Abstract
Microalgae as the photosynthetic organisms offer enormous promise in a variety of industries, such as the generation of high-value byproducts, biofuels, pharmaceuticals, environmental remediation, and others. With the rapid advancement of gene editing technology, CRISPR/Cas system has evolved into an effective tool that revolutionised the genetic engineering of microalgae due to its robustness, high target specificity, and programmability. However, due to the lack of robust delivery system, the efficacy of gene editing is significantly impaired, limiting its application in microalgae. Nanomaterials have become a potential delivery platform for CRISPR/Cas systems due to their advantages of precise targeting, high stability, safety, and improved immune system. Notably, algal-mediated nanoparticles (AMNPs), especially the microalgae-derived nanoparticles, are appealing as a sustainable delivery platform because of their biocompatibility and low toxicity in a homologous relationship. In addition, living microalgae demonstrated effective and regulated distribution into specified areas as the biohybrid microrobots. This review extensively summarised the uses of CRISPR/Cas systems in microalgae and the recent developments of nanoparticle-based CRISPR/Cas delivery systems. A systematic description of the properties and uses of AMNPs, microalgae-derived nanoparticles, and microalgae microrobots has also been discussed. Finally, this review highlights the challenges and future research directions for the development of gene-edited microalgae.
Graphical Abstract
Keywords: Algal-mediated nanoparticles, CRISPR/Cas system, Delivery system, Microalgae application, Gene editing
Introduction
Following the urgent demand for energy and food, it is critical to discover an alternative source to alleviate these issues. Due to simple cultivation conditions, rapid growth rate and high photosynthetic efficiency, microalgae have emerged as the promising renewable energy resource. They also have a significant potential to construct a productive biorefinery which is the process of converting carbon dioxide into value-added compounds, for example proteins, vitamins, fatty acids, carotenoids, and nucleic acids. Microalgae are recognised as the third generation of biofuels with the potential for high-density cultivation system designs without compete with food or agricultural crops [1]. Additionally, microalgae are now capable of producing wide range of fuels, including biodiesel [2], hydrogen [3], syngas [4], etc. They may also serve as a raw material for functional food, natural dyes, and pharmaceutical drugs [5]. Studies on microalgae have primarily concentrated on developing integrative process and culture techniques, such as photobioreactor designs [6], harvesting approaches in downstream processes [7] and extraction techniques for high-value compounds [8]. However, this field still has some drawbacks, such as the low carbon fixation efficiency, low lipid accumulation rate and long cultivation period, which need to be overcome with more robust technology.
Many microalgal genomes have been sequenced to date, providing a definite genetic background and a compelling argument for genetic modification. Zinc finger nuclease (ZFN) [9], transcription activator-like effector-based nuclease (TALEN) [10] and the clustered regularly interspaced short palindromic repeats-associated system (CRISPR/Cas system) are examples of gene editing tools that offer an useful approach to deal with microalgal issues and achieve mass production. ZFN and TALEN are now severely constrained due to low editing efficiency, costly and laborious vector construction. The CRISPR/Cas system, in contrast, has become a reliable genome editing platform for gene correction, transcriptional regulation, disease modelling, and nucleic acids imaging. Its advantages include simplicity of target design, high editing efficiency, multiplex knock-in/out ability, low cost, and a quick cycle time [11]. Since then, CRISPR/Cas systems have been used as the treatment of infectious and metabolic diseases, creating sustainable techniques to produce chemicals and fuel as well as enhance the features of food crops. The disadvantages of CRISPR/Cas systems are off-target effects [12], variable efficiency, and inactive mutant [13, 14], despite the implementation of some strategies, for example rational design and modification of sgRNA, application of Cas variants, and improvement of the repair efficiency of HDR pathway.
An ideal delivery vector is essential for the efficient intracellular distribution of CRISPR components. Due to their superior biocompatibility in a homologous relationship, algal-mediated nanoparticles (AMNPs) have a tremendous potential to provide the CRISPR/Cas system for microalgal genetic editing. Furthermore, AMNPs provide a variety of properties, including anti-bacterial, anti-fungal, anti-cancer, anti-fouling, bioremediation, and biosensing activities [15]. This, green synthesis of AMNPs has drawn a lot of interest because it is safe, simple, sustainable, cost-effective, and eco-friendly. This review provides a thorough summary of the CRISPR/Cas system's delivery status using nanoparticles (NPs), with a focus on providing an in-depth description of AMNPs from the perspectives of their synthesis, features, classifications, and numerous applications. Some strategies, like as the use of a different Cas nuclease, better codon harmonisation, the addition of introns, and proper removal of the cell wall, have been suggested to tackle the limitations of CRISPR/Cas system in microalgae [16]. This review also examines the prospects of the AMNPs-delivered CRISPR/Cas system in microalgae, which offers a clear and crucial roadmap for advancing the use of gene editing in microalgae.
Research status of microalgae gene editing
Although ZFN and TALEN have been applied in diatoms Phaeodactylum tricornutum and Chlamydomonas reinhardtii [17, 18], both technologies have not been widely used due to the limitations of laborious design steps, low editing efficiency and high off-target events. In comparison to ZFN and TALEN, the CRISPR/Cas system is regarded as a more sophisticated editing tool with several benefits, including the ease of design, increased effectiveness, and capacity to introduce mutations in many genes simultaneously [19]. In the past ten years, the CRISPR/Cas system has evolved from a groundbreaking genome-editing tool used in bacteria to a significant gene-editing tool utilised in plants, animals, and humans [20]. As a result, CRISPR technologies have advanced innovation by changing numerous eukaryotic genomes [21]. In the past, the CRISPR/Cas editing technology has been used to modify a number of microalgae species, including Nannochloropsis oceanica [22], P. tricornutum [23], Thalassiosira pseudonana [24], Chlorella vulgaris [25], and other microalgae species [26–40]. The editing process, transformation process, editing effectiveness, and experimental outcomes have all been explained in depth, as seen in Table 1 [41].
Table 1.
Application status of CRISPR/Cas system in microalgae
| Microalgae species | Microalgae subtypes | Editing method | Transformation method | Editing efficiency | Experimental results | References |
|---|---|---|---|---|---|---|
| C. Reinhardtii | C. reinhardtii CC-503 | One plasmid-driven CRISPR/Cas9 system (C. reinhardtii codon-optimized Cas9) | Electroporation | 46.7% | First successful transient expression of Cas9 and sgRNA in C. reinhardtii | [23] |
| C. reinhardtii CC-124 | CRISPR/Cas9 RNPs | Electroporation | 0.17–40% | Mutagenesis of MAA7, CpSRP43 and ChlM gene | [24] | |
| / | CRISPR/Cas9 RNPs | Electroporation | 0.56% | CpFTSY and ZEP gene knockout, increases photosynthetic productivity | [25] | |
| C. reinhardtii CC-400 | Two plasmid-driven CRISPRi/dCas9-KRAB system | Glass beads | 94% | Downregulating the expression level of CrPEPC1 gene to increase lipid synthesis | [26] | |
| C. reinhardtii CC-4349 | CRISPR/Cas9 RNPs | Natural transformation | / | Knock-out the zeaxanthin epoxidase gene to stop the formation of lutein | [27] | |
| / | CRISPR/Cas12a RNPs | Electroporation | / | Exploring the mechanisms of single-strand templated DNA repair at CRISPR/Cas12a-induced DSBs in C. reinhardtii | [28] | |
| C. Reinhardtii CC-4349, CC-124, and CC-503 | CRISPR/Cas9 RNPs | Electroporation | 30% | Knock-in antibiotics resistance gene and YFP | [29] | |
| Synechococcus elongatus | Synechococcus elongatus PCC 7942 | One plasmid-driven CRISPRi/dCas9 system | Natural transformation | 99% | The glgc gene was downregulated to increase succinate titer level | [30] |
| Synechococcus elongatus UTEX 2973 | Two plasmid-driven CRISPR/Cas9 system | Natural transformation | 30–100% | Knock-out the nblA gene | [31] | |
| Synechococcus sp. | Synechococcus sp. PCC 7002 | Genomic integration CRISPRi/dCas9 system | Natural transformation | 30–90% | Increasing central carbon flux by reducing carboxysome expression level | [32] |
| Synechococcus sp. PCC7942 | One plasmid-driven CRISPR/Cas9 system | Natural transformation | 23–57% | Improvement of succinate synthesis using glgc gene knock-out and gltA/ppc gene knock-in | [33] | |
| Synechocystis sp. PCC 6803 | One plasmid-driven CRISPRi/dCas9 system | Natural transformation | 50–95% | Prevent the synthesis of carbon storage compounds | [34] | |
| N. oceanica | N. oceanica CCMP1779 | One plasmid-driven CRISPR/Cas9 system | Electroporation | 45–90% | Generating non-transgenic marker-free nitrate reductase knock-out lines | [35] |
| N. oceanica IMET1 | CRISPR/Cas9 RNPs | Electroporation | 93% | FnCas12a as the best performer for genome editing in Nannochloropsis oceanica IMET1 | [36] | |
| Chlorella | / | One plasmid-driven CRISPR/Cas9 system | Electroporation | 67% | Enhancing lipid accumulation | [37] |
| C. vulgaris UTEX395 | CRISPR/Cas9 RNPs | Electroporation | / | Successful genome editing in C. vulgaris UTEX395 with CRISPR/Cas9 system | [38] | |
| T. pseudonana | / | One plasmid-driven CRISPR/Cas9 system | Biolistic bombardment | 61.5% | Two sgRNAs are used to induce a precise deletion in the urease gene of T. pseudonana | [39] |
| P. tricornutum | / | One plasmid-driven CRISPR/Cas9 system (C. reinhardtii codon-optimized Cas9) | Biolistic bombardment | 25–63% | Mutagenesis of the CpSRP54 gene to increase the sensitivity to high intensity light | [40] |
The problem of ineffective intracellular delivery with gene editing in microalgae persists despite varied success rates. An ideal transformation strategy and delivery vector are required to transport foreign DNA into cells. Among the transformation methods, such as electroporation [42], glass beads [43], particle bombardment [44], and Agrobacterium tumefaciens-mediated transformation [45], electroporation is the preferred method for microalgae transformation because it is rapid and highly effective in the generation of transformants (approximately 0.4 ~ 3 × 103 transformants per µg of exogenous DNA) [42, 46]. There are few delivery carriers in CRISPR systems, such as viral vectors [47], extracellular vehicles [48], cell-penetrating peptides [49], and etc. AMNPs have a greater potential to deliver CRISPR/Cas components in microalgae gene editing due to better biocompatibility in a homologous relationship [50] than other delivery carriers in CRISPR systems. Their detailed delivery in microalgae have been described systematically as follows.
NP-based delivery system for CRISPR/Cas components
Characteristics of different delivery systems
Given the existing challenges of the CRISPR/Cas system, including as off-target effects [51], poor delivery efficiency [52], and unintended adverse effects [53], various techniques have been proposed to overcome these concerns [14, 51, 54]. Choosing the best delivery system has become the most important step, with strict requirements to boost not just loading efficiency, but also correct delivery to the specified location [55].
So far, several delivery mechanisms have sprung up, resulting in an exponential expansion in the distribution of CRISPR/Cas components (Table 2). Delivery mechanisms consists of three types which are viral vectors, non-viral vectors, and physical and chemical approaches [48, 56–60]. Viral vectors including adenovirus, adeno-associated virus and lentivirus [61–63], and physical methods like electroporation [64], microinjection [65], hydrodynamic injection [66] and ultrasound [67], are relatively well-established strategies for delivering CRISPR/Cas systems [68]. Using these two techniques, the delivery efficiency of CRISPR/Cas components was enhanced to around 98% [60, 69–71], signifying a substantial achievement in the field of gene editing. Viral vectors have become the most evident method for delivering genes owing to excellent transfer efficiency and stable gene expression [72, 73].
Table 2.
Summary of common delivery strategies for CRISPR/Cas9 systems
| Delivery systems | CRISPR/Cas9 formats | Delivery efficiency | Advantages | Disadvantages | Applications |
|---|---|---|---|---|---|
| Viral delivery | |||||
| AAV | pDNA | + + | Low immunogenicity, non-pathogenic source, broad cell tropism | Limited packaging capacity, prolonged Cas9 expression, pre-existing neutralizing antibodies, high cost | In vivo |
| LV | pDNA | + + + | Large packaging capacity, low immunogenicity, broad cell tropism | Potential for insertional mutagenesis, prolonged Cas9 expression | In vitro and ex vivo |
| AV | pDNA | + + | Large packaging capacity, minimal genomic integration risk | High immunogenicity, difficult for large-scale production, pre-existing neutralizing antibodies | In vivo |
| Physical delivery | |||||
| Electroporation | pDNA, mRNA and RNP | + + + | Suitable for multiple cell types, simple operation | Severe cell damage, nonspecific transfection | In vitro and ex vivo |
| Microinjection | pDNA, mRNA and RNP | + + + | Highly specific and reproducible | Severe cell damage, low throughput, requirement of sophistication and manual skills | In vitro and ex vivo |
| Hydrodynamic injection | pDNA and RNP | + | Feasible for in vivo gene editing in small animals, highly efficient for liver transduction | Not suitable for large animals and clinical applications, non-specific and low efficiency, traumatic to tissues | In vivo |
| Non-viral delivery | |||||
| Lipid NPs | pDNA, mRNA and RNP | + | Good biocompatibility, minimal immunogenicity, feasible for large-scale production, temporal release of CRISPR-Cas9, all-in-one delivery, low toxicity | Lower delivery efficiency | In vitro and in vivo |
| Polymeric NPs | pDNA, mRNA and RNP | + | Minimal immunogenicity, feasible for large-scale production, relatively flexible functionalization, good pharmacokinetic control, large packaging capacity, all-in-one delivery | Lower delivery efficiency, variable biocompatibility and toxicity | In vitro and in vivo |
| Gold nanomaterials | pDNA and RNP | + + | Higher delivery efficiency, good biocompatibility, unique optical properties and photothermal effect, relatively flexible functionalization | Potential toxicity in vivo at high concentrations | In vitro and in vivo |
| EVs | RNP | + | Excellent biocompatibility and negligible immunogenicity, high biostability relatively flexible functionalization, low toxicity | Tedious preparation process | In vitro and in vivo |
+ denotes low; + + denotes medium; + + + denotes high
Despite ongoing improvements in viral vectors [74], their applications in the delivery of CRISPR/Cas system are still hindered by the issues with immunogenicity [75], mutagenesis [76], limited packaging capacity as well as scale-up manufacturing [77, 78]. Combinatorial distribution of multiple components is a critical challenge that may prevent widespread and flexible implementation in the future [79]. Despite its great effectiveness in the laboratory, physical delivery was less practicable for in vivo distribution due to its low scalability [80, 81]. They are more suitable for in vitro applications than clinical translations because they can be conducted at the cellular level rather than the organism level [82]. Further, the implementation of physical delivery in non-specialist laboratories and high-throughput applications is difficult because to the expensive equipment and requirement of high skilled manipulation. Improper and unoptimized physical techniques may cause inevitable cell damage or even cell death [83, 84]. Alternately, it is desirable for the CRISPR/Cas system to deliver the product by changing it directly rather than causing alterations in target cells [80]. In view of this, chemical carriers provide a novel perspective due to their low immunogenicity, adjustable cargo size, and mass-production feasibility [85]. More importantly, the ease of alteration of chemical carriers may answer future issues in terms of targeting, biosafety, loading capacity, and spatiotemporal controllability, which are desirable options for in vivo precise gene editing delivery [86, 87]. Among the many chemical vectors, nanomaterial is an efficient platform to deliver small molecule medications, genes, peptides, and diagnostic agents [88, 89]. CRISPR/Cas components could be supplied via designed NPs with advantages such as precise targeting [90], high stability [91], safety [68], enhanced immune escape [92], and combinatorial delivery of several components [60].
NPs delivery mechanism
The journey of NPs in vivo is difficult and accompanied with numerous obstacles, including disturbance of ‘protein corona’ [93], clearance by reticuloendothelial system, as well as the barriers of vasculature, extracellular matrix and endosome. In particular, the intravenous injection of NPs would face all of the difficulties of beginning from scratch, with the most challenging obstacle being the efficient access to organelles at the subcellular level [94]. The most important process for almost all successful NP-based carriers is endocytosis, which is mediated by target cells [95]. By controlling and mediating numerous signal pathways, NPs-based carriers were engulfed into endocytic vesicles through the invasion of plasma membrane [96–98].
