Abstract
Per- and polyfluoroalkyl substances (PFASs) are synthetic chemicals that are ubiquitous in environmental and biological systems, including human serum. PFASs are used in many products and industrial processes and are tied to numerous health effects. Due to multiple sources and exposure pathways, methods are needed to identify PFAS sources in communities to develop targeted interventions. We assessed effectiveness of three source apportionment methods (UNMIX, positive matrix factorization [PMF], and principal component analysis - multiple linear regression [PCA-MLR]) for identifying contributors to human serum PFAS concentrations in two highly exposed populations in Colorado and North Carolina where drinking water was contaminated via upstream sources, including a Space Force base and a fluorochemical manufacturing plant. UNMIX and PMF models extracted three to four potential PFAS exposure sources in the Colorado and North Carolina cohorts while PCA-MLR classified two in each cohort. No sources were characterized in NHANES (National Health and Nutrition Examination Study). Results suggest that these three methods can successfully identify sources in highly exposed populations. Future PFAS exposure research should focus on analyzing serum for an expanded PFAS panel, identifying cohorts with other distinct point source exposures, and combining biological and environmental data to better understand source apportionment results in the context of PFASs toxicokinetic behavior.
Keywords: Source Apportionment, Receptor Models, PFAS, Perfluoroalkyl Substances
Graphical Abstract
1. Introduction:
Per- and polyfluoroalkyl substances (PFASs) are a class of thousands of chemicals that have many uses from stain and water-repellent products to aqueous film forming foams (AFFF)1. PFASs have a range of mechanisms of action in the human body and varied behavior in the environment.1,2 Though few PFASs have been studied extensively, exposure to these substances at occupational and environmental levels has been associated with an array of adverse health effects.2,3 Since production began in the 1930s, PFASs have become globally pervasive due to their widespread use, persistence and mobility.3–7 Because of this, and varied exposure pathways, it is imperative that investigators identify key sources of exposure to develop targeted interventions.
Receptor-based methods for source apportionment are often used for identifying contamination sources and source contributions in environmental media. Some of these models (i.e. chemical mass balance [CMB]) require known source profiles,8,9 which are rarely available for PFAS-contaminated communities. Models have been developed to resolve sources in mixtures without a priori knowledge of sources.8,10,11 Such models include United States Environmental Protection Agency (USEPA)-developed UNMIX, positive matrix factorization (PMF), and principal component analysis-multiple linear regression (PCA-MLR).8,11–16 These models have been successfully used for source apportionment of PFASs in environmental media.17–23 While many source apportionment studies have used receptor models to determine source contributions from environmental media, the models have rarely been used for biological media.24,25,26 Many studies in exposed communities lack a priori knowledge of all exposure sources and/or do not have access to environmental samples from the time of the contamination. This means that biological measurements are often the best source for reconstructing prior exposure. Therefore, an understanding of model utility for biological matrices is important. This work uses multivariate receptor models–UNMIX, PMF and PCA-MLR–to evaluate putative sources of PFASs measured in serum samples from cohorts in Colorado (CO) and North Carolina (NC), United States (U.S.), along with a population representative of U.S. background exposures from 2015–2016. While some sources of contamination are known in these communities, many other PFAS concentrations in the blood samples are elevated relative to the expected background exposure, indicating multiple unknown sources of contamination. Measurements of the water sources for both the NC and CO cohorts also indicate exposure sources beyond the known sources of PFAS exposure.27–32 These cohorts lacked sufficient environmental samples or information on important exposure metrics like diet and, therefore, using source apportionment methods on biological samples may help reconstruct prior exposure. The overall objective of this work was to assess the utility of multiple source apportionment methods while elucidating source contributions using serum samples from populations exposed to varying PFAS sources. We expected to identify at least one source that aligns with known sources of PFAS contamination in these populations and additional sources that represent currently unidentified sources of exposure. This allows us to estimate the exposure contributions, the total contribution to overall exposure, of the known sources of PFASs and an indication of the magnitude of the other unidentified sources.
