Summary
Growing antibiotic resistance has encouraged the revival of phage-inspired antimicrobial approaches. On the other hand, photodynamic therapy (PDT) is considered a very promising research domain for the protection against infectious diseases. Yet, very few efforts have been made to combine the advantages of both approaches in a modular, retargetable platform. Here, we foster the M13 bacteriophage as a multifunctional scaffold, enabling the selective photodynamic killing of bacteria. We took advantage of the well-defined molecular biology of M13 to functionalize its capsid with hundreds of photo-activable Rose Bengal sensitizers and contemporarily target this light-triggerable nanobot to specific bacterial species by phage display of peptide targeting moieties fused to the minor coat protein pIII of the phage. Upon light irradiation of the specimen, the targeted killing of diverse Gram(−) pathogens occurred at subnanomolar concentrations of the phage vector. Our findings contribute to the development of antimicrobials based on targeted and triggerable phage-based nanobiotherapeutics.
Subject areas: Bacteriology, Biotechnology, Microbiology, Virology
Graphical abstract
Highlights
-
•
Novel antimicrobial approach based on triggerable nanobiotherapeutics
-
•
Antimicrobial photodynamic therapy (PDT) mediated by engineered phage vectors
-
•
M13 phages engineered to target specifically diverse Gram-negative bacteria in PDT
-
•
Lethal photosensitization of specific targets only upon light irradiation
Bacteriology; Biotechnology; Microbiology; Virology
Introduction
The incessant increase in bacterial antibiotic resistance, together with the shortening of new alternatives in the pharma R&D pipelines, has revived interest in phage-inspired antimicrobial approaches against WHO priority I and II pathogens, including many Gram(−) bacteria. Classic phage therapy, envisaging the use of natural lytic phages to control bacterial infections, provides an interesting and useful tool for the control of antibiotic-resistant infections (recently reviewed in1), and reports from the majority of studies on their application to treat infections by Staphylococcus aureus, Enterococcus faecium, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., and Escherichia coli—all belonging to the ESKAPEE group of pathogens—are encouraging to date.2,3,4,5 As an alternative to classical approach, the combination of phage therapy with antibiotics administration has been shown to enhance the antimicrobial activity exerted on bacterial infections of various pathogens.5,6,7
However, despite the recent and past successes, classic phage therapy is hampered by several drawbacks, including a generally narrow host range, which needs to be tackled by strain- and patient-tailored precision approaches.4 Moreover, the likelihood of rapid appearance and spreading of resistance against phage infection and propagation is quite high.8 An alternative approach is to take advantage of the well-defined molecular biology of several simple model phages (e.g., M13, fd, MS2) for the assembly of genetically retargeted nanobioscaffolds, overcoming the narrow host range of natural phages. These vectors may be armed with (triggerable) antimicrobial functions, providing a modular platform for the treatment of difficult-to-treat infections.9,10
Photodynamic therapy (PDT) is likewise considered a very promising therapeutic modality for the eradication of drug-resistant bacteria.11 PDT is approved for the clinical treatment of several types of diseases, including cancer,12,13 dermatological pathologies, and microbial infections.14,15 In PDT, a compound with photosensitizing properties (sensitizer) is accumulated in, or concentrated at, the target cells. The subsequent triggering of the sensitizer by light generates highly reactive oxygen species (ROS), whose local burst exerts lethal effects on the target cancer or microbial cells.15 Antimicrobial photodynamic therapy (aPDT) already finds a wide use for topical, dental, and soft tissue infections, either alone or in combination with photosensitizers.16,17,18,19 Moreover, aPDT can stimulate the immune system to improve bacterial clearance.20 The combination of antibiotic therapy with PDT has also been proven efficient in eliminating drug-resistant bacteria in patients carrying microflora resistant to a previous antibiotic therapy,21,22 whereas studies on animals have reported little evidence of host tissue damage following aPDT.17,23
Compared with conventional antibiotic therapies, antimicrobial PDT presents several interesting assets: (1) the burst of ROS affects multiple biological informational molecules at once (e.g., DNA, RNA, proteins, lipids, etc.) and disrupts membrane integrity, thus providing physical damage to cells, which less likely engenders resistance; (2) PDT is also effective, for the same reasons, on metabolically inactive, dormant, and antibiotic-resistant cells; (3) PDT can be applied locally, exploiting the possibility to focus the triggering irradiation at the desired site of action; and (4) it presents an instant onset of action.
However, a major limitation of photodynamic (as well as sonodynamic and photothermal) antimicrobial agents is that the reactive oxygen species (or the heat) generated during irradiation can harm bystander host cells. Accordingly, there is extraordinary interest in developing methods for specifically targeting the sensitizers to the infectious organism.24 The advantage of targeting the photosensitizer is that ROS production is generated only in immediate proximity of the pathogen, allowing for lower irradiation and sensitizer dosages needed to achieve lethal photosensitization. Several studies have shown the effectiveness and reduced toxicity of PDT, in conjunction with sensitizer targeting systems, including proteins (ConcanvalinA-RB25), antibodies, chemical modification, or nanoarchitectonics of vector particles24,26,27,28,29,30 and phages.31
Owing to the distinct cell surfaces, aPDT can have very different effects on Gram-positive and Gram-negative bacteria. PSs can penetrate through the cell walls of Gram-positive bacteria with higher ease, whereas Gram-negative bacteria display various degrees of resistance to PS permeabilization, due to the presence of the outer membrane, acting as a barrier to diverse drug molecules.32 To solve this problem, outer-membrane-disrupting agents or the chemical manipulation of bacterial surface structures has been proposed.33 Nevertheless, such pre-treatments may be accompanied by potential cytotoxicity of the agents used.
Despite the respective advantages provided by phage-derived therapies and PDT, only a handful of phage applications for antimicrobial PDT have been reported to date, principally involving the use of natural phages decorated with chlorine e6.34 Moreover, a systematic retargeting of photosensitizer-decorated phages through specific bacteria-targeting moieties (antibodies and peptides) has never been reported, except for one study in which chimeric M13 phages were conjugated to gold nanoparticles for antimicrobial photothermal therapy.35 This leaves a wide avenue open for scientific discovery and methodological improvements.
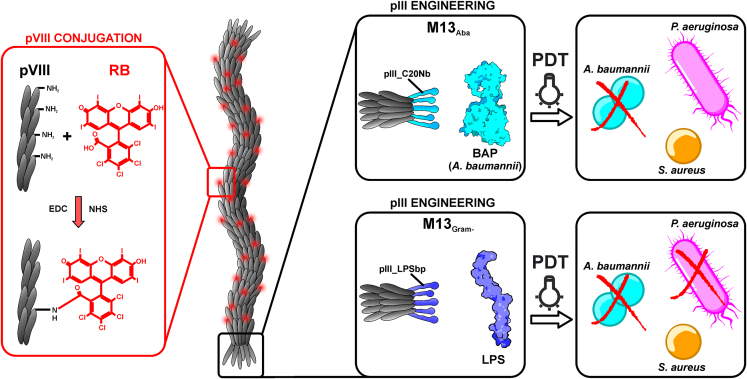
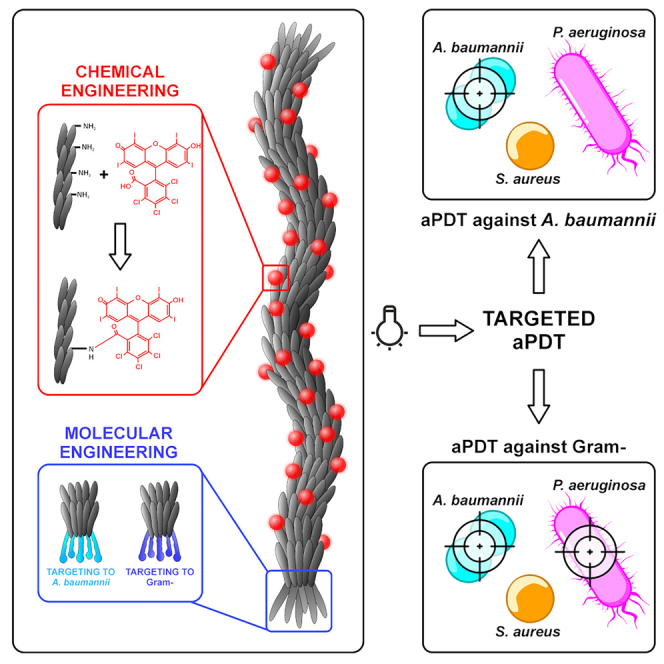
Filamentous bacteriophages such as M13 represent a harmless and effective delivery platform. They can undergo extremely flexible genetic engineering, allowing for a plethora of targeting designs.9 M13 can provide high avidity for the target due to its multivalent display of specific targeting moieties and a multitude of functionalization sites resulting in a high payload of effectors.36 In particular, the possibility to conjugate M13 with hundreds of photosensitizers37,38,39 makes them interesting vectors for antimicrobial PDT applications (Figure 1).
Figure 1.
Orthogonal engineering of M13 phage for selective PDT
Phages with targeted tropism against Acinetobacter baumannii (light blue) and against the gram-negative Acinetobacter baumannii and Pseudomonas aeruginosa cells (dark blue) were generated. M13Aba is retargeted to the biofilm activating protein (BAP) of Acinetobacter baumannii through the specific display on the minor coat protein pIII of the C20 nanobody (C20Nb), whereas M13Gram− targets the lipopolysaccharides of the Gram-negative outer membrane through the display on the pIII protein of KNYSSSISSIRAC-binding peptide (LPSbp). The amino group of M13Aba and M13Gram− major capsid protein pVIII were chemically conjugated with hundreds of RB for the production of ROS, which are highly antimicrobial, upon light irradiation, in the cell proximity.
Here we present a flexible and robust M13-based platform for antimicrobial PDT against Gram(−) bacterial pathogens. M13 phages with specific tropism against A. baumannii were generated by phage display of a specific nanobody (C20) on the minor coat protein g3p. Likewise, the fusion of an LPS-binding peptide to g3p enabled to expand the target range to different Gram(−) pathogens, not targeted by wild-type M13. These engineered vectors were orthogonally functionalized with hundreds of Rose Bengal (RB) sensitizers on the major capsid protein g8p, generating nanobioconjugates able to selectively kill their specific targets only upon light irradiation. RB was chosen as it is a widely available and biocompatible photoreactive fluorescein derivative, approved by the FDA as an ocular diagnostic stain and orphan anticancer drug.40 RB contains multiple halogens (i.e., chlorine, iodine) with high atomic number substituents, which enhance the intersystem crossing rate, generating a triplet excited state underlying effective generation of ROS.25,40 Following RB conjugation, the M13-RB phage nanobioconjugates were shown to promote photodynamic generation of ROS and mediate selective killing of their specific targets only upon light irradiation.
Results
Conjugation of Rose Bengal to the capsomers of M13 produces an ROS-generating phage PDT platform
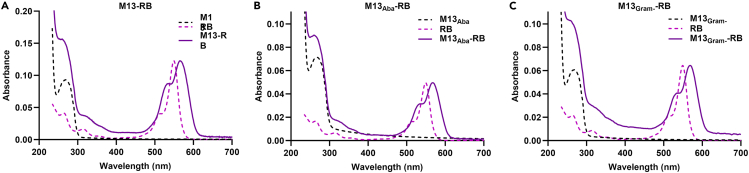
To prove that M13 can be engineered into a valuable vector platform for PDT applications, we set out to conjugate RB to the capsomers of the M13K07 phage (hereafter referred to as M13). A shifted and broader peak at 560 nm in the absorption spectrum was observed following conjugation with M13 (Figure 2). These spectral changes confirmed that RB was covalently linked to the M13 capsid.41 Considering the initial M13 concentration and the molar extinction coefficients of RB, 971, 405, and 562 RB molecules were attached to M13, M13Aba, and M13Gram−, respectively (Table 1).
Figure 2.
Conjugation of RB to M13 phage
(A–C) Chemical conjugation was performed on (A) M13, (B) M13Aba, and (C) M13Gram- phages. Absorption spectra of phages (dashed black line), RB (dashed pink line), and phage conjugated with RB (purple line).
Table 1.
