Abstract
Inflammation is a nonspecific immune response against injury caused by a harmful agent that strives to restore tissue function and homeostasis. Dodonaea angustifolia L.f. (Sapindaceae) is a medium-sized shrub used to treat a variety of diseases in traditional medicine. In the current study, integrated network-pharmacology and molecular docking approaches were used to identify the active constituents, their possible targets, signaling pathways, and anti-inflammatory effects of flavonoids from D.angustifolia. D. angustifolia active ingredients were acquired from the Indian Medicinal Plants, Phytochemistry and Therapeutics (IMPPAT), and Traditional Chinese Medicine System Pharmacology (TCMSP) databases. The screening included the ten most prevalent D. angustifolia components, and the SwissTargetPrediction database was utilized to anticipate the targets of these compounds. Anti-inflammatory genes were found using the GeneCards database. The 175 overlapping genes were discovered as prospective D. angustifolia anti-inflammatory targets. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed that the overlapped targets were closely related to the major pathogenic processes linked to inflammation, such as response to organonitrogen compound, protein kinase activity, phosphotransferase activity, pI3k-Akt signaling pathway, metabolic pathways, and chemical carcinogenesis. Compound–target–pathway, and protein–protein interaction networks revealed 6-Methoxykaempferol and 5-Hydroxy-7,8 dimethoxyflavone as key compounds, and AKT1, VEGFA, and EGFR as key targets. Furthermore, molecular docking followed by molecular dynamic (MD) simulation of D. angustifolia active ingredients with core proteins fully complemented the binding affinity of these compounds and indicated stable complexes at the docked site. These findings reveal D. angustifolia 's multi-target, multi-compound, and multi-pathway strategies against inflammation. Our study paved the way for further research into the mechanism for developing D. angustifolia -based natural products as alternative therapies for inflammation.
Keywords: Dodonea angustifolia, Anti-inflammatory, Network pharmacology, Flavonoids, KEGG, Molecular docking
1. Introduction
Inflammation is a complicated pathological and physiological process (Pant et al., 2014). It controls a variety of pathological and physiological processes in the body by influencing a number of cells and microenvironmental elements (Korniluk et al., 2017). Inflammation activates immune cells and tissue stromal cells, allowing proteins and cells from the vascular system to enter infected or injured tissues and promote repairing (Mraz and Haluzik, 2014, Owens, 2015). Inflammation often develops in stages, beginning with a quick induction phase that triggers a pro-inflammatory response, then gradually progressing to a resolution phase. Although self-limiting inflammation is physiological and essential for killing germs, it is harmful to the systemic responses of the affected organs and adjacent organs if it persists (Raucci et al., 2019). Several studies revealed that inflammation controls the onset and progression of a wide range of complex disorders. Acute inflammation and chronic inflammation are two different types of inflammation. The acute inflammation subsides quickly and typically benefits the host. Chronic inflammation occurs when inflammation persists for an extended period of time and can lead to a number of chronic diseases such as diabetes, obesity, arthritis, cardiovascular, pancreatitis, metabolic, neurological, and some types of cancer.
Investigators have focused on the mechanisms of the inflammation process in inflammatory bowel disease (IBD), psoriasis, arthritis, depression, and atherosclerotic disease, with higher levels of inflammatory mediators observed at the sites of the lesion. The inflammation aggravates the progression of the disease, which exacerbates the inflammation, producing a vicious cycle that makes therapy challenging. Hence, we must emphasize that inflammation plays a key role in the onset and progression of disease, and substances with anti-inflammatory properties are the direction to search for therapeutic drugs (Peng et al., 2021). Many related drugs including steroidal anti-inflammatory drugs (SAIDs) and non-steroidal anti-inflammatory drugs (NSADIDs) have been identified in the marker (Osborn and Hunt, 2007, Islam et al., 2018). However, the drugs have certain drawbacks because long-term usage causes unfavorable reactions in numerous organs (Narsinghani and Sharma 2014).
Flavonoids, a diverse group of phytochemicals, have been extensively studied for their health benefits, including their anti-inflammatory properties. Numerous studies have suggested that flavonoids possess potent anti-inflammatory activities, such as the suppression of proinflammatory cytokines and the inhibition of key enzymes involved in the inflammatory process, like cyclooxygenase, lipoxygenase, and inducible nitric oxide synthase (Kim et al., 2004, Amic et al., 2007). Recent research has shown that regulatory enzymes and transcription factors which are important in regulating inflammatory mediators can be inhibited by flavonoid derivatives. In fact, in vitro and animal models suggested that flavonoids have the potential to inhibit the onset as well as the development of inflammatory diseases (Maleki, Crespo and Cabanillas 2019).
Dodonaea, also known as hop-bush or sand olive, is a genus with approximately 70 species including D. angustifolia (Shepherd et al., 2007). D. angustifolia L. f. (Sapindaceae), also called as Ketketa in Ethiopia, is a 3 m tall shrub. It also occurs naturally in various forms from southern Africa to Arabia, as well as in New Zealand and Australia. D. angustifolia plant decoctions are frequently used as a cure for many ailments, injuries, infections, dental pain, and jungle fever (de Oliveira et al., 2012). The plant is said to have antifungal (Pirzada et al., 2010), antibacterial (Teffo, Aderogba and Eloff 2010), anti-inflammatory (Getie et al., 2003), antidiabetic (Veerapur et al., 2010), and antidiarrheal properties (Rajamanickam et al., 2010). Particularly for D. angustifolia, flavonoids have been reported as the primary active components. Their potential benefits in inflammation and other health conditions have been emphasized in the literature (Tadeg et al., 2005).