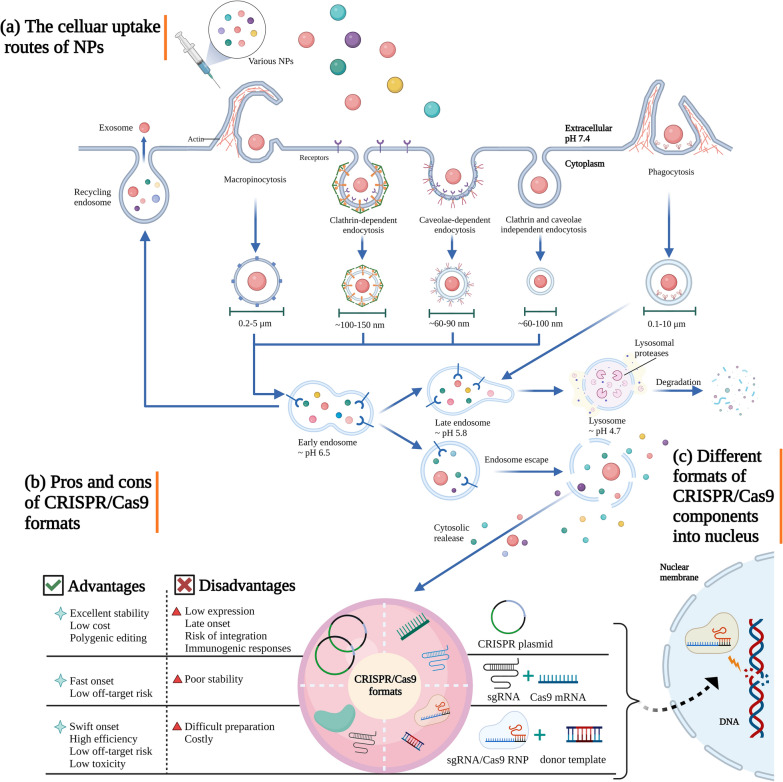
Theoretically, these pathways are subdivided into five categories based on the specific lipid and transport protein types they contain, such as surface receptors, membrane lipids, and adaptor proteins. Phagocytosis, caveolin-dependent endocytosis, clathrin- and caveolin-independent endocytosis, and macropinocytosis are all types of endocytosis [99, 100] (Fig. 1a). The fate of NPs within the cells can be determined by many types of endocytosis, which depends on the factors including cell type, size, charge, and stiffness of NPs, as well as receptor interaction [101]. Among these mechanisms, the majority of receptor-bound NPs could be translocated into the cells by clathrin-mediated and caveolin-mediated endocytosis, while non-targeting NPs in small or large size will be taken up non-specifically by macropinocytosis or phagocytosis [102]. However, a certain lipid composition (mostly cholesterol) was required for clathrin- and caveolin-independent endocytosis, which was thought to be a direct entrance route [101]. However, the uptake mechanism of each item is distinct and has not yet been thoroughly investigated. When NPs become entrapped in an endosome during endocytosis, they are unable to perform their intended biological and therapeutic roles due to a lack of quick access to cytoplasm or cellular organelles. Endosomes were frequently acidified during ageing processes, with pH variations ranging from pH 6.5–6.8 to pH 5.2–6.0. Eventually, the acidic pH and enzymatic breakdown caused the cargoes to be destroyed [103]. As a result, the inability to exert biological effects is currently a key hindrance to nanomedicine efforts, necessitating the urgent development of endosome escape to increase the effectiveness of NPs delivery.
Fig. 1.
Delivery mechanism of NPs into cells. a The cellular uptake routes of NPs. b Pros and cons of CRISPR/Cas9 formats. c Different formats of CRISPR/Cas9 components into nucleus
When NPs are subjected to the harsh environment of lysosome, endosome escape is a critical step in retaining the integrity of their cargoes and exerting their effects. According to the data, the greatest distribution effectiveness of NPs made of polymers and lipids was just 1–2%, and that the remaining 98% of NPs were useless because they were trapped in endosomes [104] or recycled back to the extracellular space [105]. It is frequently regarded as the rate-limiting step in the intracellular delivery of NPs-based systems [106, 107]. Therefore, an endosome escape technique could be used to transfer CRISPR/Cas components effectively without degrading them. However, various contentious ideas have been offered as endosome escape routes, such as proton sponge effect [108], lipid fusion with endosomal membrane [109, 110], nanoparticle swelling [111], membrane destabilisation [112], and cationic lipids induced hexagonal HII conformation [113]. These mechanisms can be categorised into three types: membrane disruption [114], membrane fusion [115], and surface modification [116]. However, it is well known that all mechanisms with endosomal pH variation will be a strong indicator of endosomal escape, which is advantageous for the development of high-efficiency gene editing. Thus, investigating pH-responsive nanomaterials, such as NPs expansion [111, 117], polymer depolymerization [118, 119], and pH buffering materials [120, 121], could be a future study topic. In addition to endocytosis, NPs can be directly internalized into cytoplasm by passive membrane fusion or pore formation, which bypasses the endosome engulfment [110, 122]. However, the understanding of the trafficking of NPs is still evolving, thus additional efforts need to be done to confirm the precise mechanism. Developing novel methods and tools to dynamically observe membrane perturbation events and real-time cargo releases through visualisation of intracellular trafficking will be crucial for emerging biomedical applications to gain an in-depth understanding of cellular uptake process. Aside from the direct progressive approaches, much effort has been invested on the use of endocytosis inhibitors to promote and encourage advancement in the field.
Selection of CRISPR/Cas formats
The ideal CRISPR/Cas format is essential for meeting the requirements for various applications, including plasmid DNA (pDNA), mRNA, and Cas9/sgRNA ribonucleoproteins (RNPs), to achieve high-efficiency gene editing [123]. When viewed in the context of NPs formulations and clinical or research application, each format offers unique characteristics [124, 125] (Fig. 1b). Due to its outstanding stability, affordable, and ease of preparation, CRISPR/Cas pDNA has become the most popular format. Additionally, it is practical to perform polygenic editing simultaneously at several sites using different sgRNA designs. However, the drawbacks of pDNA limited the applicability of the editing system. These drawbacks included the relatively low expression, delayed responsiveness, induction of immunogenic responses, and difficulty of encapsulating large molecules. Even worse, because Cas9 proteins are expressed for a longer period of time, on-target effects would be compromised and there would be a higher chance of uncontrolled plasmid sequence integration [126–129]. The delivery of mRNA or RNP complex demonstrated a rapid onset of gene editing in comparison to the pDNA format, bypassing the restrictions of pDNA and achieving functional complementation. The mRNA format can prevent undesirable insertional mutagenesis and makes it simpler to generate transient expression in the cytoplasm, which lowers the off-target effects [130–132]. However, Cas9 mRNA distribution is difficult because of the low stability which results from its fragile single-stranded structure and is susceptible to in vivo RNAse degradation [133]. Thus, attempts should be taken to improve its stability using different vectors or chemical modifications.
RNP complexes are the simplest delivery format, and they have the least amount of intracellular processing that can evade the transcription and translation process (Fig. 1c). To accomplish this, they equip components with a rapid onset and short duration, which results in substantially fewer potential side effects and cellular toxicity as well as high gene editing efficiency [134, 135]. Nevertheless, as a specific obstacle of RNP, exogenous risk and big molecule size would cause immunogenicity and difficulty in efficient nuclear entry, limiting the broad uses of the CRISPR/Cas system [136]. Additionally, the preparation of Cas9 proteins is time-consuming, complicated, and costly [137]. The compatibility between the cargo and the carrier is also important to consider, along with the reasonable choice of cargoes. Designing NPs should not compromise other qualities, such as loaded-cargo protection, targeting, and effective transfection, in order to accommodate a specific CRISPR/Cas system format [81]. In conclusion, every delivery metod hin a variety of formats has disadvantages of its own. Depending on the compatibility of the cargo and the carrier, as well as the requirements for diverse purposes, selecting the best CRISPR/Cas format is therefore essential for effective editing.
NPs-based CRISPR/Cas delivery systems
The development of delivery carriers is currently trailing behind with regard to the distribution of CRISPR/Cas systems. Hence, there is an urgent need for sophisticated delivery systems with high efficiency, targeting, controllability, and safety characteristics. Synthetic nano-delivery systems, for instance lipid NPs (LNPs) [97], polymeric NPs (PNPs) [138], and gold NPs (AuNPs) [139], showed significant potential in enhancing the editing efficacy for CRISPR/Cas systems. These systems have the advantages of transient expression patterns, feasibility for mass-production at lower cost, all-in-one delivery, and low immunogenicity risk. These systems are also more flexible to deliver various cargoes for different purposes, encompassing RNPs, pDNA, mRNA, and donor DNA [140, 141]. In conclusion, nanotechnology holds the promise of overcoming the drawbacks of traditional delivery systems through cell-specific targeting, precise molecular transport to certain organelles, and other innovative strategies.
Lipid NPs
LNPs have been extensively investigated as the cutting-edge delivery platform for the drugs [142], CRISPR/Cas components [143], and vaccines [144]. They normally consist of four main components, i.e., key cationic or ionizable lipids complexed with negatively charged genetic material, phospholipids for particle structure, cholesterol for stability and membrane fusion, and PEGylated lipids to increase stability and circulation [145, 146]. The structure and formulation of LNPs protect the cargoes from enzymatic degradation and immunological responses, which subsequently facilitated their transportation into host cells for genome editing [147]. Ionizable lipids eventually replace permanently charged lipids due to their greater transfection efficiency and reduced cytotoxicity. Uncharged ionizable lipids are likely to adhere to the cell surface when the pH is neutral due to hydrophobic interactions or receptor-mediated endocytosis [148]. Ionizable lipids change to cationic form at lower pH to enable endosome escape and promote cargo release [113]. LNPs also have various advantages such as large cargo packaging capacity, good biocompatibility and bioavailability, low cell toxicity and immunogenicity, and mature industrial manufacturing technology. However, due to low drug loading and non-targeted biodistribution, it is challenging to achieve safe, efficient, and targeted administration of CRISPR/Cas components in vivo using LNPs [149]. The rational design or modulation of LNP formulations with the aim of obtaining optimum safety profile and efficient nucleic acid delivery would be a feasible technique to achieve the desired effects mentioned above [150, 151].
Developing new LNPs composition, such as NTLA-2001 LNPs [152] and 306-O12B-LNPs [153], can achieve targeting delivery of the CRISPR/Cas system with excellent editing efficiency and safety. In the case of treating transthyretin amyloidosis, the use of NTLA-2001 LNPs, an intravenous formulation made using a patented ionizable lipid, has demonstrated liver-targeted delivery. These LNPs display a consistent editing effect that is dosage dependent, safe, and long lasting, with maximum suppression seen after 12 months. The editing efficacy of transthyretin reached up to 93.7%, resulting in 87% decrease in protein concentration. In comparison to the gold standard LNPs (MC-3 LNPs, 14.6%), 306-O12B LNPs had a more effective liver-targeted delivery efficacy of around 38.5% [154], which offered the possibility of liver-specific administration in therapeutic applications. The high liver-targeted delivery efficiency could be attributed to the strong liver tropism caused by the active-targeting mechanism mediated by apolipoprotein E (ApoE) [113, 155]. Plasma ApoE attaches to the surface of LNPs and then interacts with the low-density lipoprotein receptor to achieve active endocytosis via ligand-receptor transport. Given this, modifying LNPs with appropriate ligands or targeting moieties may be an effective method for delivering drugs to specific organs [139, 141]. Similar to this, adding functional components to LNPs can reduce the risks associated with immunogenicity. TCL053-LNPs might preferentially transfer Cas9 mRNA and sgRNA into skeletal muscle due to the pH-dependent ionizable lipid TCL053. This approach could treat skeletal muscle disorders by giving repeated intramuscular injections because of its low immunogenicity. Nevertheless, each TCL053-LNPs injection could result in a long-lasting restoration of the dystrophin protein for at least a year [156].
Additionally, altering the proportion of LNPs formulation can achieve targeted and secure delivery for the CRISPR/Cas system. Multiple CRISPR cargoes could be selectively delivered to the targeted organs using a new technique called selective organ targeting (SORT), which involves changing the biodistribution of SORT LNPs [157]. The delivery to hepatocytes attained the highest specific transfection rate of 93% by including a supplementary component to the initial LNPs formulation. By constantly controlling the SORT molecule proportion, this system offered a reliable and designable platform for extrahepatic delivery, such as lung and spleen, which is also applicable to different NPs systems for targeted delivery. The proportion of various lipids in LNPs was then optimised using SORT and MC-3 LNP nanotechnology to create a lung-targeted LNPs-mediated CRISPR/Cas13d delivery system [158]. The data showed that, when compared to control groups, this system significantly decreased mRNA and protein levels of cathepsin L only in mouse lungs and reduced lung virus infectivity by two orders of magnitude, indicating an excellent strategy with exceptional efficacy, specificity, and safety. Since it is impossible to completely eliminate off-target events, efforts must be taken to address safety issues with the CRISPR/Cas system in order to build a perfect genome editing tool [60]. These efforts include sgRNA preoptimization, transient expression of the Cas9 protein, and targeted delivery to the intended tissue.
Polymeric NPs
Using multivalent charge interactions, PNPs condensed charged genome editing cargoes into nano-sized packages, shielding them from deactivation and promoting intracellular transit for genome editing [159, 160]. Different PNPs such as polyethylenimine (PEI), poly(lactic-co-glycolic-acid) (PLGA), poly(β-amino ester) (PBAE), poly(ethylene–glycol) (PEG), and amine-terminated polyamidoamine dendrimers have become very popular to deliver CRISPR/Cas components due to their excellent pharmacokinetic control, high cargo encapsulation, minimal immunogenicity, relatively flexible functionalization, and high bioavailability [78, 161]. More significantly, PNPs were able to accurately adjust the loading effectiveness and release kinetics to perform the editing for various objectives by varying their composition, stability, responsiveness, and surface charge characteristics [162]. Increasing the effectiveness of gene editing has increasingly become the significant route for the CRISPR/Cas system. PEI-based formulations, including branching PEI 25 kDa, are frequently utilised as the carriers for efficient delivery of CRISPR/Cas plasmids into the appropriate target cells when using PNP alone. In this technique, guide RNA and Cas9 are both expressed simultaneously, producing indel efficiencies (24.4%) that are comparable to Lipofectamine 2000 (27.9%) [163]. In contrast, PLGA-NPs offered higher 38.4% indel efficacy and 70% PLGA encapsulation efficacy, making them a more effective and secure delivery method for CRISPR components. Copolymer strategy by combination of multiple PNPs could be used as an effective method for CRISPR/Cas system. For instance, PEG-b-PLGA based copolymer with a cationic lipid-assisted NPs achieved direct modulating to immune cells such as neutrophils [164], macrophages [165], and DCs [166], which induced editing efficiency about 25.5–32.7% in vitro. Based on the PEG-b-PLGA combination, PEI-coated PEG-b-PLGA produced 80% selective genome editing in endothelial cells with a 40–50% efficiency [167]. It is evident that the distribution technique of copolymer had a more pronounced editing efficacy than employing PNPs alone. Notably, the helper components, such as cationic lipids or PEI coatings, are crucial to the delivery system that encourages cellular absorption and permits endosomal escape [168]. In-depth research into their interaction in copolymers may be a future direction for PNPs to explore since our understanding of high-efficiency copolymer delivery strategies is still evolving.
Aside from improving editing effectiveness, PNPs may easily implement their multifunctionality because they have effective cargo encapsulation, good modification potential, and interesting stimuli-responsiveness. Due of the exceptional ability of cargo encapsulation, co-delivery strategies that combine medicines and CRISPR components into a single polymer carrier have a beneficial effect in a variety of applications [169, 170]. For instance, the dendrimer-based lipid NPs might co-encapsulate and deliver CRISPR components, including Cas9 mRNA, sgRNA, and donor DNA, as an all-in-one nanocarrier [171]. The findings demonstrated that more than 91% of all cells were altered in vitro with 56% HDR efficiency. In addition, PNPs were frequently used to modify existing delivery vehicles in order to provide them exceptional biocompatibility, stimuli-responsiveness, loading capacity, and other properties. As an illustration, bifunctionalized aminoguanidine-PEGylated periodic mesoporous organosilica NPs successfully delivered RNPs with 40% effectiveness rate for gene editing. In this study, PEG coatings provided the carriers with protection against opsonization, aggregation, and phagocytosis as well as many biofunctions, including exceptional storage stability, permeability, and long-lasting blood circulation of nanocarriers [172, 173]. Numerous stimuli-responsive PNPs delivery systems have been created to extend therapeutic efficacy at a lower dose frequency. These systems can remotely initiate the release of bioactive molecules in response to internal and external stimulation, such as second near-infrared light (NIR II) and pH change [174, 175]. CRISPR/Cas components can be remotely released by NIR-light trigger depending on the specified semiconducting polymer brush. Controllable release of delivery system might be assisted by photothermal conversion, which increased their editing efficiency (around 35%) [176]. Additionally, pH-responsive PNPs provide a novel method of delivering Cas9 RNPs and donor DNA to selected organs. Local administration at various sites can simultaneously provide targeted distribution to desired organs such as intravenous, intratracheal, and intramuscular delivery to the liver, lung, and skeletal muscle in mice, respectively [177]. As a result, the administration method may offer a viable strategy for overcoming the barrier of limited biodistribution at intended areas. Designing delivery systems with a low amount of positive charge or shielding the positive charge reversibly would be a promising avenue to develop a secure and effective platform in the future because most cationic polymeric materials may cause high cytotoxicity.
Gold NPs
Inorganic gold nanocarriers have been utilised extensively to deliver imaging agents, nucleic acids, and proteins because of their distinctive physicochemical qualities that are favourable size, minimal toxicity, optical properties, superior biocompatibility, and photothermal action [178, 179]. Among these, the most unique quality that triggers cargo release to control the expression and activity of Cas9 proteins is the photothermal effect provided by AuNPs core [180–182]. Converted heat promoted an endonuclear change of heat-shock factor (HSF) from inactive monomer to active trimer through external NIR laser irradiation. The combination transfection of AuNPs and Cas9 plasmid under the influence of active HSF could achieve 90% GFP-positive cells, which was significantly greater than the result of lipofectamine 2000 [183]. Similarly, after hybridising protective DNA-modified gold nanorod (GNR) with sgRNA, heat damaged the hybrids and released sgRNA into cells under regulated NIR laser irradiation [184]. Furthermore, the unique photothermal activity of AuNPs makes them an appealing candidate for disease treatment, owing to their synergistic photothermal/gene therapy effects. By way of illustration, the mCas9-sGNR nanocarrier not only used to transport CRISPR components needed for gene therapy, but it also lowered the tolerance of cancer cells to heat through photothermal therapy, which led to superadditive synergistic anticancer effects [185]. The development of AuNPs-based CRISPR-Cas12a/Cas13a systems as visual biosensors for large-scale population screening and nucleic acid bioimaging, including smartphone-based diagnostics [186–188]. Compared to earlier detection technology, these devices offer quick, ultrasensitive, specific, and on-site biosensing methods. AuNPs have also been thoroughly studied for tumour visualisation, cancer detection, and bioimaging due to their remarkable photoluminescent characteristics, with the ultimate goal of obtaining precision targeted therapy [189].