Description and main characteristics of conjugated phages
| Name | Targeting moiety | Target | [Phage] (v/mL) | [Phage] (nM) | [RB] (μM) | Ratio (RB/phage) |
|---|---|---|---|---|---|---|
| M13 | pIII | F' pilus of E. coli | 8.43E+12 | 14.0 | 13.6 | 971 |
| M13Aba | C20 nanobody | BAP protein of A. baumannii | 1.02E+13 | 17.0 | 6.89 | 405 |
| M13Gram− | Li005 peptide | LPS of gram-negative bacteria | 9.57E+12 | 15.9 | 8.94 | 562 |
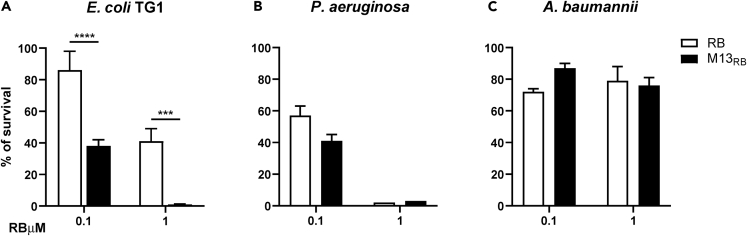
Specific photokilling of the natural E. coli host by a wild-type M13-RB bioconjugate
Next, the M13-RB conjugates were tested for their photodynamic antimicrobial activity on E. coli TG1 and two other Gram-negative bacteria, P. aeruginosa PA14 and A. baumannii ATCC 19606 (Aba). TG1 is a natural host for M13, expressing the F′ pilus, representing the primary receptor bound by the wild-type g3p phage protein promoting infection. On the contrary, P. aeruginosa and A. baumannii lack the F′ pilus and are not efficiently targeted by wild-type M13.42 Free Rose Bengal was used as control to benchmark phage-mediated PDT against the non-targeted approach. All bacterial strains tested exhibited variable sensitivity to the photodynamic treatment, presumably due to biochemical, metabolic, or genetic differences (Figure 3). In particular, E. coli TG1 viability was only partly (13.6% ± 12.3%) affected after incubation with 1 μM free RB equivalents, whereas P. aeruginosa PA14 suffered extensive (42.8% ± 6.6%) killing under the same conditions. Finally, after irradiation, A. baumannii proved to be the least sensitive to free RB, remaining essentially unaffected by PDT with 1 μM free RB. Despite these differences, M13-mediated photodynamic treatment increased the killing only toward the TG1 host strain, starting at 100 nM RB equivalents (Figure 3A). Stronger, dose-dependent photodynamic effects were recorded at the highest sensitizer concentration tested (1 μM), where the M13-RB platform increased PDT killing of TG1 cells by more than five orders of magnitude with respect to free RB alone. Remarkably, the M13-RB vector was not able to induce a significant increase in mortality (compared with free RB) on Aba and PA14. The lack of specificity toward these strains indicates that wild-type M13 can prime the accumulation of sufficient RB photosensitizers only toward its natural host, expressing the F′ pilus. These results show that the M13 phage is an excellent platform for implementing selective antimicrobial PDT.
Figure 3.
Antimicrobial PDT of M13-RB bioconjugates
(A–C) The survival rate of bacterial cells incubated with increasing concentration of either RB (white bars) or M13-RB (black bars) and irradiated with a white led amp for 45 min. PDT assays were performed on (A) E. coli, (B) P. aeruginosa, and (C) A. baumannii. Data are represented as mean ± SD. ∗∗∗ = p < 0.001, ∗∗∗∗ = p < 0.0001.
Retargeting of M13 against Acinetobacter baumannii
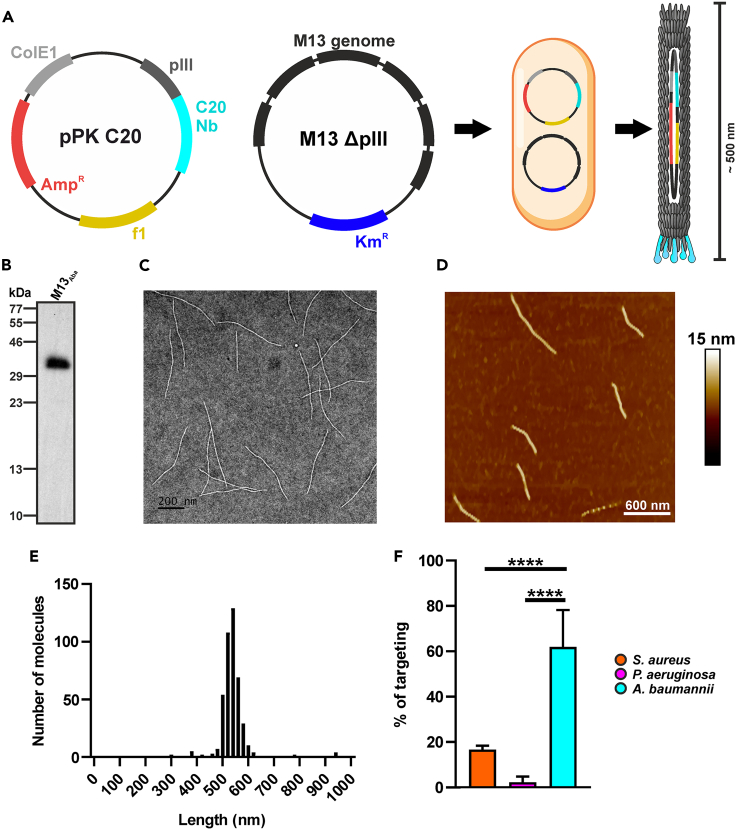
To investigate whether the phage PDT platform could be engineered to target different bacteria from the native E. coli F+ host strains, M13 was retargeted to bind to A. baumannii cells. The coding sequence of the C20 single domain antibody (sdAb or nanobody) recognizing the A. baumannii biofilm-associated protein (BAP) was fused to the C-terminal domain of the pIII (g3p) minor coat protein in the phage. This choice was driven by the high specificity of C20 for A. baumanni BAP along with a low cross-reactivity toward other bacterial species.43
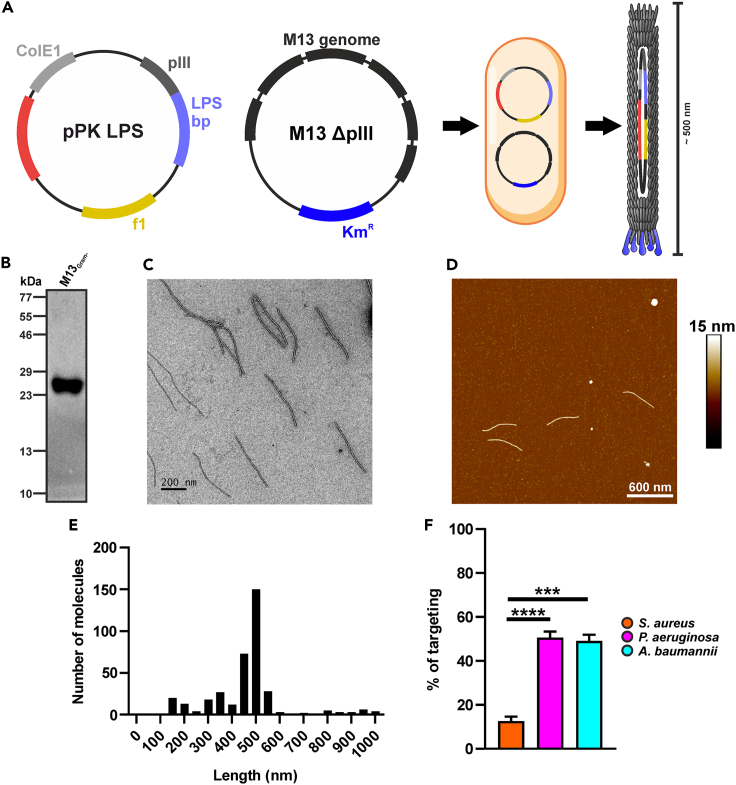
The pPK_C20 phagemid carrying the C20-pIII fusion was co-transformed with the hyperphage plasmid lacking both the native pIII cistron as well as the F1 phage origin. This strategy allowed to voiding the undesired residual packaging of the helper phage, resulting in uniform and stable recombinant virions of about 500 nm length, packaging only the C20-pIII phagemid, as validated by TEM (Figure 4C) and atomic force microscopy (AFM, Figures 4D and 4E). Immunoblotting with a pIII-C-term-specific antibody demonstrated the correct phage display of a 36.9 kDa polypeptide compatible with the expected molecular weight of the C20-pIII (C-term) fusion, absent from the wild-type M13 phages (Figure 4B). This strategy allowed to overcome occasional wild-type pIII recycling from the superinfection of helper phages, resulting in M13Aba virions displaying only the C20-pIII(C-term) fusion.
Figure 4.
Characterization of the anti-A. baumannii phage M13Aba
(A) Schematic representation of the M13 genetic engineering through co-transformation in E. coli of ppK_C20 phagemid and hyperphage genome.
(B–D) (B) Immunoblotting of the antiBAP_Nb-pIII fusion (M13Aba), demonstrating incorporation in the purified virion. The integrity of purified M13Aba phages was visualized through (C) TEM (scale bar = 200 nm) and (D) AFM (scale bar = 600 nm).
(E) M13Aba length distribution analyzed by AFM.
(F) Specificity and selectivity of engineered M13Aba phages targeting A. baumannii: the binding was determined by SyberGreen real-time PCR using an oligonucleotide that anneals on the pIII minor-coat-protein-coding gene. M13Aba phages showed significant binding to A. baumannii (cyan), whereas poor binding was observed to S. aureus (orange) and P. aeruginosa (magenta). Data are represented as mean ± SD. ∗∗∗∗ = p < 0.0001.
The specific retargeting of M13Aba to A. baumannii was demonstrated by qPCR. S. aureus and P. aeruginosa were used as negative controls, respectively for Gram(+) and Gram(−) pathogens, as they lack orthologues of the BAP protein recognized by the C20 sdAb fusion. After incubation, washing, and pelleting of the bacteria, the great majority (61.9% ± 16.7%) of M13Aba phages were bound to A. baumannii cells, whereas a significantly lower fraction of phages interacted with both S. aureus (16.6% ± 1.7%) and P. aeruginosa (2.2% ± 2.4% <3%) cells (Figure 4F). Taken together, these results demonstrate that phage display of the C20 sdAb on pIII allows the selective retargeting of M13 to A. baumannii cells.
Potent phage-mediated PDT against Acinetobacter baumannii
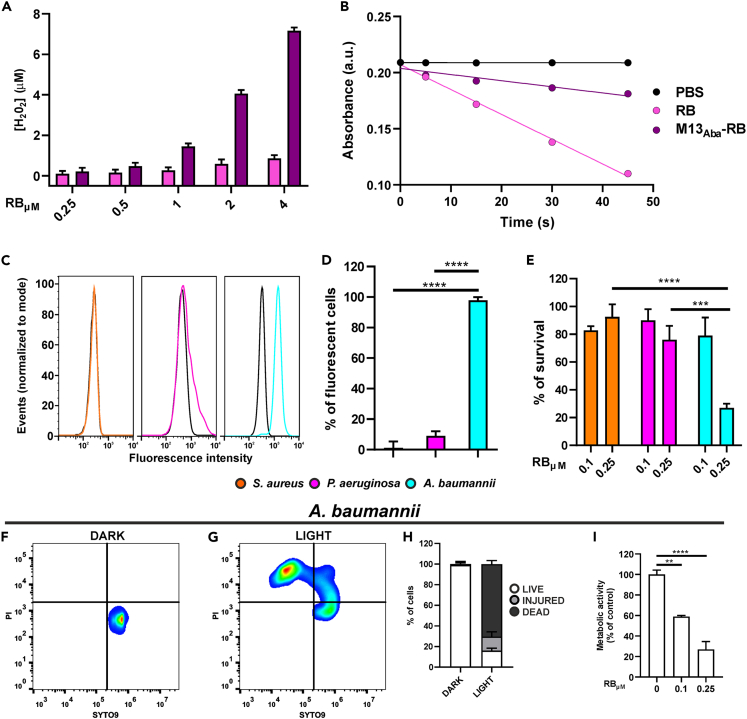
Next, the recombinant M13Aba vector was armed with hundreds of RB molecules taking advantage of the extremely high multiplicity of the pVIII capsomer, providing thousands of amine groups for the chemical conjugation of the RB sensitizer on the vector capsid. After conjugation and removal of unreacted RB, the M13Aba-RB vector was tested for the generation of ROS. The ability of phage conjugates to generate ROS, upon irradiation with visible white light, was evaluated using the Amplex Red and ABMDMA assays, respectively, to detect peroxides and 1O2. Results indicated that the conjugation of RB to phage improved the generation of peroxide (8-fold increase) over singlet oxygen production (4-fold decrease, φΔ decrease from 0.76 to 0.20) with respect to free RB, suggesting a photoactivation switch favoring type I over type II mechanisms after PS bioconjugation with proteins (Figures 5A and 5B).44,45 The results clearly showed that M13 virions can be conjugated to several hundreds of RB sensitizer molecules, without quenching the ability to produce cytotoxic ROS, providing a high sensitizer payload.