Recognizing the potential of phytoconstituents, Hopkins (Hopkins, 2007) formulated an integrative computational approach “network pharmacology”. Network pharmacology transitioned the paradigm that single gene disordered required single target drugs while complicated diseases where gene network is involved required more holistic multiple-targeted therapies (Noor et al., 2023, Noor et al., 2022). Thus, network pharmacology has now been emerged as an asset in the process of drug development, contributing significantly to the reinvigoration of traditional knowledge (Noor et al., 2022).
Explorations into the anti-inflammatory properties and mechanisms of action of D. angustifolia are in their nascent stages. Therefore, uncovering its pivotal bioactive components and potential targets will considerably augment our understanding of D. angustifolia's anti-inflammatory activity. In our present study, we have implemented an integrative approach involving network-pharmacology-based screening, molecular docking, and comprehensive molecular dynamic simulation to analyze the anti-inflammatory effects of flavonoids extracted from D. angustifolia. This methodology offers a thorough and nuanced understanding of the plant's therapeutic potential against inflammatory conditions.
2. Materials and methods
2.1. Virtual screening of active phytomolecules
The active phytomolecules of D. angustifolia were retrieved from the Traditional Chinese Medicine System Pharmacology Database (TCMSP) database (Ru et al., 2014) and the Indian Medicinal Plants, Phytochemistry and Therapeutics (IMPPAT) (Mohanraj et al., 2018). From the PubChem database, the structures of prevalent phytochemicals were downloaded in the SMILE and 3D SDF formats (Kim et al., 2019).
2.2. Screening of Anti-Inflammatory targets
The anti-inflammatory gene targets were identified by scanning the GeneCards database (The Human Gene Database) for the keywords “Inflammatory” (Safran et al., 2010). An Excel spreadsheet was used for saving all related genes for further study.
2.3. Screening of D. Angustifolia potential target genes
Potential targets of active constituents from D. angustifolia were obtained using the SwissTargetPrediction database (Daina, Michielin and Zoete 2019). For further research, only prospective targets with a probability score higher than 0 were chosen.
2.4. Identification of overlapping genes
Target intersections between anti-inflammatory targets and putative active phytomolecules targets of D. angustifolia were found employing the Venny 2.1.0 online database (Oliveros 2007). These overlapping targets were recognized as potential anti-inflammatory targets.
2.5. Protein-Protein interaction analysis
The above-mentioned potential anti-inflammatory targets were assessed using the STRING database for protein–protein interaction (PPI) analysis with a confidence score greater than 0.4 and a species limitation of “Human sapiens” (Szklarczyk et al., 2023). In order to identify possible anti-inflammatory core targets, the findings of the string PPI analysis were then loaded into the Cytoscape software (Kohl, Wiese and Warscheid 2011).
2.6. Network construction
The network construction process, which serves as the backbone of our investigation, was executed using the Cytoscape program (Kohl, Wiese and Warscheid 2011). Cytoscape is a powerful open-source bioinformatics software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations, gene expression profiles, and other state data. In this study, Cytoscape was utilized to develop an integrated network of active phytomolecules from D. angustifolia and their anti-inflammatory targets. Specifically, the active phytomolecules of D. angustifolia, identified through our earlier screening process, were imported into Cytoscape along with the associated key and core anti-inflammatory targets. These targets were identified based on their roles in the inflammation process, as per our Gene Ontology and KEGG enrichment analyses.
By using Cytoscape, these various components were connected based on their interactions and relationships. Nodes in this network represent either the active phytomolecules or the anti-inflammatory targets, and the edges represent the potential interactions between them. This complex network, thus visualized, offers a comprehensive view of how the multiple active compounds in D. angustifolia might interact with different anti-inflammatory targets, thereby hinting at the plant's multi-target, multi-compound, and multi-pathway strategies against inflammation. Such a network not only aids in comprehending the molecular mechanisms underlying the anti-inflammatory properties of D. angustifolia but also serves as a robust tool for further investigation and potential drug discovery.
2.7. Enrichment and pathway analysis
The ShinyGO database was used to perform the enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and GO functional on potentially anti-inflammatory key targets (Ge, Jung and Yao 2020). Three groups of GO terms were formed: biological process (BP), molecular function(MF), and cellular component (CC). The top 20 KEGG pathways and GO analysis data (BP, CC, and MF) were exhibited in the form of an enrichment dot bubble by uploading the data to the Bioinformatics platform (Zhao et al., 2022). Statistical significance was evaluated using the conventional hypergeometric test. The Benjamini-Hochberg method was used to control the false discovery rate (FDR) for multiple hypothesis testing, and the corrected p-value of 0.05 was then used as the significant threshold (Khan and Lee 2022).
2.8. Molecular docking
The two active chemicals; 6-Methoxykaempferol and 5-Hydroxy-7,8 dimethoxyflavone from D. angustifolia were sourced from the NCBI Pubchem database. This database provides three-dimensional (3D) structures of these molecules in a Spatial Data File (SDF) format, offering detailed molecular structure information crucial for the subsequent docking process. The crystal structures of the top three potential anti-inflammatory core targets, which are proteins believed to have significant roles in anti-inflammatory activity (AKT1 PDB ID: 3o96, EGFR PDB ID: 7jxq, VEGFA PDB ID: 2vpf), were downloaded from the Protein Data Bank (PDB) in PDB format (Berman et al., 2000). These protein structures act as docking sites for the active phytomolecules. Upon acquiring these protein structures, we removed any water molecules and ligands - compounds that may bind to the protein - embedded within the protein crystal structure complexes. This step is necessary to prevent any interference with the docking process and was achieved using the BIOVIA Discovery Studio Visualizer 2021 (Studio 2008) tool. The docking process was then initiated, wherein each phytomolecule was virtually 'fitted' into its respective protein target. This was conducted individually for each compound to ensure accurate and interference-free results. We used Autodock Vina, a well-regarded software integrated within the PyRx virtual screening tool for this purpose (Dallakyan and Olson 2015).