Microalgae-based delivery systems
AMNPs features
Nanotechnology has grown exponentially as an interdisciplinary field due to its exceptional size and properties. NPs proved to be an excellent material than other counterparts and may serve as the main target in pharmacology, biosensors, and medicine [190, 191]. Synthetic metallic NPs, which exhibit a range of unprecedented physical, chemical, and optoelectronic capabilities, have become effective tools in various sectors over the past ten years. The synthetic procedure is the most important component in evaluating their applicability. Although various cutting-edge techniques were used to boost performance of NPs, such as reliability and practicality, the consensus view is that traditional synthesis of NPs will have a significant negative influence on human well-being because their synthesis was not eco-friendly. Several hazardous reagents, including reducing agents, organic solvents, and stabilisers, are introduced during the synthesis of NPs. These reagents led to significant toxicity issues as well as undesirable byproducts. These synthetic techniques nevertheless have several drawbacks, including high cost, poor efficacy, and requirement of skilled manipulation. The tendency to use sustainable, affordable, and environmental friendly NPs, such as silver, gold, copper, and other metals, would be further increased as a result of the expanding momentum of green chemistry [192]. With regard to biosynthetic mediation, there are the remarkable capabilities to consume, accumulate, and eventually remodel metal ions into NPs, which reduce the toxicity of metal ions and minimise adverse environmental effects through the resistance mechanism by organisms [193]. Biosynthesized NPs have previously been used in antimicrobials, pathogen detection or associated protein identification, and cargo delivery [194].
Bacteria, algae, fungi, and their extracts are emerging as attractive biocatalysts for NPs synthesis. Algae also contain large amounts of natural biocompounds such phytochemicals, carotenoids, vitamins, and pigments. All of which could act as reducing and stabilising substances to hyper-accumulate metal ions and transform them into more flexible forms at the nanoscale [195]. Algae gradually emerged as the best candidate for green synthetic NPs due to their capacity for bioreduction or biosorption. A new term, phyconanotechnology, which combines phycology with nanotechnology, is proposed to describe this phenomenon [196, 197].
Although there has been substantial development in AMNPs, there are still several obstacles existing, such as the strict criteria for homogeneous NP size and shape, reduced kinetics, and difficulties in large-scale manufacture. Managing the NPs development, stability, and aggregation is essential for obtaining uniform NPs and this involves optimisation of the variables, including pH, temperature, concentration, and others [198]. Notably, multidimensional characterizations of generated NPs are a necessary step to evaluate their uses in the interdisciplinary field, as they could provide specific criteria that make NP management easier. Moreover, generating genetic modified strains or screening high-producing algae strains are attractive strategies to synthesis NPs at large scale. Importantly, due to their exceptional biocompatibility in a homologous relationship, AMNPs represent a viable substitute for the administration of medicines, bioactive macromolecules, and gene-editing components, particularly in algae. Thus, future research on the size distribution and aggregation of NPs, as well as their surface properties, morphology, and dissolution rate, will shed light on how to regulate their release for gene editing.
AMNPs biosynthesis
Top-down and bottom-up approaches are typically used in classical NPs synthesis. The former refers to the transformation of bulk material into thinner crystallites, whilst the later refers to the production of particles by assembling ultra-small building blocks. In terms of effectiveness, tunability, and environmental friendliness, bottom-up technique is best for green synthesis of NPs [199, 200]. Following the natural bio-mineralization process, the major step in synthesising AMNPs is to capture target metal ions from the surrounding environment and enzymatically reduce them to nanoscale. AMNPs can be formed intracellularly or extracellularly, depending on the origins. The intracellular pathway is a very complex and dynamic biological environment in which positively charged metal ions are absorbed by electrostatic interactions to negatively charged cell walls. They were then transformed into metallic NPs by intracellular enzymes [201]. Algae-based metabolic pathways eliminate the need for pre-treatment and give AMNPs high colloidal stability, which benefite from increased biocompatibility and steric stabilisation of bioactive compounds [202, 203]. However, downstream separation of nanomaterials originated from the living cells is still challenging [204].
Extracellular NP synthesis, on the other hand, avoids the additional downstream processing steps and is thus more promising for a variety of applications [205, 206]. Typically, the biosynthesis of NPs comprised two steps: the preparation of algae extracts and metal precursor solution, followed by their incubation for the synthesis reaction [207]. Following incubation, AMNPs have a tendency to aggregate, giving them a thermodynamic stability with different sizes and shapes [207, 208]. The successful synthesis of NPs is indicated by the colour changes of cultured mixture. Despite extracellular synthesis of metallic NPs is easy to harvest in large-scale production, it may grow slower than that of intracellular biosynthesis under the circumstance of cell-free extract. The primary reason may be due to the binding of the active proteins with intracellularly produced NPs rather than extracellularly, which suggests that the active proteins have a special function distinct from cell-free synthesis. As a result, algae have been recognised as the ideal bio-based substrate for the extracellular synthesis of NPs due to their abundance of bioactive components [209]. Despite the fact that NPs have a wide range of uses, efficient, environmental friendly, and easy to scale up, but their synthesis mechanisms are not well understood [210].
Applications of microalgae-based NPs in delivery
The ideal drug delivery systems (DDSs) primarily use nanocarriers with the requirements of safety, effectiveness, and optimal bioavailability to improve the selectivity and targeting of various cargoes into specific cells [211]. However, the shortcomings of conventional NPs, such as their poor stability, high cost, time-consuming, and toxicity, are becoming increasingly apparent in tandem with the increasing demand for nano-DDSs [212, 213]. Many marine bioactive compounds have been employed as reducing and stabilising agents based on the extracellular synthesis of NPs to create metallic nano-DDSs with remarkable biocompatibility, biodegradability, minimal immunogenicity, and non-toxicity [214, 215] (Fig. 2b). Marine carbohydrates have been shown to be the suitable substrates for the construction of AMNPs as DDSs due to considerable advantages in terms of biodiversity, diverse biological activities, and ease of preparation [216, 217]. Bioinspired multifunctional DDSs have been synthesized through extracellular synthesis using the carbohydrates found in marine algae, such as fucoidan [218], resveratrol [219], porphyrin [220], carrageenan oligosaccharide [221], and chitosan [222, 223]. These carbohydrate-based NPs have an exceptional 60%–92% drug loading efficiency, enabling a promising use in the delivery of the CRISPR/Cas system. Nonetheless, the number and type of algae-derived carbohydrates NPs are limited only to a few metals, like AuNPs and AgNPs. Therefore, additional research should be done to determine the other active components from algae that could be used for various types of cargo [15]. In contrast to metallic NPs, marine carbohydrates-based DDSs may be created as carriers through self-assembly or covalent crosslinking and combine the benefits of nanoscale systems with the characteristics of carbohydrates, such as targeted delivery, high biocompatibility, structural modularity, and biodegradability [224].
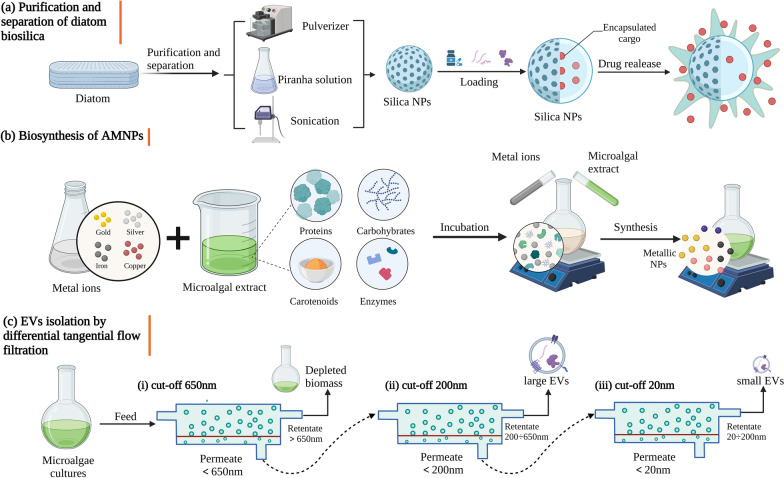
Fig. 2.
Preparation of microalgae-based nanomaterials. a Purification and separation of diatom biosilica. b Biosynthesis of AMNPs. c EVs isolation by differential tangential flow filtration
Aside from that, extracellular vesicles (EVs) generated from microalgae and diatom biosilica are also efficient biological delivery nanomaterials. Diatom-derived biosilica has high specific surface area, excellent drug-holding capacities and biocompatibility, and tailorable surface functionalization for tunable properties because it is encased in a porous 3D nanopatterned silica structure made from biosilica that self-assembled into intricate porous shells [225]. As a result, biosilica can be employed as a sophisticated microcarrier for targeted distribution for therapeutic and medical imaging purposes which is a cost effective and environmental friendly process [226–228]. Biosilica has been created using pulverisation and the strong oxidising capabilities of piranha solution, with the advantages of being cost-effective, easy, quick, and environmental friendly [227] (Fig. 2a). Cargoes could be loaded both on the surface and inside the biosilica NPs, which have a substantial impact on the releasing kinetics of active compounds due to their hierarchical 3D silica porous architectures [229]. More importantly, increased surface modification activities will aid in the targeted delivery of cargoes and the synthesis of multifunctional smart biosilica NPs [230]. For example, chitosan molecules grafted biosilica demonstrated consistent pH responsiveness and notable drug load efficiency with about 90% [231]. EVs, on the other hand, have been widely exploited as the potent tools for cargoes delivery due to their intercellular communication capabilities [232]. Microalgae are an abundant and sustainable source of natural EVs with diverse bioactivities, therefore they are positioned to become competitive competitors in innovative delivery systems [233, 234] (Fig. 2c).
Microalgae-based biohybrid microrobots
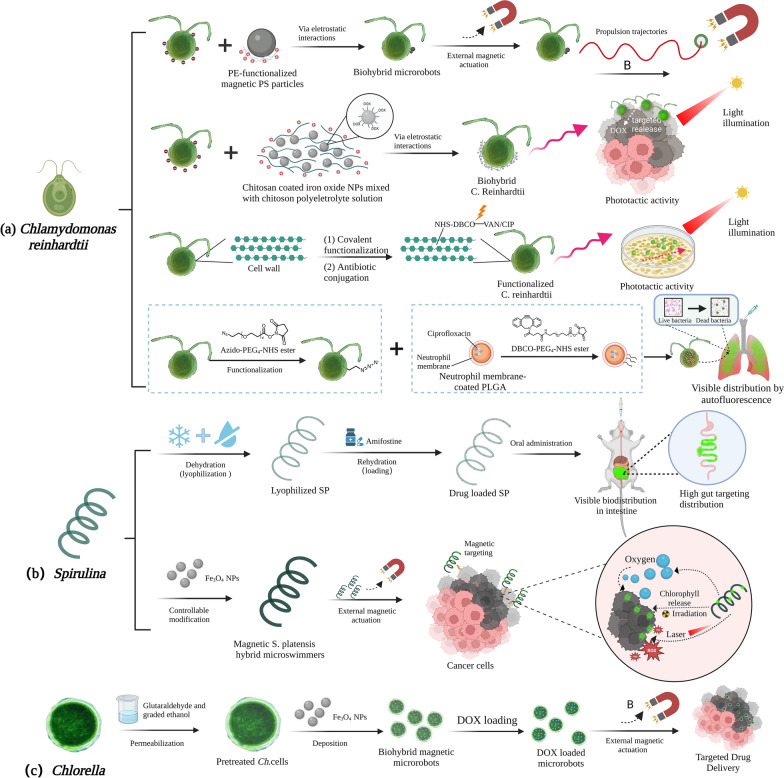
Aside from the previously described microalgae-based nanocarriers, living microalgae-based delivery systems have also exhibited impressive delivery capabilities [235]. Microalgae-based biohybrid microrobots have a number of benefits over conventional synthetic NPs, such as effective and controllable drug delivery, improved stability and bioavailability of therapeutic agents, decreased toxicity and side effects, customizability, eco-friendliness, as well as cost-effectiveness [236, 237]. These microrobots were propelled by magnetic fields or light beams and had high propulsion speeds and phototactic guidance skills, allowing for precise navigation to tissues and difficult-to-reach bodily cavities [238]. As illustrated in Fig. 3, Chlamydomonas reinhardtii, Spirulina, and Chlorella are gaining popularity for use in the construction of microrobots [239, 240]. The modifications and cargo loading of microrobots have negligible influence on their motion, thereby paving the way for the development of living delivery vehicles with enormous potential. Through external magnetically actuated control [241–245], Fe3O4 magnetized Chlorella microrobot demonstrated a greater drug loading efficiency of 98.2% and excellent targeting delivery capabilities (90% target cell death) [246]. The superior drug loading efficiency may be attributed to unique permeation pretreatment and highly negative zeta potential, providing new insights into the preparation of other microalgae-based microrobots.
Fig. 3.
Preparation and application of microalgae-based biohybrid microrobots. a–c represents the preparation process and application of microalgae-based biohybrid microrobots, including the Chlamydomonas reinhardtii (a), Spirulina (b), and Chlorella (c) etc.
Additionally, light-driven microalgal microrobots have shown great potential due to their rapid phototaxis response. Numerous tests have shown that biohybrid microrobots are resilient and can be directed towards light stimuli with powerful and precise motion. Although microalgal microrobots can be loaded with drugs with a high manufacturing yield of approximately 90% [247], the drug release effectiveness of microalgal microrobots is inadequate, ranging only from 5 to 10% [248]. Thus, improving drug release efficiency is a critical step towards expanding the use of microalgal microrobots. Microalgal microrobots have also displayed excellent delivery capabilities in local tissues without external targeted actuation. An example is the modification of Chlamydomonas reinhardtii with neutrophil membrane-coated and drug-loaded PNPs which allowed antibiotics to achieve about 90% drug release efficiency in the lungs over the first 20 h [235]. Once the issue of low drug release efficiency is resolved, microalgal microrobots will be able to deliver drugs in vivo in an effective, targeted, and safe ways. Furthermore, microalgal microrobots have outstanding fluorescence imaging capabilities that enable non-invasive tracking and real-time monitoring in vivo. They can also overcome hypoxia in tissue engineering and generate cytotoxic reactive oxygen through photodynamic treatment [249]. In summary, the development of microalgal microrobots has opened up new opportunities as a flexible platform for targeted distribution, fluorescence imaging, photothermal therapy, radiotherapy, and other biotechnology applications.
Current issues and future prospects
Current drawbacks of gene editing in microalgae
Although CRISPR/Cas systems have demonstrated great promise in the field of microalgae, their application may be hampered by a lack of understanding of microalgal biology, inefficient delivery of genetic materials, decreased expression of Cas9 due to unknown factors, low repair efficiency of the homology-directed repair (HDR) pathway, and other factors [250]. (1) Microalgae bioengineering is still in its infancy, with only a few fully sequenced genome species [251]. In the field of microalgal research, artificial intelligence (AI) has gained popularity and can be used to predict gene sequencing and editing. In this way, artificial intelligence (AI) technology fills the gap between commercial microalgae applications and genetic engineering [252, 253]. (2) It is believed that the cell wall of microalgal cells is the major barrier that reduces the delivery effectiveness of genetic materials during the introduction of macromolecules [254]. In comparison to conventional physical pretreatments like ultrasonication, high-pressure homogenization, bead milling, cryogenic grinding, and pulsed electric field, or mechanical pretreatments like acid, alkaline, and strong oxidant [255], some cutting-edge techniques, such as zinc oxide nanowire array microdevice system and optimised droplet electroporation, are significantly more effective for algal cells with rigid cell walls [256]. It is also necessary to use the right and appropriate delivery vector to increase the effectiveness of genetic materials' transport, and AMNPs have a particularly high potential for microalgae genetic editing with improved biocompatibility [15]. (3). Many approaches have been suggested to combat off-target effect of CRISPR/Cas system, for example reasonable design of sgRNA using different computational tools, like CRISPR-P 2.0 [257], E-CRISP [258], and CasFinder [259], further modification of sgRNA, application of anti-CRISPR proteins as well as appropriate editing conditions (time and temperature). Meanwhile, the cytotoxic effects of Cas9 nuclease could be mitigated by substituting it with Cas12a (4) The expression of Cas9 proteins could be reduced by unknown circumstances. Inserting introns into the Cas9 coding region could improve the efficacy of gene editing, which would significantly improve Cas9 expression [260]. (5) Due to competition from the NHEJ pathway, the HDR pathway is typically less effective in microalgae genome editing for DSBs repair in the host [261]. Given this, various approaches to improving HDR pathway repair efficiency have been suggested, including inhibition of NHEJ pathway [262], regulation of HDR-related factors [263], cell cycle synchronization [264], design of donor DNA template [265], and proximity of CRISPR component and donor DNA template [266].
Natural ingredient improvement
Currently, microalgae are potential sources of valuable bioactive compounds that can be used in different sectors, such as pharmaceutical, cosmetic, and nutraceutical industries [267]. Microalgae could replace higher plants due to its rapid growth, high biomass yield, minimal water use, daily harvest, lack of seasonal restrictions, and ease of culture in in environments with significant climate change [252]. However, most of the biomolecules are produced in relatively small amounts, making it difficult to meet industrial demands [268]. The CRISPR/Cas system is a promising way to boost the production of bioactive molecules with the benefits of pinpoint accuracy, high editing efficiency, multiplex knock-in/out capability, low cost, and short cycle [268, 269] (Fig. 4a). It is ideal to combine genetic and metabolic engineering with omic technologies (transcriptome analysis, proteomics, and liquid chromatography mass spectroscopy) for the purpose of identifying the dynamic molecular regulatory mechanisms and genetic networks involved in the various metabolic pathways in microalgae [256, 270]. Inducing targeted regulation and modification in microalgal cells could be used to precisely control the metabolic pathway and acquire the strains that can produce high-yield targeted compounds that are competitive with present commercial sources.
Fig. 4.