2. Methods:
2.1. Colorado and North Carolina Communities and Contamination Sources
Data from two on-going research studies, occurring in PFAS-exposed communities in Colorado (CO) and North Carolina (NC), were used to assess the utility of these models in determining exposure contributions of serum PFAS. The CO site is located in El Paso County, where approximately 80,000 people were exposed to high concentrations of perfluorooctanesulfonic acid (PFOS), perfluorooctanoic acid (PFOA), perfluorohexanesulfonic acid (PFHxS) and other PFASs originating from AFFF use at nearby Peterson Space Force Base (formerly Peterson Air Force Base).33 The NC site is in New Hanover County, where approximately 200,000 people were exposed to PFAS-contaminated drinking water sourced from the Cape Fear River.27 This PFAS contamination was made up in part by perfluoroalkyl acids (PFAAs; e.g., PFOS, PFOA, and PFHxS) from currently unidentified sources upstream of the drinking water intake and in part by PFASs known as novel fluoroethers which were released from a fluorochemical manufacturing facility;27 fluoroethers are a newer class of PFASs that have the traditional perfluoroalkyl carbon chains characteristic of PFAAs, such as PFOA, but interrupted by ether oxygen(s). Fluoroethers, including hexafluoropropylene oxide dimer acid (HFPO-DA, a.k.a GenX), and perfluoro-3,5,7,9-tetraoxadecanoic acid (PFO4DA), were discharged into the Cape Fear River by a fluorochemical facility 80 miles upriver from the public water utility intake.28,30,31,34–36
Select characteristics from each cohort are presented in Supplemental Table 1. In Supplemental Figure 1 distributions of select serum PFAS concentrations from both studies are displayed along with U.S. national reference range concentrations measured in the 2015–2016 National Health and Nutrition Examination Survey (NHANES).37 NHANES is an annual survey that collects information from a representative sample to assesses the health of the U.S population.37
2.2. CO Dataset
The CO study population, design, and procedures are described in detail elsewhere.33 Briefly, in spring 2018, 213 non-smoking adults who resided for at least two years in an area with AFFF-contaminated drinking water were recruited for the study. Blood samples were collected, and a questionnaire administered. Participants relied on a PFAS-impacted private well (N=16 participants) or resided in one of three PFAS-impacted water districts (Fountain, Security, or Widefield) (N=197 participants). PFAS water concentrations in the water districts displayed a north to south gradient (Security > Widefield > Fountain) moving away from the AFFF contamination source, as did certain PFAS (i.e., PFHxS) serum concentrations.33 While AFFF in drinking water is a known source of exposure in this cohort, the contribution of the AFFF has not been quantified and additional samples to characterize other local exposures (e.g., local diet, indoor and outdoor air) have not been collected. Blood samples were analyzed for 48 PFASs at the Colorado School of Mines. Details of the laboratory methods are described elsewhere.38
Eleven PFASs were detected in the serum of ≥50% of CO study participants (Table 1). Four PFASs (PFOA, PFHxS, PFOS, and perfluorononanoic acid (PFNA)) were detected in ≥98% of participants. Other PFASs detected in ≥50% of participants included: Perfluoro-n-heptanoic acid (PFHpA), perfluorodecanoic acid (PFDA), perfluoroundecanoic acid (PFUdA), perfluoropentanesulfonic acid (PFPeS), perfluoroheptanesulfonic acid (PFHpS), 2-(N-Methylperfluorooctanesulfonamido) acetic acid (MeFOSAA), and the tentatively identified unsaturated perfluorooctanesulfonic acid (U-PFOS), which contains two fewer fluorines and one double bond within the perfluorinated chain.38
Table 1:
PFASs detected in greater than 50% of participants for each cohort used as a starting point for inclusion in the three models. Not all PFASs listed below were included in the final selected modelsa.
| PFAS | Full Name | DTXIDb | CO Cohort |
NC Cohort |
||
|---|---|---|---|---|---|---|
| % Detected | Detection Limitc (ng/mL) | % Detected | Detection Limit c (ng/mL) | |||
| Perfluoroalkyl carboxylic acids (PFCAs) | ||||||
| PFHpA | Perfluoroheptanoic acid | 1037303 | 56 | 0.01–0.10 | 63 | 0.1–0.3 |
| PFOA | Perfluorooctanoic acid | 8031865 | 100 | 0.01–0.10 | 100 | 0.1–0.5 |
| PFNA | Perfluorononanoic acid | 8031863 | 98 | 0.01–0.20 | 97 | 0.1–0.9 |
| PFDA | Perfluorodecanoic acid | 3031860 | 85 | 0.01–0.20 | -- | -- |
| PFUdA | Perfluoroundecanoic acid | 8047553 | 66 | 0.01–0.20 | -- | -- |
|
Perfluorosulfonic acids (PFSAs) | ||||||
| PFPeS | Perfluoropentanesulfonic acid | 8062600 | 81 | 0.01–0.20 | -- | -- |
| PFHxS | Perfluorohexanesulfonic acid | 7040150 | 100 | 0.11–1.0 | 98 | 0.1–1.8 |
| PFHpS | Perfluoroheptanesulfonic acid | 8059920 | 99 | 0.01–0.04 | -- | -- |
| U-PFOS | Unsaturated perfluorooctanesulfonic acid | NA | 89 | 0.01–0.20 | -- | -- |
| PFOS | Perfluorooctanesulfonic acid | 3031864 | 100 | 0.10–2.0 | 99 | 0.1–0.5 |
| MeFOSAA | 2-(N-Methylperfluorooctanesulfonamido)acetic acid | 10624392 | 52 | 0.01–0.20 | -- | -- |
|
Novel fluoroethers | ||||||
| Nafionbp2 | Perfluoro-2-{[perfluoro-3-(perfluoroethoxy)-2-propanyl]oxy}ethanesulfonic acid | 10892352 | -- | -- | 99 | 0.1–0.12 |
| PFO4DA | Perfluoro-3,5,7,9-tetraoxadecanoic acid | 90723993 | -- | -- | 98 | 0.1–0.11 |
| PFO5DoA | Perfluoro-3,5,7,9,11-pentaoxadodecanoic acid | 50723994 | -- | -- | 88 | 0.1 |
UNMIX and positive matrix factorization (PMF) allow user to add and delete species to evaluate how these changes impact the fit of the resulting solution.