However, before we started the docking, the phytomolecules were energy minimized. This process, performed using the OpenBable tool in PyRx, optimizes the molecular structure for docking by finding its lowest energy conformation. Following this, the molecules were converted into pdbqt format, which is compatible with the Autodock Vina software. Finally, we visualized and analyized the docking results using BIOVIA Discovery Studio Visualizer 2021 and Pymol programs.
2.9. Molecular dynamic (MD) simulation
MD simulation is a computer technique that uses explicit modeling of each bond and atom in a system, enabling a highly thorough analysis of molecular dynamics. This method entails solving the equations of motion for each atom in the system based on the interatomic potentials that define their interactions. MD simulations of docked complexes were performed using GROMACS 2018 software and the OPLS-AA/L force field. The protein's 3D structure served as the starting point for the simulations, and the DockPrep program was used for additional optimization (Van Der Spoel et al., 2005). Docked complexes of ligand molecules with the target protein with the highest binding affinity were used as the starting point for the MD simulation. The SwissParam website was used to parameterize the ligand molecule (Zoete et al., 2011). Following earlier research, 20 ns of MD simulations were run later (Fatima et al., 2022). General MD simulation parameters for each complex such as radius of gyration (RoG), root mean square deviation (RMSD), and root mean square fluctuation (RMSF) were examined.
3. Results
3.1. D. angustifolia compounds target and Anti-Inflammatory disease target screening
This study involved a comprehensive analysis of various phytomolecules, each characterized by unique structural attributes and belonging to various subclasses of flavonoids. These natural compounds, sourced from different plant species, were scrutinized for their potential pharmacological and health-related properties. The investigation encompassed ten specific phytomolecules: Pinocembrin, a flavanone with a molecular weight of 256.25; 5-Hydroxy-7,8-dimethoxyflavone, a dimethoxyflavone with a molecular weight of 298.29; Luteolin, a tetrahydroxyflavone with a molecular weight of 286.24; Isokaempferide, a flavonol with a molecular weight of 300.26; Santin, an O-methylated flavonol with a molecular weight of 344.3; Kumatakenin, an O-methylated flavonol with a molecular weight of 314.29; Rhamnazin, an O-methylated flavonol with a molecular weight of 330.29; Rhamnocitrin, a monomethoxyflavone with a molecular weight of 300.26; Retusin, an O-methylated flavonol with a molecular weight of 358.3; and 6-Methoxykaempferol, a flavonoid with a specific structure, 3,4′,5,7-Tetrahydroxy-6-methoxyflavone, and a molecular weight of 316.26 (Table 1). These phytomolecules have been extensively studied due to their potential health benefits and diverse applications. These findings provide valuable insights into their chemical diversity and properties, offering a solid foundation for future research aimed at harnessing their therapeutic potential.
Table 1.
List of D. angustifolia’s active phytomolecules.
| PubChem CIDs | Phytomolecule name | Phytomolecule class | Phytomolecules structure | Molecular weight |
|---|---|---|---|---|
| 68,071 | Pinocembrin | Flavonoids (flavanone) |  |
256.25 |
| 188,316 | 5-Hydroxy-7,8-dimethoxyflavone | Flavonoids (dimethoxyflavone) |  |
298.29 |
| 5,280,445 | Luteolin | Flavonoids (tetrahydroxyflavone) |  |
286.24 |
| 5,280,862 | Isokaempferide | Flavonoids (flavonol) |  |
300.26 |
| 5,281,695 | Santin | Flavonoidas(trimethoxyflavone) |  |
344.3 |
| 5,318,869 | Kumatakenin | Flavonoids (O-methylated flavonol) |  |
314.29 |
| 5,320,945 | Rhamnazin | Flavonoids (O-methylated flavonol) | 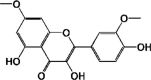 |
330.29 |
| 5,320,946 | Rhamnocitrin | Flavonoids (monomethoxyflavone) |  |
300.26 |
| 5,352,005 | Retusin | Flavonoids (O-methylated flavonol) |  |
358.3 |
| 5,377,945 | 6-Methoxykaempferol | Flavonoids (3,4′,5,7-Tetrahydroxy-6-methoxyflavone) |  |
316.26 |
Each of the ten distinct compounds investigated generated a set of 100 target predictions, resulting in a total of 1000 potential targets. To prioritize biologically significant targets for further scrutiny, we employed a stringent probability threshold of greater than 0.4 (Basavarajappa et al., 2023). This threshold ensured that only targets with a greater probability of interaction with the compounds of interest were selected for subsequent analysis, enhancing the biological relevance of our findings. Hence, 193 pharmacological targets were identified. Additionally, a total of 11,448 anti-inflammatory targets were retrieved from the GeneCards database. Venn diagram was constructed to identify common genes between pharmacological targets and compounds associated with diseases. The Venn diagram showed that a total of 175 compounds were found to be common, which later be considered as potential anti-inflammatory targets for D. angustifolia. The 175 overlapping genes were recognized as prospective D. angustifolia anti-inflammatory targets when both D. angustifolia and anti-inflammatory targets were imported into a Venn diagram (Fig. 1).
Fig. 1.
Intersecting targets between anti-inflammatory -related targets and potential targets of D. angustifolia active phytomolecules.