Extensive applications of gene editing microalgae. a Enhancing the production of natural ingredient in microalgae. b Extending the expression profile of microalgae systems. c Exploration of microalgae biofuel. d Enhancing the removal efficiency for wastewater and reduction in carbon dioxide emissions
Extending expression profile of microalgae systems
Production of recombinant biopharmaceuticals, including vaccines, antibodies, enzymes, hormones, antibiotics, cytokines, thrombolytic agents, and other proteins with medicinal value, is a rapidly growing market for disease prevention and treatment [271] (Fig. 4b). Typically, these proteins are produced by bacteria, yeast, fungus, mammalian cells, and insect, but they have drawbacks, such as expensive, poor yields, prone to contamination, fragile, and trigger immunogenic effects [272]. Eukaryotic microalgae, in contrast, have a shorter culture cycle and are less impacted by seasonal weather conditions in addition to being able to complete complex protein folding and modification to produce active proteins [271]. Both the nucleus and the chloroplast of microalgae can be genetically modified at the same time to produce biopharmaceuticals, providing a large-scale platform for recombinant protein production and industrial use [273].
Antibiotics
Antibiotic overuse and regulatory limits are driving an increase in research for antibiotic alternatives, particularly for antibiotic-resistant bacteria. Small oligopeptides known as antimicrobial peptides (AMPs) are present in plants, animals, and bacteria and serve as a general defence against pathogenic microorganisms [274, 275]. In comparison to other systems, the microalgae expression system can manufacture synthetic antibiotics that have the benefit of being flexible while protecting recombinant peptides within their cell walls from digestion [276]. More importantly, microalgae can effectively suppress the growth of bacterial strains through the interaction with their excretion products, such as fatty acids. [277]. Hence, C. reinhardtii and H. pluvialis have expressed additional antimicrobial and antifungal proteins (ToAMP4 or antimicrobial peptide piscidin-4 gene). Some of them were produced with yields up to 0.32% of total host soluble proteins with more potent antibacterial action against E. coli, S. enteritidis, S. aureus, and B. subtilis, despite the fact that expression had no effect on algal growth [278]. The future green synthesis of antibiotics with microalgae is obviously considerably facilitated by the use of gene editing techniques.
Human proteins
Recombinant human proteins could be created using the microalgal nucleus as a new expression system for the purpose of treating diseases where these proteins are deficient or where it is necessary to overexpress specific molecules, such as cytokines, growth factors, and human interferon [279, 280]. Aside from that, more health-promoting proteins, like the human growth hormone, growth factors hVEGF-165, hPDGF-B, and hSDF-1, have been successfully produced in the chloroplast system of C. reinhardtii, [281–283]. Chloroplast systems have higher levels of recombinant protein production when compared to nucleus systems. Additionally, the chloroplast system has the ability to express multiple transgenes, precise integration of foreign DNA to any locus, and the absence of gene silencing effects [275]. As a result, more algal-derived human proteins will be synthesised in the future, which could be employed as safe and economical biological agents to treat a variety of illness.
Vaccines and antigens
There are many benefits of using microalgae as an expression system for manufacturing antigens and antibodies, including their low cost, the capacity to perform post-translational modifications, and elimination of the need for protein extraction and purification. For instance, a direct vaccination against WSSV infection in shrimps could be created using transgenic Dunaliella salina that expresses the viral envelope protein VP28 [284]. In fact, the recombinant protein-based vaccinations are not just limited to aquaculture and have the potential to develop into animal feed as well as human food. For instance, biopharmaceuticals synthesize from microalgae can be employed in immunotherapy for diseases like cancer [285] and SARS-COV-2 [286]. A chimeric protein to treat breast cancer was expressed in the microalgae Schizochytrium sp. by fusing B-cell epitopes from the tumour related antigens, human epidermal growth factor receptor-2, mucin-like glycoprotein 1, Wilms' tumour antigen, and mammaglobin [287]. These epitopes serve as immunotherapy targets and aid in the synthesis of antibodies and CD8+/CD4+T cell, which kill the malignant cells [288–290]. Berndt et al. [291] use C. reinhardtii to produce recombinant SARS-CoV-2 spike RBD proteins that have a comparable affinity to those expressed in mammalian cells. The findings revealed that it is feasible to use algae to produce functional proteins [286]. In short, antigens and antibodies produced by microalgae could serve a wide variety of applications in the future, including the development of diagnostic tools and the treatment of human, plant, and animal diseases.
Exploration of microalgae biofuel
Microalgae are a viable resource for biofuel generation and the generation of biodiesel using microalgae biomass is 100 times more efficient than that using higher plants as feedstocks [292] (Fig. 4c). Furthermore, microalgae-based biofuels are compatible with existing gasoline engines, eliminating the need for engine modifications [293]. The viability of biofuel as an alternative to traditional fossil fuels will be determined primarily by its economic benefits. Unfortunately, the actual costs of microalgal biofuels are still deemed higher than conventional fossil oils, owing to limited storage lipids and carbohydrates yield in microalgae mass [294]. Numerous initiatives have been launched in the past to increase the production of microalgae biomass and biofuels, including the selection of suitable microalgae species [267], pretreatment of biomass [295] and application of nanotechnology [296]. However, this field still has several constraints that must be addressed with more robust editing tool, such as low carbon fixation efficiency, low lipid accumulation rate, and long cultivation period [297]. The CRISPR/Cas technology presents the best option for resolving the aforementioned problems in microalgae. Microalgae metabolic pathways, for example, can be altered by raising acetyl coenzyme A carboxylase, lowering enzymatic activity of phospholipase A2, or boosting lipid and carbohydrate production. Omic technologies can be used to identify the underlying dynamic molecular regulatory mechanisms and genetic networks involved in the metabolic pathways [298]. Gene-edited microalgal variations might be used in the future to produce more environmentally friendly and clean energy.
Wastewater treatment
An increasing number of pollutants found in wastewater effluents, such as pharmaceuticals, endocrine disruptors, industrial chemicals, heavy metals, pesticides, etc., are posing threat to human and environmental health [299]. Activated charcoal, flocculation, chemical precipitation, reverse osmosis, ultraviolet disinfection, ultrafiltration, electro-coagulation, and ion exchange are few examples of methods to treat wastewater [300]. However, these methods are not cost-effective since these methods demand a lot of energy and labour and they are unable to eliminate a variety of emerging pollutants in wastewater completely [301]. Microalgal-bioremediation systems have thus been shown to be an effective strategy in wastewater treatment processes to overcome the limitations of such methods. The advantages of microalgal-bioremediation [302] (Fig. 4d) are that it may effectively remove N and P through photosynthetic processes and CO2 sequestration. In the meantime, oxygen is released into the water, increasing the amount of dissolved oxygen [303]. Additionally, microalgae increase the pH and dissolved oxygen in the broth as a result of their photosynthetic activities, which reduces pathogen survival [304]. Microalgae have a variety of appealing advantages, but there are still several obstacles existing which are high turbidity, contamination by other microbes, and difficulties in harvesting microalgae biomass. These issues can be resolved using the methods including genomics, computational biology, proteomics, bioinformatics, molecular modelling, molecular dynamics simulation, and a specialised algorithm for pathways prediction [305]. These combined approaches may provide a fast understanding of pollutant binding, degradation, and absorption [306]. Based on this, more robust microalgal strains could be created using gene editing by altering the related genes that are responsible for the biosynthesis of specific enzymes, increasing the adaptability and biosorption ability. This would further improve the efficiency of wastewater treatment and lower operating costs.
CO2 sequestration
Large-scale human activity and excessive greenhouse gas emissions have caused enormous changes in the environment, including the greenhouse effect. These changes have resulted in global warming, climate change, and changes in other environmental aspects [307]. Critical climate change has sparked initiatives worldwide to cut greenhouse gas emissions, especially CO2, with the primary goal of keeping rises in global temperature to 2 ℃ [308]. CO2 sequestration is critical for managing climate stability and balancing CO2 level in the atmosphere [309]. CO2 sequestration by microalgae is the most promising approach for mitigating the consequences of greenhouse emissions [310] (Fig. 4d). Microalgae capture CO2 and provide various economic and ecological advantages over conventional terrestrial plants, such as rapid growth and sustainability [311], high photosynthetic efficiency, and cooperation with CO2 sequestration and wastewater treatment [312]. Additionally, the resulting microalgae biomass can be used to synthesis high-value biomolecules, such as carbohydrates and lipids to produce byproducts for example biofuels and as chemical precursors [313]. A better understanding of the microalgae metabolic pathway with the establishment of ideal genome editing platform has enabled researchers to increase the photosynthetic efficiency of microalgae chassis cells, optimise the microalgae culture conditions, and increase microalgae biomass and yield [314]. Engineered microalgal cell factories based on synthetic biology methods have made it feasible to increase economic efficiency, lessen the greenhouse effect, and protect the environment.
Conclusion
Target-specific gene editing methods have substantially helped the microalgae industry, enabling improvements in the fields of natural ingredients enhancement, expression profile extension, wastewater treatment, CO2 sequestration, and other related fields. Even though this field has made significant advancements, there are still some problems that prevent the widespread application of this cutting-edge technology. These problems include the difficulty of delivering editing components, lack of knowledge about microalgae biology, ambiguous molecular regulatory mechanisms and genetic networks associated with microalgae, and the ineffectiveness of gene editing tools. In this review, a variety of efficient approaches aimed at resolving those problems have been thoroughly discussed. The synthesis and use of novel AMNPs has the potential to increase the effectiveness of gene editing component delivery while minimising damage to microalgae cells. Due to their intrinsic anti-bacterial, anti-cancer, anti-viral, anti-pollution, and bio-remediation capabilities, AMNPs confer additional competitiveness in the gene editing of microalgae in the realms of biomedicine and environmental remediation. Following extensive study on AMNPs, gene editing in microalgae would offer a plethora of opportunities in a variety of industries.
Acknowledgements
Not applicable.
Abbreviations
- AMNPs
Algal-mediated nanoparticles
- CRISPR/Cas system
Clustered regularly interspaced short palindromic repeats-associated system
- ZFN
Zinc finger nuclease
- TALEN
Transcription activator-like effector-based nuclease
- NPs
Nanoparticles
- NP
Nanoparticle
- pDNA
Plasmid DNA
- RNPs
Ribonucleoproteins
- LNPs
Lipid NPs
- PNPs
Polymeric nanoparticles
- AuNPs
Gold nanoparticles
- ApoE
Apolipoprotein E
- SORT
Selective organ targeting
- PEI
Polyethylenimine
- PLGA
Poly (lactic-co-glycolic-acid)
- PBAE
Poly (β-amino ester)
- PEG
Poly (ethylene–glycol)
- NIR
Near-infrared
- GFP
Green fluorescent protein
- HSF
Heat-shock factor
- GNR
Gold nanorod
- DDSs
Drug delivery systems
- EVs
Extracellular vehicles
- PE
Polyelectrolyte
- DOX
Doxorubicin
- NHS
N-hydroxysuccinimide
- DBCO
Dibenzocyclooctyne
- HDR
Homology-directed repair
- AI
Artificial intelligence
- NHEJ
Nonhomologous end joining
- AMPs
Antimicrobial peptides
- GHGs
Greenhouse gases
Author contributions
All authors took part in writing, reviewing, and editing the manuscript. SYF, XX, and JJL wrote the manuscript. AFL, QQW and DDG prepared the figures. SXL and ZLW created the table. TG and YLL collected and organized the literature. JZ modified the paper. PLS and DYYT reviewed and edited the paper. All authors reviewed the manuscript and approved it for publication.
Funding
This work was supported by the National Natural Science Foundation of China (No. U1804112), the Basic Research Project of the Key Research Program of Colleges and Universities in Henan Province (23ZX005), and the Natural Science Foundation of Henan Province (232300421164).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shuying Feng, Xin Xie and Junjie Liu, contributed equally to this work.
Contributor Information
Shuying Feng, Email: fsy@hactcm.edu.cn.
Tao Guo, Email: gt010010@163.com.
Jin Zhou, Email: zhou.jin@sz.tsinghua.edu.cn.
Pau Loke Show, Email: PauLoke.Show@ku.ac.ae.
References
- 1.Su H, Feng J, Lv J, Liu Q, Nan F, Liu X, et al. Molecular mechanism of lipid accumulation and metabolism of oleaginous chlorococcum sphacosum gd from soil under salt stress. Int J Mol Sci. 2021;22:1304. doi: 10.3390/ijms22031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Da Ros PC, Silva CS, Silva-Stenico ME, Fiore MF, De Castro HF. Assessment of chemical and physico-chemical properties of cyanobacterial lipids for biodiesel production. Mar Drugs. 2013;11:2365–2381. doi: 10.3390/md11072365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giri DD, Dwivedi H, Khalaf DAA, Pal DB, Otaibi AA, Areeshi MY, et al. Sustainable production of algae-bacteria granular consortia based biological hydrogen: new insights. Bioresour Technol. 2022;352:127036. doi: 10.1016/j.biortech.2022.127036. [DOI] [PubMed] [Google Scholar]
- 4.Harahap BM, Ahring BK. Acetate production from syngas produced from lignocellulosic biomass materials along with gaseous fermentation of the syngas: a review. Microorganisms. 2023;11:995. doi: 10.3390/microorganisms11040995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayona-Morcillo PJ, Gomez-Serrano C, Gonzalez-Lopez CV, Massa D, Jimenez-Becker S. Effect of the application of hydrolysate of chlorella vulgaris extracted by different techniques on the growth of pelargonium x hortorum. Plants. 2022;11:191. doi: 10.3390/plants11172308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsvetanova F, Yankov D. Bioactive compounds from red microalgae with therapeutic and nutritional value. Microorganisms. 2022;10:2290. doi: 10.3390/microorganisms10112290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallick N, Bagchi SK, Koley S, Singh AK. Progress and challenges in microalgal biodiesel production. Front Microbiol. 2016;7:1019. doi: 10.3389/fmicb.2016.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkata Subhash G, Rajvanshi M, Navish Kumar B, Govindachary S, Prasad V, Dasgupta S. Carbon streaming in microalgae: extraction and analysis methods for high value compounds. Bioresour Technol. 2017;244:1304–1316. doi: 10.1016/j.biortech.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Hlavova M, Turoczy Z, Bisova K. Improving microalgae for biotechnology–from genetics to synthetic biology. Biotechnol Adv. 2015;33:1194–1203. doi: 10.1016/j.biotechadv.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Kurita T, Moroi K, Iwai M, Okazaki K, Shimizu S, Nomura S, et al. Efficient and multiplexable genome editing using platinum talens in oleaginous microalga, nannochloropsis oceanica nies-2145. Genes Cells. 2020;25:695–702. doi: 10.1111/gtc.12805. [DOI] [PubMed] [Google Scholar]
- 11.Wang JY, Doudna JA. Crispr technology: a decade of genome editing is only the beginning. Science. 2023;379:eadd8643. doi: 10.1126/science.add8643. [DOI] [PubMed] [Google Scholar]
- 12.Coelho MA, De Braekeleer E, Firth M, Bista M, Lukasiak S, Cuomo ME, et al. Crispr guard protects off-target sites from cas9 nuclease activity using short guide rnas. Nat Commun. 2020;11:4132. doi: 10.1038/s41467-020-17952-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng H, Zhang F, Ding Y. Crispr/cas9 delivery system engineering for genome editing in therapeutic applications. Pharmaceutics. 2021;13:1649. doi: 10.3390/pharmaceutics13101649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng S, Wang Z, Li A, Xie X, Liu J, Li S, et al. Strategies for high-efficiency mutation using the crispr/cas system. Front Cell Dev Biol. 2021;9:803252. doi: 10.3389/fcell.2021.803252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhary R, Nawaz K, Khan AK, Hano C, Abbasi BH, Anjum S. An overview of the algae-mediated biosynthesis of nanoparticles and their biomedical applications. Biomolecules. 2020;10:1498. doi: 10.3390/biom10111498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerken HG, Donohoe B, Knoshaug EP. Enzymatic cell wall degradation of chlorella vulgaris and other microalgae for biofuels production. Planta. 2013;237:239–253. doi: 10.1007/s00425-012-1765-0. [DOI] [PubMed] [Google Scholar]
