ABSTRACT
Ecuador had substantial COVID-19-mortality during 2020 despite early implementation of non-pharmaceutical interventions (NPIs). Resource-limited settings like Ecuador have high proportions of informal labour which entail high human mobility, questioning efficacy of NPIs. We performed a retrospective observational study in Ecuador’s national reference laboratory for viral respiratory infections during March 2020–February 2021 using stored respiratory specimens from 1950 patients, corresponding to 2.3% of all samples analysed within the Ecuadorian national surveillance system per week. During 2020, detection of SARS-CoV-2 (Pearson correlation; r = −0.74; p = 0.01) and other respiratory viruses (Pearson correlation; r = −0.68; p = 0.02) by real-time RT–PCR correlated negatively with NPIs stringency. Among respiratory viruses, adenoviruses (Fisher’s exact-test; p = 0.026), parainfluenzaviruses (p = 0.04), enteroviruses (p < 0.0001) and metapneumoviruses (p < 0.0001) occurred significantly more frequently during months of absent or non-stringent NPIs (characterized by <55% stringency according to the Oxford stringency index data for Ecuador). Phylogenomic analyses of 632 newly characterized SARS-CoV-2 genomes revealed 100 near-parallel SARS-CoV-2 introductions during early 2020 in the absence of NPIs. NPI stringency correlated negatively with the number of circulating SARS-CoV-2 lineages during 2020 (r = −0.69; p = 0.02). Phylogeographic reconstructions showed differential SARS-CoV-2 dispersion patterns during 2020, with more short-distance transitions potentially associated with recreational activity during non-stringent NPIs. There were also fewer geographic transitions during strict NPIs (n = 450) than during non-stringent or absent NPIs (n = 580). Virological evidence supports that NPIs had an effect on virus spread and distribution in Ecuador, providing a template for future epidemics in resource-limited settings and contributing to a balanced assessment of societal costs entailed by strict NPIs.
KEYWORDS: Non-pharmaceutical interventions, Ecuador, COVID-19, SARS-CoV-2, public health, respiratory infections
Introduction
Latin America is a COVID-19 hot spot, accumulating 20% of reported cases and 32% of deaths worldwide in 2020 [1], albeit hosting only 8% of the world’s population. Latin America and the Caribbean suffered substantial economic constrains due to the pandemic, exemplified by the decline of 9.4% in gross domestic product in 2020 (https://www.imf.org/en/Publications/WEO/Issues/2020/06/24/WEOUpdateJune2020). Most of the economic losses were associated with compulsory non-pharmaceutical interventions (NPIs), i.e. policies restricting human contact, movement, and shutting down public and private services to avoid SARS-CoV-2 transmission. However, in resource-limited settings like Latin America, informal labour exceeds 80%, leading to significant human mobility that could potentially undermine NPIs effectiveness (International Labour Organization; https://ilostat.ilo.org/data/). Even in affluent settings, the use of NPIs in containing COVID-19 mortality and SARS-CoV-2 spread is debated. On the one hand, early research in China and Brazil suggested that NPIs decreased SARS-CoV-2 transmission rates [2] and reproduction number [3], and a time series meta-analysis of 149 countries showed a decrease of COVID-19 incidence rates by 13% after NPIs implementation [4]. On the other hand, a modelling study analysing European countries found no efficacy of less stringent NPIs in lowering SARS-CoV-2 transmission apart from complete lockdown [5], and another study in Europe found that closure of businesses and stay-at-home orders were unlikely to lower COVID-19 incidence [6]. Within Latin America, Ecuador was one of the earliest epicentres of the pandemic, with one of the highest COVID-19-associated mortality rates worldwide reaching 8.5% in late 2020 [7] and an 80% increase in excess deaths of baseline annual mortality [7] despite strict and early implementation of NPIs. Here, we use molecular data gathered in Ecuador’s national reference laboratory to analyse the efficacy of NPIs in a prototypic resource-limited setting in Latin America.
Materials and methods
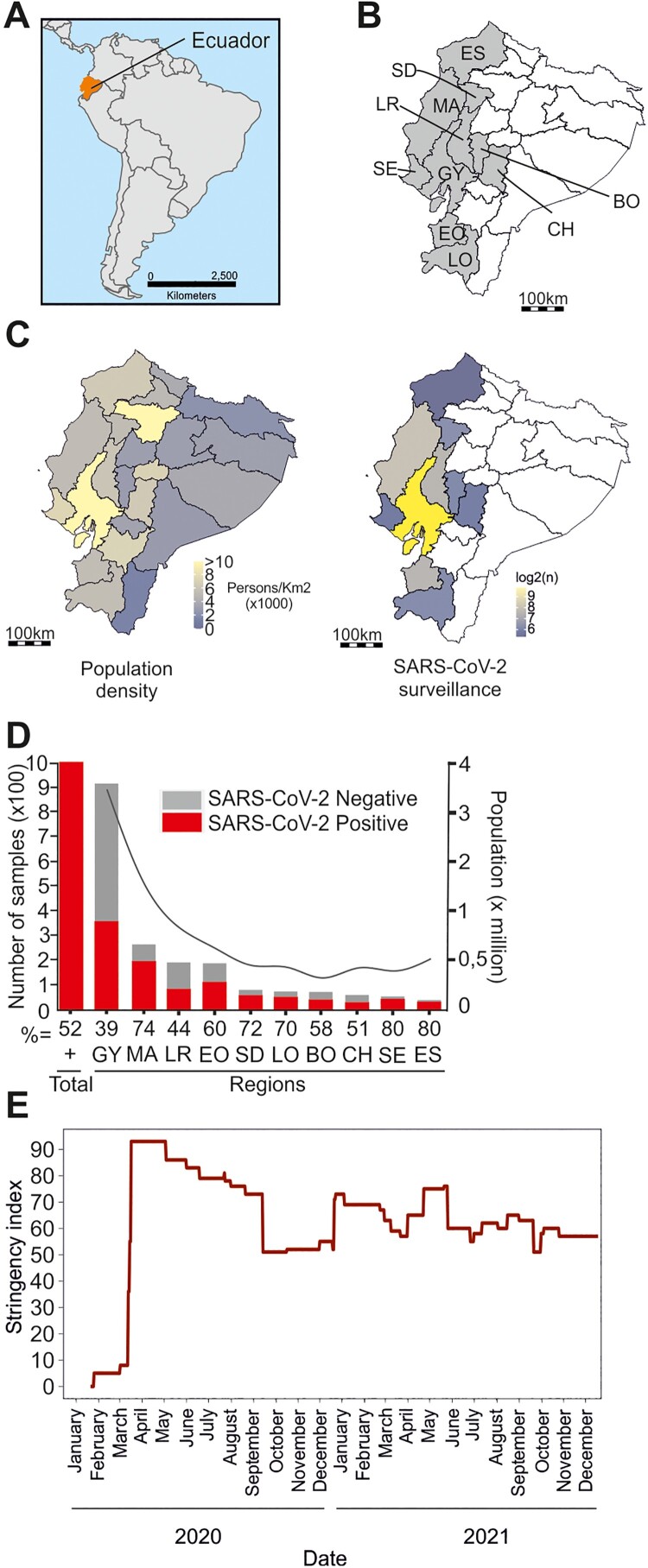
Study design. Samples originated mainly from coastal Ecuador, home to most of the Ecuadorian population (Figure 1(A–C)). The coastal region of Ecuador was selected due to severe early COVID-19 impact, mainly in the city of Guayaquil. This region is home to the Instituto Nacional de Investigación en Salud Pública Dr. Leopoldo Izquieta Pérez (INSPI), the Ecuadorian SARS-CoV-2 reference laboratory. The decision to focus on this region was also influenced by limited sample availability in other Ecuadorian laboratories. We used 1950 oro-nasopharyngeal swabs and sputum samples retrospectively collected during March 2020 to February 2021 from routine disease surveillance from the Ecuadorian national surveillance system (Figure 1(C)) right. The samples correspond to 2.3% of all the samples analysed by the INSPI per week (subsample data in Figures S1, S2 and in Table S1). The median age of the subpopulation was 40 years (interquartile range: 29–55), corresponding to 48.5% (946) female and 51.5% (1004) male patients (Figure S1 and Table S1). The number of samples gathered per province was comparable with the total population (Figure 1(D)).