DTXSID is a unique substance identifier used in the U.S. EPA CompTox Chemistry Dashboard (Williams et al. 2017).
Multiple analytical runs were used to analyze sample sets, causing some run-to-run variation in detection limits. The range of detection limits for each compound is provided.
2.3. NC Dataset
The NC study population, design, and procedures are also described in detail elsewhere.27 Briefly, 344 residents of New Hanover County, ages 6 years and older, living in a home served with Cape Fear Public Utility Authority (CFPUA) drinking water for at least 12 months prior to November 2017, were recruited for the study. Blood samples were collected, and a questionnaire administered. Samples were analyzed at U.S. Environmental Protection Agency (EPA) in Research Triangle Park, North Carolina. Eight PFASs were detected in ≥50% of NC study participants (Table 1). Two fluoroethers (Nafion byproduct 2 and PFO4DA) and four PFAAs (PFOS, PFOA, PFHxS, and PFNA) were found in ≥97% participants. Other PFASs detected in ≥50% of participants included perfluoro-3,5,7,9,11-pentaoxadodecanoic acid (PFO5DoA) and PFHpA. Samples representative of exposure to fluoroethers via this population’s drinking water were not available because the source of contamination was shut off prior to the, and levels of fluoroethers have dropped substantially since then.27,29,30,34 Zhang et al. (2019) provides a snapshot of the presence of novel fluoroethers in the Cape Fear River prior to discharge control.34
2.4. Receptor Modeling
The three multivariate receptor models used in this analysis (UNMIX, PMF, and PCA-MLR) were run for each cohort individually.16,39,40 The CO serum results were also stratified by water district of residence (PMF only) to evaluate if exposure contribution varied when residential water was sourced further from the primary water contamination source (i.e., AFFF released from Peterson Space Force Base). The model objectives are to identify the number of sources, composition of each source, and exposure contributions for chemical constituents in each sample. For each model, an analyte inclusion requirement of ≥50% detection (above the limit of detection or LOD) within each cohort was used to ensure accuracy, and ½ the LOD was substituted for PFAS measurements below the LOD as established in similar works.17,20,22 Other methods that can be used for censored environmental data are maximum likelihood estimation, survival analysis, and non-parametric approaches. 41 While substitution remains the most common, these approaches can be much more robust, especially when data with less than 50% detects is included. 42 With this requirement, 11 PFASs were included for CO, 8 PFASs were included for NC, and 5 PFASs were included for NHANES. PFASs included in the initial models and their abbreviations are displayed in Table 1. Unmix and PMF both have built in methods to assess the robustness and stability of each model including bootstrapping, rotation, and assessment of influential points. Table 2 compares the assumptions across each model and selection criteria used for determining the most appropriate solution using each method.15,16,39,40
Table 2.
Model assumptions and selection criteria.
| Model Assumptions | UNMIXa | PMFb | PCA-MLRc |
|---|---|---|---|
| Chemical Mass Balance | x | x | |
| Linear Correlation | x | ||
| Normality | x | ||
| Source Composition Approximately Constant | x | x | |
| Positive Contributions | x | x | |
|
| |||
| Selection Criteria | |||
|
| |||
| High Model R2 | x | x | x |
| High Compound R2 | x | x | |
| High S/N | x | x | |
| User Discretion on Set of Included Compounds | x | x | x |
| Identification and Adjustment of Influential Outliers | x | x | |
| Runs Fall Within IQR of Bootstraps | x | x | |
| No Significant Negative Bias | x | ||
| No Errors, Significant Changes in Q, or Significant Swaps in Sources During Bootstrap-Displacement | x | ||
| No Source Swaps During Bootstrap-Displacement | x | ||
| Close to 1 | x | ||
| Close to 100 Bootstrap Runs for 100 Feasible Results | x | ||
| Primary Goodness-of-fit Metric | S/N | Q | R2 |
Abbreviations: PMF, positive matrix factorization; PCA-MLR, Principal component analysis-multiple linear regression; S/N, Signal to noise ratio; IQR, interquartile range;
Norris et al. 2007
Norris et al. 20014
Thurston and Spengler, 1985
2.5. UNMIX Model
USEPA’s UNMIX Model software version 6.0 was employed for this analysis.12,39 UNMIX assumes all concentrations are positive and species do not degrade or react with one another thus conserving mass, an appropriate assumption for many PFAAs, such as those presented in Table 143. These assumptions allow for a mass balance calculation using Equation 1.