- 17.Hao X, Luo L, Jouhet J, Rebeille F, Marechal E, Hu H, et al. Enhanced triacylglycerol production in the diatom phaeodactylum tricornutum by inactivation of a hotdog-fold thioesterase gene using talen-based targeted mutagenesis. Biotechnol Biofuels. 2018;11:312. doi: 10.1186/s13068-018-1309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sizova I, Greiner A, Awasthi M, Kateriya S, Hegemann P. Nuclear gene targeting in chlamydomonas using engineered zinc-finger nucleases. Plant J. 2013;73:873–882. doi: 10.1111/tpj.12066. [DOI] [PubMed] [Google Scholar]
- 19.Vazquez-Dominguez I, Garanto A, Collin RWJ. Molecular therapies for inherited retinal diseases-current standing, opportunities and challenges. Genes. 2019;10:654. doi: 10.3390/genes10090654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghribi M, Nouemssi SB, Meddeb-Mouelhi F, Desgagne-Penix I. Genome editing by crispr-cas: a game change in the genetic manipulation of chlamydomonas. Life. 2020;10:295. doi: 10.3390/life10110295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Baltasar R, Garcia-Torralba A, Nieto-Romero V, Page A, Molinos-Vicente A, Lopez-Manzaneda S, et al. Efficient and fast generation of relevant disease mouse models by in vitro and in vivo gene editing of zygotes. CRISPR J. 2022;5:422–434. doi: 10.1089/crispr.2022.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naduthodi MIS, Sudfeld C, Avitzigiannis EK, Trevisan N, Van Lith E, Alcaide Sancho J, et al. Comprehensive genome engineering toolbox for microalgae nannochloropsis oceanica based on crispr-cas systems. ACS Synth Biol. 2021;10:3369–3378. doi: 10.1021/acssynbio.1c00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nymark M, Sharma AK, Sparstad T, Bones AM, Winge P. A crispr/cas9 system adapted for gene editing in marine algae. Sci Rep. 2016;6:24951. doi: 10.1038/srep24951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopes A, Nekrasov V, Kamoun S, Mock T. Editing of the urease gene by crispr-cas in the diatom thalassiosira pseudonana. Plant Methods. 2016;12:49. doi: 10.1186/s13007-016-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J, Chang KS, Lee S, Jin E. Establishment of a genome editing tool using crispr-cas9 in chlorella vulgaris utex395. Int J Mol Sci. 2021;22:480. doi: 10.3390/ijms22020480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farasat I, Kushwaha M, Collens J, Easterbrook M, Guido M, Salis HM. Efficient search, mapping, and optimization of multi-protein genetic systems in diverse bacteria. Mol Syst Biol. 2014;10:731. doi: 10.15252/msb.20134955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin SE, Lim JM, Koh HG, Kim EK, Kang NK, Jeon S, et al. Crispr/cas9-induced knockout and knock-in mutations in chlamydomonas reinhardtii. Sci Rep. 2016;6:27810. doi: 10.1038/srep27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baek K, Kim DH, Jeong J, Sim SJ, Melis A, Kim JS, et al. DNA-free two-gene knockout in chlamydomonas reinhardtii via crispr-cas9 ribonucleoproteins. Sci Rep. 2016;6:30620. doi: 10.1038/srep30620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao PH, Ng IS. Crispri mediated phosphoenolpyruvate carboxylase regulation to enhance the production of lipid in chlamydomonas reinhardtii. Bioresour Technol. 2017;245:1527–1537. doi: 10.1016/j.biortech.2017.04.111. [DOI] [PubMed] [Google Scholar]
- 30.Baek K, Yu J, Jeong J, Sim SJ, Bae S, Jin E. Photoautotrophic production of macular pigment in a chlamydomonas reinhardtii strain generated by using DNA-free crispr-cas9 rnp-mediated mutagenesis. Biotechnol Bioeng. 2018;115:719–728. doi: 10.1002/bit.26499. [DOI] [PubMed] [Google Scholar]
- 31.Ferenczi A, Chew YP, Kroll E, Von Koppenfels C, Hudson A, Molnar A. Mechanistic and genetic basis of single-strand templated repair at cas12a-induced DNA breaks in chlamydomonas reinhardtii. Nat Commun. 2021;12:6751. doi: 10.1038/s41467-021-27004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, Lee S, Baek K, Jin E. Site-specific gene knock-out and on-site heterologous gene overexpression in chlamydomonas reinhardtii via a crispr-cas9-mediated knock-in method. Front Plant Sci. 2020;11:306. doi: 10.3389/fpls.2020.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Shen CR, Huang CH, Sung LY, Wu MY, Hu YC. Crispr-cas9 for the genome engineering of cyanobacteria and succinate production. Metab Eng. 2016;38:293–302. doi: 10.1016/j.ymben.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Wendt KE, Ungerer J, Cobb RE, Zhao H, Pakrasi HB. Crispr/cas9 mediated targeted mutagenesis of the fast growing cyanobacterium synechococcus elongatus utex 2973. Microb Cell Fact. 2016;15:115. doi: 10.1186/s12934-016-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon GC, Korosh TC, Cameron JC, Markley AL, Begemann MB, Pfleger BF. Crispr interference as a titratable, trans-acting regulatory tool for metabolic engineering in the cyanobacterium Synechococcus sp. Strain pcc 7002. Metab Eng. 2016;38:170–179. doi: 10.1016/j.ymben.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang CH, Shen CR, Li H, Sung LY, Wu MY, Hu YC. Crispr interference (crispri) for gene regulation and succinate production in cyanobacterium s. Elongatus pcc 7942. Microb Cell Fact. 2016;15:196. doi: 10.1186/s12934-016-0595-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao L, Cengic I, Anfelt J, Hudson EP. Multiple gene repression in cyanobacteria using crispri. ACS Synth Biol. 2016;5:207–212. doi: 10.1021/acssynbio.5b00264. [DOI] [PubMed] [Google Scholar]
- 38.Poliner E, Takeuchi T, Du ZY, Benning C, Farre EM. Nontransgenic marker-free gene disruption by an episomal crispr system in the oleaginous microalga, nannochloropsis oceanica ccmp1779. ACS Synth Biol. 2018;7:962–968. doi: 10.1021/acssynbio.7b00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Lu Y, Xin Y, Wei L, Huang S, Xu J. Genome editing of model oleaginous microalgae nannochloropsis spp. By crispr/cas9. Plant J. 2016;88:1071–1081. doi: 10.1111/tpj.13307. [DOI] [PubMed] [Google Scholar]
- 40.Liu X, Zhang D, Zhang J, Chen Y, Liu X, Fan C, et al. Overexpression of the transcription factor atlec1 significantly improved the lipid content of chlorella ellipsoidea. Front Bioeng Biotechnol. 2021;9:626162. doi: 10.3389/fbioe.2021.626162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vingiani GM, De Luca P, Ianora A, Dobson ADW, Lauritano C. Microalgal enzymes with biotechnological applications. Mar Drugs. 2019;17:459. doi: 10.3390/md17080459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Yang L, Wen X, Chen Z, Liang Q, Li J, et al. Rapid and high efficiency transformation of chlamydomonas reinhardtii by square-wave electroporation. Biosci Rep. 2019;39:BSR20181210. doi: 10.1042/BSR20181210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nouemssi SB, Ghribi M, Beauchemin R, Meddeb-Mouelhi F, Germain H, Desgagne-Penix I. Rapid and efficient colony-pcr for high throughput screening of genetically transformed chlamydomonas reinhardtii. Life. 2020;10:186. doi: 10.3390/life10090186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moosburner MA, Gholami P, Mccarthy JK, Tan M, Bielinski VA, Allen AE. Multiplexed knockouts in the model diatom phaeodactylum by episomal delivery of a selectable cas9. Front Microbiol. 2020;11:5. doi: 10.3389/fmicb.2020.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolanos-Martinez OC, Mahendran G, Rosales-Mendoza S, Vimolmangkang S. Current status and perspective on the use of viral-based vectors in eukaryotic microalgae. Mar Drugs. 2022;20:434. doi: 10.3390/md20070434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown LE, Sprecher SL, Keller LR. Introduction of exogenous DNA into chlamydomonas reinhardtii by electroporation. Mol Cell Biol. 1991;11:2328–2332. doi: 10.1128/mcb.11.4.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee S, Kim YY, Ahn HJ. Systemic delivery of crispr/cas9 to hepatic tumors for cancer treatment using altered tropism of lentiviral vector. Biomaterials. 2021;272:120793. doi: 10.1016/j.biomaterials.2021.120793. [DOI] [PubMed] [Google Scholar]
- 48.Yao X, Lyu P, Yoo K, Yadav MK, Singh R, Atala A, et al. Engineered extracellular vesicles as versatile ribonucleoprotein delivery vehicles for efficient and safe crispr genome editing. J Extracell Vesicles. 2021;10:e12076. doi: 10.1002/jev2.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of cas9 protein and guide rna. Genome Res. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren X, Wei C, Yan Q, Shan X, Wu M, Zhao X, et al. Optimization of a novel lipid extraction process from microalgae. Sci Rep. 2021;11:20221. doi: 10.1038/s41598-021-99356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manghwar H, Li B, Ding X, Hussain A, Lindsey K, Zhang X, et al. Crispr/cas systems in genome editing: Methodologies and tools for sgrna design, off-target evaluation, and strategies to mitigate off-target effects. Adv Sci. 2020;7:1902312. doi: 10.1002/advs.201902312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tong S, Moyo B, Lee CM, Leong K, Bao G. Engineered materials for in vivo delivery of genome-editing machinery. Nat Rev Mater. 2019;4:726–737. doi: 10.1038/s41578-019-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shivram H, Cress BF, Knott GJ, Doudna JA. Controlling and enhancing crispr systems. Nat Chem Biol. 2021;17:10–19. doi: 10.1038/s41589-020-00700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu R, Liang L, Freed EF, Gill RT. Directed evolution of crispr/cas systems for precise gene editing. Trends Biotechnol. 2021;39:262–273. doi: 10.1016/j.tibtech.2020.07.005. [DOI] [PubMed] [Google Scholar]
- 55.Kaczmarek JC, Kowalski PS, Anderson DG. Advances in the delivery of rna therapeutics: from concept to clinical reality. Genome Med. 2017;9:60. doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov. 2021;20:101–124. doi: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hatit MZC, Lokugamage MP, Dobrowolski CN, Paunovska K, Ni H, Zhao K, et al. Species-dependent in vivo mrna delivery and cellular responses to nanoparticles. Nat Nanotechnol. 2022;17:310–318. doi: 10.1038/s41565-021-01030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Zhang H, Gu J, Zhang J, Shi H, Qian H, et al. Engineered extracellular vesicles for cancer therapy. Adv Mater. 2021;33:e2005709. doi: 10.1002/adma.202005709. [DOI] [PubMed] [Google Scholar]
- 59.Wang D, Zhang F, Gao G. Crispr-based therapeutic genome editing: strategies and in vivo delivery by aav vectors. Cell. 2020;181:136–150. doi: 10.1016/j.cell.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilbie D, Walther J, Mastrobattista E. Delivery aspects of crispr/cas for in vivo genome editing. Acc Chem Res. 2019;52:1555–1564. doi: 10.1021/acs.accounts.9b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu W, Mookherjee S, Chaitankar V, Hiriyanna S, Kim JW, Brooks M, et al. Nrl knockdown by aav-delivered crispr/cas9 prevents retinal degeneration in mice. Nat Commun. 2017;8:14716. doi: 10.1038/ncomms14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, et al. Generation of gene-modified cynomolgus monkey via cas9/rna-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 63.Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, et al. Crispr-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan Y, Zhu X, Yu Y, Li C, Zhang Z, Wang F. Nanotechnology strategies for plant genetic engineering. Adv Mater. 2021;24:e2106945. doi: 10.1002/adma.202106945. [DOI] [PubMed] [Google Scholar]
- 65.Chaverra-Rodriguez D, Macias VM, Hughes GL, Pujhari S, Suzuki Y, Peterson DR, et al. Targeted delivery of crispr-cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat Commun. 2018;9:3008. doi: 10.1038/s41467-018-05425-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, et al. Genome editing with cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pu Y, Yin H, Dong C, Xiang H, Wu W, Zhou B, et al. Sono-controllable and ros-sensitive crispr-cas9 genome editing for augmented/synergistic ultrasound tumor nanotherapy. Adv Mater. 2021;33:e2104641. doi: 10.1002/adma.202104641. [DOI] [PubMed] [Google Scholar]
- 68.Wang HX, Li M, Lee CM, Chakraborty S, Kim HW, Bao G, et al. Crispr/cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem Rev. 2017;117:9874–9906. doi: 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- 69.Seki A, Rutz S. Optimized rnp transfection for highly efficient crispr/cas9-mediated gene knockout in primary t cells. J Exp Med. 2018;215:985–997. doi: 10.1084/jem.20171626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fajrial AK, He QQ, Wirusanti NI, Slansky JE, Ding X. A review of emerging physical transfection methods for crispr/cas9-mediated gene editing. Theranostics. 2020;10:5532–5549. doi: 10.7150/thno.43465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song X, Liu C, Wang N, Huang H, He S, Gong C, et al. Delivery of crispr/cas systems for cancer gene therapy and immunotherapy. Adv Drug Deliv Rev. 2021;168:158–180. doi: 10.1016/j.addr.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 72.Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, Mcconkey ME, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using crispr-cas9 genome editing. Nat Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Russell S, Bennett J, Wellman JA, Chung DC, Yu ZF, Tillman A, et al. Efficacy and safety of voretigene neparvovec (aav2-hrpe65v2) in patients with rpe65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390:849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 75.Shirley JL, De Jong YP, Terhorst C, Herzog RW. Immune responses to viral gene therapy vectors. Mol Ther. 2020;28:709–722. doi: 10.1016/j.ymthe.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baum C, Kustikova O, Modlich U, Li Z, Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum Gene Ther. 2006;17:253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- 77.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 78.Chen F, Alphonse M, Liu Q. Strategies for nonviral nanoparticle-based delivery of crispr/cas9 therapeutics. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2020;12:e1609. doi: 10.1002/wnan.1609. [DOI] [PubMed] [Google Scholar]
- 79.Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, et al. Therapeutic genome editing by combined viral and non-viral delivery of crispr system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Glass Z, Lee M, Li Y, Xu Q. Engineering the delivery system for crispr-based genome editing. Trends Biotechnol. 2018;36:173–185. doi: 10.1016/j.tibtech.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the crispr-cas9 gene-editing system for therapeutic applications. J Control Release. 2017;266:17–26. doi: 10.1016/j.jconrel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li L, Hu S, Chen X. Non-viral delivery systems for crispr/cas9-based genome editing: challenges and opportunities. Biomaterials. 2018;171:207–218. doi: 10.1016/j.biomaterials.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Canatella PJ, Karr JF, Petros JA, Prausnitz MR. Quantitative study of electroporation-mediated molecular uptake and cell viability. Biophys J. 2001;80:755–764. doi: 10.1016/S0006-3495(01)76055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bak RO, Dever DP, Reinisch A, Cruz Hernandez D, Majeti R, Porteus MH. Multiplexed genetic engineering of human hematopoietic stem and progenitor cells using crispr/cas9 and aav6. Elife. 2017;6:e27873. doi: 10.7554/eLife.27873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li L, He ZY, Wei XW, Gao GP, Wei YQ. Challenges in crispr/cas9 delivery: potential roles of nonviral vectors. Hum Gene Ther. 2015;26:452–462. doi: 10.1089/hum.2015.069. [DOI] [PubMed] [Google Scholar]
- 86.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 87.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 88.Patra JK, Das G, Fraceto LF, Campos EVR, Rodriguez-Torres MDP, Acosta-Torres LS, et al. Nano based drug delivery systems: recent developments and future prospects. J Nanobiotechnology. 2018;16:71. doi: 10.1186/s12951-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klochkov SG, Neganova ME, Nikolenko VN, Chen K, Somasundaram SG, Kirkland CE, et al. Implications of nanotechnology for the treatment of cancer: recent advances. Semin Cancer Biol. 2021;69:190–199. doi: 10.1016/j.semcancer.2019.08.028. [DOI] [PubMed] [Google Scholar]
- 90.Rajasekaran D, Srivastava J, Ebeid K, Gredler R, Akiel M, Jariwala N, et al. Combination of nanoparticle-delivered sirna for astrocyte elevated gene-1 (aeg-1) and all-trans retinoic acid (atra): An effective therapeutic strategy for hepatocellular carcinoma (hcc) Bioconjug Chem. 2015;26:1651–1661. doi: 10.1021/acs.bioconjchem.5b00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liang Y, Iqbal Z, Wang J, Xu L, Xu X, Ouyang K, et al. Cell-derived extracellular vesicles for crispr/cas9 delivery: engineering strategies for cargo packaging and loading. Biomater Sci. 2022;10:4095–4106. doi: 10.1039/D2BM00480A. [DOI] [PubMed] [Google Scholar]