Figure 1.
Study setting. (A) Map of South America. Ecuador, orange. (B) Sampled Ecuadorian provinces (gray). ES = Esmeraldas, MA = Manta, SD = Santo Domingo, LR = Los Rios, SE = Santa Elena, GY = Guayas, BO = Bolivar, CH = Chimborazo, EO = El Oro, LO = Loja. (C) Population density, Ecuador (left). Spatial distribution of samples retrospectively tested for SARS-CoV-2 (right). (D) Sample number (left axis) and population in millions (right axis) of sampled provinces. Abbreviations as in (B). (E) OxCGRT stringency index; Ecuador.
Laboratory analyses. Nucleic acid purification was performed using the MagNA Pure 96 DNA and Viral NA small-volume kit following the manufacturer’s instructions. (Roche, Penzberg, Germany). SARS-CoV-2 testing was performed using the SarbecoV E-gene and RdRP real time RT–PCR-based kits (TIBMolbiol, Germany) [8]. Multiplexed testing for common respiratory viruses was performed using multiplex real-time RT–PCR kits (TIB Molbiol) targeting the four endemic human coronaviruses (HCoV)-OC43, -NL63, -229E, -HKU1; Human adenoviruses, metapneumovirus, parechovirus, influenza A/B virus, respiratory syncytial viruses A/B, enteroviruses and parainfluenza viruses 1–4 (https://shop.tib-molbiol.de/cgi-bin/WebObjects/TIB-MOLBIOL.woa/5/wo/7JgBubYtXT9F5y0V0Qvnrw/17.0.0.11.1.13.9.1.5.7.1.9.20.1.1#target). Whole-genome amplification of SARS-CoV-2 was done using the Artic V3 PCR-based protocol (https://artic.network/ncov-2019). Library preparation was done using the KAPA Frag kit and KAPA Hyper Prep kit (Roche Molecular Diagnostics, Switzerland) and sequencing was done using MiSeq reagent v2 chemistry (Illumina, USA) according to the manufacturers’ protocols. Genome assembly was done by mapping MiSeq reads to the Wuhan-Hu-1 SARS-CoV-2 reference sequence (GenBank accession number: NC045512) using the Python-based CoVpipe pipeline (https://gitlab.com/RKIBioinformaticsPipelines/ncov_minipipe). COVID-19 lineage assignment was performed using a dynamic lineage classification method called Phylogenetic Assignment of Named Global Outbreak LINeages (PANGOLIN) version 3.1.16 [9].
Evolutionary analyses. Sequences were aligned using mafft v7.445 [10]. An approximately-maximum likelihood (ML) phylogeny encompassing all SARS-CoV-2 sequences from this study and all available Ecuadorian GISAID genomes until September 1, 2021 was reconstructed using the program fasttree 2.1.10 [11] with a GTR + CAT substitution model. To explore the temporal signal of SARS-CoV-2, an ancestral state inference in an ML framework was calculated in TreeTime 0.7.6 [12], using a fixed clock rate calculated previously for SARS-CoV-2 of 8 × 10−4 substitutions per site per year and a standard deviation of 4 × 10−4 (https://docs.nextstrain.org/projects/ncov/en/latest/reference/configuration.html;Nextstrain version:8fdf1932). Time-stamped phylogenies were estimated under a HKY + I nucleotide substitution model and a strict molecular clock in BEAST v.1.10.4 [3,13]. Phylogeographic inference was calculated using the relaxed random walk (RRW) diffusion model implemented in BEAST v1.10.4 [13] using a dataset comprising only SARS-CoV-2 B.1.1 Pango lineage sequences to limit biases from different transmission dynamics between lineages; and because B.1.1 was the only lineage detected throughout the whole study period. This model accommodates branch-specific variation in rates of dispersal with a Cauchy distribution. Briefly, latitude and longitude were attributed to a point within the patient’s province of residence. Each Markov chain was run for 1 × 108 generations and sampled every 1000 generations. Functions available in the R package “seraphim” [14] were used to extract spatio-temporal information embedded within the whole posterior trees dataset, after a 25% burn in and visualize the continuous phylogeographic reconstructions using three discrete variables: No NPIs, strict NPIs and relaxed NPIs. We calculated the 80% highest posterior density (HPD) interval and depicted the uncertainty of the phylogeographic estimates for each node. We also used the patristic wavefront distance, which calculates the sum of geographical distances associated with each branch connecting a given node to the root, also available in the R package “seraphim” [14].
Statistical analyses and modelling. Chi-square tests of proportions and Fisher’s exact tests were used to compare categorical variables, t-tests and Pearson correlation tests to compare continuous variables using R (v. 4.0.3). Kernel density estimations (KDE), used to visualize the distribution of continuous variables in time were performed in R (v. 4.0.3). KDE is employed to estimate the probability density function (PDF) of a random variable. Using a chosen kernel function centred at each data point over time, a smooth curve contributes to overall density estimation. Kernel width and shape are set by a bandwidth parameter. Summing up individual kernel curves yields the overall density estimate, representing the variable’s PDF. This non-parametric method provides a smooth distribution estimation. Georeferencing was performed in R (v. 4.0.3) using open source maps (https://www.diva-gis.org/gdata; https://gadm.org/; https://hub.worldpop.org/geodata/summary?id=46031; DOI:10.5258/SOTON/WP00675). The OxCGRT stringency index was gathered from Our World in Data [15] (Figure 1(E)). The OxCGRT stringency index was classified as a discrete variable for statistical analyses, months with >55% stringency classified as high, and <55% as low based on highest and lowest stringency during the year (highest 93 and lowest 50) (Figure 1(E)). Human mobility data from Ecuador was gathered from Google Community Mobility Reports (https://www.google.com/covid19/mobility/) which aggregates movement trends over time by geography, across categories of places such as retail and recreation, groceries and pharmacies, parks, transit stations, workplaces, and residential areas.