Figure 5.
Photodynamic properties of M13Aba-RB
(A) Amplex Red assay for the generation of peroxides using different concentrations of RB (pink) and M13Aba-RB (purple).
(B) Determination of 1O2 generation following the decrease of ABMDMA absorbance over the irradiation time under white light irradiation for RB (pink line), M13Aba-RB (purple line), and PBS (black line).
(C) Flow cytometry analysis of refactored and RB-conjugated M13Aba-RB targeting to S. aureus (orange), P. aeruginosa (magenta), and A. baumannii (cyan). The fluorescence, detected in the PE channel, is related to RB.
(D–G) (D) Percentage of fluorescent cells at the flow cytometer. (E) Selective photodynamic killing of S. aureus (orange), P. aeruginosa (magenta), and A. baumannii (cyan) after 45 min of light irradiation following incubation with 0.1 and 0.25 μM (RB equivalents) of photoactive M13Aba-RB. Live/dead assays were performed on A. baumannii cells preincubated with M13Aba-RB and (F) kept in dark conditions or (G) irradiated.
(H) percentage of live, injured, and dead cells measured in the live/dead assay, 20 min post-irradiation.
(I) Metabolic activity of A. baumannii cells after phage-mediated aPDT. Data are represented as mean ± SD. ∗∗ = p < 0.01, ∗∗∗ = p < 0.001, ∗∗∗∗ = p < 0.0001.
To exclude impairment of vector targeting after conjugation with the sensitizer, the specificity of M13Aba-RB for A. baumannii cells was evaluated by flow cytometry, taking advantage of the intrinsic fluorescence of RB. Again, S. aureus and P. aeruginosa were used as negative controls. As a result, 97.9% of fluorescent events were detected for an A. baumannii culture incubated with the M13Aba-RB phage conjugate, confirming excellent binding even after conjugation with the photosensitizer. On the contrary, M13Aba-RB bound only a minor fraction of S. aureus and P. aeruginosa cells, confirming the lack of affinity for non-specific targets (Figures 5C and 5D). These results demonstrate that the tropism of M13 can be successfully modified to target a specific bacterial species and that the retargeted M13 virions withhold specificity even after conjugation with hundreds of photosensitizers.
To test the efficacy of the M13Aba-RB bioconjugate, antimicrobial PDT assays were performed. Interestingly, the M13Aba-RB bioconjugate showed no antimicrobial effects without irradiation, whereas photostimulation of M13Aba-RB led to the killing of more than 75% of A. baumannii cells at 0.25 μM RB equivalents (Figure 5E). Considering the phage functionalization rate (Table 1), this value corresponds to a picomolar (pM) effective vector concentration. The bacterial photokilling proved phage concentration-dependent, as indicated by increased Acinetobacter survival rates at 0.1 μM RB equivalents, whereas free RB sensitizer molecules (not conjugated to phage) showed no significant effect at the same concentrations (data not shown). The absence of photodynamic effects was also observed for S. aureus and P. aeruginosa, proving the specific killing mediated by the engineered phage (Figure 5E). These results demonstrate a potent and light-dependent antimicrobial effect of the engineered M13 platform, only toward the specific target pathogen. To further validate the photodynamic killing of A. baumannii and provide first insights into the possible killing activity triggered by the phage vector, live/dead and metabolic assays were carried out. After incubation with the M13Aba-RB bioconjugates, A. baumannii cells showed a strong light-dependent shift in viability in cytofluorometry: under dark conditions all cells stained positive for SYTO9 only, a hallmark of live cells (Figure 5F). Shortly after irradiation (20 min post-irradiation) a large fraction of the population stained positive for propidium iodide (PI, Fig, 5G), comprehending a minor fraction of injured cells (SYTO9/PI positive) and more than 70% of dead cells (PI positive, SYTO9 negative) (Figure 5H). This result demonstrates that the fast onset of phage-mediated aPDT is accompanied by a rapid permeabilization of the cell membrane, as most of the bacterial population becomes positive to the otherwise non-permeant PI stain. Metabolic assays showed a 70% decrease in the capacity to reduce tetrazolium violet following irradiation (Figure 5I), consistent with the observed decrease in vitality (Figures 5E and 5H). These results indicate that the rapid permeabilization of A. baumannii membranes is followed by a marked drop in the reducing power of the bacterium. Judging from the similar decrease in replication rates (Figure 5E) and increase in numbers of dead cells (Figure 5H), this drop is likely irreversible.
Retargeting of the M13 platform against Gram-negative bacteria
We next investigated whether the tropism of the phage vector platform could be broadened to target not a single species such as A. baumannii, but Gram(−) bacteria in general. To this sake, a new phagemid was constructed (pPK_LPS), allowing for the multivalent display of a lipopolysaccharide (LPS)-binding peptide (KNYSSSISSIRAC) in fusion with the C-terminus of the pIII protein. LPS represents a major component of the outer membrane of Gram-negative bacteria and is a proven key virulence factor for P. aeruginosa and A. baumannii species.46,47 The KNYSSSISSIRAC peptide exhibits two basic residues, but it is generally hydrophilic and was selected because it has a known high binding affinity to LPS.48
After co-transformation with the ΔpIII ΔF1ori helper phage plasmid, M13Gram- virions were produced and purified (Figure 6A). Immunoblotting with a pIII-C-term-specific antibody demonstrated the correct phage display of the LPS-binding peptide with the C-terminal domain of pIII (Figure 6B). Moreover, AFM analysis showed that the engineered M13Gram− virions are stable and uniform in their shortened size, packaging only the pPK_LPS phagemid (Figures 6D and 6E).
Figure 6.
Characterization of the anti-Gram phage M13Gram−
(A) Schematic representation of the M13 genetic engineering through co-transformation in E. coli of pPK_LPS phagemid and hyperphage genome.
(B–F) (B) Immunoblotting of the KNYSSSISSIRAC-pIII fusion (M13Gram−) demonstrating incorporation in the purified virion. Visualization of purified M13Gram− phage through (C) TEM (scale bar = 200 nm) and (D) AFM (scale bar = 600 nm). (E) M13Gram− length distribution analyzed by AFM. (F) Selective tropism of engineered M13Gram− to Gram-negative bacteria was proved through targeting assays performed on S. aureus (orange), P. aeruginosa (magenta), and A. baumannii (cyan). Data are represented as mean ± SD. ∗∗∗ = p < 0.001, ∗∗∗∗ = p < 0.0001.
As for the M13Aba vector, the effective retargeting of M13Gram- virions was investigated by qPCR on the same panel of bacterial pathogens used before, including P. aeruginosa and A. baumannii as Gram-negatives, as well as S. aureus as Gram(+) control. M13Gram− proved to bind significantly better in targeting Gram(−) bacteria, with 50.6% ± 2.6% of the virions binding to P. aeruginosa and 49.1% ± 2.8% to A. baumannii, whereas significantly less phages (12.6% ± 2.0%) bound to S. aureus. These data indicate a broadened specificity of the retargeted M13Gram− phage for Gram(−) bacteria expressing LPS on their surface (Figure 6F).
Potent phage-mediated PDT against antibiotic-resistant Gram(−) bacteria
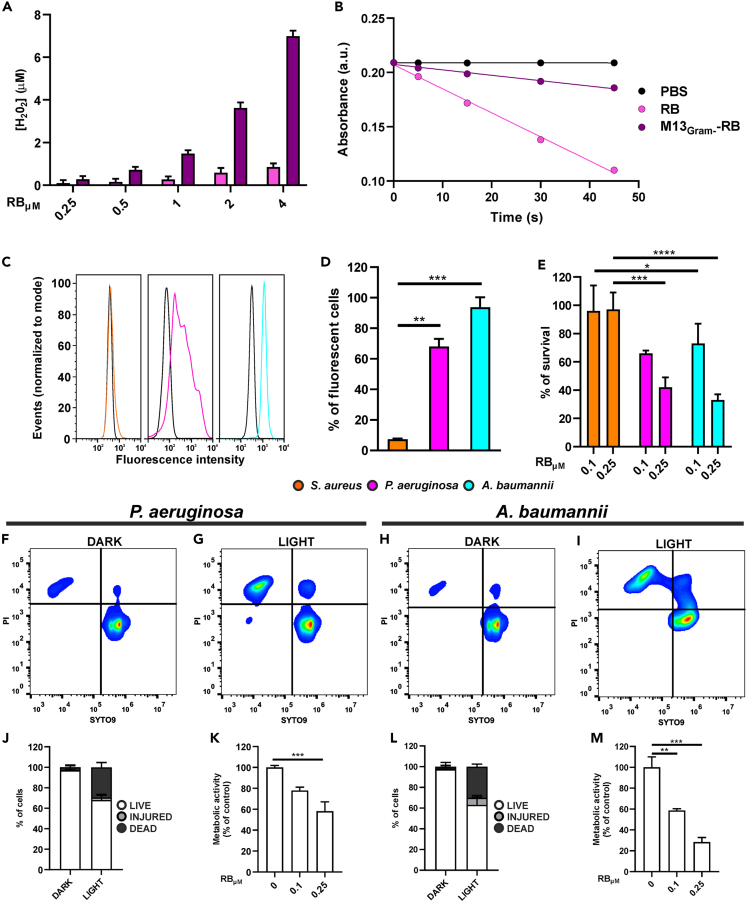
The retargeted M13Gram− phages were then conjugated with RB, and purified from unreacted sensitizer molecules, as previously described. Again, the generation of peroxide of the bioconjugate increased 8-fold, whereas the production of the singlet oxygen decreased 4-fold (φΔ decrease from 0.76 to 0.20), compared with free RB.
These results indicate that the LPS-binding phages retain similar ROS generating rates to their M13 and M13Aba counterparts (Figures 7A and 7B), testifying reproducible results of the vector platform after retargeting.
Figure 7.
Photodynamic properties of the M13Gram− phage vector
(A) Amplex Red assay for the generation of peroxides using different concentrations of RB (pink) and M13Gram−-RB (purple).
(B) Determination of 1O2 generation following the decrease of ABMDMA absorbance over the irradiation time under white light irradiation for RB (pink line), M13Gram−-RB (purple line), and PBS (black line).
(C) Flow cytometry analysis of retargeted and RB-conjugated M13Gram− phages directed to the lipopolysaccharides of Gram-negative bacteria: dark lines correspond to the control cells treated with PBS, whereas colored ones highlight binding of the engineered M13Gram−-RB to S. aureus (orange), P. aeruginosa (magenta), and A. baumannii (cyan).
(D) Percentage of bacterial fluorescent cells measured by flow cytometry analysis.
(E–I) (E) Percentage of bacteria survived to light irradiation after preincubation with M13Gram−-RB phages. The activity of phages was tested at two concentrations, 0.1 and 0.25 μM, of equivalent RB. Live/dead assay was performed on P. aeruginosa and A. baumannii preincubated with M13Gram−-RB and (F–H) kept in dark or (G–I) irradiated.
(J–M) Percentage of live, injured, and dead cells measured in live/dead assay 20 min after irradiation. Metabolic activity of (K) P. aeruginosa and (M) A. baumannii after phage-mediated aPDT. Data are represented as mean ± SD. ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001, ∗∗∗∗ = p < 0.0001.