- 92.Ebeid K, Meng X, Thiel KW, Do AV, Geary SM, Morris AS, et al. Synthetically lethal nanoparticles for treatment of endometrial cancer. Nat Nanotechnol. 2018;13:72–81. doi: 10.1038/s41565-017-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gunawan C, Lim M, Marquis CP, Amal R. Nanoparticle-protein corona complexes govern the biological fates and functions of nanoparticles. J Mater Chem B. 2014;2:2060–2083. doi: 10.1039/c3tb21526a. [DOI] [PubMed] [Google Scholar]
- 94.Pan L, He Q, Liu J, Chen Y, Ma M, Zhang L, et al. Nuclear-targeted drug delivery of tat peptide-conjugated monodisperse mesoporous silica nanoparticles. J Am Chem Soc. 2012;134:5722–5725. doi: 10.1021/ja211035w. [DOI] [PubMed] [Google Scholar]
- 95.Degors IMS, Wang C, Rehman ZU, Zuhorn IS. Carriers break barriers in drug delivery: endocytosis and endosomal escape of gene delivery vectors. Acc Chem Res. 2019;52:1750–1760. doi: 10.1021/acs.accounts.9b00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yaron PN, Holt BD, Short PA, Losche M, Islam MF, Dahl KN. Single wall carbon nanotubes enter cells by endocytosis and not membrane penetration. J Nanobiotechnology. 2011;9:45. doi: 10.1186/1477-3155-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rouet R, Thuma BA, Roy MD, Lintner NG, Rubitski DM, Finley JE, et al. Receptor-mediated delivery of crispr-cas9 endonuclease for cell-type-specific gene editing. J Am Chem Soc. 2018;140:6596–6603. doi: 10.1021/jacs.8b01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33:941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Donahue ND, Acar H, Wilhelm S. Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Adv Drug Deliv Rev. 2019;143:68–96. doi: 10.1016/j.addr.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 100.De Sousa Almeida M, Susnik E, Drasler B, Taladriz-Blanco P, Petri-Fink A, Rothen-Rutishauser B. Understanding nanoparticle endocytosis to improve targeting strategies in nanomedicine. Chem Soc Rev. 2021;50:5397–5434. doi: 10.1039/D0CS01127D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Behzadi S, Serpooshan V, Tao W, Hamaly MA, Alkawareek MY, Dreaden EC, et al. Cellular uptake of nanoparticles: journey inside the cell. Chem Soc Rev. 2017;46:4218–4244. doi: 10.1039/C6CS00636A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Khalil IA, Kogure K, Futaki S, Hama S, Akita H, Ueno M, et al. Octaarginine-modified multifunctional envelope-type nanoparticles for gene delivery. Gene Ther. 2007;14:682–689. doi: 10.1038/sj.gt.3302910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu S, Olenyuk BZ, Okamoto CT, Hamm-Alvarez SF. Targeting receptor-mediated endocytotic pathways with nanoparticles: rationale and advances. Adv Drug Deliv Rev. 2013;65:121–138. doi: 10.1016/j.addr.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, et al. Image-based analysis of lipid nanoparticle-mediated sirna delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 105.Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, et al. Efficiency of sirna delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pei D. How do biomolecules cross the cell membrane? Acc Chem Res. 2022;55:309–318. doi: 10.1021/acs.accounts.1c00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 108.Wojnilowicz M, Glab A, Bertucci A, Caruso F, Cavalieri F. Super-resolution imaging of proton sponge-triggered rupture of endosomes and cytosolic release of small interfering rna. ACS Nano. 2019;13:187–202. doi: 10.1021/acsnano.8b05151. [DOI] [PubMed] [Google Scholar]
- 109.Yuba E, Kanda Y, Yoshizaki Y, Teranishi R, Harada A, Sugiura K, et al. Ph-sensitive polymer-liposome-based antigen delivery systems potentiated with interferon-gamma gene lipoplex for efficient cancer immunotherapy. Biomaterials. 2015;67:214–224. doi: 10.1016/j.biomaterials.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 110.Akita H, Kudo A, Minoura A, Yamaguti M, Khalil IA, Moriguchi R, et al. Multi-layered nanoparticles for penetrating the endosome and nuclear membrane via a step-wise membrane fusion process. Biomaterials. 2009;30:2940–2949. doi: 10.1016/j.biomaterials.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 111.Hu Y, Litwin T, Nagaraja AR, Kwong B, Katz J, Watson N, et al. Cytosolic delivery of membrane-impermeable molecules in dendritic cells using ph-responsive core-shell nanoparticles. Nano Lett. 2007;7:3056–3064. doi: 10.1021/nl071542i. [DOI] [PubMed] [Google Scholar]
- 112.Manganiello MJ, Cheng C, Convertine AJ, Bryers JD, Stayton PS. Diblock copolymers with tunable ph transitions for gene delivery. Biomaterials. 2012;33:2301–2309. doi: 10.1016/j.biomaterials.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for sirna delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 114.Nakase I, Kobayashi S, Futaki S. Endosome-disruptive peptides for improving cytosolic delivery of bioactive macromolecules. Biopolymers. 2010;94:763–770. doi: 10.1002/bip.21487. [DOI] [PubMed] [Google Scholar]
- 115.Yang J, Bahreman A, Daudey G, Bussmann J, Olsthoorn RC, Kros A. Drug delivery via cell membrane fusion using lipopeptide modified liposomes. ACS Cent Sci. 2016;2:621–630. doi: 10.1021/acscentsci.6b00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Selby LI, Cortez-Jugo CM, Such G, Such G, Johnston APR. Nanoescapology: Progress toward understanding the endosomal escape of polymeric nanoparticles. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2017;9:e1452. doi: 10.1002/wnan.1452. [DOI] [PubMed] [Google Scholar]
- 117.Mura S, Nicolas J, Couvreur P. Stimuli-responsive nanocarriers for drug delivery. Nat Mater. 2013;12:991–1003. doi: 10.1038/nmat3776. [DOI] [PubMed] [Google Scholar]
- 118.Massignani M, Lopresti C, Blanazs A, Madsen J, Armes SP, Lewis AL, et al. Controlling cellular uptake by surface chemistry, size, and surface topology at the nanoscale. Small. 2009;5:2424–2432. doi: 10.1002/smll.200900578. [DOI] [PubMed] [Google Scholar]
- 119.Su X, Fricke J, Kavanagh DG, Irvine DJ. In vitro and in vivo mrna delivery using lipid-enveloped ph-responsive polymer nanoparticles. Mol Pharm. 2011;8:774–787. doi: 10.1021/mp100390w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu H, Zhu L, Torchilin VP. Ph-sensitive poly(histidine)-peg/dspe-peg co-polymer micelles for cytosolic drug delivery. Biomaterials. 2013;34:1213–1222. doi: 10.1016/j.biomaterials.2012.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao ZX, Gao SY, Wang JC, Chen CJ, Zhao EY, Hou WJ, et al. Self-assembly nanomicelles based on cationic mpeg-pla-b-polyarginine(r15) triblock copolymer for sirna delivery. Biomaterials. 2012;33:6793–6807. doi: 10.1016/j.biomaterials.2012.05.067. [DOI] [PubMed] [Google Scholar]
- 122.Dimitrov DS. Virus entry: Molecular mechanisms and biomedical applications. Nat Rev Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Givens BE, Naguib YW, Geary SM, Devor EJ, Salem AK. Nanoparticle-based delivery of crispr/cas9 genome-editing therapeutics. AAPS J. 2018;20:108. doi: 10.1208/s12248-018-0267-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the crispr-cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using crispr-cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, et al. Somatic crispr/cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015;6:7391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mali P, Yang L, Esvelt KM, Aach J, Guell M, Dicarlo JE, et al. Rna-guided human genome engineering via cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using staphylococcus aureus cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, et al. Genome-wide crispr screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, et al. Efficient genome modification by crispr-cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 131.Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, et al. Genome editing with rna-guided cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, et al. Generation of gene-modified mice via cas9/rna-mediated gene targeting. Cell Res. 2013;23:720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M, et al. Rapid and highly efficient mammalian cell engineering via cas9 protein transfection. J Biotechnol. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 134.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient rna-guided genome editing in human cells via delivery of purified cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, et al. Chemically modified guide rnas enhance crispr-cas genome editing in human primary cells. Nat Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, et al. DNA-free genome editing in plants with preassembled crispr-cas9 ribonucleoproteins. Nat Biotechnol. 2015;33:1162–1164. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- 137.Crudele JM, Chamberlain JS. Cas9 immunity creates challenges for crispr gene editing therapies. Nat Commun. 2018;9:3497. doi: 10.1038/s41467-018-05843-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang S, Shen J, Li D, Cheng Y. Strategies in the delivery of cas9 ribonucleoprotein for crispr/cas9 genome editing. Theranostics. 2021;11:614–648. doi: 10.7150/thno.47007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lee B, Lee K, Panda S, Gonzales-Rojas R, Chong A, Bugay V, et al. Nanoparticle delivery of crispr into the brain rescues a mouse model of fragile x syndrome from exaggerated repetitive behaviours. Nat Biomed Eng. 2018;2:497–507. doi: 10.1038/s41551-018-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wei T, Cheng Q, Farbiak L, Anderson DG, Langer R, Siegwart DJ. Delivery of tissue-targeted scalpels: Opportunities and challenges for in vivo crispr/cas-based genome editing. ACS Nano. 2020;14:9243–9262. doi: 10.1021/acsnano.0c04707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wei T, Cheng Q, Min YL, Olson EN, Siegwart DJ. Systemic nanoparticle delivery of crispr-cas9 ribonucleoproteins for effective tissue specific genome editing. Nat Commun. 2020;11:3232. doi: 10.1038/s41467-020-17029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Riley RS, June CH, Langer R, Mitchell MJ. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discov. 2019;18:175–196. doi: 10.1038/s41573-018-0006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Han JP, Kim M, Choi BS, Lee JH, Lee GS, Jeong M, et al. In vivo delivery of crispr-cas9 using lipid nanoparticles enables antithrombin gene editing for sustainable hemophilia a and b therapy. Sci Adv. 2022;8:6901. doi: 10.1126/sciadv.abj6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Miao L, Zhang Y, Huang L. Mrna vaccine for cancer immunotherapy. Mol Cancer. 2021;20:41. doi: 10.1186/s12943-021-01335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cheng X, Lee RJ. The role of helper lipids in lipid nanoparticles (lnps) designed for oligonucleotide delivery. Adv Drug Deliv Rev. 2016;99:129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 146.Kowalski PS, Rudra A, Miao L, Anderson DG. Delivering the messenger: advances in technologies for therapeutic mrna delivery. Mol Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Liu Q, Chen F, Hou L, Shen L, Zhang X, Wang D, et al. Nanocarrier-mediated chemo-immunotherapy arrested cancer progression and induced tumor dormancy in desmoplastic melanoma. ACS Nano. 2018;12:7812–7825. doi: 10.1021/acsnano.8b01890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN, et al. Targeted delivery of rnai therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fenton OS, Olafson KN, Pillai PS, Mitchell MJ, Langer R. Advances in biomaterials for drug delivery. Adv Mater. 2018;20:e1705328. doi: 10.1002/adma.201705328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fonseca-Santos B, Gremiao MP, Chorilli M. Nanotechnology-based drug delivery systems for the treatment of alzheimer's disease. Int J Nanomedicine. 2015;10:4981–5003. doi: 10.2147/IJN.S87148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sercombe L, Veerati T, Moheimani F, Wu SY, Sood AK, Hua S. Advances and challenges of liposome assisted drug delivery. Front Pharmacol. 2015;6:286. doi: 10.3389/fphar.2015.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gillmore JD, Gane E, Taubel J, Kao J, Fontana M, Maitland ML, et al. Crispr-cas9 in vivo gene editing for transthyretin amyloidosis. N Engl J Med. 2021;385:493–502. doi: 10.1056/NEJMoa2107454. [DOI] [PubMed] [Google Scholar]
- 153.Qiu M, Glass Z, Chen J, Haas M, Jin X, Zhao X, et al. Lipid nanoparticle-mediated codelivery of cas9 mrna and single-guide rna achieves liver-specific in vivo genome editing of angptl3. Proc Natl Acad Sci USA. 2021;118:e2020401118. doi: 10.1073/pnas.2020401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Akinc A, Maier MA, Manoharan M, Fitzgerald K, Jayaraman M, Barros S, et al. The onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 155.Bottger R, Pauli G, Chao PH, Al Fayez N, Hohenwarter L, Li SD. Lipid-based nanoparticle technologies for liver targeting. Adv Drug Deliv Rev. 2020;154–155:79–101. doi: 10.1016/j.addr.2020.06.017. [DOI] [PubMed] [Google Scholar]
- 156.Kenjo E, Hozumi H, Makita Y, Iwabuchi KA, Fujimoto N, Matsumoto S, et al. Low immunogenicity of lnp allows repeated administrations of crispr-cas9 mrna into skeletal muscle in mice. Nat Commun. 2021;12:7101. doi: 10.1038/s41467-021-26714-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA, Siegwart DJ. Selective organ targeting (sort) nanoparticles for tissue-specific mrna delivery and crispr-cas gene editing. Nat Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cui Z, Zeng C, Huang F, Yuan F, Yan J, Zhao Y, et al. Cas13d knockdown of lung protease ctsl prevents and treats sars-cov-2 infection. Nat Chem Biol. 2022;18:1056–1064. doi: 10.1038/s41589-022-01094-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of crispr/cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zou L, Chen F, Bao J, Wang S, Wang L, Chen M, et al. Preparation, characterization, and anticancer efficacy of evodiamine-loaded plga nanoparticles. Drug Deliv. 2016;23:908–916. doi: 10.3109/10717544.2014.920936. [DOI] [PubMed] [Google Scholar]
- 161.Wang Y, Ma B, Abdeen AA, Chen G, Xie R, Saha K, et al. Versatile redox-responsive polyplexes for the delivery of plasmid DNA, messenger rna, and crispr-cas9 genome-editing machinery. ACS Appl Mater Interfaces. 2018;10:31915–31927. doi: 10.1021/acsami.8b09642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen F, Zhang J, Wang L, Wang Y, Chen M. Tumor ph(e)-triggered charge-reversal and redox-responsive nanoparticles for docetaxel delivery in hepatocellular carcinoma treatment. Nanoscale. 2015;7:15763–15779. doi: 10.1039/C5NR04612B. [DOI] [PubMed] [Google Scholar]
- 163.Ryu N, Kim MA, Park D, Lee B, Kim YR, Kim KH, et al. Effective pei-mediated delivery of crispr-cas9 complex for targeted gene therapy. Nanomedicine. 2018;14:2095–2102. doi: 10.1016/j.nano.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 164.Liu Y, Cao ZT, Xu CF, Lu ZD, Luo YL, Wang J. Optimization of lipid-assisted nanoparticle for disturbing neutrophils-related inflammation. Biomaterials. 2018;172:92–104. doi: 10.1016/j.biomaterials.2018.04.052. [DOI] [PubMed] [Google Scholar]
- 165.Luo YL, Xu CF, Li HJ, Cao ZT, Liu J, Wang JL, et al. Macrophage-specific in vivo gene editing using cationic lipid-assisted polymeric nanoparticles. ACS Nano. 2018;12:994–1005. doi: 10.1021/acsnano.7b07874. [DOI] [PubMed] [Google Scholar]
- 166.Zhang Y, Shen S, Zhao G, Xu CF, Zhang HB, Luo YL, et al. In situ repurposing of dendritic cells with crispr/cas9-based nanomedicine to induce transplant tolerance. Biomaterials. 2019;217:119302. doi: 10.1016/j.biomaterials.2019.119302. [DOI] [PubMed] [Google Scholar]
- 167.Zhang X, Jin H, Huang X, Chaurasiya B, Dong D, Shanley TP, et al. Robust genome editing in adult vascular endothelium by nanoparticle delivery of crispr-cas9 plasmid DNA. Cell Rep. 2022;38:110196. doi: 10.1016/j.celrep.2021.110196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu Q, Zhao K, Wang C, Zhang Z, Zheng C, Zhao Y, et al. Multistage delivery nanoparticle facilitates efficient crispr/dcas9 activation and tumor growth suppression in vivo. Adv Sci. 2019;6:1801423. doi: 10.1002/advs.201801423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li Q, Lv X, Tang C, Yin C. Co-delivery of doxorubicin and crispr/cas9 or rnai-expressing plasmid by chitosan-based nanoparticle for cancer therapy. Carbohydr Polym. 2022;287:119315. doi: 10.1016/j.carbpol.2022.119315. [DOI] [PubMed] [Google Scholar]
- 170.Wan T, Pan Q, Ping Y. Microneedle-assisted genome editing: a transdermal strategy of targeting nlrp3 by crispr-cas9 for synergistic therapy of inflammatory skin disorders. Sci Adv. 2021;7:eabe2888. doi: 10.1126/sciadv.abe2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Farbiak L, Cheng Q, Wei T, Alvarez-Benedicto E, Johnson LT, Lee S, et al. All-in-one dendrimer-based lipid nanoparticles enable precise hdr-mediated gene editing in vivo. Adv Mater. 2021;33:e2006619. doi: 10.1002/adma.202006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Park J, Fong PM, Lu J, Russell KS, Booth CJ, Saltzman WM, et al. Pegylated plga nanoparticles for the improved delivery of doxorubicin. Nanomedicine. 2009;5:410–418. doi: 10.1016/j.nano.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Faure AC, Dufort S, Josserand V, Perriat P, Coll JL, Roux S, et al. Control of the in vivo biodistribution of hybrid nanoparticles with different poly(ethylene glycol) coatings. Small. 2009;5:2565–2575. doi: 10.1002/smll.200900563. [DOI] [PubMed] [Google Scholar]
- 174.Rao NV, Ko H, Lee J, Park JH. Recent progress and advances in stimuli-responsive polymers for cancer therapy. Front Bioeng Biotechnol. 2018;6:110. doi: 10.3389/fbioe.2018.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang S, Liu Q, Li L, Urban MW. Recent advances in stimuli-responsive commodity polymers. Macromol Rapid Commun. 2021;42:e2100054. doi: 10.1002/marc.202100054. [DOI] [PubMed] [Google Scholar]