| (Equation 1) |
Where is the concentration of the ith species in the jth sample, is the ith species concentration from the nth source, is the contribution from the nth source to the jth sample and is the error term.8–10,15
Unlike other receptor models (e.g., PMF), UNMIX does not require additional inputs beyond chemical species and concentrations. UNMIX employs singular value decomposition to reduce dimensionality of the data space, normalizes the data, and uses an algorithm to identify “edges” in the data to distinguish sources.12 To determine the best convergent model, UNMIX largely relies on the minimum R2 and the minimum signal to noise ratio (S/N) to determine results.12
2.6. PMF Model
An in-depth explanation of the PMF algorithm can be found in the PMF 5.0 handbook and Reff et al.15,40 The algorithm underlying PMF differs greatly from UNMIX, and criteria for selecting one over the other in the event that the results disagree can be found in a review by Henry and Christensen, where they conclude that Unmix is more appropriate when edges in the data are distinct and PMF functions better when there are many zeros in the loading and score matrices.44.Like UNMIX, the PMF model is based on the CMB equation (Equation 1).45 Unlike UNMIX, the number of sources in a PMF solution are user defined. Thus, the user can test solutions with a varying number of sources to determine an appropriate number for optimizing the diagnostic criteria. Another practical difference between PMF and UNMIX is that PMF includes uncertainty estimates for each data point. The uncertainty estimates are used in an equation to identify the value of the parameter , a goodness-of-fit parameter:
| (Equation 2) |
Where, is the uncertainty of the jth species concentration in sample , is the number of samples, and m is the number of species.15 Three different values are generated: is equal to (number of non-weak, a user selected label, data values in the data set (X)) -(numbers of elements in the factor contributions (G) and factor profiles (F), taken together), is the goodness-of-fit parameter calculated including all points, and is the goodness-of-fit parameter calculated excluding points not fit by the model, defined as samples for which the uncertainty-scaled residual is greater than four.40 Along with other criteria (Table 2) these calculation can, in part, be used to choose the best model. Depending on the data available, various equations may be used to estimate uncertainties.15 is commonly applied and was used in this analysis.15,44
2.7. PCA-MLR
A comprehensive explanation of the use of PCA-MLR for source apportionment can be found in Thurston and Spengler.16 The PCA-MLR model relies on the same underlying principles as the two models described above, however the PCA-MLR model does not rely on the CMB equation and therefore does not impose any positive constraints on the model. For the PCA, the data were normalized via log-transformation The PCA was run with varimax rotation and varimax factors with an eigenvalue >1 were used in the analysis as done in previous works on the subject.16,46–48 Following identification of factors, MLR was employed where the factors identified in the PCA were modelled as independent variables and the sum of the measured pollutant concentrations was the dependent variable. The regression coefficients from the MLR were then used to determine the exposure contribution, in percent, of each source with the following equation:
| (Equation 5) |
where is the beta coefficient for a given factor.16
3. Results and Discussion:
The overarching goal of this study was to quantify source contributions of PFAS exposure in human serum in two highly exposed communities and assess the utility of three different source apportionment methods in human serum across these communities and a broader reference population (NHANES). UNMIX, PMF and PCA-MLR were run on CO and NC serum datasets with results shown in Figures 1, 2 and 3. For the CO and NC cohorts, the UNMIX and PMF models described three to four major sources while the PCA-MLR method characterized two sources. Each cohort contained at least one distinct source of contamination that was by these models, but none of the sources elucidated were similar across the two cohorts. This is expected due to the overall lack of similarity in likely PFAS exposure source across these two cohorts. The NHANES analysis did not result in feasible solutions or solutions that met diagnostic criteria (Table 2) for any of the three models (results not shown).
Figure 1.