To demonstrate that the tropism toward Gram(−) bacteria was not altered after conjugation with RB, flow cytometry experiments with M13Gram−-RB were carried out (Figures 7C and 7D). Clearly, the phage vector bound to both A. baumannii and P. aeruginosa, whereas no fluorescence shift was observed for S. aureus. In particular, the totality (100%) of A. baumannii cells was labeled and detected after incubation with M13Gram−-RB; slightly lower rates were recorded for P. aeruginosa (70%), whereas only 7% of S. aureus cells were targeted by the phage, suggesting an intact specificity of M13Gram− for Gram(−) bacteria, even after chemical conjugation.
Photodynamic antimicrobial assays were eventually set up to investigate the specificity of M13Gram−-RB in killing preferentially Gram(−) bacteria. Bacteria were treated with different concentrations of the phage bioconjugate and irradiated with the same settings used for M13Aba. Again, M13Gram−-RB vector showed no antimicrobial effects when bacteria were kept in the dark, whereas irradiation provoked a significant reduction in survival rates only for A. baumannii and P. aeruginosa (Figure 7E). In both cases, a dose-dependent effect could be observed at pM phage concentrations, with higher vector concentrations and RB equivalents correlating with increased light-dependent antimicrobial effects. On the contrary, neither of the two vector concentrations tested showed effective in antimicrobial photodynamic therapy (aPDT) on S. aureus, suggesting that M13Gram−-RB is harmless against Gram(+) cells. As for M13Aba-RB (Figure 5), additional live/dead and metabolic assays were carried out to validate the photodynamic proprieties of M13Gram−-RB on P. aeruginosa and A. baumannii. Both species showed a consistent shift in the live-dead staining after incubation with M13Gram−-RB and irradiation (Figures 7F and 7G for P. aeruginosa; Figures 7H and 7I for A. baumannii), albeit at lower permeabilization rates (30%–40% PI positive cells) (respectively, Figures 7J and 7L), suggesting weaker photodynamic performances of M13Gram−-RB with respect to M13Aba-RB (>70% PI positive cells; Figure 5H). The metabolic assays confirmed this trend both for P. aeruginosa (Figure 7K) as well as for A. baumannii (Figure 7M) with a more pronounced decrease of the metabolic activity for the latter. These results indicate that the phage- and light-dependent permeabilization of membranes is followed by a pursuant drop in the metabolic activity of bacteria targeted by the vector.
Discussion
Our results demonstrate that engineered M13 phages can serve as potent vectors for antimicrobial PDT and that their tropism can be conveniently tweaked to target specifically diverse Gram(−) bacterial pathogens. Light-activated killing of microorganisms represents a promising alternative to treat antibiotic-resistant infections.
Phages, in their own right, represent eclectic antibacterial solutions. Classic phage therapy with natural phages has provided a useful tool before the advent of antibiotics and is receiving renewed attention for treating antibiotic-resistant pathogens. However, the intimate co-evolution of phages with specific bacterial strains results in intrinsic therapeutic limitations. Natural phages generally sport a very restricted host range, variable antibacterial (lytic) efficacy, and frequent emergence of host resistance against phage infection and propagation.8,49 The lack of full biological characterization of many natural phage preparations may present further risks due to the possible transduction of virulence genes.50 In addition, the complex and varying pharmacokinetics and pharmacodynamics of different phages constitute a major challenge for clinical translation.8,51,52,53
As such, there is a growing consensus over the idea that designer strategies, including synthetic biology and nanotechnology approaches on well-studied model phages, could contribute to overcome some constraints of classic phage therapy, for example, providing engineered phages with new (modular) functions, tailored host ranges, improved safety profiles, and a more reproducible pharmacological behavior.54,55,56,57,58
M13 represents an excellent platform for aPDT, with significant advantages over other active targeting schemes (recently reviewed in24). Taking advantage of the long-standing and straightforward molecular toolkit available for this simple phage,59 targeting moieties such as peptides and antibodies can be genetically displayed on g3p at one tip of the recombinant virion, much alike the bits of a modular screwdriver. This leaves the large number of g8p capsomers available for functionalization with hundreds of sensitizers. The exposed domain of g8p (pVIII) harbors accessible and reactive amino acids such as the lysine residues, the aromatic groups of tyrosine, and carboxylic acid groups of acidic residues, which can be combined with functional derivatives in simple processes including EDC/NHS activation, maleimide-thiol coupling, diazonium reagents, and click chemistry.60,61
The pentavalent display of the targeting moiety on g3p together with the high sensitizer payload allows respectively to (1) exploit possible cooperative binding effects of the targeting moieties displayed on g3p and (2) increase ROS generation only in close proximity of the pathogen, reaching lethal Gram(−) photosensitization at sub-micromolar concentrations of the vector, without losing specificity (Figures 5 and 7). This result indicates that the ROS produced extracellularly by the M13 platform in aPDT can be very effective without need of photosensitizer internalization. The dispensability of sensitizer internalization was similarly determined for a study on photobactericidal activity of porphyrin–cellulose nanocrystals (CNC-Por) against A. baumannii.62 However, the use of a phage scaffold allowed to attain similar killing effects with >100-fold reduction in sensitizer concentration as compared with CNC-Por. Also, for the other Gram(−) bacteria tested in this study, lethal photosensitization occurred at sub-micromolar RB concentrations when the sensitizer was conjugated to the M13 vector (Figures 3 and 7, respectively for E. coli and P. aeruginosa). By contrast, significantly higher concentrations of RB (10–100 μM) were needed to observe photoinhibition effects when the bare sensitizer was administered to planktonic Gram(−) bacteria or biofilms (63,64, and data not shown). These results are particularly interesting when we also consider that Gram(−) bacteria, due to different cell wall structure, are more resistant to aPDT than Gram(+) bacteria.65 The effectiveness at lower sensitizer concentrations may be important to reduce side effects but also to overcome the limitations of suboptimal irradiation intensities deriving from the poor penetration of light (see below).
These results add to the evidence that important improvements can be obtained in aPDT when routing the sensitizers with bacteriophage-sensitizer nanoconjugates, likely because of the higher sensitizer payload of the phage scaffolds. A pioneering study in which natural phages were conjugated with chlorine e6 demonstrated for the first time that a phage can be used to deliver photosensitizers to a target organism/cell, resulting in enhanced photosensitization and killing of S. aureus.34 This approach relied on natural phages, limiting the host range to specific species or strains. More recently, a difunctional bacteriophage, displaying a scFv for specific binding to Candida albicans and armed with pheophorbide (PPA), a chlorophyll-based photosensitizer, was used for fungal photodynamic inactivation. The PPa-JM-phage nanobioconjugate was shown to trigger light-dependent killing of C. albicans through a caspase-dependent (apoptotic) pathway.35,66
To date, the best evidence for the potential of designer phage nanobioconjugates in photodynamic antimicrobial treatments is represented by M13 phages conjugated to gold nanorods or nanoparticles.35,66 The latter can be excited by light in the near-infrared (NIR) window, translating the light energy into heat and creating extremely high local temperatures useful for photothermal therapy (PTT).67,68 In one study, M13 phanorods were retargeted to bind selectively P. aeruginosa through pIII-display of the orthologous receptor-binding domain of the P. aeruginosa-specific filamentous PF1 phage. Following their functionalization with gold nanorods, the conjugates were shown to eradicate P. aeruginosa biofilms after NIR irradiation, provoking only minor cytotoxicity to bystander mammalian cells, co-cultured with the bacteria. Notably, the irradiation also destroyed the phage vector, reducing potential risks of phage replication and horizontal gene transfer associated with classic phage therapy.35 More recently, the derivatization of phanorods with Zn2+-releasing peptides allowed to counter antibiotic-resistant P. aeruginosa wound infections in vivo, showing no detectable toxicity or systemic effects in mice following wound irradiation.69 As for phanorods, the proficient targeting and negligible side effects of the M13-RB bioconjugates need to be confirmed in an appropriate animal infection model, to validate the applicability for in vivo bacterial control.
Our results further add to these evidences, highlighting the advantage of phage-based targeting in photodynamic antimicrobial therapies. This potential is boosted by the possibility to rationally modify (change, narrow, or broaden) their tropism: we show that the targeting moieties, achieving specificity against different bacterial hosts, are not limited to the repertoire of characterized receptor-binding domains of natural phages, but also include simple peptides and stable monomeric sdAb (or scFv) scaffolds, expanding the choice to a wide portfolio of peptides, immunoglobulin (Ig) folds, and other non-Ig domains able to bind to bacteria. In particular, the broadening of the target spectrum, achieved by genetic display of an LPS-binding peptide (Figure 6), may be important if several Gram(−) pathogens need to be targeted at once or in cases where more specific targeting moieties are not available. Similarly, the receptor-binding domains of several phages with broad host range70 could serve for this purpose.
Among the advantages of antimicrobial photodynamic therapy is that the killing mechanism relies on physical damage of the biomolecules, thereby acting also on metabolically inactive or even antibiotic-resistant cells.22 ROS produced by aPDT also affect and impair lipopolysaccharide,71 leading to an immediate onset of membrane damage25 (as also evident from the live/dead assays in Figures 5G, 7G, and 7I). As such, aPDT seems exempt from the selection of resistant strains, even after repeated cycles of treatment.72 One study reported that initial exposure to low doses of antibacterial blue light (aBL, 405 nm) eventually increased the tolerance of methicillin-susceptible S. aureus to higher doses of violet light, possibly because of upregulation of bacterial stress responses.73 However, a second set of experiments showed that repeated sub-lethal aBL irradiations did not trigger photodynamic tolerance in S. aureus.74 Moreover, the dual daily administration of Indocyanine green (ICG), triggered with near-infrared 810 nm light for aPDT, combined with 405 nm aBL resulted in a sustained antibacterial effect on S. aureus.75 These results speak in favor of limited risks for developing resistance against aPDT.
On the other hand, aPDT may be affected by low specificity for target cells, whereas the photosensitizers could have low bioavailability per se in the absence of carriers (e.g., poor solubility, dispersion). Our results indicate that engineered M13 virions are not damaged by extensive conjugation with the photosensitizers. This was not predictable as the conjugated molecules may alter the solubility, monodispersion, or charge of the phage, which in turn could impair target recognition and specificity. Moreover, as the phage acts as a suicide vector, with the only scope to vehiculate sensitizers to the target cells, the downstream evolution of resistance mechanisms against phage replication and cell lysis, which frequently plagues classic phage therapy, becomes irrelevant.
For non-invasive applications, the penetration of light into tissue is highly dependent on the wavelength. Therefore, aPDT is currently best suited for topic applications, directly accessible to irradiation sources that can produce light intensities sufficient to activate the photosensitizers. Building on the improvement of irradiation and optical fiber systems, also employed in anti-cancer PDT approaches,76,77 aPDT could in principle be suitable to treat deeper infections and abscesses. For example, the use of sensitizers activable with laser light in the near-infrared (NIR) window allows to extend PDT approaches to subcutaneous settings, up to 3 cm deep in the tissues.78 Ultrasound has also been used to activate certain photosensitizers, including Rose Bengal.79 Antimicrobial sonodynamic therapy (aSDT) with RB-conjugated phages could therefore present several advantages to achieve lethal sensitization even deeper in the body than visible light or NIR.80,81
In general, photodynamic therapy seems to be a viable alternative for treating infections caused by ESKAPEE bacteria, especially for topical use. We anticipate that engineered model phages and self-assembling (virus-like) nanoparticles will provide excellent platforms for delivering sensitizers as well as antimicrobial drugs for targeted treatments. Furthermore, synergistic effects can be obtained by aPDT when combined with antibiotics, antimicrobial peptides, nanoparticles, or efflux pump inhibitors. This study shows that M13 can be engineered from an evolving biological entity with a limited host-range into a controlled, nanoparticle-like reagent with a high sensitizer payload.
In principle this nanobot may be retargeted to any cell if a targeting peptide or moiety can be fused to the g3p protein. In this scheme the possible mechanisms to develop resistance are likely to be constrained to mutations in the chosen receptor or in the acquisition of exceptionally robust ROS detoxification mechanisms.