Results
Implementation of NPIs in Ecuador
On 29 February 2020, the Ministry of Public Health in Ecuador confirmed the first case of SARS-CoV-2 infection and implemented strict NPIs about 2 weeks later [16]. In general, the population adhered to the implemented NPIs due to the fear of COVID-19, shown as the culprit of the public health emergency with bodies lying openly in the streets [17]. The implemented NPIs increased in severity and included closing of national borders on March 16, followed by banning social gatherings, a curfew, closing of public spaces, commerce, schools, and stay-at-home orders by March 23, corresponding to a high NPIs stringency level of 93% according to the Oxford stringency index [15] (Figure 1(E)). The stringency index is the mean score of nine metrics: school closures; workplace closures; cancellation of public events; restrictions on public gatherings; closures of public transport; stay-at-home requirements; public information campaigns; restrictions on internal movements; and international travel controls. In Ecuador, the NPI stringency index decreased 3–7% per month until mid-September 2020, time in which Ecuador central government lifted stay-at-home orders, travel restrictions, permitted public gatherings, and gave political power to local governments to decide on NPIs. Afterwards, the stringency index was approximately 50% for the rest of the year (Figure 1(E)). Of note, the obligatory use of masks in indoor and outdoor spaces was implemented on April 6, 2020 and lifted only during April 2022.
Modified circulation of SARS-CoV-2 and other respiratory viruses
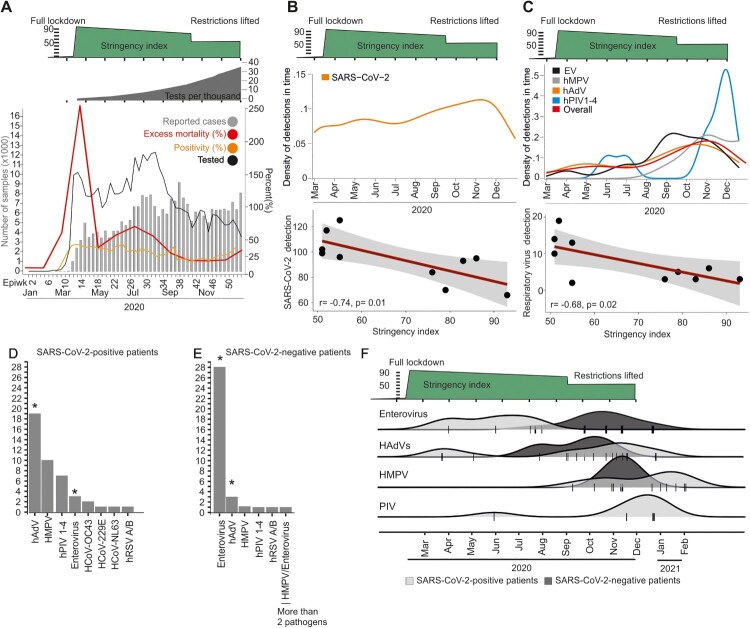
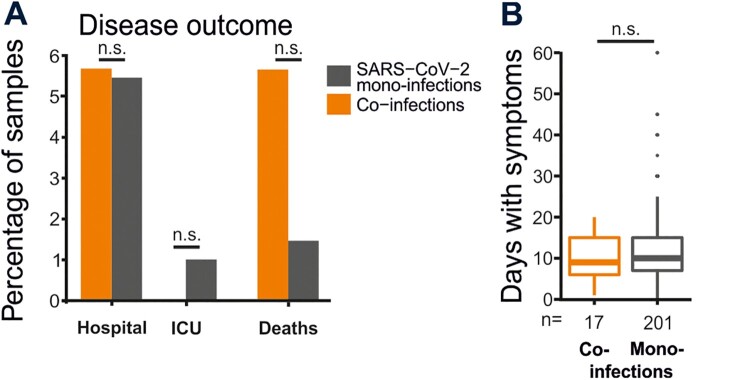
During the onset of the pandemic, Ecuador’s laboratory testing capacity was below all other South American countries, reaching 40 tests per 1000 inhabitants as of December, 2020 [1], and lack of reagents challenged homogenous testing over time (total number of tests performed in Ecuador is shown in dark grey in Figure 2(A)) (https://www.theguardian.com/world/live/2020/jun/12/coronavirus-live-news-markets-fall-over-fears-of-long-us-recovery-as-brazil-cases-top-800000). Therefore, we re-tested 1,950 samples stored at Ecuador’s reference laboratory continuously during the first year of the COVID-19 pandemic correspondint to a mean 47.8 samples per week (range: 27.5–60). In total, 52% (1017/1950) of those samples were SARS-CoV-2 positive (Figure 1(D)). The high RT–PCR detection rate is consistent with the Ecuadorian testing algorithm, focusing on symptomatic cases and contacts [18]. The SARS-CoV-2 RT–PCR detection per month correlated significantly with lower stringency of NPIs, implying increased virus circulation before NPIs were implemented and after NPIs were relaxed (Pearson correlation test; r = −0.74; p = 0.01) (Figure 2(B)). Consistent with those data relying on re-tested samples, the positivity rate reported from Ecuador was approximately 40% in March 2020, decreased subsequently and turned to increase to approximately 40% during late 2020, soon after NPIs were relaxed and following the reported increase of excess mortality (Figure 2(A), orange and red lines). The overall detection rate of all other respiratory viruses in our sample was low at 4.7% (91/1950; CI: 3.9–5.8) (Figure 2(C)). Among common respiratory viruses, adenoviruses (Fisher’s exact test; p = 0.026), parainfluenzaviruses (Fisher’s exact-test; p = 0.04), enteroviruses (Fisher’s exact; p < 0.0001) and metapneumoviruses (Fisher’s exact; p < 0.0001) occurred significantly more frequently during months of non-stringent NPIs (Figure 2(D,E)). Similar to SARS-CoV-2, the detection of those common respiratory viruses significantly correlated with less stringent NPIs (Pearson correlation; r = −0.68; p = 0.02) (Figure 2(C)), reminiscent of the reduced circulation of SARS-CoV-2 and suggesting modified respiratory virus circulation according to NPI stringency (Figure 2(F)). Therefore, there was reduced respiratory virus circulation in comparison to pre-pandemic detection rates in Ecuador [19]. Co-infection between SARS-CoV-2 and other respiratory viruses was detected in 4.4% of SARS-CoV-2 positive samples (44/1017; CI: 6.1–9.4), and only in a single additional sample by respiratory viruses other than SARS-CoV-2 (Figure 2(D,E)). The detected rates of co-infection were considerably lower than co-infection rates found in pre-pandemic studies from tropical settings, reaching more than 10% using similar methodology [20]. Overall decreased virus circulation was likely associated with stringent NPIs and included the complete absence of influenza viruses, consistent with a molecular study from the United Kingdom [21]. Finally, co-infections neither affected COVID-19 severity, nor duration (Figure 3(A,B)).
Figure 2.