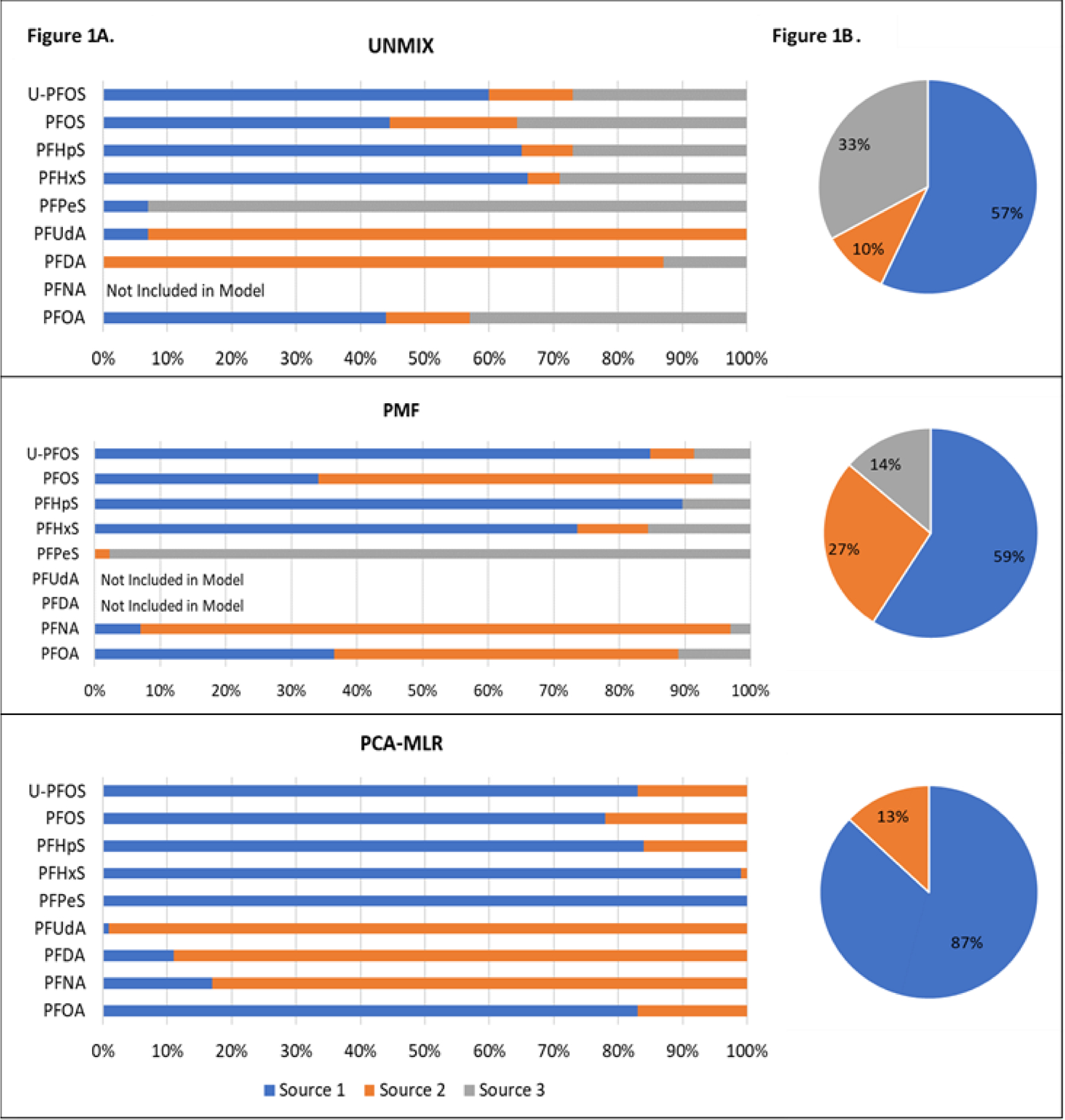
A (Left Side) Source compositions for the CO cohort from the three different models. B (Right Side) Exposure contributions for the CO from the three different models.
Figure 2.
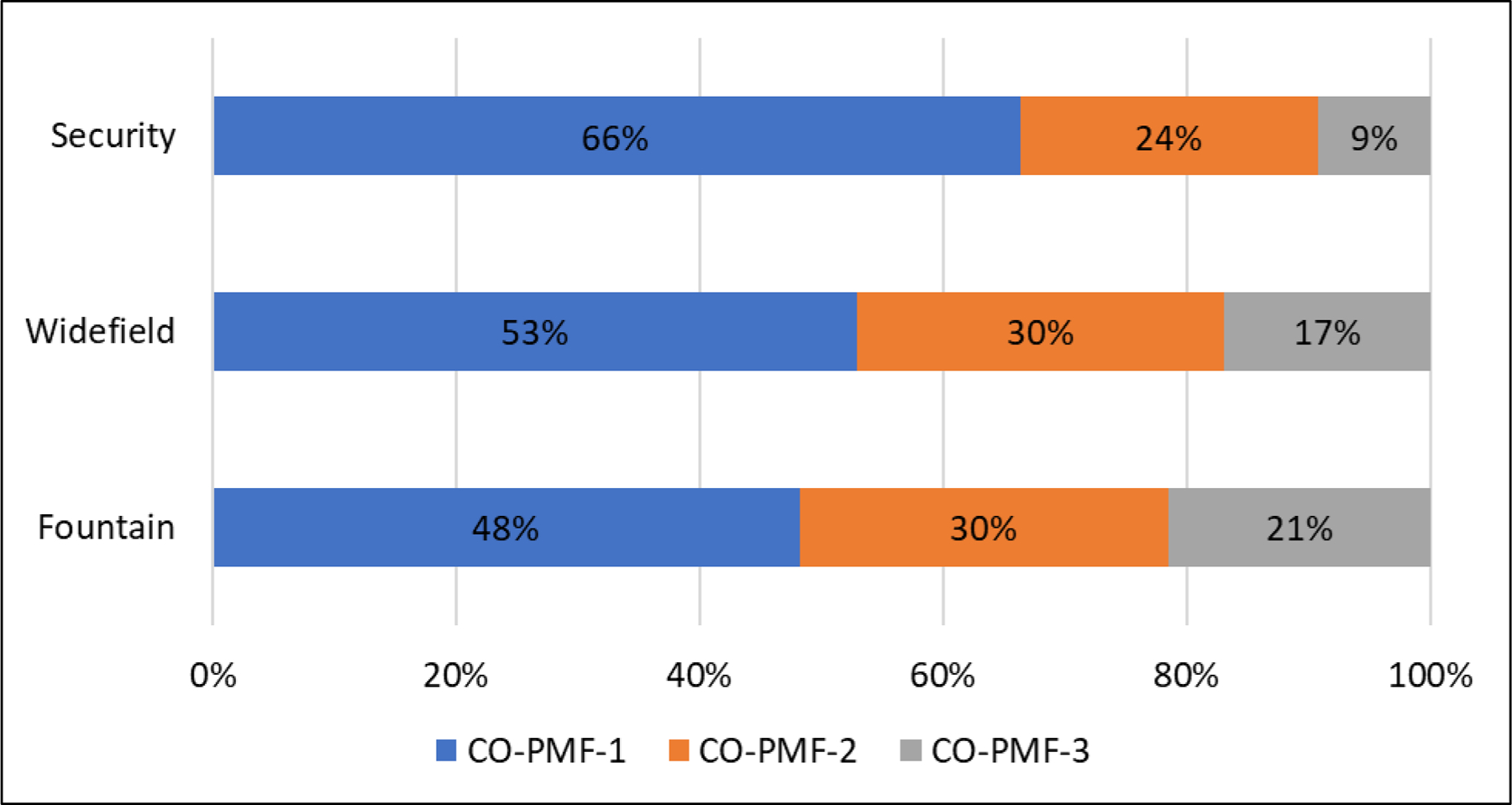