Limitations of the study
Our study presents a novel modular phage vector platform for the application of antimicrobial photodynamic therapy against Gram-negative bacterial pathogens. However, there are a number of limitations to our study to be pointed out. First, a potential limitation of targeting M13 against A. baumannii by expressing the C20 nanobody fused with the pIII protein is the variability of the BAP protein among all A. baumannii strains. Although the gene is conserved in nearly all strains of A. baumannii, variations in the coding sequence, such as insertions, deletions, or mutations, could alter the binding epitope and hinder efficient phage targeting.82 To overcome this challenge, M13Gram− can be employed, as it has the ability to target broadly Gram(−) bacteria, including A. baumannii. Alternatively, the modular nature of our platform can be exploited by displaying other targeting moieties specific for A. baumannii, in fusion with the pIII protein of M13. As a proof of concept in this study, both wild-type M13 phage and engineered phages were conjugated with the photosensitizer Rose Bengal. However, this does not exclude the possibility that other photosensitizers may exhibit improved efficiency in generating ROS and subsequent increased antimicrobial photodynamic therapy (aPDT) effects on pathogens. Considering the wide range of available sensitizers to choose from, the effective potential on phage-mediated aPDT may be further validated and expanded.83,84,85 The selection of a specific sensitizer can also be based on therapeutic needs. For instance, the use of sensitizers excitable with infrared lasers allows for deeper tissue penetration of light, enabling the treatment of subcutaneous infections.85 Lastly, our investigations were limited to in vitro experiments, in order to provide a first proof-of-principle of the technology and did not involve in vivo animal models. To fully comprehend the potential of engineered phages rigged for aPDT in targeting bacterial infections, it will be important to explore the challenges associated with animal treatments. This would involve investigating different modes of phage administration, such as intraperitoneal, intravenous, or local injection, to determine the optimal approach for targeting the infections. In addition, the efficacy of phage administration may vary depending on the bacterial species and the region of infection. Therefore, further research is needed to evaluate the applicability of our platform in diverse infection models.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-M13 g3p monoclonal antibody | New England BioLabs | Cat#E8033; RRID:AB_1559734 |
| HRP-conjugated IgG goat anti-mouse IgG | Jackson | Cat#115-035-166; RRID:AB_2338511 |
| Bacterial and virus strains | ||
| E. coli Tg1 | DSMZ | 6056 |
| E. coli DH5α | DSMZ | 6897 |
| Acinetobacter baumannii ATCC 19606 | American Type Culture Collection | 19606 |
| Pseudomonas aeruginosa PA14 | DSMZ | 19882 |
| Staphylococcus aureus ATCC 29213 | American Type Culture Collection | 29213 |
| M13KO7 Helper Phage | New England BioLabs | NO315S |
| Hyperphage M13 KO7ΔpIII | PROGEN | PRHYPE |
| Chemicals, peptides, and recombinant proteins | ||
| Rose Bengal (RB) | Merck | 330000 |
| EDC | Merck | 03450 |
| Tetrazolium Violet | Merck | T0138 |
| NHS | Merck | 130672 |
| ABMDMA | Merck | 75068 |
| Amplex Red | Merck | 90101 |
| HRP | Merck | P6782 |
| Critical commercial assays | ||
| PowerUp™ SYBR™ Green Master Mix for qPCR | Thermo Fisher Scientific | A25742 |
| LIVE/DEAD™ BacLight™ Bacterial Viability Kit | Thermo Fisher Scientific | L7007 |
| Deposited data | ||
| pPK_LPS | This study | GenBank: OQ803482 |
| pPKC20 | This study | GenBank: OQ803483 |
| M13KO7Δg3pΔoriF1 | This study | GenBank: OQ803484 |
| Plasmids and phagemids | ||
| pComb3XSS | Addgene | 63890 |
| pPK_LPS | This study | GenBank: OQ803482 |
| pPK_C20 | This study | GenBank: OQ803483 |
| M13KO7Δg3pΔoriF1 | This study | GenBank: OQ803484 |
| Oligonucleotides | ||
| Primer: forward amplification C20 (AD251) GTTTTGAGCTCCAGGTGCAG |
This study | N/A |
| Primer: reverse amplification C20 (AD252) GTTTTACTAGTACTAACGGTAACCTGG |
This study | N/A |
| Primer: forward amplification Li005 (AD300) CAAAAACTATTCGAGCTCGATTAGCAGCATTCGTGCGTGCA |
This study | N/A |
| Primer: reverse amplification Li005 (AD301) CTAGTGCACGCACGAATGCTGCTAA TCGAGCTCGAATAGTTTTTGAGCT |
This study | N/A |
| Primer: forward M13 F1ori (AD046) AGGCTGCACCGGTGCGTCAG |
This study | N/A |
| Primer: reverse M13 F1ori (AD047) ACACCCGCCGCGCTTAATGCG |
This study | N/A |
| Primer: forward qPCR (AD211) TCCGGCCTTGCTAATGGTAA |
This study | N/A |
| Primer: reverse qPCR (AD212) CGCAAAGACACCACGGAATA |
This study | N/A |
| Software and algorithms | ||
| BLAST | https://blast.ncbi.nlm.nih.gov/BlastAlign.cgi | v 2.6.0 |
| MUSCLE | https://www.ebi.ac.uk/Tools/msa/muscle/ | v 3.8.31 |
| ImageJ | GitHub | v 1.43t |
| NanoScope Analysis | Bruker corporation | v 1.80 |
| MATLAB | Mathworks | v 9.7 R2019b |
| CytExpert | Beckman | v 2.4 |
| FlowJo | BD Biosciences | v 10 |
| Prism | GraphPad | v 10 |
Resource availability
Lead contact
Further inquiries and request for data, strains and resources should be directed to the lead contact Alberto Danielli, alberto.danielli@unibo.it.
Materials availability
Materials generated in this study are available upon request from the lead contact.
Experimental model and study participant details
Bacterial strains used in this study are listed in the key resources table. All the bacterial strains used in this study were routinely cultured in solid LB agar plates or LB liquid medium at 37°C.
Method details
Bacterial cultures
Acinetobacter baumannii ATCC 19606, Pseudomonas aeruginosa PA14 (clinical isolate), Escherichia coli TG1 and Staphylococcus aureus ATCC 29213 were routinely cultured in solid LB agar plates or LB liquid medium. E. coli TG1 cells transformed with plasmids or phagemids were grown solid or liquid LB supplemented with appropriate antibiotic as described below.
Phagemid cloning and phage preparation
The translational fusion of the anti-A. baumannii (C20) nanobody or the LPS-binding peptide at the N-terminus of the minor capsid protein pIII was performed by cloning the coding sequences of interest in pPK3DsbA a modified version of the pComb3XSS phagemid86 obtained from Addgene (pComb3XSS was a gift from Carlos Barbas - Addgene plasmid # 63890; http://n2t.net/addgene:63890; RRID: Addgene_63890). Briefly, to generate pPK3DsbA, the ompA leader sequence found in the original pComb3XSS vector was replaced with a dsbA leader sequence, while the SacI-SpeI insert encompassing the original light and heavy chain stuffers was replaced by a multiple cloning site (MCS), in frame with the C-terminal domain of the pIII protein. The gene encoding for the anti-BAP nanobody C2043 was synthesized by Eurofins Genomics. Amplification of the synthetic C20 gene with oligonucleotides AD0251 (SacI overhang) and AD0252 (SpeI overhang), generated amplicons that were further purified and digested with SacI/SpeI restriction endonucleases (NEB, New England Biolabs). and then purified through PCR clean-up. The LPS binding peptide coding sequence (KNYSSSISSIRAC,48) was obtained by annealing of oligonucleotides AD0300 (CAAAAACTATTCGAGCTCGATTAGCAGCATTCGTGCGTGCA) and AD0301 (CTAGTGCACGCACGAATGCTGCTAATCGAGCTCGAATAGTTTTTGAGCT), resulting in SacI and SpeI overhangs. Both inserts were directionally cloned into the SacI-SpeI linearized pPK3DsbA vector, generating phagemids pPK_LPS (GenBank: OQ803482) and pPK_C20 (GenBank: OQ803483). Positive clones were validated by sequencing of purified the constructs.
Engineered phages (M13Aba and M13Gram-) were generated by co-transforming E. coli TG1 with pPK_LPS or pPK_C20 phagemids and a Hyperphage helper derivative (ProGen), M13KO7Δg3pΔoriF1 (GenBank: OQ803484), in which the oriF1 was removed by inverse PCR with oligos AD0046 (AGGCTGCACCGGTGCGTCAG) and AD0047 (ACACCCGCCGCGCTTAATGCG) followed by re-ligation of the amplicon. Thereby, uniform virions (about 600 nm in length), packaging only the pPK phagemids containing the f1 ori where produced. Cotransformants were grown for 24 h in LB medium supplemented with Ampicillin (100 mg L−1), Kanamicin (25 mg L−1) and 0.4 mM IPTG to induce expression of the fusion construct. Purification of phage was performed as previously described.38 Briefly, the bacterial culture was pelleted for 30 min at 12.000g to separate bacterial cells from phages present in the supernatant. PEG 8000 (4% w/v) and NaCl (3% w/v) were added to the supernatant and incubated for 90 min at 4°C. Next, the solution was centrifuged for 15 min at 12.000g to precipitate virions. The phage pellet was then resuspended in PBS 1x and further purified by isoelectric point (IEP) precipitation.87 Purified phages were finally resuspended in PBS 1x pH 7.4. Phage concentrations were determined by measurement of the absorbance at 269 nm wavelength in a UV-Vis spectrophotometer (extinction coefficient ε = 3.84 cm2 mg−1) using a 320 nm wavelength spectrum as baseline.
Transmission electron microscopy (TEM) analysis
A drop of M13 bacteriophages solution (1x1011 v/mL) was deposited on an oxygen plasma cleaned TEM grid (formvar-coated copper 200 mesh, Ted Pella, Redding, CA, USA) for 30 s and then removed with filter paper. The negative staining was performed using 1% uranyl acetate, and the TEM analysis was performed using a JEOL JEM 1400 microscope (JEOL, Tokyo, Japan) operating at 120 kV. The TEM micrographs were analyzed by using ImageJ.
AFM analysis of engineered phages
AFM analysis was performed with a Multimode 8 AFM (Bruker, U.S.A.). Aliquots of concentration-adjusted phage solutions diluted PBS buffer were deposited on the freshly-cleaved muscovite mica (Electron Microscopy Sciences, U.S.A.) surface let to absorb for 2 min. The AFM fluid cell was mounted and flowed with a small volume of ultrapure water (milliQ, Millipore, U.S.A.). Images were taken in PeakForce Tapping in liquid using ScanAsyst Fluid+ probes (Bruker, U.S.A.), and processed with NanoScope Analysis software (ver. 1.80) by only flattening the micrographs. The contour space coordinates of hundreds phages were digitized semi-automatically from the micrographs using a custom-developed algorithm implemented in MATLAB (Mathworks, U.S.A.).88,89
Immunoblotting
M13Gram- and M13C20_Nb purified phages at a concentration of 1010 pfu μL−1 were resolved using a 12% (w/v) SDS gel and transferred to a PVDF membrane (GVS). After treatment with blocking solution (5% milk in PBS pH 7.4 with 0.05% Tween), the membrane was incubated with Anti-M13 g3p Monoclonal Antibody (New England BioLabs) diluted 1: 5000 in blocking solution for 1 h at room temperature. After three washes in PBS Tween 0.05%, the membrane was incubated with HRP-conjugated IgG anti-mouse secondary antibody (Jackson) diluted 1:10 000 and incubated for 1 h at room temperature. The membrane development was performed using LightWave Plus - ECL HRP reagent (GVS). Images were acquired via ChemiDoc Imaging System (Bio-Rad).
Validation of phage retargeting by qPCR
Engineered or wild type M13 phages were incubated with exponential phase P. aeruginosa, A. baumannii, and E. coli cells (106 CFU·mL−1) for 15 min at room temperature under gentle shaking. Unbound phages were removed by centrifugation and washing, twice in PBS. The bacterial pellet containing bound phages was the resuspended in PBS 1x, collected and store at −20°C. Samples were boiled at 98°C for 15 min, to release the viral genome, and then 2 μL were used as template for the qPCR reaction (PowerUp Green Master Mix; ThermoFisher Scientific) using primers AD211 (TCCGGCCTTGCTAATGGTAA) and AD212 (CGCAAAGACACCACGGAATA), annealing to the C-terminal domain of the pIII gene. Commercial M13K07 (NEB) was used as a standard to set the calibration curve.