3.2. Enrichment analysis
Pathway enrichment and GO analysis of overlapping proteins were carried out to ascertain their biological properties using ShinyGO resources. In Fig. 2, the top 20 KEGG pathways and the top 20 GO analysis data (BP, CC, and MF) are displayed. According to the findings of the GO enrichment analysis, D. angustifolia anti-inflammatory targets are engaged in a number of biological processes, including response to oxygen containing compound, response to organonitrogen compound, and regulation of programmed cell death, etc (Fig. 2A). Similarly, anti-inflammatory targets of D. angustifolia are mostly receptor complex, plasma membrane region, membrane raft, perinuclear region of cytoplasm, and axon in the cellular component category (Fig. 2B). The same is applicable for enhanced molecular function ontologies, which predominate protein kinase activity, phosphotransferase activity, ATP binding, etc (Fig. 2C). The anti-inflammatory targets of D. angustifolia appear to be predominantly implicated in pathways in cancer, pI3k-Akt signaling pathway, metabolic pathways, chemical carcinogenesis, endocrine resistance, etc., according to the KEGG pathway enrichment data. (Fig. 2D).
Fig. 2.
GO enrichment analysis of 175 Anti-Inflammatory core targets (A) Biological process, (B) Cellular components, (C) Molecular function, and (D) KEGG pathway Analysis.
3.3. Protein-Protein interaction analysis and network construction
The 175 anti-inflammatory targets of D. angustifolia were imported into STRING Database 11.5 to create the PPI network. In terms of the PPI enrichment, the local clustering coefficient, average node degree, and p-value were 0.528, 4.48, and 1.0e-16, respectively (Fig. 3A). A network exhibiting the 10 active chemicals operating on the 175 anti-inflammatory targets of D. angustifolia was created using Cytoscape Software 3.9.0 Each edge indicates the link between an active substance and D. angustifolia anti-inflammatory target (Fig. 3B).
Fig. 3.
(A) The PPI network of 175 potential anti-inflammatory key targets constructed by employing STRING database. (B) The binding interaction network of D. angustifolia active phytochemicals with potential anti-inflammatory -related targets. (C) The binding interaction network of 6-Methoxykaempferol and 5-Hydroxy-7,8 dimethoxyflavone and with potential anti-inflammatory -related targets.
Only two active chemicals 6-Methoxykaempferol and 5-Hydroxy-7,8 dimethoxyflavone were found to interact with D. angustifolia anti-inflammatory targets. So these two active chemicals were considered for further research. The binding interaction network of these two active chemicals with potential anti-inflammatory -related targets is represented in Fig. 3C.
To acquire the PPI network, the PPI analysis findings were exported as a simple textual data format (.tsv) file and uploaded into Cytoscape software 3.9.0. The network is depicted in Fig. 4A for the top 20 effect targets, based on the calculation of degree of centrality, as a series of colored circles with each node's color representing its degree, ranging from yellow (lowest) to red (highest).
Fig. 4.
(A) Top 20 potential anti-inflammatory key targets, and (B) The top five potential anti-inflammatory core-targets constructed using the Cytoscape software based on degree centrality (DC). The color of each node changes from gradually from red (highest) to yellow (lowest) as its degree decreases. (D) The five anti-inflammatory core-targets ranked by DC > average value of (62.8). (E) Table shown the details of five anti-inflammatory core-targets ranked by DC.
The five nodes that satisfied the degree of centrality (DC) criterion with an average value of around ≥ 70.00 were retrieved and are considered of as anti-inflammatory core targets Fig. 4B. Fig. 4C shows a bar graph of the top ten anti-inflammatory core targets as determined by DC. The five anti-inflammatory core targets include AKT1, VEGFA, EGFR, ESR1, and SRC. Details of these targets were mentioned in Fig. 4D.
3.4. Molecular docking
The top three anti-inflammatory core targets (AKT1, VEGFA, and EGFR) were molecularly docked with the two active compounds (5-Hydroxy-7,8 dimethoxyflavone and 6-Methoxykaempferol) of D. angustifolia. Molecular docking was performed to determine the binding ability of D. angustifolia bioactive components and the anti-inflammatory genes. These active chemicals exhibit strong binding ability towards these genes. The binding energy scores for 5-Hydroxy-7,8 dimethoxyflavone and 6-Methoxykaempferol against AKT1 were −9.2 and −8.9 kcal/mol, respectively (Fig. 5A). In term of interaction, 5-Hydroxy-7,8 dimethoxyflavone formed stable hydrogen bonds with Asn54 and Thr82 within the active site of AKT1 (Fig. 5B). Similarly, 6-Methoxykaempferol interact with Gln79, Thr82 and Asp292 via hydrogen bonds at the active site chamber (Fig. 5C).
Fig. 5.
(A) Docking and interaction mechanism of Ligands with AKT1. Amino acid interactions with (B) 5-Hydroxy-7–8-dimethoxyflavone and (C) 6-Methoxykaempferol within the active site of AKT1. (D) Docking and interaction mechanism of Ligands with VEGFA. Amino acid interactions with (E) 5-Hydroxy-7–8-dimethoxyflavone and (F) 6-Methoxykaempferol within the active site of VEGFA. (G) Docking and interaction mechanism of Ligands with EGFR. (H) 5-Hydroxy-7–8-dimethoxyflavone and (I) 6-Methoxykaempferol against EGFR.
On the other hand, The binding energy scores for 5-Hydroxy-7,8 dimethoxyflavone and 6-Methoxykaempferol against VEGFA were −6.0 and −6.6 kcal/mol, respectively. The interaction mechanism of compounds with VEGFA is represented in Fig. 5D. Both ligands adapted similar binding modes in the binding site cavity. 5-Hydroxy-7,8 dimethoxyflavone formed hydrogen bonds with Cys61 and Asn62 while 6-Methoxykaempferol interact with Asp64 and Arg224 at the active site chamber (Fig. 5E & F).Furthermore, the results of docking the ligands with EGFR showed that the binding energy scores for 5-Hydroxy-7,8 dimethoxyflavone and 6-Methoxykaempferol against EGFR were −8.0 and −8.9 kcal/mol, respectively. interact with identical catalytic residues at substrate-binding pockets (Fig. 5G). In details, the 6-Hydroxy-7,8 dimethoxyflavone was involved in a single hydrogen bond with Lys745 and many stabilized via several homophobic forces with hydrophobic residues such as Leu777 and Leu858 (Fig. 5H). Similarly, 6-Methoxykaempferol interacted mainly with hydrophobic residues at the active site chamber (Fig. 5I).