Epidemiology of SARS-CoV-2 and other respiratory viruses, Ecuador. (A) Reported number of cases and all-cause excess mortality (www.salud.gob.ec), tested samples and positivity rate from Ecuador’s reference laboratory INSPI. Top: Total number of tests per thousand inhabitants (Our World in Data) and summarized Oxford stringency index. (B) Kernel density estimations of SARS-CoV-2 detections; correlation of NPI stringency index versus number of SARS-CoV-2 detections per month. (C) Kernel density estimations of other respiratory viruses; correlation of NPI stringency index versus number of respiratory virus detections per month. Viruses with less than two detections are not shown for graphical reasons. Human adenoviruses (hAdV), metapneumovirus (hMPV), enteroviruses (EV) and parainfluenza viruses 1–4 (hPIV 1–4). Epiwk: Epidemiological week. (D) and (E) Overall detection of other respiratory viruses. Asterisk denotes statistically significant difference between SARS-CoV-2 confirmed- and negative-patients (Fisher’s exact test; p < 0.01). Abbreviations as in (C). (F) Kernel Density Estimation (KDE) of individual pathogen detection in SARS-CoV-2-confirmed and -negative patients over time. Densities are stacked on top of each other for clarity of presentation. Abbreviations as in (C).
Figure 3.
COVID-19 course of SARS-CoV-2 co-infected versus mono-infected patients. (A) Percentage of co-infected and SARS-CoV-2 mono-infected patients that were hospitalized, admitted to the intensive care unit (ICU) and reported mortality. (B) Mean days with symptoms of co-infected and SARS-CoV-2 mono-infected patients (t-test with Bonferroni correction; p = 0.08). Dots represent outliers. N.S. not significant.
Travel restrictions were implemented too late to prevent parallel SARS-CoV-2 introductions
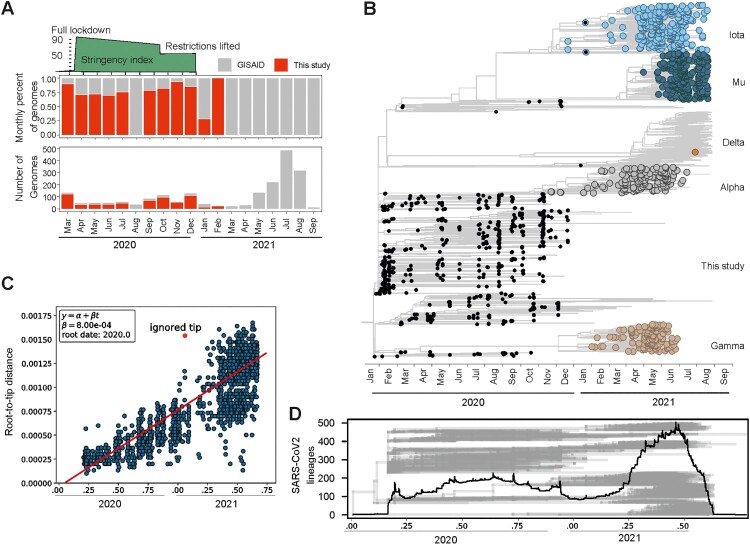
Since there is a lack of SARS-CoV-2 genomic data from the onset of the COVID-19 pandemic in Ecuador [22], we sequenced 62.1% (632/1017) of the SARS-CoV-2-positive samples during 2020, (Figure 4(A)) including the earliest SARS-CoV-2 genome in Ecuador to date, from the 16th of March 2020. We additionally downloaded 1431 complete, high-coverage and high-quality SARS-CoV-2 sequences from Ecuador in GISAID until collection date September 01, 2021. The full dataset comprised 2,063 sequences, 30% of which were newly generated for this study (Figure 4(A)). Notably, sequences from this study increased by more than 60% the early genomic information in 2020, leading to a more comprehensive dataset to assess the SARS-CoV-2 evolutionary dynamics (Figure 4(A)). Phylogenomic reconstructions using the complete dataset (Figure 4(B)) revealed that Ecuadorian SARS-CoV-2 sequences shared a common ancestor (TMRCA) projected to January 1st, 2020 in a root-to-tip regression analysis (Figure 4(C)). Additionally, the estimates of the SARS-CoV-2 TMRCA in Ecuador were projected within the tree to the 14th of January 2020 (median; 90% of the posterior probability distribution, December 2019 to March 2020) (Figure 4(B)). Those reconstructions were consistent with the likely time of emergence and rapid global spread of SARS-CoV-2 and with estimates from Brazil, the first country in Latin America detecting SARS-CoV-2 [3]. Close to 100 separate SARS-CoV-2 introduction events into Ecuador were suggested by detection of multiple closely related viruses at the end of January, 2020 (Figure 4(B,D)). Although Ecuador was among the first Latin American countries to implement travel restrictions by March 16, 2020, those NPIs were apparently implemented too late to prevent multiple parallel introduction events. For comparison, similarly high numbers of parallel SARS-CoV-2 introductions were reported during the onset of COVID-19 in Brazil [3].
Figure 4.
Evolution of SARS-CoV-2 in Ecuador. (A) Monthly proportion and number of genomes analysed for this study. Red, genomes sequenced for this study. Gray, available genomes in GISAID. (B) Time-resolved maximum clade credibility phylogeny of SARS-CoV-2 in Ecuador. Black, sequences generated for this study. (C) Root-to-tip versus time regression analysis reconstructing the time of SARS-CoV-2 introduction to Ecuador. (D) Number of SARS-CoV-2 tips per time in the time-stamped phylogenetic tree (shown in gray).