Bioconjugation and purification of M13-RB, M13Aba and M13Gram- phages
Rose Bengal (RB) (Merck, Cat. No. 330000) was conjugated to the capsomers of the M13, M13Aba and M13Gram- phage vectors by EDC/NHS cross-coupling reaction between the amine groups of M13 capsomers and the activated carboxylic group of RB, as previously reported.38 A solution 10 mM of RB in DMSO (Merck, Cat. No. 472301) was prepared. The activation of the RB carboxylic group was carried out, under stirring condition, by adding EDC (Merck, Cat. No. 03450) and NHS (Merck, Cat. No. 130672) to the RB solution, with a final concentration of 10 mM and 15 mM, respectively. The solution was incubated for 3 h at 25°C under constant shaking at 700 rpm (ThermoMixer HC, S8012-0000; STARLAB, Hamburg, Germany). 50 μL of the activated RB solution was then mixed with 1 mL of the M13, M13Aba or M13Gram- phage solution at a concentration of 2.4 × 101³ virions mL⁻1 (40 nM) in PBS, resulting in a final RB concentration of 500 μM. The mixture was incubated overnight at 25°C and 700 rpm. The bioconjugates were purified using a PD-10 Desalting Column (containing 8.3 mL of Sephadex G-25 Medium, Merck, Cat. No. GE17-0851-01) to remove unreacted RB and the byproducts of the crosslinking reaction using PBS as elution solvent.
Singlet oxygen assay
The production of singlet oxygen (1O2) generated by light irradiation was detected by monitoring the decline of absorbance of 9,10-Anthracenediyl-bis(methylene) dimalonic acid (ABMDMA) (Merck, Cat. No. 75068), at 401 nm, due to the reaction of 1O2 with ABMDMA to give the corresponding endoperoxide. Iso-absorbing solutions of RB, M13Gram--RB, and M13Aba-RB were prepared in deuterated PBS (10 mM, pH 7.4, Merck, Cat. No. 151882). 500 μL of each solution, containing a concentration of 15 μM of RB and 25 μM of ABMDMA, were irradiated with a visible light lamp (Valex cold white LED, with an irradiance of 2.4 mW cm−2, measured with the photo-radiometer Delta Ohm LP 471 RAD) and maintained under vigorous stirring. The singlet oxygen quantum yield (φΔ) of RB in deuterated PBS is 0.7690 and was used as a reference to determine the φΔ of M13Gram--RB and M13Aba-RB using the following equation: φΔS = kS/kR × φΔR, where kS and kR are the slopes of the photodegradation rate of ABMDMA of the samples (M13Gram--RB and M13Aba-RB) and of the reference (RB), and φΔS and φΔR are the singlet oxygen quantum yields of the samples and the reference, respectively.
Peroxides assay
Amplex Red assay was used to quantify the peroxides generated by light irradiation. Amplex Red (Merck, Cat. No. 90101) is a colorless molecule that reacts with peroxides in the presence of horseradish peroxidase (HRP, Merck, Cat. No. P6782) to produce resorufin, a fluorescent product. The concentration of peroxides was determined by measuring the difference in the fluorescence intensity of the irradiated samples and non-irradiated references. A working solution (WS) was prepared by mixing 10 μL of HRP (0.4 mg mL−1) in PBS with 1 mL of Amplex Red (500 μM) in phosphate buffer (50 mM, pH 7.4). Solutions of RB, M13Gram--RB, and M13Aba-RB were prepared at concentrations of 0.0 μM, 0.25 μM, 0.5 μM, 1.0 μM, 2.0 μM, and 4.0 μM of RB in a 96-well plate. The sample solutions were irradiated for 45 min with visible light (white LED Valex 30 W lamp, irradiation power density on the cell plate = 20 mW cm−2; measured with the photo-radiometer Delta Ohm LP 471 RAD). After irradiation, 10 μL of WS was added to each well. The solutions were then incubated for 30 min at room temperature in the dark. Three technical replicates were performed for each sample. An identical plate was prepared and kept in the dark. To calculate the concentration of peroxides in the samples from the fluorescence signal, a calibration curve was made using standard solutions of H2O2. The fluorescence emission was measured at 590 nm (λex = 530 nm) using a EnSpire Multimode plate reader (PerkinElmer).
Validation of phage retargeting by flow cytometry
RB-conjugated phages were incubated with 106 exponential phase bacterial cells, with a virion:bacteria ratio of to 100:1, for 15 min at room temperature and under gently shacking. Bacterial cells were centrifuged at 6000g for 10 min and washed twice with PBS, to remove unbound phages, prior to fixation with a solution of paraformaldehyde (PFA) 4% in PBS for 15 min. Fixed cells were centrifuged and washed three times with PBS to remove excess PFA, resuspended in PBS and analyzed with a CytoFLEX S cytofluorimeter (Beckam Coulter). Events present in the gated population (gating performed on forward (FSC) and side (SSC) scatter) were analyzed for the fluorescence in the FL1 channel (emission filter 585/42 nm). At least 10.000 events were recorded for each sample. Results are presented as percentage of fluorescent cells in comparison to the control (untreated bacterial cells). Data analysis was processed using CytExpert (Beckman) and FlowJo.
Antimicrobial PDT assays
The photodynamic effect of RB-phage bioconjugates was assessed on bacteria grown to exponential phase in planktonic culture. Free Rose Bengal was used as a further control to verify the phage vector platform’s targeting efficacy. Bacterial cultures were grown to exponential phase (OD600 = 0.4–0.7), pelleted twice by centrifugation for 10 min at 5000g and resuspended in PBS, with a target concentration of 106 cfu mL−1 50 μL of this suspension was seeded in triplicate together with the phage conjugates in the wells of two 96 well u-bottom plates (Sarstedt), reaching a final volume of 100 μL. Both plates were preincubated in the dark for 15 min under gentle agitation to allow phage-bacteria interaction. Unbound phages were removed by washing twice in PBS. Then, one of the two plates was exposed to white light (white LED Valex 30 W lamp, irradiation power density on the cell plate = 20 mW cm−2; measured with the photo-radiometer Delta Ohm LP 471 RAD) at room temperature, for 45 min. The second plate, representing the negative control, was kept in the dark at room temperature for the same time. After irradiation, bacteria were gently mixed and transferred to fresh growth medium (LB broth, dilution 1:10) in a 96-well flat bottom plate (Sarstedt). Growth rates of live bacteria were then monitored with an EnSpire Multimode plate reader (PerkinElmer), by measuring the increase of absorbance (OD600) every 10 min for 15 h at 37°C in each well. Absorbance OD600 values for each time point were averaged and plotted on graphs mirroring the kinetics (recovery) of growth after irradiation. The antimicrobial activity of PDT was inferred by measuring the delay in time (=number of generations) needed to reach the onset of the exponential phase of growth (normalized OD600 = 0.125/0.15), relative to the control sample without sensitizer kept in the dark. Bacterial survival rates of the different strains were normalized with respect to light exposure without RB conjugated phages or free RB, therefore considering the different susceptibility of these strains to light. At least three biological replicates were performed for each sample. Live/dead assay was performed on bacterial cells 20 min after aPDT treatment using the commercial LIVE/DEAD BacLight Bacterial Viability Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Metabolic activity after aPDT treatment was evaluated as described before.91 Briefly, immediately after PDT treatment, cultures were diluted 1:2 in LB medium supplemented with 0.02% Tetrazolium violet (Merck) and then incubated for 16 h at 37°C. The reduction of tetrazolium violet by metabolically active bacteria was estimated by measuring the absorbance at 550 nm using the multimode plate reader EnSpire (PerkinElmer).
Quantification and statistical analysis
All statistical inference tests were performed by using GraphPad Prism 10. Tests used and corresponding p values are reported along with each result as appropriate.
Acknowledgments
This research was supported by a CARISBO grant (Ricerca Medica e Alta Tecnologia 2021), AIRC under the MFAG 2019 ID. 22894 project (P.I.: M.C.), and EU funding within the NextGeneration EU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT) and from the Horizon Pathfinder Open programme under Grant Agreement N10104678ECLipse. A.D. was supported by a Proof-of-Concept grant by the Italian Ministry of Economic Development and the University of Bologna. The support from the Italian MUR (PRIN2022, PhageTarget project 2022K452FJ) is gratefully acknowledged.
Author contributions
Investigation, Data Curation, Formal Analysis: A.P., R.S., F.S., E.S., L.U., L.B., A.C., A.M., S.K.Z., G.Z., Z.P., M.D.G., and P.E.C. Conceptualization, Formal Analysis, Funding Acquisition, Methodology, Supervision: L.P. P.P.P., P.E.C, M.C., and A.D. Writing: all authors.
Declaration of interests
There are no conflicts to declare.
Published: September 27, 2023
Contributor Information
Paolo Emidio Costantini, Email: paolo.costantini4@unibo.it.
Matteo Calvaresi, Email: matteo.calvaresi3@unibo.it.
Alberto Danielli, Email: alberto.danielli@unibo.it.