The details of amino acids involved in stabilizing the two compounds at protein active sites are given in Table 2. Overall, the molecular docking outcomes agreed with the network pharmacology-based screening results, indicating that network pharmacology tools were effective in this study.
Table 2.
Docking and amino acid interactions of 5-Hydroxy-7–8-dimethoxyflavone and 6-Methoxykaempferol against AKT1, VEGFA and EGFR.
| Protein | Phytochemical |
|||
|---|---|---|---|---|
|
5-Hydroxy-7–8-dimethoxyflavone |
6-Methoxykaempferol |
|||
| H- bonds | Hydrophobic | H- bonds | Hydrophobic | |
| AKT1 | Asn54, Thr82 | Trp80 | Asn54, Gln79, Thr82, Val271, Tyr272, Asp292 | Trp80, Val270 |
| VEGFA | Ser50, Cys61, Asn62, Asp63 | Asp34, Glu64 | Asp63, Arg224 | Ile46, Glu64 |
| EGFR | Lys745 | Leu777, Leu788, Leu858, Phe856 | – | Leu759, Met766, Leu788, Leu858, |
3.5. MD simulation
GROMACS was used to execute all-atom MD simulations for 20 ns to evaluate the interaction of AKT1, VEGFA, and EGFR protein, and the stability of the ligand molecules. MD simulation of each complex was performed to get the RMSD, RMSF, and RoG values, which ultimately helped determine the docked complex's stability.
Protein-ligand RMSD (Root Mean Square Deviation) is a metric commonly used to assess the similarity between two structures of a protein–ligand complex. It measures the deviation between the positions of the atoms in the protein–ligand complex structures after the structures have been aligned based on a common reference frame. RMSD is calculated by taking the square root of the average of the squared differences between the corresponding atoms in the two structures. In the case of a protein–ligand complex, the RMSD is calculated only for the ligand atoms, while the protein atoms are held fixed. A low RMSD value indicates that the two structures are similar, while a high RMSD value indicates that they are different. Typically, a cutoff value of 2 Å is used to define a “good” ligand pose, although the acceptable RMSD value can vary depending on the application and the accuracy of the computational method used. In Fig. 6A protein in complex with both ligands was stable and no deviation was observed during 20 ns and 1000 frames. In Fig. 6B the RMSD of the ligands was observed and it was concluded that both ligands during 20 ns simulations were stable at their best binding poses, no deviation was observed.
Fig. 6.
MD Simulation analysis of 5-Hydroxy-7–8-dimethoxyflavone and 6-Methoxykaempferol in complex with AKT1.
Similarly, RMSD in Fig. 7A was observed and it was revealed that protein was stable during 20 ns simulation after binding with both ligands throughout 1000 frames. An average deviation 0.01 ± 0.001 nm was observed and a minor deviation of 0.01 was seen at 18 ns. In Fig. 7B both ligands with the respective protein were strongly stable till 8 ns but after 8 ns 0.01 nm deviation was observed which was in the range of protein–ligand strong binding affinity. In Fig. 8A, the protein with respective both ligands was stable and the mean deviation was calculated which was 0.001 nm and this was an acceptable range. In Fig. 8B, RMSD showed that 5-Hydroxy-7-8-dimethoxyflavone was stable during the whole simulation time but 6-Methoxykaempferol was not stable till 9 ns of simulation but after 9 ns simulation this ligand was also stable. Herein, results showed that protein–ligand complexes were strongly stable during 20 ns in a range of 2 Å.
Fig. 7.
MD Simulation analysis of 5-Hydroxy-7–8-dimethoxyflavone and 6-Methoxykaempferol in complex with EGFR.
Fig. 8.
MD Simulation analysis of 5-Hydroxy-7–8-dimethoxyflavone and 6-Methoxykaempferol in complex with VEGFA.
Protein root mean square fluctuation (RMSF) is a measure of the flexibility or mobility of a protein's structure. It calculates the average deviation of each atom in a protein from its mean position over a given period of time of simulation. The RMSF can provide insights into protein dynamics and functional changes, such as protein–ligand binding or conformational changes. Typically, the RMSF values are plotted as a function of the protein's residue number to visualize regions of high flexibility or mobility in the protein structure. High RMSF values usually correspond to more flexible regions, such as loop regions or exposed surface areas, while low RMSF values correspond to more rigid or stable regions, such as alpha-helices or beta-sheets. In Fig. 6D, Fig. 7D and Fig. 8D the RMSF of protein respective to all ligands revealed that there were no higher conformational changes in protein structure during 20 ns and the RMSF values were in the acceptable range of 2 Å.
In the case of a protein–ligand complex, the radius of gyration is used to characterize the overall size and shape of the complex. However, it is important to note that the ligand is typically much smaller than the protein, and therefore its contribution to the overall radius of gyration is relatively small. In general, the radius of gyration of a protein–ligand complex depends on the size and shape of both the protein and the ligand, as well as the specific interactions between them. Therefore, it can be a useful parameter for characterizing the stability and binding affinity of the complex, as well as for guiding drug design efforts. In Fig. 6C, Fig. 7C, and Fig. 8C, all the complexes showed strong stability because their Rg values are in the range of acceptance. All the ligands showed an acceptable range of radius of gyration.
4. Discussion
For centuries, medicinal plants have been used to cure a variety of diseases, including inflammation. Many plants contain anti-inflammatory bioactive chemicals, making them ideal candidates for the creation of new anti-inflammatory drugs (Aslam et al., 2021, Noor et al., 2022, Rehman et al., 2022). D.angustifolia is a plant species native to Australia and other areas of the world. The plant is well-known for its traditional medical applications in treating a number of diseases, including inflammation (Getie et al., 2003). D.angustifolia extracts and compounds may have anti-inflammatory potential but more research is required to fully understand their modes of action and possible therapeutic applications.