Implementation of NPIs likely slowed the emergence of new SARS-CoV-2 lineages
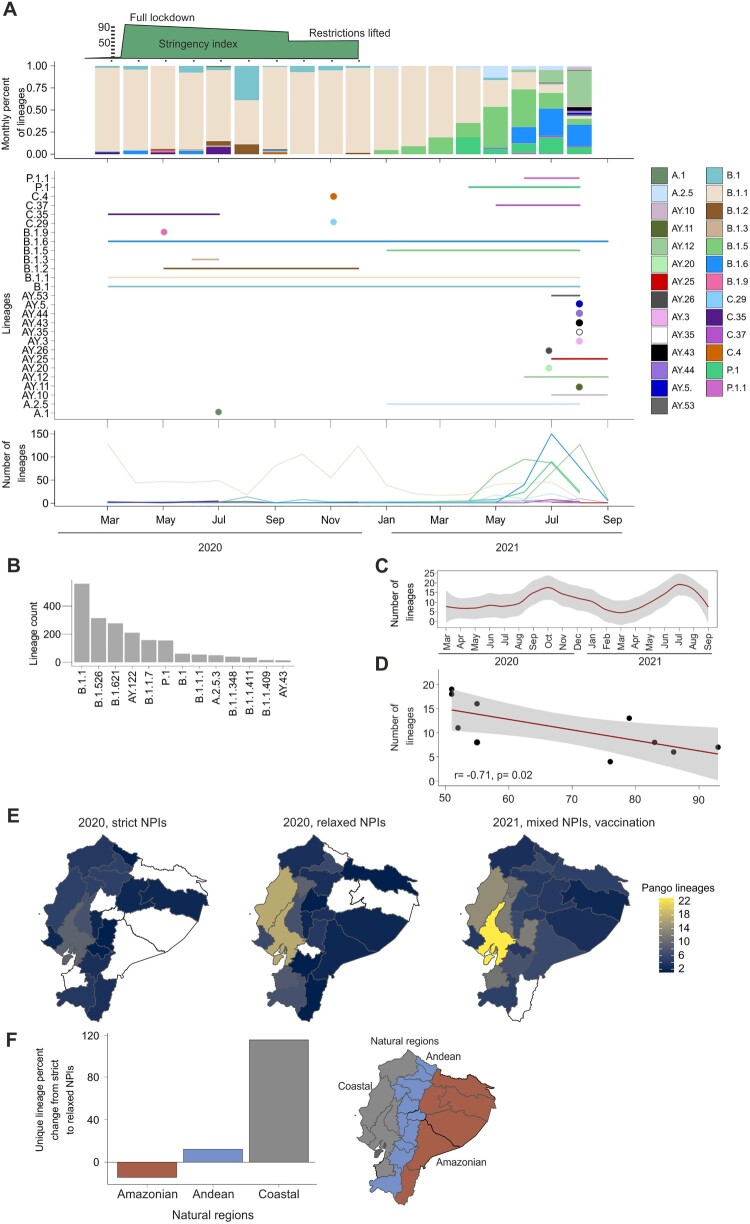
The SARS-CoV-2 strains circulating in Ecuador during 2020 and 2021 were assigned to 81 different lineages. Newly sequenced genomes from this study (shown in red in Figure 4(A) and in black in Figure 4(B)) were classified mainly as B.1.1 (436 in this study and 115 from GISAID), and its sub-lineages; B.1.1 representing 45.2% of the total genomic data from this study (Figure 5(A,B)). That distribution was in concordance to continent-wide South American estimates showing 37% of all SARS-CoV-2 strains to be B.1.1 during early 2020 [23]. Both B.1 and B.1.1. lineages were detected during February 2020 in Europe and to a lesser extent in North America and Asia [24], and in Ecuador during March 2020, compatible with travel-aided SARS-CoV-2 introduction events into Ecuador and elsewhere in Latin America. Moreover, the B.1.1 lineage was detected across the whole timespan (Figure 5(B)). In our dataset, a mean of 7.3 (range: 4–12; CI: 4.4–10.8) SARS-CoV-2 Pango lineages were detected during March to August 2020 (Figure 5(C)). Once NPIs were relaxed from September to December 2020, almost twice as many lineages circulated despite the shorter time span (mean, 15.5; range: 11–19; CI: 10.4–21.6) (Figure 5(C)). Moreover, the number of circulating lineages was negatively correlated with the stringency of NPIs, suggesting accelerated viral evolution due to increased transmission and introduction of previously non-endemic lineages once NPIs were relaxed (Figure 5(D)) Pearson correlation test; (r = −0.69; p = 0.02). The geographical distribution of the Pango lineages varied with the stringency of NPIs (Figure 5(E,F)). The coastal region of Ecuador, corresponding to the highest densely populated natural region in Ecuador and where the metropolis Guayaquil is located (Figure 1(C)), showed a 114.8% increase of unique Pango lineage detection in months with relaxed NPIs versus strict NPI months (Figure 5(F)). The Andean region also showed a 12.2% increase, where Quito, the capital, is located. For the least densely populated area, the amazon region, there was a decrease of −14.2% of unique Pango lineage detection during relaxed NPI months. The geographical distribution of distinct Pango lineages varied according to the stringency of NPIs during 2020–2021, with most distinct lineages emerging in coastal Ecuador during relaxed NPIs (Figure 5(E)). During 2020, the coastal region of Ecuador, corresponding to the most densely populated natural region in Ecuador containing the metropolis Guayaquil (Figure 1(C)), showed a 114.8% increase of unique Pango lineages in months with relaxed NPIs versus strict NPI months (Figure 5(F)). Higher numbers of newly emerging variants in 2021 (Figure 5(E)) are in concordance with global estimates of increased lineage detection [23,25] potentially associated with heterogeneous levels of NPIs following the onset of vaccination and population-level immune responses enhancing coronavirus antigenic drift [26]. Notably, only two of our newly generated sequences belonged to a former variant of concern termed “iota” (Figure 4(B)).
Figure 5.
Modified SARS-CoV-2 lineage emergence in Ecuador according to strictness of non-pharmaceutical interventions. (A) Total number of lineages in the complete dataset. Lineages with less than 10 genomes are not shown for graphical reasons. (B) Distribution of lineages and number across time in the whole dataset. (C) Number of distinct Pango lineages over time. (D) Pearson correlation of stringency index versus number of lineages per month. Gray in F and G, 95% confidence intervals. (E) Number of distinct Pango lineages across natural climatic regions. (F) Percent change of distinct Pango lineages from strict to relaxed NPIs in Ecuadorian natural regions during 2020.
NPIs modified the dispersion of SARS-CoV-2
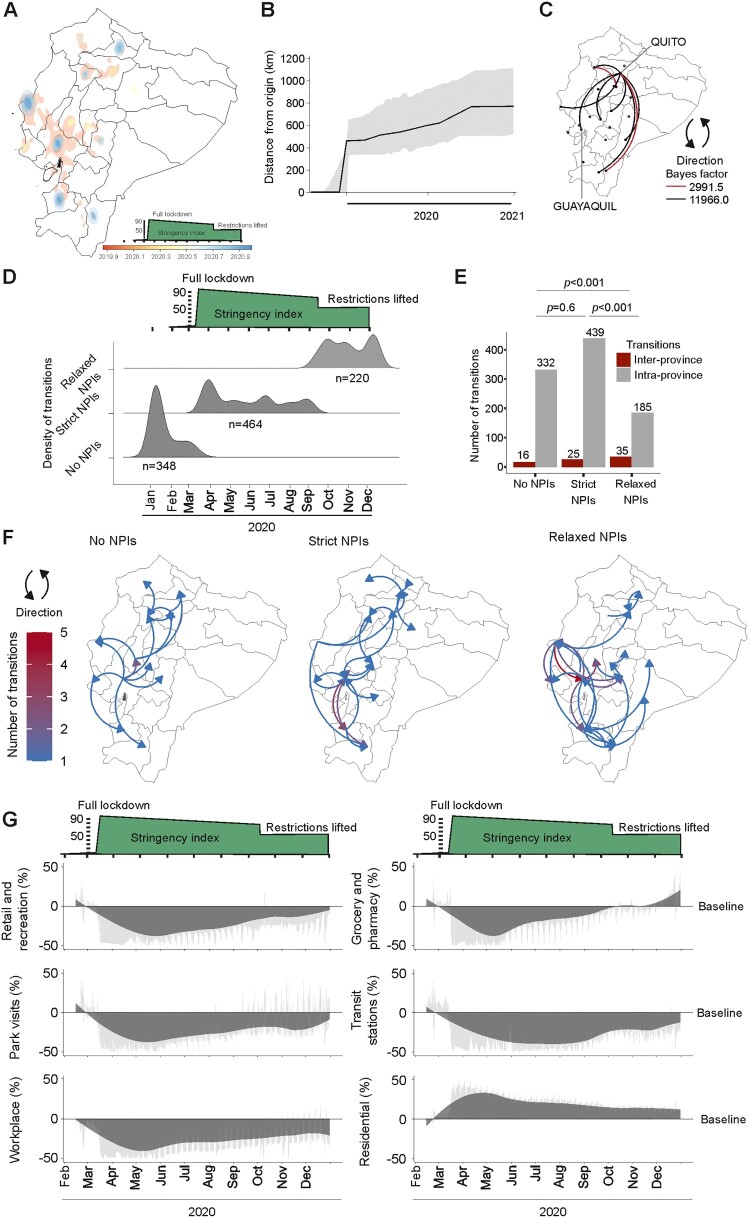
To mitigate biases arising from distinct transmission dynamics among lineages, and as B.1.1 was the sole lineage identified throughout the study duration, we exclusively employed SARS-CoV-2 lineage B.1.1 (n = 517 viral genomes) for mapping its dispersion patterns in 2020 (Figure 5(A,B)). The dataset comprised 292 SARS-CoV-2 sequences detected during months of strict NPIs and 225 during months of relaxed NPIs (X2 = 0.034; p = 0.85365), suggesting a comparable sequence population overtime. Using the extracted spatio-temporal information of randomly sampled 5% (500/10,000) of phylogenetic trees, and the maximum clade credibility (MCC) tree (Figure 6(A)), HPD analyses suggested differential SARS-CoV-2 dispersion patterns during 2020. Notably, a larger dispersion area probability was detected during months preceding the implementation of NPIs (red colour in Figure 6(A)), consistent with intense viral spread. Furthermore, SARS-CoV-2 exhibited a more localized distribution outside main cities during stringent NPIs months (yellow colour in Figure 6(A)), shifting back to city centres during relaxed NPIs periods (blue colour in Figure 6(A)). Intense viral spread at the onset of the pandemic in Ecuador was consistent with peak movements exceeding 400 km in early 2020 in Bayesian analyses of maximal patristic wavefront distance (MPWD) (Figure 6(B)). The 268 km straight-line distance between Quito and Guayaquil, Ecuador’s largest cities, highlights that various virus movements likely contributed to the 2020 MPWD spanning 500–700 km (Figure 6(B)). Long-distance transitions were confirmed using Bayes factor analyses, showing multiple highly supported transitions between Northern Andean and coastal provinces, including Quito and Guayaquil (Figure 6(C)).
Figure 6.