Data and code availability
-
•
Sequences of plasmids and phagemids generated in this study have been deposited to GenBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All the other data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Strathdee S.A., Hatfull G.F., Mutalik V.K., Schooley R.T. Phage therapy: From biological mechanisms to future directions. Cell. 2023;186:17–31. doi: 10.1016/J.CELL.2022.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uyttebroek S., Chen B., Onsea J., Ruythooren F., Debaveye Y., Devolder D., Spriet I., Depypere M., Wagemans J., Lavigne R., et al. Safety and efficacy of phage therapy in difficult-to-treat infections: a systematic review. Lancet Infect. Dis. 2022;22:e208–e220. doi: 10.1016/S1473-3099(21)00612-5. [DOI] [PubMed] [Google Scholar]
- 3.Pirnay J.P., Kutter E. Bacteriophages: it’s a medicine, Jim, but not as we know it. Lancet Infect. Dis. 2021;21:309–311. doi: 10.1016/S1473-3099(20)30464-3. [DOI] [PubMed] [Google Scholar]
- 4.Schooley R.T., Biswas B., Gill J.J., Hernandez-Morales A., Lancaster J., Lessor L., Barr J.J., Reed S.L., Rohwer F., Benler S., et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berryhill B.A., Huseby D.L., McCall I.C., Hughes D., Levin B.R. Evaluating the potential efficacy and limitations of a phage for joint antibiotic and phage therapy of Staphylococcus aureus infections. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/PNAS.2008007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Nieuwenhuyse B., Van der Linden D., Chatzis O., Lood C., Wagemans J., Lavigne R., Schroven K., Paeshuyse J., de Magnée C., Sokal E., et al. Bacteriophage-antibiotic combination therapy against extensively drug-resistant Pseudomonas aeruginosa infection to allow liver transplantation in a toddler. Nat. Commun. 2022;13:5725–5812. doi: 10.1038/s41467-022-33294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordillo Altamirano F.L., Kostoulias X., Subedi D., Korneev D., Peleg A.Y., Barr J.J. Phage-antibiotic combination is a superior treatment against Acinetobacter baumannii in a preclinical study. EBioMedicine. 2022;80 doi: 10.1016/J.EBIOM.2022.104045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dąbrowska K., Abedon S.T. Pharmacologically aware phage therapy: pharmacodynamic and pharmacokinetic obstacles to phage antibacterial action in animal and human bodies. Microbiol. Mol. Biol. Rev. 2019;83:e00012–e00019. doi: 10.1128/MMBR.00012-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang R., Li H.D., Cao Y., Wang Z.Y., Yang T., Wang J.H. M13 phage: a versatile building block for a highly specific analysis platform. Anal. Bioanal. Chem. 2023;415:3927–3944. doi: 10.1007/S00216-023-04606-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henry K.A., Arbabi-Ghahroudi M., Scott J.K. Beyond phage display: non-traditional applications of the filamentous bacteriophage as a vaccine carrier, therapeutic biologic, and bioconjugation scaffold. Front. Microbiol. 2015;6:755. doi: 10.3389/fmicb.2015.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sperandio F.F., Huang Y.-Y., Hamblin M.R. Antimicrobial photodynamic therapy to kill Gram-negative bacteria. Recent Pat. Anti-Infect. Drug Discov. 2013;8:108–120. doi: 10.2174/1574891X113089990012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi H., Sadler P.J. How promising is phototherapy for cancer? Br. J. Cancer. 2020;123:871–873. doi: 10.1038/s41416-020-0926-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolmans D.E.J.G.J., Fukumura D., Jain R.K. Photodynamic therapy for cancer. Nat. Rev. Cancer. 2003;3:380–387. doi: 10.1038/NRC1071. [DOI] [PubMed] [Google Scholar]
- 14.Yin R., Agrawal T., Khan U., Gupta G.K., Rai V., Huang Y.Y., Hamblin M.R. Antimicrobial photodynamic inactivation in nanomedicine: small light strides against bad bugs. Nanomedicine. 2015;10:2379–2404. doi: 10.2217/NNM.15.67. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Li X., Lovell J.F., Yoon J., Chen X. Clinical development and potential of photothermal and photodynamic therapies for cancer. Nat. Rev. Clin. Oncol. 2020;17:657–674. doi: 10.1038/s41571-020-0410-2. [DOI] [PubMed] [Google Scholar]
- 16.Cieplik F., Deng D., Crielaard W., Buchalla W., Hellwig E., Al-Ahmad A., Maisch T. Antimicrobial photodynamic therapy - what we know and what we don’t. Crit. Rev. Microbiol. 2018;44:571–589. doi: 10.1080/1040841X.2018.1467876. [DOI] [PubMed] [Google Scholar]
- 17.Hu X., Huang Y.Y., Wang Y., Wang X., Hamblin M.R. Antimicrobial photodynamic therapy to control clinically relevant biofilm infections. Front. Microbiol. 2018;9:1299. doi: 10.3389/FMICB.2018.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomb R.M., White T.A., Coia J.E., Anderson J.G., MacGregor S.J., Maclean M. Review of the comparative susceptibility of microbial species to photoinactivation using 380-480 nm violet-blue Light. Photochem. Photobiol. 2018;94:445–458. doi: 10.1111/PHP.12883. [DOI] [PubMed] [Google Scholar]
- 19.Wozniak A., Grinholc M. Combined antimicrobial activity of photodynamic inactivation and antimicrobials-state of the art. Front. Microbiol. 2018;9:930. doi: 10.3389/FMICB.2018.00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka M., Mroz P., Dai T., Huang L., Morimoto Y., Kinoshita M., Yoshihara Y., Nemoto K., Shinomiya N., Seki S., Hamblin M.R. Photodynamic therapy can induce a protective innate immune response against murine bacterial arthritis via neutrophil accumulation. PLoS One. 2012;7 doi: 10.1371/JOURNAL.PONE.0039823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barra F., Roscetto E., Soriano A.A., Vollaro A., Postiglione I., Pierantoni G.M., Palumbo G., Catania M.R. Photodynamic and antibiotic therapy in combination to fight biofilms and resistant surface bacterial infections. Int. J. Mol. Sci. 2015;16:20417–20430. doi: 10.3390/IJMS160920417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcez A.S., Nuñez S.C., Hamblim M.R., Suzuki H., Ribeiro M.S. Photodynamic therapy associated with conventional endodontic treatment in patients with antibiotic-resistant microflora: a preliminary report. J. Endod. 2010;36:1463–1466. doi: 10.1016/J.JOEN.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Takasaki A.A., Aoki A., Mizutani K., Schwarz F., Sculean A., Wang C.Y., Koshy G., Romanos G., Ishikawa I., Izumi Y. Application of antimicrobial photodynamic therapy in periodontal and peri-implant diseases. Periodontology. 2009;51:109–140. doi: 10.1111/J.1600-0757.2009.00302.X. [DOI] [PubMed] [Google Scholar]
- 24.Thomas-Moore B.A., del Valle C.A., Field R.A., Marín M.J. Recent advances in nanoparticle-based targeting tactics for antibacterial photodynamic therapy. Photochem. Photobiol. Sci. 2022;21:1111–1131. doi: 10.1007/S43630-022-00194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cantelli A., Piro F., Pecchini P., Di Giosia M., Danielli A., Calvaresi M. Concanavalin A-Rose Bengal bioconjugate for targeted Gram-negative antimicrobial photodynamic therapy. J. Photochem. Photobiol., B. 2020;206:111852. doi: 10.1016/J.JPHOTOBIOL.2020.111852. [DOI] [PubMed] [Google Scholar]
- 26.Cao B., Xu H., Yang M., Mao C. Virus-based cancer therapeutics for targeted photodynamic therapy. Methods Mol. Biol. 2018;1776:643–652. doi: 10.1007/978-1-4939-7808-3_41. [DOI] [PubMed] [Google Scholar]
- 27.Cohen B.A., Bergkvist M. Targeted in vitro photodynamic therapy via aptamer-labeled, porphyrin-loaded virus capsids. J. Photochem. Photobiol., B. 2013;121:67–74. doi: 10.1016/J.JPHOTOBIOL.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Ariga K., Leong D.T., Mori T. Nanoarchitectonics for hybrid and related materials for bio-oriented applications. Adv. Funct. Mater. 2018;28 doi: 10.1002/ADFM.201702905. [DOI] [Google Scholar]
- 29.Kalarical Janardhanan S., Narayan S., Abbineni G., Hayhurst A., Mao C. Architectonics of phage-liposome nanowebs as optimized photosensitizer vehicles for photodynamic cancer therapy. Mol. Cancer Therapeut. 2010;9:2524–2535. doi: 10.1158/1535-7163.MCT-10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deken M.M., Kijanka M.M., Beltrán Hernández I., Slooter M.D., de Bruijn H.S., van Diest P.J., van Bergen en Henegouwen P.M.P., Lowik C.W.G.M., Robinson D.J., Vahrmeijer A.L., Oliveira S. Nanobody-targeted photodynamic therapy induces significant tumor regression of trastuzumab-resistant HER2-positive breast cancer, after a single treatment session. J. Contr. Release. 2020;323:269–281. doi: 10.1016/J.JCONREL.2020.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulfo L., Costantini P.E., Di Giosia M., Danielli A., Calvaresi M. EGFR-targeted photodynamic therapy. Pharmaceutics. 2022;14:241. doi: 10.3390/PHARMACEUTICS14020241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia H.R., Zhu Y.X., Chen Z., Wu F.G. Cholesterol-assisted bacterial cell surface engineering for photodynamic inactivation of Gram-positive and Gram-negative Bacteria. ACS Appl. Mater. Interfaces. 2017;9:15943–15951. doi: 10.1021/ACSAMI.7B02562. [DOI] [PubMed] [Google Scholar]
- 33.Jia H.R., Zhu Y.X., Liu Y., Guo Y., Sayed S.M., Zhu X.Y., Cheng X., Wu F.G. Direct chemical editing of Gram-positive bacterial cell walls via an enzyme-catalyzed oxidative coupling reaction. Explorations. 2022;2 doi: 10.1002/EXP.20220010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Embleton M.L., Nair S.P., Heywood W., Menon D.C., Cookson B.D., Wilson M. Development of a novel targeting system for lethal photosensitization of antibiotic-resistant strains of Staphylococcus aureus. Antimicrob. Agents Chemother. 2005;49:3690–3696. doi: 10.1128/AAC.49.9.3690-3696.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng H., Borg R.E., Dow L.P., Pruitt B.L., Chen I.A. Controlled phage therapy by photothermal ablation of specific bacterial species using gold nanorods targeted by chimeric phages. Proc. Natl. Acad. Sci. USA. 2020;117:1951–1961. doi: 10.1073/pnas.1913234117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J.-W., Song J., Hwang M.P., Lee K.H. Nanoscale bacteriophage biosensors beyond phage display. Int. J. Nanomed. 2013;8:3917–3925. doi: 10.2147/IJN.S51894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gandra N., Abbineni G., Qu X., Huai Y., Wang L., Mao C. Bacteriophage bionanowire as a carrier for both cancer-targeting peptides and photosensitizers and its use in selective cancer cell killing by photodynamic therapy. Small. 2013;9:215–221. doi: 10.1002/SMLL.201202090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ulfo L., Cantelli A., Petrosino A., Costantini P.E., Nigro M., Starinieri F., Turrini E., Zadran S.K., Zuccheri G., Saporetti R., et al. Orthogonal nanoarchitectonics of M13 phage for receptor targeted anticancer photodynamic therapy. Nanoscale. 2022;14:632–641. doi: 10.1039/d1nr06053h. [DOI] [PubMed] [Google Scholar]
- 39.Bortot B., Apollonio M., Baj G., Andolfi L., Zupin L., Crovella S., di Giosia M., Cantelli A., Saporetti R., Ulfo L., et al. Advanced photodynamic therapy with an engineered M13 phage targeting EGFR: Mitochondrial localization and autophagy induction in ovarian cancer cell lines. Free Radic. Biol. Med. 2022;179:242–251. doi: 10.1016/J.FREERADBIOMED.2021.11.019. [DOI] [PubMed] [Google Scholar]
- 40.Demartis S., Obinu A., Gavini E., Giunchedi P., Rassu G. Nanotechnology-based rose Bengal: a broad-spectrum biomedical tool. Dyes Pigments. 2021;188 doi: 10.1016/J.DYEPIG.2021.109236. [DOI] [Google Scholar]
- 41.Moczek Ł., Nowakowska M. Novel water-soluble photosensitizers from chitosan. Biomacromolecules. 2007;8:433–438. doi: 10.1021/bm060454+. [DOI] [PubMed] [Google Scholar]
- 42.Russel M., Whirlow H., Sun T.P., Webster R.E. Low-frequency infection of F- bacteria by transducing particles of filamentous bacteriophages. J. Bacteriol. 1988;170:5312–5316. doi: 10.1128/JB.170.11.5312-5316.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Payandeh Z., Rasooli I., Mousavi Gargari S.L., Rajabi Bazl M., Ebrahimizadeh W. Immunoreaction of a recombinant nanobody from camelid single domain antibody fragment with Acinetobacter baumannii. Trans. R. Soc. Trop. Med. Hyg. 2014;108:92–98. doi: 10.1093/TRSTMH/TRT114. [DOI] [PubMed] [Google Scholar]
- 44.Di Giosia M., Bomans P.H.H., Bottoni A., Cantelli A., Falini G., Franchi P., Guarracino G., Friedrich H., Lucarini M., Paolucci F., et al. Proteins as supramolecular hosts for C60: a true solution of C60 in water. Nanoscale. 2018;10:9908–9916. doi: 10.1039/C8NR02220H. [DOI] [PubMed] [Google Scholar]