Exposure contributions by water district of residence for CO cohort, results from PMF model.
Figure 3.
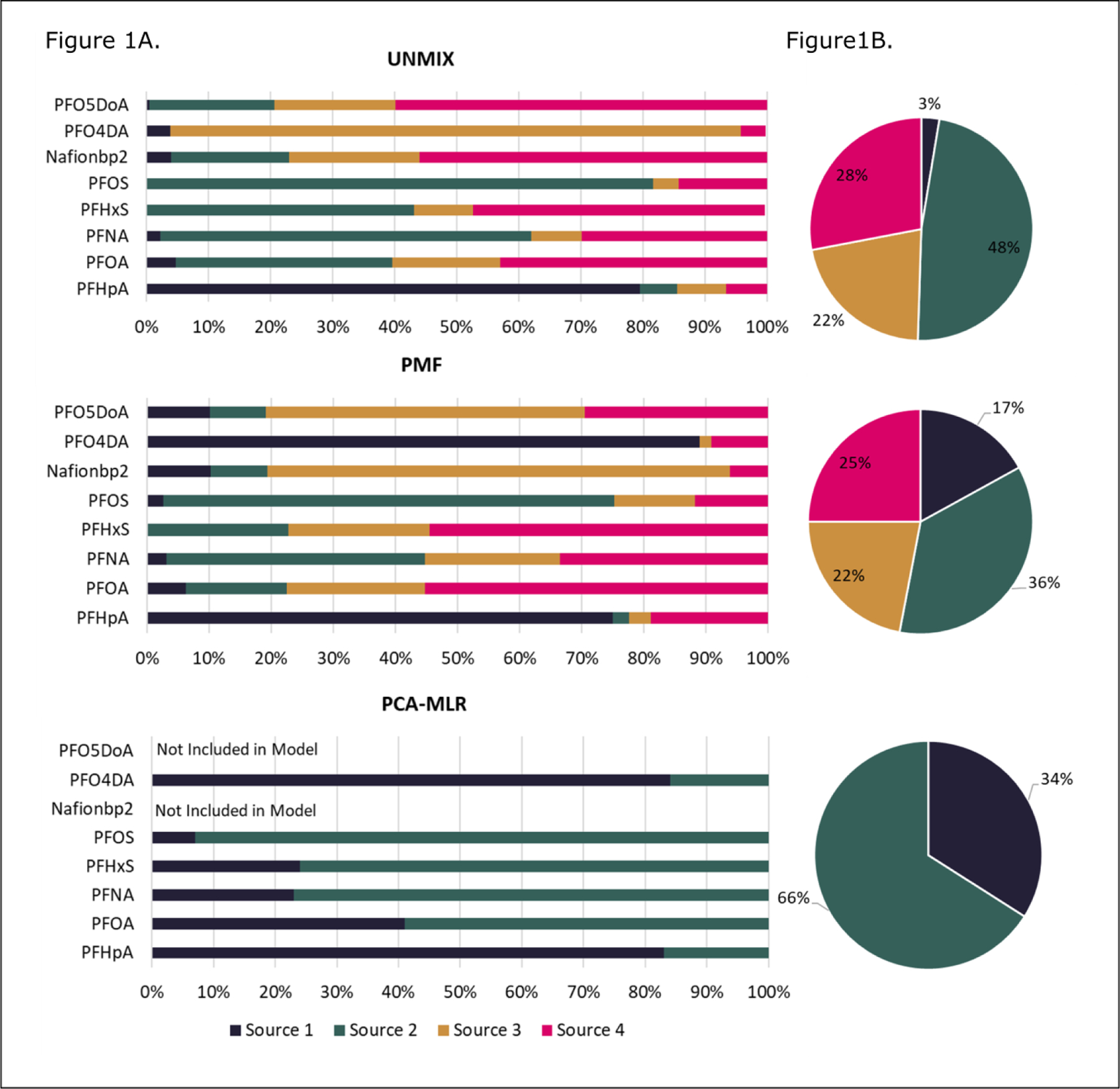
A (Left Side) Source compositions for the NC cohort from the three different models. B (Right Side) Exposure contributions for the NC cohort from the three different models.