- 176.Li L, Yang Z, Zhu S, He L, Fan W, Tang W, et al. A rationally designed semiconducting polymer brush for nir-ii imaging-guided light-triggered remote control of crispr/cas9 genome editing. Adv Mater. 2019;31:e1901187. doi: 10.1002/adma.201901187. [DOI] [PubMed] [Google Scholar]
- 177.Xie R, Wang X, Wang Y, Ye M, Zhao Y, Yandell BS, et al. Ph-responsive polymer nanoparticles for efficient delivery of cas9 ribonucleoprotein with or without donor DNA. Adv Mater. 2022;34:e2110618. doi: 10.1002/adma.202110618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Luther DC, Huang R, Jeon T, Zhang X, Lee YW, Nagaraj H, et al. Delivery of drugs, proteins, and nucleic acids using inorganic nanoparticles. Adv Drug Deliv Rev. 2020;156:188–213. doi: 10.1016/j.addr.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Goddard ZR, Marin MJ, Russell DA, Searcey M. Active targeting of gold nanoparticles as cancer therapeutics. Chem Soc Rev. 2020;49:8774–8789. doi: 10.1039/D0CS01121E. [DOI] [PubMed] [Google Scholar]
- 180.Nihongaki Y, Kawano F, Nakajima T, Sato M. Photoactivatable crispr-cas9 for optogenetic genome editing. Nat Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 181.Wirth J, Garwe F, Meyer R, Csaki A, Stranik O, Fritzsche W. Plasmonically enhanced electron escape from gold nanoparticles and their polarization-dependent excitation transfer along DNA nanowires. Nano Lett. 2014;14:3809–3816. doi: 10.1021/nl5009184. [DOI] [PubMed] [Google Scholar]
- 182.Zhang Z, Wang L, Wang J, Jiang X, Li X, Hu Z, et al. Mesoporous silica-coated gold nanorods as a light-mediated multifunctional theranostic platform for cancer treatment. Adv Mater. 2012;24:1418–1423. doi: 10.1002/adma.201104714. [DOI] [PubMed] [Google Scholar]
- 183.Chen X, Chen Y, Xin H, Wan T, Ping Y. Near-infrared optogenetic engineering of photothermal nanocrispr for programmable genome editing. Proc Natl Acad Sci USA. 2020;117:2395–2405. doi: 10.1073/pnas.1912220117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Peng H, Le C, Wu J, Li XF, Zhang H, Le XC. A genome-editing nanomachine constructed with a clustered regularly interspaced short palindromic repeats system and activated by near-infrared illumination. ACS Nano. 2020;14:2817–2826. doi: 10.1021/acsnano.9b05276. [DOI] [PubMed] [Google Scholar]
- 185.Huang L, Zhou M, Abbas G, Li C, Cui M, Zhang XE, et al. A cancer cell membrane-derived biomimetic nanocarrier for synergistic photothermal/gene therapy by efficient delivery of crispr/cas9 and gold nanorods. Adv Healthc Mater. 2022;11:e2201038. doi: 10.1002/adhm.202201038. [DOI] [PubMed] [Google Scholar]
- 186.Ma L, Yin L, Li X, Chen S, Peng L, Liu G, et al. A smartphone-based visual biosensor for crispr-cas powered sars-cov-2 diagnostics. Biosens Bioelectron. 2022;195:113646. doi: 10.1016/j.bios.2021.113646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lopez-Valls M, Escalona-Noguero C, Rodriguez-Diaz C, Pardo D, Castellanos M, Milan-Rois P, et al. Cascade: Naked eye-detection of sars-cov-2 using cas13a and gold nanoparticles. Anal Chim Acta. 2022;1205:339749. doi: 10.1016/j.aca.2022.339749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang WS, Pan J, Li F, Zhu M, Xu M, Zhu H, et al. Reverse transcription recombinase polymerase amplification coupled with crispr-cas12a for facile and highly sensitive colorimetric sars-cov-2 detection. Anal Chem. 2021;93:4126–4133. doi: 10.1021/acs.analchem.1c00013. [DOI] [PubMed] [Google Scholar]
- 189.Fan M, Han Y, Gao S, Yan H, Cao L, Li Z, et al. Ultrasmall gold nanoparticles in cancer diagnosis and therapy. Theranostics. 2020;10:4944–4957. doi: 10.7150/thno.42471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhang P, Guo Z, Ullah S, Melagraki G, Afantitis A, Lynch I. Nanotechnology and artificial intelligence to enable sustainable and precision agriculture. Nat Plants. 2021;7:864–876. doi: 10.1038/s41477-021-00946-6. [DOI] [PubMed] [Google Scholar]
- 191.Wu D, Zhou J, Creyer MN, Yim W, Chen Z, Messersmith PB, et al. Phenolic-enabled nanotechnology: Versatile particle engineering for biomedicine. Chem Soc Rev. 2021;50:4432–4483. doi: 10.1039/D0CS00908C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mousavi SM, Hashemi SA, Ghasemi Y, Atapour A, Amani AM, Savar Dashtaki A, et al. Green synthesis of silver nanoparticles toward bio and medical applications: review study. Artif Cells Nanomed Biotechnol. 2018;46:S855–S872. doi: 10.1080/21691401.2018.1517769. [DOI] [PubMed] [Google Scholar]
- 193.Zhang D, Ma XL, Gu Y, Huang H, Zhang GW. Green synthesis of metallic nanoparticles and their potential applications to treat cancer. Front Chem. 2020;8:799. doi: 10.3389/fchem.2020.00799. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 194.Alnadhari S, Al-Enazi NM, Alshehrei F, Ameen F. A review on biogenic synthesis of metal nanoparticles using marine algae and its applications. Environ Res. 2021;194:110672. doi: 10.1016/j.envres.2020.110672. [DOI] [PubMed] [Google Scholar]
- 195.Fawcett D, Verduin JJ, Shah M, Sharma SB, Poinern GEJ. A review of current research into the biogenic synthesis of metal and metal oxide nanoparticles via marine algae and seagrasses. J Nanosci. 2017;2017:8013850. [Google Scholar]
- 196.Jacob JM, Ravindran R, Narayanan M, Samuel SM, Pugazhendhi A, Kumar G. Microalgae: a prospective low cost green alternative for nanoparticle synthesis. Curr Opin Environ Sci Health. 2021;20:100163. doi: 10.1016/j.coesh.2019.12.005. [DOI] [Google Scholar]
- 197.Pal S, Tak YK, Song JM. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the gram-negative bacterium escherichia coli. Appl Environ Microbiol. 2007;73:1712–1720. doi: 10.1128/AEM.02218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Makarov VV, Love AJ, Sinitsyna OV, Makarova SS, Yaminsky IV, Taliansky ME, et al. "Green" nanotechnologies: Synthesis of metal nanoparticles using plants. Acta Naturae. 2014;6:35–44. doi: 10.32607/20758251-2014-6-1-35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tran TV, Nguyen DTC, Kumar PS, Din ATM, Jalil AA, Vo DN. Green synthesis of zro2 nanoparticles and nanocomposites for biomedical and environmental applications: a review. Environ Chem Lett. 2022;20:1309–1331. doi: 10.1007/s10311-021-01367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mohd Yusof H, Mohamad R, Zaidan UH, Abdul Rahman NA. Microbial synthesis of zinc oxide nanoparticles and their potential application as an antimicrobial agent and a feed supplement in animal industry: a review. J Anim Sci Biotechnol. 2019;10:57. doi: 10.1186/s40104-019-0368-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hulkoti NI, Taranath TC. Biosynthesis of nanoparticles using microbes-a review. Colloids Surf B Biointerfaces. 2014;121:474–483. doi: 10.1016/j.colsurfb.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 202.Dahoumane SA, Mechouet M, Wijesekera K, Filipe CDM, Sicard C, Bazylinski DA, et al. Algae-mediated biosynthesis of inorganic nanomaterials as a promising route in nanobiotechnology-a review. Green Chem. 2017;19:552–587. doi: 10.1039/C6GC02346K. [DOI] [Google Scholar]
- 203.Rahman A, Kumar S, Bafana A, Lin J, Dahoumane SA, Jeffryes C. A mechanistic view of the light-induced synthesis of silver nanoparticles using extracellular polymeric substances of chlamydomonas reinhardtii. Molecules. 2019;24:3506. doi: 10.3390/molecules24193506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhang ZW, Chen J, Yang QL, Lan K, Yan ZY, Chen JQ. Eco-friendly intracellular microalgae synthesis of fluorescent cdse qds as a sensitive nanoprobe for determination of imatinib. Sensor Actuat B-Chem. 2018;263:625–633. doi: 10.1016/j.snb.2018.02.169. [DOI] [Google Scholar]
- 205.Gahlawat G, Choudhury AR. A review on the biosynthesis of metal and metal salt nanoparticles by microbes. RSC Adv. 2019;9:12944–12967. doi: 10.1039/C8RA10483B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Patil MP, Kim GD. Marine microorganisms for synthesis of metallic nanoparticles and their biomedical applications. Colloids Surf B Biointerfaces. 2018;172:487–495. doi: 10.1016/j.colsurfb.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 207.Sharma A, Sharma S, Sharma K, Chetri SPK, Vashishtha A, Singh P, et al. Algae as crucial organisms in advancing nanotechnology: a systematic review. J Appl Phycol. 2016;28:1759–1774. doi: 10.1007/s10811-015-0715-1. [DOI] [Google Scholar]
- 208.Prasad TNVKV, Kambala VSR, Naidu R. Phyconanotechnology: synthesis of silver nanoparticles using brown marine algae cystophora moniliformis and their characterisation. J Appl Phycol. 2013;25:177–182. doi: 10.1007/s10811-012-9851-z. [DOI] [Google Scholar]
- 209.Siddiqi KS, Husen A. Fabrication of metal and metal oxide nanoparticles by algae and their toxic effects. Nanoscale Res Lett. 2016;11:363. doi: 10.1186/s11671-016-1580-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Alijani HQ, Pourseyedi S, Torkzadeh Mahani M, Khatami M. Green synthesis of zinc sulfide (zns) nanoparticles using stevia rebaudiana bertoni and evaluation of its cytotoxic properties. J Mol Struct. 2019;1175:214–218. doi: 10.1016/j.molstruc.2018.07.103. [DOI] [Google Scholar]
- 211.Yun YH, Lee BK, Park K. Controlled drug delivery: Historical perspective for the next generation. J Control Release. 2015;219:2–7. doi: 10.1016/j.jconrel.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bozzuto G, Molinari A. Liposomes as nanomedical devices. Int J Nanomedicine. 2015;10:975–999. doi: 10.2147/IJN.S68861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Svenson S. The dendrimer paradox–high medical expectations but poor clinical translation. Chem Soc Rev. 2015;44:4131–4144. doi: 10.1039/C5CS00288E. [DOI] [PubMed] [Google Scholar]
- 214.Sun Y, Ma XL, Hu H. Marine polysaccharides as a versatile biomass for the construction of nano drug delivery systems. Mar Drugs. 2021;19:345. doi: 10.3390/md19060345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chen XY, Zhao X, Gao YY, Yin JQ, Bai MY, Wang FH. Green synthesis of gold nanoparticles using carrageenan oligosaccharide and their in vitro antitumor activity. Mar Drugs. 2018;16:277. doi: 10.3390/md16080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Manivasagan P, Oh J. Marine polysaccharide-based nanomaterials as a novel source of nanobiotechnological applications. Int J Biol Macromol. 2016;82:315–327. doi: 10.1016/j.ijbiomac.2015.10.081. [DOI] [PubMed] [Google Scholar]
- 217.Cardoso MJ, Costa RR, Mano JF. Marine origin polysaccharides in drug delivery systems. Mar Drugs. 2016;14:34. doi: 10.3390/md14020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Manivasagan P, Bharathiraja S, Bui NQ, Jang B, Oh YO, Lim IG, et al. Doxorubicin-loaded fucoidan capped gold nanoparticles for drug delivery and photoacoustic imaging. Int J Biol Macromol. 2016;91:578–588. doi: 10.1016/j.ijbiomac.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 219.Tomoaia G, Horovitz O, Mocanu A, Nita A, Avram A, Racz CP, et al. Effects of doxorubicin mediated by gold nanoparticles and resveratrol in two human cervical tumor cell lines. Colloids Surf B Biointerfaces. 2015;135:726–734. doi: 10.1016/j.colsurfb.2015.08.036. [DOI] [PubMed] [Google Scholar]
- 220.Venkatpurwar V, Shiras A, Pokharkar V. Porphyran capped gold nanoparticles as a novel carrier for delivery of anticancer drug: in vitro cytotoxicity study. Int J Pharm. 2011;409:314–320. doi: 10.1016/j.ijpharm.2011.02.054. [DOI] [PubMed] [Google Scholar]
- 221.Chen X, Han W, Zhao X, Tang W, Wang F. Epirubicin-loaded marine carrageenan oligosaccharide capped gold nanoparticle system for ph-triggered anticancer drug release. Sci Rep. 2019;9:6754. doi: 10.1038/s41598-019-43106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Salem DS, Sliem MA, El-Sesy M, Shouman SA, Badr Y. Improved chemo-photothermal therapy of hepatocellular carcinoma using chitosan-coated gold nanoparticles. J Photochem Photobiol B. 2018;182:92–99. doi: 10.1016/j.jphotobiol.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 223.Manivasagan P, Bharathiraja S, Bui NQ, Lim IG, Oh J. Paclitaxel-loaded chitosan oligosaccharide-stabilized gold nanoparticles as novel agents for drug delivery and photoacoustic imaging of cancer cells. Int J Pharm. 2016;511:367–379. doi: 10.1016/j.ijpharm.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 224.George A, Shah PA, Shrivastav PS. Natural biodegradable polymers based nano-formulations for drug delivery: a review. Int J Pharm. 2019;561:244–264. doi: 10.1016/j.ijpharm.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 225.Hussein HA, Abdullah MA. Anticancer compounds derived from marine diatoms. Mar Drugs. 2020;18:356. doi: 10.3390/md18070356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tang F, Li L, Chen D. Mesoporous silica nanoparticles: synthesis, biocompatibility and drug delivery. Adv Mater. 2012;24:1504–1534. doi: 10.1002/adma.201104763. [DOI] [PubMed] [Google Scholar]
- 227.Uthappa UT, Brahmkhatri V, Sriram G, Jung HY, Yu J, Kurkuri N, et al. Nature engineered diatom biosilica as drug delivery systems. J Control Release. 2018;281:70–83. doi: 10.1016/j.jconrel.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 228.Delasoie J, Zobi F. Natural diatom biosilica as microshuttles in drug delivery systems. Pharmaceutics. 2019;11:537. doi: 10.3390/pharmaceutics11100537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Aw MS, Simovic S, Yu Y, Addai-Mensah J, Losic D. Porous silica microshells from diatoms as biocarrier for drug delivery applications. Powder Technol. 2012;223:52–58. doi: 10.1016/j.powtec.2011.04.023. [DOI] [Google Scholar]
- 230.Delasoie J, Rossier J, Haeni L, Rothen-Rutishauser B, Zobi F. Slow-targeted release of a ruthenium anticancer agent from vitamin b12 functionalized marine diatom microalgae. Dalton Trans. 2018;47:17221–17232. doi: 10.1039/C8DT02914H. [DOI] [PubMed] [Google Scholar]
- 231.Sasirekha R, Sheena TS, Sathiya Deepika M, Santhanam P, Townley HE, Jeganathan K, et al. Surface engineered amphora subtropica frustules using chitosan as a drug delivery platform for anticancer therapy. Mater Sci Eng C Mater Biol Appl. 2019;94:56–64. doi: 10.1016/j.msec.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 232.Fu P, Zhang J, Li H, Mak M, Xu W, Tao Z. Extracellular vesicles as delivery systems at nano-/micro-scale. Adv Drug Deliv Rev. 2021;179:113910. doi: 10.1016/j.addr.2021.113910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Paterna A, Rao E, Adamo G, Raccosta S, Picciotto S, Romancino D, et al. Isolation of extracellular vesicles from microalgae: a renewable and scalable bioprocess. Front Bioeng Biotechnol. 2022;10:836747. doi: 10.3389/fbioe.2022.836747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Adamo G, Fierli D, Romancino DP, Picciotto S, Barone ME, Aranyos A, et al. Nanoalgosomes: Introducing extracellular vesicles produced by microalgae. J Extracell Vesicles. 2021;10:e12081. doi: 10.1002/jev2.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zhang F, Zhuang J, Li Z, Gong H, De Avila BE, Duan Y, et al. Nanoparticle-modified microrobots for in vivo antibiotic delivery to treat acute bacterial pneumonia. Nat Mater. 2022;21:1324–1332. doi: 10.1038/s41563-022-01360-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sun L, Yu Y, Chen Z, Bian F, Ye F, Sun L, et al. Biohybrid robotics with living cell actuation. Chem Soc Rev. 2020;49:4043–4069. doi: 10.1039/D0CS00120A. [DOI] [PubMed] [Google Scholar]
- 237.Chen QW, Qiao JY, Liu XH, Zhang C, Zhang XZ. Customized materials-assisted microorganisms in tumor therapeutics. Chem Soc Rev. 2021;50:12576–12615. doi: 10.1039/D0CS01571G. [DOI] [PubMed] [Google Scholar]
- 238.Xin H, Zhao N, Wang Y, Zhao X, Pan T, Shi Y, et al. Optically controlled living micromotors for the manipulation and disruption of biological targets. Nano Lett. 2020;20:7177–7185. doi: 10.1021/acs.nanolett.0c02501. [DOI] [PubMed] [Google Scholar]
- 239.Dawiec-Lisniewska A, Podstawczyk D, Bastrzyk A, Czuba K, Pacyna-Iwanicka K, Okoro OV, et al. New trends in biotechnological applications of photosynthetic microorganisms. Biotechnol Adv. 2022;59:107988. doi: 10.1016/j.biotechadv.2022.107988. [DOI] [PubMed] [Google Scholar]
- 240.Zhang D, Zhong D, Ouyang J, He J, Qi Y, Chen W, et al. Microalgae-based oral microcarriers for gut microbiota homeostasis and intestinal protection in cancer radiotherapy. Nat Commun. 2022;13:1413. doi: 10.1038/s41467-022-28744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Li M, Wu J, Lin D, Yang J, Jiao N, Wang Y, et al. A diatom-based biohybrid microrobot with a high drug-loading capacity and ph-sensitive drug release for target therapy. Acta Biomater. 2022;154:443–453. doi: 10.1016/j.actbio.2022.10.019. [DOI] [PubMed] [Google Scholar]
- 242.Liu L, Wu J, Chen B, Gao J, Li T, Ye Y, et al. Magnetically actuated biohybrid microswimmers for precise photothermal muscle contraction. ACS Nano. 2022;16:6515–6526. doi: 10.1021/acsnano.2c00833. [DOI] [PubMed] [Google Scholar]
- 243.Yan X, Zhou Q, Vincent M, Deng Y, Yu J, Xu J, et al. Multifunctional biohybrid magnetite microrobots for imaging-guided therapy. Sci Robot. 2017;2:eaaq155. doi: 10.1126/scirobotics.aaq1155. [DOI] [PubMed] [Google Scholar]
- 244.Yasa O, Erkoc P, Alapan Y, Sitti M. Microalga-powered microswimmers toward active cargo delivery. Adv Mater. 2018;30:e1804130. doi: 10.1002/adma.201804130. [DOI] [PubMed] [Google Scholar]
