and averaged.
2.5. Intact mass spectrometry and activity assay
Varying final concentrations of TEVp (21, 2.1, 0.21, or 0.021 μM) were added to samples of 21 μM Ubc9. Buffer A was used to make up the reaction volumes. The final reaction mixtures also contained 1 mM DTT and 0.5 mM EDTA. The samples were gently mixed and incubated at 4 or 25 °C. At various time points (up to 16 h for 4 °C incubation), 50 μL aliquots were taken and quenched with 0.1% final concentration formic acid, and the extent of proteolytic cleavage was evaluated using intact protein mass spectrometry. Intact protein mass spectra were acquired using an Agilent 6545 Q-TOF LC/MS mass spectrometer linked to an Agilent 1290 Infinity II HPLC system. All HPLC runs used water and acetonitrile (ACN) with 0.1% formic acid as mobile phases. Two microliters of diluted samples (4 μM) were injected into a Poroshell 300SB-C18, 2.1 × 75mm, 5-μm HPLC column and eluted with a 5–10% ACN gradient in 12 min while maintaining a flow rate of 0.5 mL/min. Agilent MassHunter Acquisition software was used to collect mass spectra, and the resulting m/z spectra were deconvoluted and analyzed with Agilent MassHunter Analysis software. The peak integration data from the mass spectra were used to quantify the relative amounts of substrate and products.
All experiments were performed at least twice unless specified.
3. Results
3.1. Protein production using NT*-fusion construct
The NT* fusion tag is promoted as a solubility enhancer for expressing aggregation-prone peptides. It has a history of usage and has demonstrated promising outcomes even in the context of folded proteins. Previously, a distinct NT* variant was shown to increase TEVp synthesis (NT*FlSp-Tev) without impairing the protease's activity (Zhong et al., 2022). Likewise, an earlier report highlighted recombinant fusion proteins of NT, in combination with green fluorescent protein or purine nucleoside phosphorylase, showcasing high expression yields and robust activity (Arndt et al., 2022). Furthermore, the anti-aggregation properties of the NT* domain were exploited in preventing protein aggregation in human cells and established that the anti-aggregation effect of NT* remained consistent, regardless of its placement within the fusion protein (Schellhaus et al., 2022).
In this study, we created an N-terminal NT*-fusion of TEVp that self-processes intracellularly to create a Gly-His7-TEVp. The glycine in position P1ʹ is a cleavage-friendly residue. Fig. 1a shows the fusion protein sequence with an oblique symbol indicating the TEVp cleavage site. Recognizing TEVp's well-known solubility challenges, we endeavored to enhance its yield by employing NT* for solubilization. We used E. coli Rosetta 2(DE3) for protein expression, and the expressed protein was entirely soluble. A single metal affinity step captured most of the protein, and His7-TEV was almost 85% pure at this stage, as determined by SDS-PAGE. The protein purity was further enhanced by size-exclusion chromatography and the final products were judged to be >95% pure by SDS-PAGE (Fig. 1b), and the protein molecular weight was confirmed by electrospray ionization mass spectroscopy (ESI-MS) (Fig. 1c). We estimated the concentrations using a molar extinction coefficient of 31,970 M−1 cm−1 derived using the Expasy ProtParam tool (Wilkins et al., 1998). The measurements were cross-verified using a standard BCA protein assay. The final yield was about 90–100 mg from 1 L LB culture. The protein estimations are shown in the Supplementary Figs. S1 and S2.
Fig. 1.
Characterization of NT*-TEVp. (a) Amino acid sequence of the NT*-TEVp. The TEVp cleaving site is indicated with an oblique sign. (b) Purification of TEVp: SDS-PAGE analysis. Lanes 1–3 display samples from the immobilized metal-affinity chromatography run (1, soluble protein or load; 2, flow-through; 3, elution fraction). Lane 4 represents the peak fraction obtained from the SEC run. Lane M indicates the molecular weight marker. Downward arrows highlight the processed TEVp and the NT* tag in lane 1. (C) Deconvoluted mass spectrum of purified TEVp. The experimental mass is as expected.