- 45.Cantelli A., Malferrari M., Soldà A., Simonetti G., Forni S., Toscanella E., Mattioli E.J., Zerbetto F., Zanelli A., Di Giosia M., et al. Human serum albumin-oligothiophene bioconjugate: a phototheranostic platform for localized killing of cancer cells by precise light activation. JACS Au. 2021;1:925–935. doi: 10.1021/JACSAU.1C00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pier G.B. Pseudomonas aeruginosa lipopolysaccharide: A major virulence factor, initiator of inflammation and target for effective immunity. Int. J. Med. Microbiol. 2007;297:277–295. doi: 10.1016/J.IJMM.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moffatt J.H., Harper M., Mansell A., Crane B., Fitzsimons T.C., Nation R.L., Li J., Adler B., Boyce J.D. Lipopolysaccharide-deficient Acinetobacter baumannii shows altered signaling through host Toll-like receptors and increased susceptibility to the host antimicrobial peptide LL-37. Infect. Immun. 2013;81:684–689. doi: 10.1128/IAI.01362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki M.M., Matsumoto M., Yamamoto A., Ochiai M., Horiuchi Y., Niwa M., Omi H., Kobayashi T., Takagi T. Molecular design of LPS-binding peptides. J. Microbiol. Methods. 2010;83:153–155. doi: 10.1016/J.MIMET.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 49.Castledine M., Padfield D., Sierocinski P., Soria Pascual J., Hughes A., Mäkinen L., Friman V.P., Pirnay J.P., Merabishvili M., De Vos D., Buckling A. Parallel evolution of Pseudomonas aeruginosa phage resistance and virulence loss in response to phage treatment in vivo and in vitro. Elife. 2022;11 doi: 10.7554/ELIFE.73679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Penadés J.R., Chen J., Quiles-Puchalt N., Carpena N., Novick R.P. Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 2015;23:171–178. doi: 10.1016/J.MIB.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 51.Hesse S., Adhya S. Phage therapy in the twenty-first century: facing the decline of the antibiotic era; is it finally time for the age of the phage? Annu. Rev. Microbiol. 2019;73:155–174. doi: 10.1146/ANNUREV-MICRO-090817-062535. [DOI] [PubMed] [Google Scholar]
- 52.Suh G.A., Lodise T.P., Tamma P.D., Knisely J.M., Alexander J., Aslam S., Barton K.D., Bizzell E., Totten K.M.C., Campbell J.L., et al. Considerations for the use of phage therapy in clinical practice. Antimicrob. Agents Chemother. 2022;66 doi: 10.1128/AAC.02071-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmalstig A.A., Freidy S., Hanafin P.O., Braunstein M., Rao G.G. Reapproaching old treatments: considerations for PK/PD studies on phage therapy for bacterial respiratory infections. Clin. Pharmacol. Ther. 2021;109:1443–1456. doi: 10.1002/CPT.2214. [DOI] [PubMed] [Google Scholar]
- 54.Peng H., Chen I.A. Phage engineering and the evolutionary arms race. Curr. Opin. Biotechnol. 2021;68:23–29. doi: 10.1016/J.COPBIO.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lavigne R., Loessner M.J. Editorial overview: Phage therapy in the 21st century - inspired by biotechnology. Curr. Opin. Biotechnol. 2021;68 doi: 10.1016/J.COPBIO.2021.04.002. vi–vii. [DOI] [PubMed] [Google Scholar]
- 56.Lenneman B.R., Fernbach J., Loessner M.J., Lu T.K., Kilcher S. Enhancing phage therapy through synthetic biology and genome engineering. Curr. Opin. Biotechnol. 2021;68:151–159. doi: 10.1016/J.COPBIO.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordillo Altamirano F.L., Barr J.J. Unlocking the next generation of phage therapy: the key is in the receptors. Curr. Opin. Biotechnol. 2021;68:115–123. doi: 10.1016/J.COPBIO.2020.10.002. [DOI] [PubMed] [Google Scholar]
- 58.Payaslian F., Gradaschi V., Piuri M. Genetic manipulation of phages for therapy using BRED. Curr. Opin. Biotechnol. 2021;68:8–14. doi: 10.1016/J.COPBIO.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 59.Smith G.P. Phage display: simple evolution in a petri dish (Nobel Lecture) Angew. Chem. Int. Ed. Engl. 2019;58:14428–14437. doi: 10.1002/ANIE.201908308. [DOI] [PubMed] [Google Scholar]
- 60.Bernard J.M.L., Francis M.B. Chemical strategies for the covalent modification of filamentous phage. Front. Microbiol. 2014;5:734. doi: 10.3389/FMICB.2014.00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang S.H., Chung W.J., McFarland S., Lee S.W. Assembly of bacteriophage into functional materials. Chem. Rec. 2013;13:43–59. doi: 10.1002/TCR.201200012. [DOI] [PubMed] [Google Scholar]
- 62.Carpenter B.L., Feese E., Sadeghifar H., Argyropoulos D.S., Ghiladi R.A. Porphyrin-cellulose nanocrystals: a photobactericidal material that exhibits broad spectrum antimicrobial activity. Photochem. Photobiol. 2012;88:527–536. doi: 10.1111/J.1751-1097.2012.01117.X. [DOI] [PubMed] [Google Scholar]
- 63.Nakonieczna J., Wolnikowska K., Ogonowska P., Neubauer D., Bernat A., Kamysz W. Rose Bengal-mediated photoinactivation of multidrug resistant Pseudomonas aeruginosa is enhanced in the presence of antimicrobial peptides. Front. Microbiol. 2018;9:1949. doi: 10.3389/FMICB.2018.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakonieczna J., Wozniak A., Pieranski M., Rapacka-Zdonczyk A., Ogonowska P., Grinholc M. Photoinactivation of ESKAPE pathogens: overview of novel therapeutic strategy. Future Med. Chem. 2019;11:443–461. doi: 10.4155/FMC-2018-0329. [DOI] [PubMed] [Google Scholar]
- 65.Malik Z., Ladan H., Nitzan Y. Photodynamic inactivation of Gram-negative bacteria: Problems and possible solutions. J. Photochem. Photobiol., B. 1992;14:262–266. doi: 10.1016/1011-1344(92)85104-3. [DOI] [PubMed] [Google Scholar]
- 66.Ngo-Duc T.T., Alibay Z., Plank J.M., Cheeney J.E., Haberer E.D. Gold-decorated M13 I-forms and S-forms for targeted photothermal lysis of bacteria. ACS Appl. Mater. Interfaces. 2020;12:126–134. doi: 10.1021/ACSAMI.9B15682. [DOI] [PubMed] [Google Scholar]
- 67.Ray P.C., Khan S.A., Singh A.K., Senapati D., Fan Z. Nanomaterials for targeted detection and photothermal killing of bacteria. Chem. Soc. Rev. 2012;41:3193–3209. doi: 10.1039/C2CS15340H. [DOI] [PubMed] [Google Scholar]
- 68.Costantini P.E., Di Giosia M., Ulfo L., Petrosino A., Saporetti R., Fimognari C., Pompa P.P., Danielli A., Turrini E., Boselli L., Calvaresi M. Spiky gold nanoparticles for the photothermal eradication of colon cancer cells. Nanomaterials. 2021;11:1608. doi: 10.3390/NANO11061608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peng H., Rossetto D., Mansy S.S., Jordan M.C., Roos K.P., Chen I.A. Treatment of wound infections in a mouse model using Zn2+-releasing phage bound to gold nanorods. ACS Nano. 2022;16:4756–4774. doi: 10.1021/acsnano.2c00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koskella B., Meaden S. Understanding bacteriophage specificity in natural microbial communities. Viruses. 2013;5:806–823. doi: 10.3390/V5030806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kömerik N., Wilson M., Poole S. The effect of photodynamic action on two virulence factors of Gram-negative bacteria. Photochem. Photobiol. 2000;72:676–680. doi: 10.1562/0031-8655(2000)072<0676:teopao>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 72.Giuliani F., Martinelli M., Cocchi A., Arbia D., Fantetti L., Roncucci G. In vitro resistance selection studies of RLP068/Cl, a new Zn(II) phthalocyanine suitable for antimicrobial photodynamic therapy. Antimicrob. Agents Chemother. 2010;54:637–642. doi: 10.1128/AAC.00603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rapacka-Zdonczyk A., Wozniak A., Pieranski M., Woziwodzka A., Bielawski K.P., Grinholc M. Development of Staphylococcus aureus tolerance to antimicrobial photodynamic inactivation and antimicrobial blue light upon sub-lethal treatment. Sci. Rep. 2019;9:9423–9518. doi: 10.1038/s41598-019-45962-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tomb R.M., Maclean M., Coia J.E., MacGregor S.J., Anderson J.G. Assessment of the potential for resistance to antimicrobial violet-blue light in Staphylococcus aureus. Antimicrob. Resist. Infect. Control. 2017;6:100–113. doi: 10.1186/s13756-017-0261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nikinmaa S., Podonyi A., Raivio P., Meurman J., Sorsa T., Rantala J., Kankuri E., Tauriainen T., Pätilä T. Daily administered dual-light photodynamic therapy provides a sustained antibacterial effect on Staphylococcus aureus. Antibiotics. 2021;10:1240. doi: 10.3390/ANTIBIOTICS10101240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moore C.M., Pendse D., Emberton M. Photodynamic therapy for prostate cancer-a review of current status and future promise. Nat. Clin. Pract. Urol. 2009;6:18–30. doi: 10.1038/NCPURO1274. [DOI] [PubMed] [Google Scholar]
- 77.Hamblin M.R., Abrahamse H. Factors affecting photodynamic therapy and anti-tumor immune response. Anti Cancer Agents Med. Chem. 2021;21:123–136. doi: 10.2174/1871520620666200318101037. [DOI] [PubMed] [Google Scholar]
- 78.Henderson T.A., Morries L.D. Near-infrared photonic energy penetration: can infrared phototherapy effectively reach the human brain? Neuropsychiatric Dis. Treat. 2015;11:2191–2208. doi: 10.2147/NDT.S78182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vanerio N., Stijnen M., De Mol B.A.J.M., Kock L.M. Biomedical applications of photo- and sono-activated Rose Bengal: a review. Photobiomodul. Photomed. Laser Surg. 2019;37:383–394. doi: 10.1089/PHOTOB.2018.4604. [DOI] [PubMed] [Google Scholar]
- 80.Qian X., Zheng Y., Chen Y. Micro/Nanoparticle-augmented Sonodynamic Therapy (SDT): breaking the depth shallow of photoactivation. Adv. Mater. 2016;28:8097–8129. doi: 10.1002/ADMA.201602012. [DOI] [PubMed] [Google Scholar]
- 81.Costley D., Nesbitt H., Ternan N., Dooley J., Huang Y.Y., Hamblin M.R., McHale A.P., Callan J.F. Sonodynamic inactivation of Gram-positive and Gram-negative bacteria using a Rose Bengal-antimicrobial peptide conjugate. Int. J. Antimicrob. Agents. 2017;49:31–36. doi: 10.1016/J.IJANTIMICAG.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brossard K.A., Campagnari A.A. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect. Immun. 2012;80:228–233. doi: 10.1128/IAI.05913-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garapati C., Boddu S.H., Jacob S., Ranch K.M., Patel C., Babu R.J., Tiwari A.K., Yasin H. Photodynamic therapy: A special emphasis on nanocarrier-mediated delivery of photosensitizers in antimicrobial therapy. Arab. J. Chem. 2023;16 doi: 10.1016/J.ARABJC.2023.104583. [DOI] [Google Scholar]
- 84.Polat E., Kang K. Natural photosensitizers in antimicrobial photodynamic therapy. Biomedicines. 2021;9:584. doi: 10.3390/BIOMEDICINES9060584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abrahamse H., Hamblin M.R. New photosensitizers for photodynamic therapy. Biochem. J. 2016;473:347–364. doi: 10.1042/BJ20150942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andris-Widhopf J., Rader C., Steinberger P., Fuller R., Barbas C.F. Methods for the generation of chicken monoclonal antibody fragments by phage display. J. Immunol. Methods. 2000;242:159–181. doi: 10.1016/S0022-1759(00)00221-0. [DOI] [PubMed] [Google Scholar]
- 87.Passaretti P., Khan I., Dafforn T.R., Goldberg Oppenheimer P. Improvements in the production of purified M13 bacteriophage bio-nanoparticle. Sci. Rep. 2020;10:18538–18539. doi: 10.1038/s41598-020-75205-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sampaolese B., Bergia A., Scipioni A., Zuccheri G., Savino M., Samorì B., De Santis P. Recognition of the DNA sequence by an inorganic crystal surface. Proc. Natl. Acad. Sci. USA. 2002;99:13566–13570. doi: 10.1073/PNAS.202471699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zuccheri G., Scipioni A., Cavaliere V., Gargiulo G., De Santis P., Samorì B. Mapping the intrinsic curvature and flexibility along the DNA chain. Proc. Natl. Acad. Sci. USA. 2001;98:3074–3079. doi: 10.1073/PNAS.051631198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Redmond R.W., Gamlin J.N. A Compilation of Singlet Oxygen Yields from Biologically Relevant Molecules. Photochem. Photobiol. 1999;70:391–475. doi: 10.1111/J.1751-1097.1999.TB08240.X. [DOI] [PubMed] [Google Scholar]
- 91.Bochner B.R., Gadzinski P., Panomitros E. Phenotype Microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
-
•
Sequences of plasmids and phagemids generated in this study have been deposited to GenBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All the other data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.