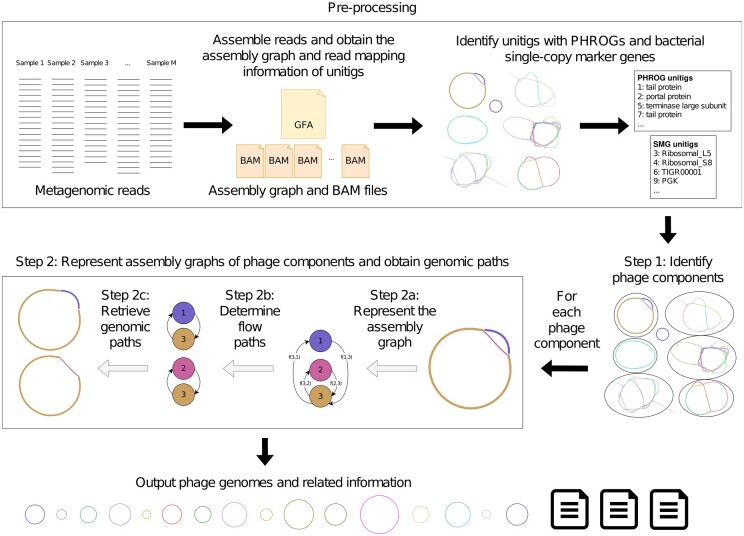
Figure 1.
Phables workflow. Preprocessing: assemble reads, obtain the assembly graph and read mapping information, and identify unitigs with PHROGs and bacterial single-copy marker genes. Step 1: Identify phage components from the initial assembly. Step 2: For each phage component, represent the assembly graph, determine the flow paths and retrieve the genomic paths. Finally, output phage genomes and related information.