3.2. TEV protease activity
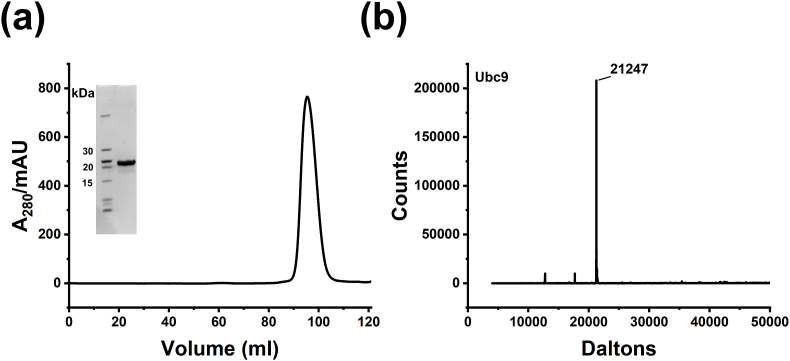
We isolated a His6-tagged Ubc9 protein with a TEVp cleavable site for use as a substrate in the activity assays. Ubc9 is an enzyme involved in the sumoylation cycle. As previously described, the plasmid pDN2405 (K48A/K49A/E54A) was used for expression and purification (Hewitt et al., 2016). The purified protein was judged to be >95% pure. The gel filtration profile and the mass data are given in Fig. 2.
Fig. 2.
Purification of substrate, His6-Ubc9. (a) SEC profile from S-200 Hiload 16/600 superdex 200 pg column run. The peak fraction analyzed by SDS-PAGE is shown in the inset. (b) Deconvoluted mass spectrum of pure protein. The experimental mass is as expected.
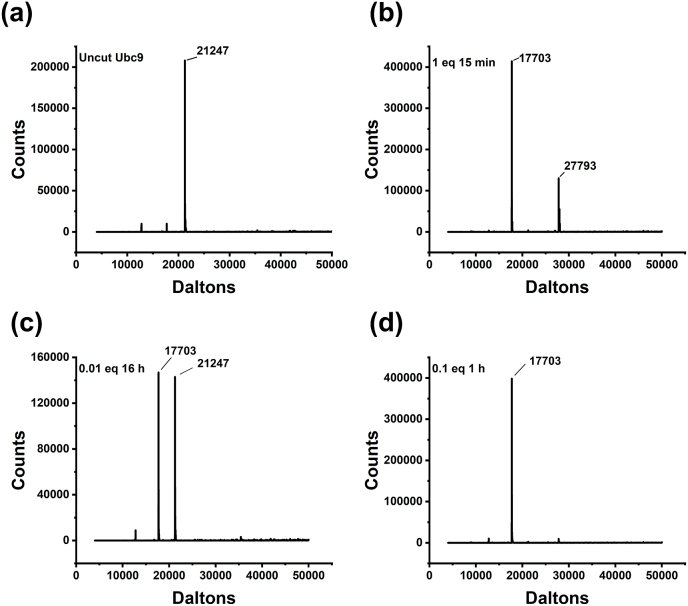
For determining the activity of TEVp, we used four different molar ratios of Ubc9: TEVp (1:1, 1:0.1, 1:0.01, and 1:0.001). The proteolytic cleavage was monitored over time using intact protein mass spectrometry (Fig. 3 and Supplementary Figs. S3 and S4). The reactions were carried out at 4 and 25 °C to account for the effects of temperature on activity. Almost complete digestion was seen at both temperatures at an equimolar ratio. At a molar ratio 1:0.1, approximately 90% of the substrate was cleaved after 4 h at 4 °C, whereas a similar amount was cleaved in 30 min at 25 °C. At 4 °C, ∼47% of the substrate was cleaved in 16 h at a molar ratio of 1: 0.01. In 60 min at the same molar ratio at 25 °C, ∼25% cleavage efficiency was seen. At a 0.001 M ratio, we observed extremely low cleavage efficiency at both temperatures. In terms of percentage, it was approximately 8% at 4 °C (16 h) and 4% at 25 °C (1 h). Fig. 3(b) and (d) show the results of an SDS-PAGE investigation of a few representative samples. These data agree well with the mass spectrometric observations (Fig. 4). Fig. 4 showcases selected mass spectrometry profiles from the digestions used to calculate the cleavage efficiencies mentioned above. The whole data set is shown in Supplementary Figs. S3 and S4. Mass spectrometry showed no evidence of non-specific cleavage or protein degradation even after incubation with TEVp in a 1:1 ratio at 4 °C for 16 h.