Spatiotemporal reconstructions of SARS-CoV-2 spread according to strictness of non-pharmaceutical interventions. (A) 80% highest posterior density regions of SARS-CoV-2 in coastal Ecuador. Colours denote the probability of SARS-CoV-2 geographical distribution in time. (B) Maximal-patristic wavefront distance calculating the sum of geographical distances associated with each branch connecting a given node to the root. (C) Bayes factor analyses of transitions. (D) Density estimations of SARS-CoV-2 transitions between geographical locations. (E) Number of intra- or inter-province transitions. Top, Chi-square significance level. (F) Spatiotemporal reconstruction of inter-province transitions of SARS-CoV-2 in Ecuador during differential non-pharmaceutical interventions (NPIs). (G) Percent change from baseline of different mobility estimates in Ecuador.
Differential dispersion patterns during different phases of the pandemic were also observed in analyses of the density and number of transitions in time. Specifically, there were fewer geographic transitions during strict NPIs (n = 450) than during non-stringent or absent NPIs (n = 580) (Figure 6(D)). Two weeks after the implementation of strict NPIs, a 54% decrease of geographic transitions entailed, followed by continuously low numbers of geographic transitions (Figure 6(D)). Most transitions occurred intra-province in all three periods analysed, suggesting predominantly local transmission (Figure 6(E)). However, intra-province transitions increased significantly during month with strict NPIs, compatible with localized viral dispersion due to movement restrictions (X2 = 20.6; p = 0.0001). Conversely, more inter-province transitions entailed during months with relaxed NPIs, compatible with long-distance human movement (X2 = 21.1; p = 0.0001; Figure 6(E)). Analysing geographical dispersion patterns using the inter-province movements, we mapped the transitions in every period analysed. At the onset of the pandemic, geographic transitions originated mainly unidirectionally outward from coastal Ecuador’s main cities to other provinces (50%; 8/16; Figure 6(F) left panel), which was in concordance to our time-stamped analyses (Figure 4(B)). Virus movements during strict NPIs occurred multidirectionally over relatively larger distances (Figure 6(F) middle panel), potentially due to commercial reactivation during mid-2020 in all of South America [27]. Key activities entailing enhanced virus transmission over shorter distances under relaxed NPIs may include retail, grocery and pharmacy and recreation as illustrated by virus transitions to the pacific coast (Figure 6(F) right panel) and according to publically available mobility data (Figure 6(G)).
Discussion
We gathered virological data describing the onset of the COVID-19 pandemic in a Latin American COVID-19 epicentre to show that NPIs could be a modifying and limiting factor of SARS-CoV-2 transmission. NPIs are a well-known part of public health responses to epidemics and have been extensively used for example during the SARS-CoV epidemic in 2003 and the Spanish flu in 1918. However, the success of NPIs in containing outbreaks varied depending on region, time of implementation, cultural background of the population, its prior experience to epidemics and civil compliance to the imposed restrictions [28,29]. For SARS-CoV-2, a comparative study analysing public health responses in developed and limited number of developing countries suggested that the success of NPIs depended on governmental monitoring and early stringent application of NPIs [30], which are particularly difficult in resource-limited settings such as Latin America due to several factors. First, testing of SARS-CoV-2 in Latin America is insufficient and assessments of base mortality are often delayed [7]. Second, monitoring and implementation of NPIs is complicated in rural settings, in which circa 20% of Latin American populations reside [31] and in which access to healthcare services is limited and COVID-19 stigmatization, i.e. delay of seeking care due to negative economic or social consequences, is frequent [31,32]. Third, compliance with NPIs by the population is affected by socio-demographic characteristics such as poverty and ethnicity [33]. Fourth and potentially most importantly, informal labour reaches more than 60% of workforce in Ecuador and other parts of Latin America [34], and stay-at-home orders cannot be followed by persons relying on daily informal income. Those factors may explain why despite strict NPIs, the death toll continued to rise in Ecuador and other regions of Latin America, and why Latin America was a priority region for rapid vaccination [35]. Whether NPIs were thus a meaningful tool to contain COVID-19 in Latin America and whether the socio-economic costs of NPIs were justified thus remained unclear [36]. In contrast, our data highlight NPIs’ significant influence on SARS-CoV-2 epidemiology. They strongly indicate that without NPIs, the first-year pandemic death toll in Latin America would have been notably higher, mirroring an India study projecting a 60% death reduction due to NPIs in 2020 [37].
The study has limitations. The respiratory samples used don’t accurately represent Ecuador’s population, compliance of NPIs over time is unknown, and unmeasured NPIs like mask usage are also impactful but not included in the indexes. Other factors influencing our data are fear of COVID-19 [17] and significant population immunity after the first wave, leading to transient population immunity [38]. The Oxford stringency index doesn’t fully consider regional factors like informal labour and local NPI enforcement. However, aggregating policy responses could be useful in situations were lots of countries have limited variation in individual policies such as during the initial pandemic year [15]. Potential biases of phylogenetic analyses include sample representativeness and a smaller genomic dataset compared to other countries like Brazil [3]. Nonetheless, our samples follow Ecuador’s testing algorithm and were collected consistently and should thus allow interpretation of the SARS-CoV-2 public health response across time. Phylogenomic analyses from an affluent setting confirmed usability of genomic data to analyse NPI efficacy [39]. The study’s strengths include comprehensive molecular, phylogenetic, and phylogeographic analyses that whose results mutually confirmed each other.