3.1. CO Cohort: Results
For the CO dataset, UNMIX and PMF resulted in similar 3-source solutions and PCA-MLR resulted in a 2-source solution (Figure 1a/b).
PFNA, PFHpA and MeFOSAA were excluded from UNMIX due to high specific variance (variance due to error >50%). The final solution had an overall R2 of 0.87, had a minimum S/N ratio of 3.24 and met all diagnostic fit criteria as shown in Table 2. Source 1 (CO-UNMIX-1) accounted for 57% of the total mass and had high loadings of sulfonates including PFHxS, PFHpS, and U-PFOS. Source 2 (CO-UNMIX-2) accounted for 10% of the total mass and had high percent contributions of PFDA and PFUdA. Source 3 (CO-UNMIX-3) accounted for 33% of the total mass and had high percent contribution of PFPeS.
PMF produced a solution after investigation of 2 through 4-source solutions. PFUdA and PFDA were excluded from the final PMF solution due to a low R2 for the observed versus predicted estimates. PFNA was flagged as weak due to a low R2. While the 4-source solution had lower parameters, it displayed a high degree of rotational ambiguity (caused by multiple similar solutions being generated when the matrices are rotated) during F-Peak rotation and had comparable values across species. Therefore, the 3-source solution was used. This solution had a of 11.7 and a of 4761. The model was selected using the diagnostic criteria and resulted in the following sources: Source 1 (CO-PMF-1) with high percent contributions of sulfonates including PFHxS, PFHpS and U-PFOS, made up 59% of the total; Source 2 (CO-PMF-2), with a high percent contribution of PFNA made up 27% of the total; and Source 3 (CO-PMF-3), with a high percent contribution of PFPeS, made up 14% of the total.
The PCA model was run with all compounds other than PFHpA and MeFOSAA because inclusion resulted in decreased overall R2 and decreased percent variance explained. Two eigenvalues ≥1 were found in the PCA, which explained 76% of dataset variation. When absolute principal component scores were fit in the MLR, source 1 (CO-PCAMLR-1) contributed 87% and source 2 (CO-PCAMLR-2) contributed 13% to the total solution identified by PCA-MLR, with a model of 0.94. CO-PCAMLR-1 had high percent contributions from the sulfonates and PFOA while CO-PCAMLR-2 had high percent contributions of PFNA, PFDA and PFUdA.
Following evaluation of source apportionment results for the CO cohort, PMF results were stratified by participant’s water district of residence due to a clear north to south gradient of PFAS concentrations in the affected area.33,38 PMF has the option to include a sampling site with data input so that different locations may be compared. Results indicate decreasing exposure contribution from CO-PMF-1 by water district with increasing distance from the known AFFF release site (Figure 2). On average, CO-PMF-1 (the sulfonate dominated source) contributed 66% of the exposure for the serum PFAS in Security water district participants, (i.e., participants living closest to drinking water contamination source); Widefield water district participants averaged 53% contribution from CO-PMF-1; and Fountain water district participants (furthest from the contamination source) averaged a 48% contribution from CO-PMF-1. Average exposure contributions from CO-PMF-2, the source with high percent contributions from PFNA, ranged from 24% in Security to 30% in Widefield and Fountain. Average exposure contributions from CO-PMF-3, the source dominated by PFPeS, ranged from 9% in Security to 21% in Fountain (Figure 2).
3.2. CO Cohort: Potential Exposure Sources
The first exposure source characterized with each modeling approach (CO-UNMIX-1, CO-PMF-1 and CO-PCAMLR-1) had high percent contributions of sulfonates, specifically PFHpS, PFHxS, and U-PFOS for CO-UNMIX-1 and CO-PMF-1. The second source in all models had high percent contributions of longer chain carboxylates: PFDA and PFUdA for CO-UNMIX-1, PFNA in CO-PMF-2, and PFNA, PFDA and PFUdA in CO-PCAMLR-2. CO-UNMIX-3 and CO-PMF-3 had high percent contributions of PFPeS. PFOS and PFOA were not helpful in distinguishing sources despite being elevated in CO serum samples. However, PFOS and PFOA did contribute approximately 80% of their mass to CO-PCAMLR-1.