This study can be utilized as a basis for early screening of flavonoids obtained from D. angustifolia, and it provides a novel therapeutic idea for further research into the plant's ability to treat inflammation. Many flavonoids have anti-inflammatory effects, according to research. Flavonoids are thought to act as anti-inflammatory agents by suppressing the generation of pro-inflammatory enzymes and cytokines, scavenging free radicals, and altering inflammatory signaling pathways. This analysis discovered 10 active components and 1000 targets in total. A total of 11,448 genes associated with inflammation were also collected from the Gencards database. Among these were 175 intersecting targets, which were related to the inflammation.
GO functional analysis showed that the anti-inflammatory targets of D. angustifolia are chiefly engaged in response to oxygen-containing compounds, response to organonitrogen compounds, and the regulation of programmed cell death. Each of these activities has notable roles within inflammatory processes. Oxygen-containing compounds include reactive oxygen species (ROS), which are known mediators of inflammation. An overproduction of ROS can lead to oxidative stress, triggering inflammatory responses and contributing to the pathogenesis of several inflammatory diseases, such as atherosclerosis and rheumatoid arthritis (Mittal et al., 2014). On the other hand, Organonitrogen compounds such as nitric oxide (NO) are recognized as key signaling molecules in various physiological processes, including inflammation. Elevated levels of NO are often observed in inflammatory conditions, playing a crucial role in vasodilation, increased vascular permeability, and leukocyte adhesion, which are hallmarks of inflammation (Bogdan 2001).
Regulation of programmed cell death or apoptosis is a vital process in controlling inflammation. During an inflammatory response, apoptotic cell death helps in the resolution of inflammation by promoting the removal of activated immune cells, thereby preventing excessive tissue damage (Serhan et al., 2015). In addition to the GO functional analysis, our KEGG pathway analysis revealed that these targets are involved in various pathways including cancer pathways, pI3k-Akt signaling pathway, metabolic pathways, chemical carcinogenesis, and endocrine resistance. Each of these pathways has established connections to inflammation.
For instance, the pI3k-Akt pathway is a key regulator of cell survival and proliferation and is often dysregulated in inflammation. The activation of this pathway leads to the production of pro-inflammatory cytokines, promoting inflammatory responses (Fruman et al., 2017). Similarly, inflammation is an integral part of cancer development and progression, with many signaling pathways shared between inflammation and cancer, such as NF-kB and STAT3 pathways (DiDonato, Mercurio and Karin 2012).
Metabolic pathways are also closely intertwined with inflammation. During inflammation, there is a metabolic shift in immune cells, with changes in glucose, lipid, and amino acid metabolism that support the immune response (O'Neill, Kishton and Rathmell 2016). Chemical carcinogenesis involves the formation of cancer due to chemical exposure, with many carcinogens causing DNA damage and triggering inflammation, thereby contributing to cancer development (Kundu and Surh 2008). Taken together, our results suggest that the anti-inflammatory actions of D. angustifolia may involve multiple biological processes and pathways that are critical in the regulation of inflammation.
Further, compound-target-pathway and PPI network identified 6 Methoxykaempferol and 5-Hydroxy-7,8 dimethoxyflavone as potential key chemicals, as well as AKT1, VEGFA, and EGFR as potential key targets. AKT1 is important for the promotion of microvascular leakage in response to inflammatory stimuli such as histamine, and consequently controls the amplitude of this reaction(Di Lorenzo et al., 2009). VEGFA (vascular endothelial growth factor A) is a protein family that has been linked to the development of vascular endothelial cells, with VEGFA being the most ubiquitous and abundantly expressed member(Vafadari, Salamian and Kaczmarek 2016). Recent research has found that VEGFA is intimately associated to the occurrence and progression of inflammatory disorders(Fatima et al., 2017). The epidermal growth factor receptor (EGFR) has been identified as a major initiator exploited by several pathogens for host survival and triggering inflammatory responses. Current research suggests that EGFR activation may have a role in inflammatory disorders(Pastore et al., 2008).
Further, molecular docking and molecular dynamic (MD) simulation of D. angustifolia active ingredients (6-Methoxykaempferol and 5-hydroxy-7,8 dimethoxyflavone) with core proteins (AKT1, VEGFA, and EGFR) was performed. Docking and simulation results supported our findings and demonstrated that these compounds bind steadily to the target genes' active pockets, highlighting the possibility that these substances could be used to treat inflammation by inhibiting the AKT1, VEGFA, and EGFR genes.
To sum up, in the context of acute inflammation, the active compounds of D. angustifolia, especially the identified flavonoids 6-Methoxykaempferol and 5-Hydroxy-7,8-dimethoxyflavone, may have a significant role. By modulating key molecular targets like AKT1, VEGFA, and EGFR, which play integral roles in cell survival, proliferation, and angiogenesis, these compounds could potentially control the excessive inflammatory response and prevent consequent tissue damage.
On the other hand, chronic inflammation, often linked to conditions like metabolic disorders, cancer, and autoimmune diseases, may also be mitigated by D. angustifolia. Our Gene Ontology and KEGG enrichment analyses suggest that the anti-inflammatory targets of D. angustifolia participate in pathways often dysregulated in chronic inflammatory conditions, such as the PI3k-Akt signaling pathway, metabolic pathways, and cancer pathways. However, the specific impacts of D. angustifolia on acute versus chronic inflammation may be influenced by several factors, including the type of inflammatory trigger, dosage and administration route of D. angustifolia, and individual patient characteristics. Hence, while our findings provide a strong basis for understanding the potential anti-inflammatory mechanisms of D. angustifolia, more focused experimental and clinical studies are warranted to explore its precise effects in different inflammatory contexts.