Our data suggest that stringent NPIs modifying human behaviour were a modifying factor on spread and circulation of COVID-19 in Ecuador, but intense SARS-CoV-2 spread occurred before their implementation. Therefore, NPIs should be rapidly implemented and sustained in immunologically naïve populations upon introduction of a highly transmissible pathogen such as SARS-CoV-2. This interpretation is in concordance with studies showing increased mortality when NPIs were introduced too late and lifted too early during the 1918 Spanish flu pandemic in the US [29] and from early travel restrictions globally reducing case numbers during the onset of the COVID-19 pandemic [40]. Finally, the emergence of antigenically distinct SARS-CoV-2 variants worldwide [41] leading to decreased vaccine effectiveness [42], together with the delayed modification of vaccines worldwide has led countries such as China, Canada and Peru to implement similarly stringent NPIs as in 2020, affecting hundreds of millions of people in 2022. Overall, our analyses provide an evidence-based way to justify rapid implementation of NPIs, providing data-driven support to stakeholders facing resurge of SARS-CoV-2 immune escape variants or future epidemics globally. Non-compliance with NPIs on the population level has been associated with low socioeconomic status, male sex and young age, among others [43,44]. Therefore, it is essential to communicate virological data in an easily apprehensible way to contribute to acceptance of NPIs. Generally, the virological benefit of NPIs must be weighed against the capacities of public health systems in managing severe COVID-19 cases, e.g. intensive care units, respirators, and oxygen, all limited in resource-limited settings like Latin America [45,46] and the high societal [47] and economic costs associated with NPIs.
Supplementary Material
Acknowledgements
We thank Arne Kühne for technical support. We thank Carlo Fischer for the critical reading of the manuscript. All patient data were managed in anonymized databases by the INSPI. Clinical symptoms were retrieved from medical charts. The study was approved by the Ecuadorian Ministry of Health (MSP-MSP-2021-0006-O-FDQ) and the Ethics committee of the Espiritu Santo University in Guayaquil, Ecuador (2022-002A). Conceptualization (AM-S, JCZ, JFD), data curation (AB, DdM, MP, JG,JG, BW, A-LS, MO, MJB, MG, AO, SS), formal analysis (AM-S, AB, DdM, MP, JG, BW, A-LS, MO, MJB, MG, AO, SS, JCZ, JFD), funding acquisition (JFD), investigation (AM-S, AB, DdM, MP, JG, BW, A-LS, MO, MJB, MG, AO, SS, JCZ, JFD), methodology (AM-S, AB, DdM, MP, JG, BW, A-LS, MO, MJB, MG, AO, SS, JCZ, JFD), project administration (AM-S, AO, JCZ, JFD), resources (AM-S, AB, DdM, MP, JG, BW, A-LS, MO, MJB, MG, AO, SS, JCZ, JFD), software (AM-S, A-LS, BW), visualization (AM-S, BW, A-LS), writing–original draft (AM-S, AB, DdM, MP, JG, BW, A-LS, MO, MJB, MG, AO, SS, JCZ, JFD), and writing–review & editing (AM-S, JFD).
Funding Statement
This work was supported by the German Federal Ministry for Economic Cooperation and Development (BMZ) through the Deutsche Gesellschaft für Internationale Zusammenarbeit (GIZ) to JFD (project number 81272349).
Disclosure statement
We have no conflicts of interest to disclose.
Data and materials availability
The SARS-CoV-2 sequence assembly can be found in ebi.ac.uk under the accession: ERP145042. All assembled genomes are available in GISAID under EPI_ISL_18283221-EPI_ISL_18283825. All the code written to perform the analyses can be found at: https://github.com/Dokandres/EcuadorNPI.
References
- 1.Roser M. Coronavirus pandemic (COVID-19). Our world in data. 2020.
- 2.Hou C, Chen J, Zhou Y, et al. The effectiveness of quarantine of Wuhan city against the Corona Virus Disease 2019 (COVID-19): a well-mixed SEIR model analysis. J Med Virol. 2020 Jul;92(7):841–848. DOI: 10.1002/jmv.25827 [DOI] [PubMed] [Google Scholar]
- 3.Candido DS, Claro IM, de Jesus JG, et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020 Sep 4;369(6508):1255–1260. DOI: 10.1126/science.abd2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Islam N, Sharp SJ, Chowell G, et al. Physical distancing interventions and incidence of coronavirus disease 2019: natural experiment in 149 countries. Br Med J. 2020 Jul 15;370:m2743. DOI: 10.1136/bmj.m2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flaxman S, Mishra S, Gandy A, et al. Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature. 2020 Aug;584(7820):257–261. DOI: 10.1038/s41586-020-2405-7 [DOI] [PubMed] [Google Scholar]
- 6.Hunter PR, Colon-Gonzalez FJ, Brainard J, et al. Impact of non-pharmaceutical interventions against COVID-19 in Europe in 2020: a quasi-experimental non-equivalent group and time series design study. Euro Surveill. 2021 Jul;26:28. DOI: 10.2807/1560-7917.ES.2021.26.28.2001401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuellar L, Torres I, Romero-Severson E, et al. Excess deaths reveal the true spatial, temporal and demographic impact of COVID-19 on mortality in Ecuador. Int J Epidemiol. 2022;51(1):54–62. DOI: 10.1093/ije/dyab163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020 Jan;25:3. DOI: 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rambaut A, Holmes EC, O’Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(11):1403–1407. DOI: 10.1038/s41564-020-0770-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katoh K, Standley DM.. Mafft multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. DOI: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price MN, Dehal PS, Arkin AP.. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010 Mar 10;5(3):e9490. DOI: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sagulenko P, Puller V, Neher RA.. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 2018 Jan;4(1):vex042. DOI: 10.1093/ve/vex042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suchard MA, Lemey P, Baele G, et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018 Jan;4(1):vey016. DOI: 10.1093/ve/vey016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dellicour S, Rose R, Faria NR, et al. SERAPHIM: studying environmental rasters and phylogenetically informed movements. Bioinformatics. 2016;32(20):3204–3206. DOI: 10.1093/bioinformatics/btw384 [DOI] [PubMed] [Google Scholar]
- 15.Hale T, Angrist N, Goldszmidt R, et al. A global panel database of pandemic policies (Oxford COVID-19 Government Response Tracker). Nat Hum Behav. 2021 Apr;5(4):529–538. DOI: 10.1038/s41562-021-01079-8 [DOI] [PubMed] [Google Scholar]