- 245.Zhong DN, Li WL, Qi YC, He J, Zhou M. Photosynthetic biohybrid nanoswimmers system to alleviate tumor hypoxia for fl/pa/mr imaging-guided enhanced radio-photodynamic synergetic therapy. Adv Funct Mater. 2020;30:1910395. doi: 10.1002/adfm.201910395. [DOI] [Google Scholar]
- 246.Gong D, Celi N, Zhang D, Cai J. Magnetic biohybrid microrobot multimers based on chlorella cells for enhanced targeted drug delivery. ACS Appl Mater Interfaces. 2022;14:6320–6330. doi: 10.1021/acsami.1c16859. [DOI] [PubMed] [Google Scholar]
- 247.Akolpoglu MB, Dogan NO, Bozuyuk U, Ceylan H, Kizilel S, Sitti M. High-yield production of biohybrid microalgae for on-demand cargo delivery. Adv Sci. 2020;7:2001256. doi: 10.1002/advs.202001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Shchelik IS, Molino JVD, Gademann K. Biohybrid microswimmers against bacterial infections. Acta Biomater. 2021;136:99–110. doi: 10.1016/j.actbio.2021.09.048. [DOI] [PubMed] [Google Scholar]
- 249.Zhong D, Zhang D, Xie T, Zhou M. Biodegradable microalgae-based carriers for targeted delivery and imaging-guided therapy toward lung metastasis of breast cancer. Small. 2020;16:e2000819. doi: 10.1002/smll.202000819. [DOI] [PubMed] [Google Scholar]
- 250.Wang Y, Huang C, Zhao W. Recent advances of the biological and biomedical applications of crispr/cas systems. Mol Biol Rep. 2022;49:7087–7100. doi: 10.1007/s11033-022-07519-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Tan FHP, Nadir N, Sudesh K. Microalgal biomass as feedstock for bacterial production of pha: advances and future prospects. Front Bioeng Biotechnol. 2022;10:879476. doi: 10.3389/fbioe.2022.879476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Sreenikethanam A, Raj S, Gugulothu P, Bajhaiya AK. Genetic engineering of microalgae for secondary metabolite production: recent developments, challenges, and future prospects. Front Bioeng Biotechnol. 2022;10:836056. doi: 10.3389/fbioe.2022.836056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Schmidt TJN, Berarducci B, Konstantinidou S, Raffa V. Crispr/cas9 in the era of nanomedicine and synthetic biology. Drug Discov Today. 2023;28:103375. doi: 10.1016/j.drudis.2022.103375. [DOI] [PubMed] [Google Scholar]
- 254.Wang D, Li Y, Hu X, Su W, Zhong M. Combined enzymatic and mechanical cell disruption and lipid extraction of green alga neochloris oleoabundans. Int J Mol Sci. 2015;16:7707–7722. doi: 10.3390/ijms16047707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Ren X, Liu Y, Fan C, Hong H, Wu W, Zhang W, et al. Production, processing, and protection of microalgal n-3 pufa-rich oil. Foods. 2022;11:1215. doi: 10.3390/foods11091215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Guiheneuf F, Khan A, Tran LS. Genetic engineering: a promising tool to engender physiological, biochemical, and molecular stress resilience in green microalgae. Front Plant Sci. 2016;7:400. doi: 10.3389/fpls.2016.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen LL. Crispr-p 2.0: an improved crispr-cas9 tool for genome editing in plants. Mol Plant. 2017;10:530–532. doi: 10.1016/j.molp.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 258.Heigwer F, Kerr G, Boutros M. E-crisp: fast crispr target site identification. Nat Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 259.Abby SS, Neron B, Menager H, Touchon M, Rocha EP. Macsyfinder: a program to mine genomes for molecular systems with an application to crispr-cas systems. PLoS ONE. 2014;9:e110726. doi: 10.1371/journal.pone.0110726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Dasgupta I, Flotte TR, Keeler AM. Crispr/cas-dependent and nuclease-free in vivo therapeutic gene editing. Hum Gene Ther. 2021;32:275–293. doi: 10.1089/hum.2021.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Liu M, Rehman S, Tang X, Gu K, Fan Q, Chen D, et al. Methodologies for improving hdr efficiency. Front Genet. 2018;9:691. doi: 10.3389/fgene.2018.00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with crispr-cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Paulsen BS, Mandal PK, Frock RL, Boyraz B, Yadav R, Upadhyayula S, et al. Ectopic expression of rad52 and dn53bp1 improves homology-directed repair during crispr-cas9 genome editing. Nat Biomed Eng. 2017;1:878–888. doi: 10.1038/s41551-017-0145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ferrari S, Jacob A, Beretta S, Unali G, Albano L, Vavassori V, et al. Efficient gene editing of human long-term hematopoietic stem cells validated by clonal tracking. Nat Biotechnol. 2020;38:1298–1308. doi: 10.1038/s41587-020-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Renaud JB, Boix C, Charpentier M, De Cian A, Cochennec J, Duvernois-Berthet E, et al. Improved genome editing efficiency and flexibility using modified oligonucleotides with talen and crispr-cas9 nucleases. Cell Rep. 2016;14:2263–2272. doi: 10.1016/j.celrep.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 266.Ma M, Zhuang F, Hu X, Wang B, Wen XZ, Ji JF, et al. Efficient generation of mice carrying homozygous double-floxp alleles using the cas9-avidin/biotin-donor DNA system. Cell Res. 2017;27:578–581. doi: 10.1038/cr.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Khan MI, Shin JH, Kim JD. The promising future of microalgae: current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb Cell Fact. 2018;17:36. doi: 10.1186/s12934-018-0879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Deguchi M, Kane S, Potlakayala S, George H, Proano R, Sheri V, et al. Metabolic engineering strategies of industrial hemp (cannabis sativa l.): a brief review of the advances and challenges. Front Plant Sci. 2020;11:580621. doi: 10.3389/fpls.2020.580621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Yi Z, Xu M, Magnusdottir M, Zhang Y, Brynjolfsson S, Fu W. Photo-oxidative stress-driven mutagenesis and adaptive evolution on the marine diatom phaeodactylum tricornutum for enhanced carotenoid accumulation. Mar Drugs. 2015;13:6138–6151. doi: 10.3390/md13106138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Fu W, Nelson DR, Mystikou A, Daakour S, Salehi-Ashtiani K. Advances in microalgal research and engineering development. Curr Opin Biotechnol. 2019;59:157–164. doi: 10.1016/j.copbio.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 271.Rosales-Mendoza S, Solis-Andrade KI, Marquez-Escobar VA, Gonzalez-Ortega O, Banuelos-Hernandez B. Current advances in the algae-made biopharmaceuticals field. Expert Opin Biol Ther. 2020;20:751–766. doi: 10.1080/14712598.2020.1739643. [DOI] [PubMed] [Google Scholar]
- 272.Kesik-Brodacka M. Progress in biopharmaceutical development. Biotechnol Appl Biochem. 2018;65:306–322. doi: 10.1002/bab.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Lingg N, Zhang P, Song Z, Bardor M. The sweet tooth of biopharmaceuticals: importance of recombinant protein glycosylation analysis. Biotechnol J. 2012;7:1462–1472. doi: 10.1002/biot.201200078. [DOI] [PubMed] [Google Scholar]
- 274.Reddy KV, Yedery RD, Aranha C. Antimicrobial peptides: Premises and promises. Int J Antimicrob Agents. 2004;24:536–547. doi: 10.1016/j.ijantimicag.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 275.Griesbeck C, Kobl I, Heitzer M. Chlamydomonas reinhardtii: a protein expression system for pharmaceutical and biotechnological proteins. Mol Biotechnol. 2006;34:213–223. doi: 10.1385/MB:34:2:213. [DOI] [PubMed] [Google Scholar]
- 276.Walmsley AM, Arntzen CJ. Plants for delivery of edible vaccines. Curr Opin Biotechnol. 2000;11:126–129. doi: 10.1016/S0958-1669(00)00070-7. [DOI] [PubMed] [Google Scholar]
- 277.Lopez-Pacheco IY, Rodas-Zuluaga LI, Cuellar-Bermudez SP, Hidalgo-Vazquez E, Molina-Vazquez A, Araujo RG, et al. Revalorization of microalgae biomass for synergistic interaction and sustainable applications: Bioplastic generation. Mar Drugs. 2022;20:601. doi: 10.3390/md20100601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Marinescu M, Popa CV. Pyridine compounds with antimicrobial and antiviral activities. Int J Mol Sci. 2022;23:5659. doi: 10.3390/ijms23105659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lai CL, Lau JY, Wu PC, Ngan H, Chung HT, Mitchell SJ, et al. Recombinant interferon-alpha in inoperable hepatocellular carcinoma: a randomized controlled trial. Hepatology. 1993;17:389–394. doi: 10.1002/hep.1840170307. [DOI] [PubMed] [Google Scholar]
- 280.Li H, Liu Q, Cui K, Liu J, Ren Y, Shi D. Expression of biologically active human interferon alpha 2b in the milk of transgenic mice. Transgenic Res. 2013;22:169–178. doi: 10.1007/s11248-012-9623-1. [DOI] [PubMed] [Google Scholar]
- 281.Jarquin-Cordero M, Chavez MN, Centeno-Cerdas C, Bohne AV, Hopfner U, Machens HG, et al. Towards a biotechnological platform for the production of human pro-angiogenic growth factors in the green alga chlamydomonas reinhardtii. Appl Microbiol Biotechnol. 2020;104:725–739. doi: 10.1007/s00253-019-10267-6. [DOI] [PubMed] [Google Scholar]
- 282.Centeno-Cerdas C, Jarquin-Cordero M, Chavez MN, Hopfner U, Holmes C, Schmauss D, et al. Development of photosynthetic sutures for the local delivery of oxygen and recombinant growth factors in wounds. Acta Biomater. 2018;81:184–194. doi: 10.1016/j.actbio.2018.09.060. [DOI] [PubMed] [Google Scholar]
- 283.Chavez MN, Schenck TL, Hopfner U, Centeno-Cerdas C, Somlai-Schweiger I, Schwarz C, et al. Towards autotrophic tissue engineering: photosynthetic gene therapy for regeneration. Biomaterials. 2016;75:25–36. doi: 10.1016/j.biomaterials.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 284.Feng S, Feng W, Zhao L, Gu H, Li Q, Shi K, et al. Preparation of transgenic dunaliella salina for immunization against white spot syndrome virus in crayfish. Arch Virol. 2014;159:519–525. doi: 10.1007/s00705-013-1856-7. [DOI] [PubMed] [Google Scholar]
- 285.Hernandez-Ramirez J, Wong-Arce A, Gonzalez-Ortega O, Rosales-Mendoza S. Expression in algae of a chimeric protein carrying several epitopes from tumor associated antigens. Int J Biol Macromol. 2020;147:46–52. doi: 10.1016/j.ijbiomac.2019.12.250. [DOI] [PubMed] [Google Scholar]
- 286.Chia WY, Kok H, Chew KW, Low SS, Show PL. Can algae contribute to the war with covid-19? Bioengineered. 2021;12:1226–1237. doi: 10.1080/21655979.2021.1910432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Aurisicchio L, Peruzzi D, Koo G, Wei WZ, La Monica N, Ciliberto G. Immunogenicity and therapeutic efficacy of a dual-component genetic cancer vaccine cotargeting carcinoembryonic antigen and her2/neu in preclinical models. Hum Gene Ther. 2014;25:121–131. doi: 10.1089/hum.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Kufe DW. Muc1-c oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32:1073–1081. doi: 10.1038/onc.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Rafiq S, Purdon TJ, Daniyan AF, Koneru M, Dao T, Liu C, et al. Optimized t-cell receptor-mimic chimeric antigen receptor t cells directed toward the intracellular wilms tumor 1 antigen. Leukemia. 2017;31:1788–1797. doi: 10.1038/leu.2016.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Soysal SD, Muenst S, Kan-Mitchell J, Huarte E, Zhang X, Wilkinson-Ryan I, et al. Identification and translational validation of novel mammaglobin-a cd8 t cell epitopes. Breast Cancer Res Treat. 2014;147:527–537. doi: 10.1007/s10549-014-3129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Lin TW, Huang PH, Liao BH, Chao TL, Tsai YM, Chang SC, et al. Tag-free sars-cov-2 receptor binding domain (rbd), but not c-terminal tagged sars-cov-2 rbd, induces a rapid and potent neutralizing antibody response. Vaccines. 2022;10:1839. doi: 10.3390/vaccines10111839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Georgianna DR, Mayfield SP. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature. 2012;488:329–335. doi: 10.1038/nature11479. [DOI] [PubMed] [Google Scholar]
- 293.Gilmour DJ. Microalgae for biofuel production. Adv Appl Microbiol. 2019;109:1–30. doi: 10.1016/bs.aambs.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 294.Wen X, Du K, Wang Z, Peng X, Luo L, Tao H, et al. Effective cultivation of microalgae for biofuel production: a pilot-scale evaluation of a novel oleaginous microalga graesiella sp. Wbg-1. Biotechnol Biofuels. 2016;9:123. doi: 10.1186/s13068-016-0541-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Blatti JL, Beld J, Behnke CA, Mendez M, Mayfield SP, Burkart MD. Manipulating fatty acid biosynthesis in microalgae for biofuel through protein-protein interactions. PLoS ONE. 2012;7:e42949. doi: 10.1371/journal.pone.0042949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Frleta R, Popovic M, Smital T, Simat V. Comparison of growth and chemical profile of diatom skeletonema grevillei in bioreactor and incubation-shaking cabinet in two growth phases. Mar Drugs. 2022;20:697. doi: 10.3390/md20110697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Ward VCA, Rehmann L. Fast media optimization for mixotrophic cultivation of chlorella vulgaris. Sci Rep. 2019;9:19262. doi: 10.1038/s41598-019-55870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Luo L, Ren H, Pei X, Xie G, Xing D, Dai Y, et al. Simultaneous nutrition removal and high-efficiency biomass and lipid accumulation by microalgae using anaerobic digested effluent from cattle manure combined with municipal wastewater. Biotechnol Biofuels. 2019;12:218. doi: 10.1186/s13068-019-1553-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Goodman JM, Boone-Heinonen J, Richardson DM, Andrea SB, Messer LC. Analyzing policies through a dohad lens: What can we learn? Int J Environ Res Public Health. 2018;15:2906. doi: 10.3390/ijerph15122906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Andrade-Guel M, Cabello-Alvarado C, Bartolo-Perez P, Medellin-Banda DI, Avila-Orta CA, Cruz-Ortiz B, et al. Surface modification of tio(2)/zno nanoparticles by organic acids with enhanced methylene blue and rhodamine b dye adsorption properties. RSC Adv. 2022;12:28494–28504. doi: 10.1039/D2RA04961A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Forootanfar H, Rezaei S, Zeinvand-Lorestani H, Tahmasbi H, Mogharabi M, Ameri A, et al. Studies on the laccase-mediated decolorization, kinetic, and microtoxicity of some synthetic azo dyes. J Environ Health Sci Eng. 2016;14:7. doi: 10.1186/s40201-016-0248-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Srivastava A, Seo SH, Ko SR, Ahn CY, Oh HM. Bioflocculation in natural and engineered systems: Current perspectives. Crit Rev Biotechnol. 2018;38:1176–1194. doi: 10.1080/07388551.2018.1451984. [DOI] [PubMed] [Google Scholar]
- 303.Savchenko O, Xing J, Yang X, Gu Q, Shaheen M, Huang M, et al. Algal cell response to pulsed waved stimulation and its application to increase algal lipid production. Sci Rep. 2017;7:42003. doi: 10.1038/srep42003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Fuchs T, Arnold ND, Garbe D, Deimel S, Lorenzen J, Masri M, et al. A newly designed automatically controlled, sterilizable flat panel photobioreactor for axenic algae culture. Front Bioeng Biotechnol. 2021;9:697354. doi: 10.3389/fbioe.2021.697354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Ranjbar S, Malcata FX. Is genetic engineering a route to enhance microalgae-mediated bioremediation of heavy metal-containing effluents? Molecules. 2022;27:1473. doi: 10.3390/molecules27051473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Goveas LC, Nayak S, Vinayagam R, Loke Show P, Selvaraj R. Microalgal remediation and valorisation of polluted wastewaters for zero-carbon circular bioeconomy. Bioresour Technol. 2022;365:128169. doi: 10.1016/j.biortech.2022.128169. [DOI] [PubMed] [Google Scholar]
- 307.Gondi R, Kavitha S, Yukesh Kannah R, Parthiba Karthikeyan O, Kumar G, Kumar Tyagi V, et al. Algal-based system for removal of emerging pollutants from wastewater: a review. Bioresour Technol. 2022;344:126245. doi: 10.1016/j.biortech.2021.126245. [DOI] [PubMed] [Google Scholar]
- 308.Ravutsov M, Mitrev Y, Shestakova P, Lazarova H, Simeonov S, Popova M. Co(2) adsorption on modified mesoporous silicas: the role of the adsorption sites. Nanomaterials. 2021;11:2831. doi: 10.3390/nano11112831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Rekker S, Ives MC, Wade B, Webb L, Greig C. Measuring corporate paris compliance using a strict science-based approach. Nat Commun. 2022;13:4441. doi: 10.1038/s41467-022-31143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Chen Y, Xu C, Vaidyanathan S. Microalgae: a robust "green bio-bridge" between energy and environment. Crit Rev Biotechnol. 2018;38:351–368. doi: 10.1080/07388551.2017.1355774. [DOI] [PubMed] [Google Scholar]
- 311.Cheah WY, Show PL, Chang JS, Ling TC, Juan JC. Biosequestration of atmospheric co2 and flue gas-containing co2 by microalgae. Bioresour Technol. 2015;184:190–201. doi: 10.1016/j.biortech.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 312.Li L, Huang J, Almutairi AW, Lan X, Zheng L, Lin Y, et al. Integrated approach for enhanced bio-oil recovery from disposed face masks through co-hydrothermal liquefaction with spirulina platensis grown in wastewater. Biomass Convers Biorefin. 2021;25:1–12. doi: 10.1007/s13399-021-01891-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Solovchenko AKhozin-Goldberg I. High-co2 tolerance in microalgae: possible mechanisms and implications for biotechnology and bioremediation. Biotechnol Lett. 2013;35:1745–1752. doi: 10.1007/s10529-013-1274-7. [DOI] [PubMed] [Google Scholar]
- 314.Choi KR, Jang WD, Yang D, Cho JS, Park D, Lee SY. Systems metabolic engineering strategies: Integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 2019;37:817–837. doi: 10.1016/j.tibtech.2019.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.