In this study, network pharmacology application was employed to elaborate the putative active components, possible targets, and key biological pathways of D.angustifolia in anti-inflammatory treatment, giving a theoretical foundation for additional experimental validation. However, due to the limitations of network pharmacology, further clarification is still needed for the pharmacological mechanism of D.angustifolia in the treatment of inflammation through in vitro and clinical studies which can facilitate the design of new drugs with a wide range of anti-inflammatory activity.
5. Conclusion
In conclusion, this study demonstrated a potential D. angustifolia mechanism in the treatment of inflammation based on a network pharmacological approach. It was discovered that 6-Methoxykaempferol and 5-Hydroxy-7,8 dimethoxyflavone played a significant role in inflammation by affecting AKT1, VEGFA, and EGFR. Additionally, our molecular docking studies demonstrated that the primary active D. angustifolia components (6-Methoxykaempferol and 5-Hydroxy-7,8 dimethoxyflavone) could dock favorably with AKT1, VEGFA, and EGFR, providing a crucial foundation for future research. However, this study has some limitations because additional clinical and pharmacological research is required to support our results.
Funding
This study is supported via funding from Prince sattam bin Abdulaziz University project number (PSAU/2023/R/1444).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The author thanks the Deanship of Scientific Research (DSR), Prince Sattam bin Abdulaziz University, Al-Kharj, Saudi Arabia for providing the funding for this research.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Mubarak A. Alamri, Email: m.alamri@psau.edu.sa.
Muhammad Tahir ul Qamar, Email: tahirulqamar@gcuf.edu.pk.
References
- Amic D., Davidovic-Amic D., Beslo D., et al. SAR and QSAR of the antioxidant activity of flavonoids. Curr. Med. Chem. 2007;14:827–845. doi: 10.2174/092986707780090954. [DOI] [PubMed] [Google Scholar]
- Aslam S., Ahmad S., Noor F., et al. Designing a multi-epitope vaccine against chlamydia trachomatis by employing integrated core proteomics, immuno-informatics and in silico approaches. Biology. 2021;10:997. doi: 10.3390/biology10100997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavarajappa G.M., Rehman A., Shiroorkar P.N., et al. Therapeutic effects of Crataegus monogyna inhibitors against breast cancer. Front. Pharmacol. 2023;14:1187079. doi: 10.3389/fphar.2023.1187079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan C. Nitric oxide and the immune response. Nat. Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47:W357–W364. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Chem. Biol.: Methods and Protocols. 2015:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- de Oliveira S.Q., de Almeida M.T.R., Maraslis F., et al. Isolation of three new ent-labdane diterpenes from Dodonaea viscosa Jacquin (Sapindaceae): preliminary evaluation of antiherpes activity. Phytochem. Lett. 2012;5:500–505. [Google Scholar]
- Di Lorenzo A., Fernández-Hernando C., Cirino G., et al. Akt1 is critical for acute inflammation and histamine-mediated vascular leakage. Proc. Natl. Acad. Sci. 2009;106:14552–14557. doi: 10.1073/pnas.0904073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato J.A., Mercurio F., Karin M. NF-κB and the link between inflammation and cancer. Immunol. Rev. 2012;246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- Fatima I., Ahmad S., Alamri M.A., et al. Discovery of Rift Valley fever virus natural pan-inhibitors by targeting its multiple key proteins through computational approaches. Sci. Rep. 2022;12:9260. doi: 10.1038/s41598-022-13267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima L., Campello R., Santos R.D., et al. Estrogen receptor 1 (ESR1) regulates VEGFA in adipose tissue. Sci. Rep. 2017;7:1–14. doi: 10.1038/s41598-017-16686-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruman D.A., Chiu H., Hopkins B.D., et al. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getie M., Gebre-Mariam T., Rietz R., et al. Evaluation of the anti-microbial and anti-inflammatory activities of the medicinal plants Dodonaea viscosa, Rumex nervosus and Rumex abyssinicus. Fitoterapia. 2003;74:139–143. doi: 10.1016/s0367-326x(02)00315-5. [DOI] [PubMed] [Google Scholar]
- Islam M.T., Ali E.S., Uddin S.J., et al. Andrographolide, a diterpene lactone from Andrographis paniculata and its therapeutic promises in cancer. Cancer Lett. 2018;420:129–145. doi: 10.1016/j.canlet.2018.01.074. [DOI] [PubMed] [Google Scholar]
- Khan S.A., Lee T.K.W. Network pharmacology and molecular docking-based investigations of Kochiae Fructus's active phytomolecules, molecular targets, and pathways in treating COVID-19. Front. Microbiol. 2022;3020 doi: 10.3389/fmicb.2022.972576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Chen J., Cheng T., et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47:D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.P., Son K.H., Chang H.W., et al. Anti-inflammatory plant flavonoids and cellular action mechanisms. J. Pharmacol. Sci. 2004;96:229–245. doi: 10.1254/jphs.crj04003x. [DOI] [PubMed] [Google Scholar]
- Kohl M., Wiese S., Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Data mining in proteomics: from standards to applications. 2011:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- Korniluk A., Koper O., Kemona H., et al. From inflammation to cancer. Irish J. Medical Sci. (1971) 2017;186:57–62. doi: 10.1007/s11845-016-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu J.K., Surh Y.-J. Inflammation: gearing the journey to cancer. Mutation Res./Rev. Mutation Res. 2008;659:15–30. doi: 10.1016/j.mrrev.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Maleki S.J., Crespo J.F., Cabanillas B. Anti-inflammatory effects of flavonoids. Food Chem. 2019;299 doi: 10.1016/j.foodchem.2019.125124. [DOI] [PubMed] [Google Scholar]
- Mittal M., Siddiqui M.R., Tran K., et al. Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signal. 2014;20:1126–1167. doi: 10.1089/ars.2012.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraj K., Karthikeyan B.S., Vivek-Ananth R., et al. IMPPAT: a curated database of Indian medicinal plants, phytochemistry and therapeutics. Sci. Rep. 2018;8:4329. doi: 10.1038/s41598-018-22631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mraz M., Haluzik M. The role of adipose tissue immune cells in obesity and low-grade inflammation. J. Endocrinol. 2014;222:R113–R127. doi: 10.1530/JOE-14-0283. [DOI] [PubMed] [Google Scholar]
- Narsinghani T., Sharma R. Lead optimization on conventional non-steroidal anti-inflammatory drugs: an approach to reduce gastrointestinal toxicity. Chem. Biol. Drug Des. 2014;84:1–23. doi: 10.1111/cbdd.12292. [DOI] [PubMed] [Google Scholar]
- Noor F., Rehman A., Ashfaq U.A., et al. Integrating network pharmacology and molecular docking approaches to decipher the multi-target pharmacological mechanism of Abrus precatorius L. Acting on Diabetes. Pharmaceuticals. 2022;15:414. doi: 10.3390/ph15040414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros, J.C., 2007. VENNY. An interactive tool for comparing lists with Venn Diagrams. http://bioinfogp.cnb.csic.es/tools/venny/index. html.