- 16.Garcia PJ, Alarcon A, Bayer A, et al. COVID-19 response in Latin America. Am J Trop Med Hyg. 2020 Nov;103(5):1765–1772. DOI: 10.4269/ajtmh.20-0765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goolsbee A, Syverson C.. Fear, lockdown, and diversion: Comparing drivers of pandemic economic decline 2020. J Public Econ. 2021 Jan;193:104311. DOI: 10.1016/j.jpubeco.2020.104311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cañizares Fuentes R, Aroca R, Blasco Carlos M.. Evaluation of COVID-19 surveillance strategy in Ecuador. Disaster Med Public Health Prep. 2020;16(1):51–54. DOI: 10.1017/dmp.2020.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caini S, de Mora D, Olmedo M, et al. The epidemiology and severity of respiratory viral infections in a tropical country: Ecuador, 2009–2016. J Infect Public Health. 2019;12(3):357–363. DOI: 10.1016/j.jiph.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Góes LGB, Zerbinati RM, Tateno AF, et al. Typical epidemiology of respiratory virus infections in a Brazilian slum [Research Support, Non-U S Gov't]. J Med Virol. 2020;92(8):1316–1321. DOI: 10.1002/jmv.25636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poole S, Brendish NJ, Clark TW.. SARS-CoV-2 has displaced other seasonal respiratory viruses: results from a prospective cohort study. J Infect. 2020 Dec;81(6):966–972. DOI: 10.1016/j.jinf.2020.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez B, Márquez S, Prado-Vivar B, et al. Genomic epidemiology of SARS-CoV-2 transmission lineages in Ecuador. Virus Evolution. 2021;7(2):veab051. DOI: 10.1093/ve/veab051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018 Dec 1;34(23):4121–4123. DOI: 10.1093/bioinformatics/bty407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alteri C, Cento V, Piralla A, et al. Genomic epidemiology of SARS-CoV-2 reveals multiple lineages and early spread of SARS-CoV-2 infections in Lombardy, Italy. Nat Commun. 2021 Jan 19;12(1):434. DOI: 10.1038/s41467-020-20688-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giovanetti M, Slavov SN, Fonseca V, et al. Genomic epidemiology of the SARS-CoV-2 epidemic in Brazil. Nat Microbiol. 2022 Sep;7(9):1490–1500. DOI: 10.1038/s41564-022-01191-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jo WK, Drosten C, Drexler JF.. The evolutionary dynamics of endemic human coronaviruses. Virus Evol. 2021 Jan;7(1):veab020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Ruitenbeek RE, Slik JS, Bhulai S.. On the relation between COVID-19, mobility, and the stock market. PLoS One. 2021;16(12):e0261381. DOI: 10.1371/journal.pone.0261381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews Pillemer F, Blendon RJ, Zaslavsky AM, et al. Predicting support for non-pharmaceutical interventions during infectious outbreaks: a four region analysis. Disasters. 2015 Jan;39(1):125–145. DOI: 10.1111/disa.12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bootsma MC, Ferguson NM.. The effect of public health measures on the 1918 influenza pandemic in U.S. cities. Proc Natl Acad Sci U S A. 2007 May 1;104(18):7588–7593. DOI: 10.1073/pnas.0611071104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xylogiannopoulos KF, Karampelas P, Alhajj R.. COVID-19 pandemic spread against countries’ non-pharmaceutical interventions responses: a data-mining driven comparative study. BMC Public Health. 2021 Sep 1;21(1):1607. DOI: 10.1186/s12889-021-11251-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreira-Soto A, Pachamora Diaz JM, Gonzalez-Auza L, et al. High SARS-CoV-2 Seroprevalence in Rural Peru, 2021: a cross-sectional population-based study. mSphere. 2021 Dec 22;6(6):e0068521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDermott JH, Newman WG.. Refusal of viral testing during the SARS-CoV-2 pandemic. Clin Med. 2020 Sep;20(5):e163–e164. DOI: 10.7861/clinmed.2020-0388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan WK, Fernandez D, Tyrovolas S, et al. Heterogeneity in the effectiveness of non-pharmaceutical interventions during the first SARS-CoV2 wave in the United States. Front Public Health. 2021;9:754696. DOI: 10.3389/fpubh.2021.754696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canelas C. Informality and poverty in Ecuador. Small Bus Econ. 2019 2019/12/01;53(4):1097–1115. DOI: 10.1007/s11187-018-0102-9 [DOI] [Google Scholar]
- 35.Bollyky TJ, Murray CJL, Reiner RC.. Epidemiology, not geopolitics, should guide COVID-19 vaccine donations. Lancet. 2021 Jul 10;398(10295):97–99. DOI: 10.1016/S0140-6736(21)01323-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knaul FM, Touchton M, Arreola-Ornelas H, et al. Punt politics as failure of health system stewardship: evidence from the COVID-19 pandemic response in Brazil and Mexico. Lancet Reg Health Am. 2021 Dec;4:100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salvatore M, Purkayastha S, Ganapathi L, et al. Lessons from SARS-CoV-2 in India: a data-driven framework for pandemic resilience. Sci Adv. 2022 Jun 17;8(24):eabp8621. DOI: 10.1126/sciadv.abp8621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall VJ, Foulkes S, Charlett A, et al. SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: a large, multicentre, prospective cohort study (SIREN). Lancet. 2021 Apr 17;397(10283):1459–1469. DOI: 10.1016/S0140-6736(21)00675-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nadeau SA, Vaughan TG, Beckmann C, et al. Swiss public health measures associated with reduced SARS-CoV-2 transmission using genome data. Sci Transl Med. 2022 Nov 8;14:eabn7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grépin KA, Ho T-L, Liu Z, et al. Evidence of the effectiveness of travel-related measures during the early phase of the COVID-19 pandemic: a rapid systematic review. BMJ Global Health. 2021;6(3):e004537. DOI: 10.1136/bmjgh-2020-004537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yewdell JW. Antigenic drift: understanding COVID-19. Immunity. 2021;54(12):2681–2687. DOI: 10.1016/j.immuni.2021.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andrews N, Stowe J, Kirsebom F, et al. Covid-19 vaccine effectiveness against the Omicron (B.1.1.529) variant. N Engl J Med. 2022 Apr 21;386(16):1532–1546. DOI: 10.1056/NEJMoa2119451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Noronha N, Moniz M, Gama A, et al. Non-adherence to COVID-19 lockdown: who are they? A cross-sectional study in Portugal. Public Health. 2022 Jul 11;211:5–13. DOI: 10.1016/j.puhe.2022.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coroiu A, Moran C, Campbell T, et al. Barriers and facilitators of adherence to social distancing recommendations during COVID-19 among a large international sample of adults. PLoS One. 2020;15(10):e0239795. DOI: 10.1371/journal.pone.0239795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benítez MA, Velasco C, Sequeira AR, et al. Responses to COVID-19 in five Latin American countries. Health Policy Technol. 2020;9(4):525–559. DOI: 10.1016/j.hlpt.2020.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reperant LA, Osterhaus A.. COVID-19 vaccination and critical care capacity: Perilous months ahead. Vaccine. 2021;39(16):2183–2186. DOI: 10.1016/j.vaccine.2021.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lichand G, Doria CA, Leal-Neto O, et al. The impacts of remote learning in secondary education during the pandemic in Brazil. Nat Hum Behav. 2022 Aug;6(8):1079–1086. DOI: 10.1038/s41562-022-01350-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.