Fig. 3.
Cleavage efficiency analyses of TEVp using mass spectrometry. Progress of the His6-Ubc9 digestion at 4 °C (a) and 25 °C (c) at four different molar ratios. The ratios (E:S) are indicated. (b) and (d) are SDS-PAGE analyses of some representative samples belonging to (a) and (c), respectively. Gel loading in (b): Lane 1, uncut His6-Ubc9; lanes 2–5 carry samples from reactions of 1 eq (15 min), 0.1 eq (4 h), 0.01 eq (16 h) and 0.001 eq (16 h), respectively. Gel loading in (d): Lanes 1–4 carry samples from reactions of 1 eq (15 min), 0.1 eq (15 min), 0.01 eq (1 h), and 0.001 eq (1 h), respectively. Lane M denotes the molecular weight marker. Cleaved products, substrate, and enzyme bands are marked in (d). The Y-axis scale indicates cleavage efficiency, where 1.0 represents 100% cleavage, and 0.0 means 0% cleavage in (a) and (c). The peak integration data were used for all calculations.
Fig. 4.
Mass spectrometric analyses of enzymatic activity. Uncut substrate (a) and TEVp cleavage reactions [(b), (c), (d)]. A few representative sample analyses are shown. Panels (b) and (c) illustrate samples incubated at 4 °C, while panel (d) depicts samples incubated at 25 °C. Deconvoluted mass spectra are displayed for all samples. The respective enzyme: substrate molar ratios and duration are indicated.
3.3. Stability and storage
Typically, we store the TEVp samples at −80 °C after snap-freezing. The concentrations ranged from 2 to 5 mg/mL. Our preparations showed no activity loss or precipitation from the stored vials when thawed. We compared the secondary structures of stored and freshly prepared TEVp samples using Circular Dichroism (CD). Generally, the two spectra were comparable (Fig. 5a). The protein samples were also subjected to analytical size exclusion chromatography (SEC). They showed identical peak elution volumes (∼17 mL). The chromatogram confirmed the lack of protein aggregation or degradation (Fig. 5b). Moreover, we show that their catalytic efficiency remained more or less similar. (Fig. 5c).
Fig. 5.
Assessment of stability and activity of stored TEVp. (a) Comparative CD spectra: Comparison of CD spectra between freshly prepared and −80 °C stored (6 months) TEVp samples reveals strong alignment. The spectra exhibit a notable similarity, indicating structural stability during storage. (b) SEC analysis: Superimposed normalized chromatograms of stored and freshly prepared TEVp samples show a close match with no alteration in peak elution volumes. (c) SDS-PAGE analysis of activity: SDS-PAGE analysis for comparison of the activity of freshly prepared vs. stored TEVp samples. The digestion was performed at 25 °C using a molar ratio of 1:0.1 (His6-Ubc9: TEVp) for up to 30 min. Lanes 1 (t = 15 min) and 2 (t = 30 min) contain products from the reaction using stored protease; lane 3 carries the uncut His6-Ubc9. Lanes 4 (t = 15 min) and 5 (t = 30 min) show products from the reaction using freshly prepared protease. Lane M indicates the molecular weight marker. Cleaved products, substrate, and enzyme bands are marked. Freshly prepared and stored TEVp samples exhibit comparable activity.