- O'Neill L.A., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn D., Hunt R. Cochrane Database Syst. Rev. CD005945. 2007 doi: 10.1002/14651858.CD005945.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens B.M. Inflammation, innate immunity, and the intestinal stromal cell niche: opportunities and challenges. Front. Immunol. 2015;6:319. doi: 10.3389/fimmu.2015.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant S., Deshmukh A., GuruMurthy G.S., et al. Inflammation and atherosclerosis—revisited. J. Cardiovasc. Pharmacol. Ther. 2014;19:170–178. doi: 10.1177/1074248413504994. [DOI] [PubMed] [Google Scholar]
- Pastore S., Gubinelli E., Leoni L., et al. Biological drugs targeting the immune response in the therapy of psoriasis. Biologics: Targets Ther. 2008;2:687–697. doi: 10.2147/btt.s2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Ao M., Dong B., et al. Anti-inflammatory effects of curcumin in the inflammatory diseases: status, limitations and countermeasures. Drug Des. Devel. Ther. 2021:4503–4525. doi: 10.2147/DDDT.S327378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirzada A., Shaikh W., Usmanghani K., et al. Antifungal activity of Dodonaea viscosa Jacq extract on pathogenic fungi isolated from super ficial skin infection. Pak. J. Pharm. Sci. 2010;23 [PubMed] [Google Scholar]
- Rajamanickam V., Rajasekaran A., Anandarajagopal K., et al. Anti-diarrheal activity of Dodonaea viscosa root extracts. Int J Pharm. Bio. Sci. 2010;1:182–185. [Google Scholar]
- Raucci F., Iqbal A.J., Saviano A., et al. IL-17A neutralizing antibody regulates monosodium urate crystal-induced gouty inflammation. Pharmacol. Res. 2019;147 doi: 10.1016/j.phrs.2019.104351. [DOI] [PubMed] [Google Scholar]
- Rehman A., Ashfaq U.A., Javed M.R., et al. The Screening of phytochemicals against NS5 Polymerase to treat Zika Virus infection: integrated computational based approach. Comb. Chem. High Throughput Screen. 2022;25:738–751. doi: 10.2174/1386207324666210712091920. [DOI] [PubMed] [Google Scholar]
- Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminf. 2014;6:1–6. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M., Dalah I., Alexander J., et al. GeneCards Version 3: the human gene integrator. Database. 2010 doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan C.N., Dalli J., Colas R.A., et al. Protectins and maresins: New pro-resolving families of mediators in acute inflammation and resolution bioactive metabolome. Biochimica et Biophysica Acta (BBA)-Mol. Cell Biol. Lipids. 2015;1851:397–413. doi: 10.1016/j.bbalip.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd, K.A., Rye, B.L., Meissner, R.A. et al., 2007. Two new Western Australian species of Dodonaea (Sapindaceae) from northern Yilgarn ironstones.
- Studio D. Discovery studio. Accelrys [2.1]. 2008 [Google Scholar]
- Szklarczyk D., Kirsch R., Koutrouli M., et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadeg H., Mohammed E., Asres K., et al. Antimicrobial activities of some selected traditional Ethiopian medicinal plants used in the treatment of skin disorders. J. Ethnopharmacol. 2005;100:168–175. doi: 10.1016/j.jep.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Teffo L.S., Aderogba M.A., Eloff J.N. Antibacterial and antioxidant activities of four kaempferol methyl ethers isolated from Dodonaea viscosa Jacq. var. angustifolia leaf extracts. S. Afr. J. Bot. 2010;76:25–29. [Google Scholar]
- Vafadari B., Salamian A., Kaczmarek L. MMP-9 in translation: from molecule to brain physiology, pathology, and therapy. J. Neurochem. 2016;139:91–114. doi: 10.1111/jnc.13415. [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Veerapur, V., Prabhakar, K., Thippeswamy, B., et al., 2010. Antidiabetic effect of Dodonaea viscosa (L). Lacq. aerial parts in high fructose-fed insulin resistant rats: a mechanism based study. [PubMed]
- Zhao J., He K., Du H., et al. Bioinformatics prediction and experimental verification of key biomarkers for diabetic kidney disease based on transcriptome sequencing in mice. PeerJ. 2022;10:e13932. doi: 10.7717/peerj.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete V., Cuendet M.A., Grosdidier A., et al. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]