4. Discussion
The high-level expression of TEVp using the novel construct is the key feature of our work. We produced approximately 100 mg of pure protein per liter of bacterial culture. According to our findings under initial rate settings, TEVp has approximately five-fold higher enzyme activity at 25 °C than at 4 °C. TEVp is also known to be less active at lower temperatures than another well-known viral protease, HRV 3Cp. Therefore, more incubation times are warranted at lower temperatures. Another practical consideration is to utilize more TEVp, which is feasible because the enzyme is highly rated for its proteolytic specificity. The specificity is probably attributed to their lower turnover rates than serine proteases, even though their Michaelis constants (KM) are similar (ranging from low to mid micromolar). In contrast to serine proteases, viral proteases exhibit catalytic rate constants (kcat) that are approximately 100 times lower. While transient recognition of non-canonical sites might occur in enzymatic activities, the sluggish turnover rates of viral proteases decrease the probability of undesired catalysis (Waugh, 2011). As a result, using more viral protease to digest the fusion protein is a viable option that researchers often exercise. In this context, the recombinant production of large amounts of protease becomes indispensable. The new construct we use to produce TEVp is significantly better and offers high purity and yield with minimal effort. Compared to an earlier reported MBP-fusion, pDZ2087 plasmid (Raran-Kurussi et al., 2017), the yield has nearly doubled, and the purified enzyme showed similar storage properties. In-house production of high-quality protease using the pSRK51 vector is much cheaper than commercial alternatives. A cost projection stemming from consumable expenses associated with our protein production, along with a cost comparison to other retail suppliers, is shown in the supplementary section (Tables T1 and T2). A recent publication emphasized NT* fusion in generating HRV 3Cp (Abdelkader and Otting, 2021). However, in our approach, we ensured intracellular cleavage before purification. Thus, all the yields presented in this study pertain exclusively to TEVp. The NT* tag may not offer any additional advantages in terms of activity; however, further assessment is needed to evaluate its effect on the storage of NT*-TEVp fusion protein.
CRediT authorship contribution statement
Pragyan P. Parida: Investigation, Writing – review & editing. Deepa Saraswathi: Investigation. Subbarao M.V. Mopidevi: Data curation, Formal analysis, Writing – review & editing. Sreejith Raran-Kurussi: Conceptualization, Investigation, Validation, Supervision, Methodology, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge the support of the Department of Atomic Energy, Government of India, under project identification number RTI. 4007. Additionally, our thanks go to Dr. Kaustubh R. Mote for his insightful discussions, and we recognize the Biophysics facility at TIFRH for their support.
Handling Editor: Dr A Wlodawer
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2023.100106.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Abdelkader E.H., Otting G. NT*-HRV3CP: an optimized construct of human rhinovirus 14 3C protease for high-yield expression and fast affinity-tag cleavage. J. Biotechnol. 2021;325:145–151. doi: 10.1016/j.jbiotec.2020.11.005. [DOI] [PubMed] [Google Scholar]
- Arnau J., Lauritzen C., Petersen G.E., Pedersen J. Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Expr. Purif. 2006;48:1–13. doi: 10.1016/j.pep.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Arndt T., Jaudzems K., Shilkova O., Francis J., Johansson M., Laity P.R., Sahin C., Chatterjee U., Kronqvist N., Barajas-Ledesma E., Kumar R., Chen G., Strömberg R., Abelein A., Langton M., Landreh M., Barth A., Holland C., Johansson J., Rising A. Spidroin N-terminal domain forms amyloid-like fibril based hydrogels and provides a protein immobilization platform. Nat. Commun. 2022;13:4695. doi: 10.1038/s41467-022-32093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommel P.G., Fox B.G. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr. Purif. 2007;55:53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita L.D., Gilis D., Robertson A.L., Dehouck Y., Rooman M., Bottomley S.P. Enhancing the stability and solubility of TEV protease using in silico design. Protein Sci. 2007;16:2360–2367. doi: 10.1110/ps.072822507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesaratto F., Burrone O.R., Petris G. Tobacco Etch Virus protease: a shortcut across biotechnologies. J. Biotechnol. 2016;231:239–249. doi: 10.1016/j.jbiotec.2016.06.012. [DOI] [PubMed] [Google Scholar]
- Dougherty W.G., Cary S.M., Dawn Parks T. Molecular genetic analysis of a plant virus polyprotein cleavage site: a model. Virology. 1989;171:356–364. doi: 10.1016/0042-6822(89)90603-X. [DOI] [PubMed] [Google Scholar]
- Fang L., Jia K.-Z., Tang Y.-L., Ma D.-Y., Yu M., Hua Z.-C. An improved strategy for high-level production of TEV protease in Escherichia coli and its purification and characterization. Protein Expr. Purif. 2007;51:102–109. doi: 10.1016/j.pep.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Hewitt W.M., Lountos G.T., Zlotkowski K., Dahlhauser S.D., Saunders L.B., Needle D., Tropea J.E., Zhan C., Wei G., Ma B., Nussinov R., Waugh D.S., Schneekloth J.S. Insights into the allosteric inhibition of the SUMO E2 enzyme Ubc9. Angew. Chem. Int. Ed. 2016;55:5703–5707. doi: 10.1002/anie.201511351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Li Q., Chen A.S., Kang C. West Nile virus protease activity in detergent solutions and application for affinity tag removal. Anal. Biochem. 2013;435:44–46. doi: 10.1016/j.ab.2012.12.015. [DOI] [PubMed] [Google Scholar]
- Kapust R.B., Tözsér J., Fox J.D., Anderson D.E., Cherry S., Copeland T.D., Waugh D.S. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. Des. Sel. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- Kronqvist N., Sarr M., Lindqvist A., Nordling K., Otikovs M., Venturi L., Pioselli B., Purhonen P., Landreh M., Biverstål H., Toleikis Z., Sjöberg L., Robinson C.V., Pelizzi N., Jörnvall H., Hebert H., Jaudzems K., Curstedt T., Rising A., Johansson J. Efficient protein production inspired by how spiders make silk. Nat. Commun. 2017;8 doi: 10.1038/ncomms15504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVallie E.R., McCoy J.M., Smith D.B., Riggs P. Enzymatic and Chemical Cleavage of Fusion Proteins. Curr. Protoc. Mol. Biol. 1994;28 doi: 10.1002/0471142727.mb1604bs28. [DOI] [PubMed] [Google Scholar]
- Matthews D.A., Smith W.W., Ferre R.A., Condon B., Budahazi G., Slsson W., Villafranca J.E., Janson C.A., McElroy H.E., Gribskov C.L., Worland S. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- Nallamsetty S., Kapust R.B., Tözsér J., Cherry S., Tropea J.E., Copeland T.D., Waugh D.S. Efficient site-specific processing of fusion proteins by tobacco vein mottling virus protease in vivo and in vitro. Protein Expr. Purif. 2004;38:108–115. doi: 10.1016/j.pep.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Nallamsetty S., Waugh D.S. A generic protocol for the expression and purification of recombinant proteins in Escherichia coli using a combinatorial His6-maltose binding protein fusion tag. Nat. Protoc. 2007;2:383–391. doi: 10.1038/nprot.2007.50. [DOI] [PubMed] [Google Scholar]
- Nam H., Hwang B.J., Choi D., Shin S., Choi M. Tobacco etch virus (TEV) protease with multiple mutations to improve solubility and reduce self‐cleavage exhibits enhanced enzymatic activity. FEBS Open Bio. 2020;10:619–626. doi: 10.1002/2211-5463.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan J., Zdanov A., Evdokimov A.G., Tropea J.E., Peters H.K., Kapust R.B., Li M., Wlodawer A., Waugh D.S. Structural basis for the substrate specificity of tobacco etch virus protease. J. Biol. Chem. 2002;277:50564–50572. doi: 10.1074/jbc.M207224200. [DOI] [PubMed] [Google Scholar]
- Raran-Kurussi S., Cherry S., Zhang D., Waugh D.S. In: Heterologous Gene Expression in E.Coli. Burgess-Brown N.A., editor. Springer New York; New York, NY: 2017. Removal of affinity tags with TEV protease; pp. 221–230. [DOI] [Google Scholar]
- Raran-Kurussi S., Sharwanlal S.B., Balasubramanian D., Mote K.R. A comparison between MBP- and NT* as N-terminal fusion partner for recombinant protein production in E. coli. Protein Expr. Purif. 2022;189 doi: 10.1016/j.pep.2021.105991. [DOI] [PubMed] [Google Scholar]
- Sanchez M.I., Ting A.Y. Directed evolution improves the catalytic efficiency of TEV protease. Nat. Methods. 2020;17:167–174. doi: 10.1038/s41592-019-0665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellhaus A.K., Xu S., Gierisch M.E., Vornberger J., Johansson J., Dantuma N.P. A spider silk-derived solubility domain inhibits nuclear and cytosolic protein aggregation in human cells. Commun. Biol. 2022;5:505. doi: 10.1038/s42003-022-03442-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Berg S., Löfdahl P.-Å., Härd T., Berglund H. Improved solubility of TEV protease by directed evolution. J. Biotechnol. 2006;121:291–298. doi: 10.1016/j.jbiotec.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Waugh D.S. An overview of enzymatic reagents for the removal of affinity tags. Protein Expr. Purif. 2011;80:283–293. doi: 10.1016/j.pep.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh D.S. Making the most of affinity tags. Trends Biotechnol. 2005;23:316–320. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Wilkins M.R., Gasteiger E., Bairoch A., Sanchez J.-C., Williams K.L., Appel R.D., Hochstrasser D.F. 2-D Proteome Analysis Protocols. Humana Press; New Jersey: 1998. Protein identification and analysis tools in the ExPASy server; pp. 531–552. [DOI] [PubMed] [Google Scholar]
- Wong J., Chen X., Truong K. Engineering a temperature sensitive tobacco etch virus protease. Protein Eng. Des. Sel. 2017;30:705–712. doi: 10.1093/protein/gzx050. [DOI] [PubMed] [Google Scholar]
- Wu X., Wu D., Lu Z., Chen W., Hu X., Ding Y. A novel method for high-level production of TEV protease by superfolder GFP tag. J. Biomed. Biotechnol. 2009;2009:1–8. doi: 10.1155/2009/591923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav D.K., Yadav N., Yadav S., Haque S., Tuteja N. An insight into fusion technology aiding efficient recombinant protein production for functional proteomics. Arch. Biochem. Biophys. 2016;612:57–77. doi: 10.1016/j.abb.2016.10.012. [DOI] [PubMed] [Google Scholar]
- Zhang D., Tözsér J., Waugh D.S. Molecular cloning, overproduction, purification and biochemical characterization of the p39 nsp2 protease domains encoded by three alphaviruses. Protein Expr. Purif. 2009;64:89–97. doi: 10.1016/j.pep.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Kumar R., Wang Y., Biverstål H., Ingeborg Jegerschöld C., J B Koeck P., Johansson J., Abelein A., Chen G. Amyloid fibril formation of arctic amyloid-β 1-42 peptide is efficiently inhibited by the BRICHOS domain. ACS Chem. Biol. 2022;17:2201–2211. doi: 10.1021/acschembio.2c00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.