Based on what is known about PFAS releases near the CO site, CO-UNMIX-1, CO-PMF-1 and CO-PCAMLR-1, representing 57%, 59% and 87% relative overall source contribution, respectively, suggests that they are theAFFF-contaminated drinking water source.33,38 Three important PFASs for this source (AFFF) across models were PFHpS, PFHxS and U-PFOS which were detected in raw drinking water samples taken in 2018 as part of the CO study.38 Further, PFHpS and PFHxS are likely derived from AFFF, and have been found at high concentrations at other AFFF release sites.49,50 PFHxS has been found at elevated concentrations in the serum of firefighters exposed to AFFF51 and both PFHxS and PFHpS have been found in the serum of residents exposed to AFFF-contaminated drinking water, and in the associated raw water samples, in a Swedish community near a military installation.52,53 While previous research from the CO site is the first to our knowledge to report U-PFOS in human serum, 36 others have detected U-PFOS in AFFF-contaminated water and products as well as in the serum of mice dosed with AFFF.38,54,55
The supposition that CO-PMF-1 may be identifying the AFFF-contaminated drinking water is further supported by the results of the stratified PMF analysis (Figure 2). When stratifying by water district of residence, the exposure contribution by CO-PMF-1 decreases monotonically moving from the northernmost water district closest to the known AFFF source (Security) to the water district furthest from the AFFF source (Fountain). This is consistent with the Barton et. al 2019 findings that water district of residence was a primary predictor of PFAS serum concentrations and McDonough et al. 2021 results showing that untreated water concentrations varied by water district in the same pattern.38
The second source described by all three models (i.e., CO-UNMIX-2, CO-PMF-2, and CO-PCAMLR-2), with exposure contributions of 10%, 27% and 13%, respectively, had high percent contributions of the longer chain carboxylates, including PFNA, PFDA, and PFUdA. This source is not likely to be associated with drinking water, as these PFASs were either not detected at all or were detected only at very low concentrations in the CO untreated water samples and concentrations did not vary significantly by water district.38 Further, though only PFNA was included in the Barton et al. analyses, it was not associated with any drinking water-related variables, like many of the other PFASs.33 Based on market basket studies (i.e., where representative diets are characterized and daily intakes of contaminants or nutrients are estimated56) conducted in several countries, it is plausible that this source should be attributed to diet.57 Long chain perfluorocarboxylates, such as those identified here, are more likely to accumulate in seafood58,59 and dairy products60 compared to shorter chain PFASs.
A third source characterized by the UNMIX and PMF models (i.e., CO-UNMIX-3 and CO-PMF-3) had relative contributions of 33% and 14%, respectively, and high percent contributions of PFPeS. Two potential candidates for this source are: 1) an AFFF-contaminated drinking water source, or 2) an outside exposure, such as consumer product exposure. It is difficult to assign this factor to a specific source as there is very limited literature on the use of PFPeS and it is not routinely measured in human serum. The Australian Department of Health found that PFPeS has been used in electroplating, antireflective coatings, carpet treatments, and is present in AFFF.61 However, none of the sources the report cites specifically tested for PFPeS, rather they tested for PFHpS and PFHxS that are structurally similar but not identical. It is possible that PFPeS is related to the AFFF-contaminated drinking water given PFPeS was found in untreated drinking water samples at the CO Site38 as well as in the blood of residents exposed to AFFF-contaminated water in Sweden.62 Further, the PCA-MLR analysis only separated out two sources, with 100% of PFPeS allocated to CO-PCAMLR-1 with the other sulfonates known to be derived from AFFF.
One potential issue with this interpretation is the lack of an expected trend for PFPeS (i.e., CO-PMF-3) in the stratified analysis (Figure 2). Further, as presented in McDonough et al 2021, in a second year of sampling at the CO site in a subset of the year one participants (N=53 in year 2 [2019]; N=213 in year 1 [2018]) there was no significant decline in PFPeS serum concentrations.38 Over this one-year period, which took place after the water systems had mitigated the AFFF-contamination, other PFASs present in the contaminated drinking water (i.e., PFHxS, PFOS, PFOA, PFHpS and UPFOS) did decline significantly (p<0.05). Given PFPeS is estimated to have a shorter elimination half-life than the other PFASs listed above, it follows that if drinking water was the primary source of PFPeS exposure and the drinking water exposure was remediated prior to the blood testing in 2018, a significant decline in serum concentrations would be expected.62 In fact, of 53 CO study participants, 16 (30%) saw an increase in PFPeS from 2018 to 2019, with an average percent decline of only 11%.34 This, coupled with the fact that both CO-UNMIX-3 and CO-PMF-3 were similarly influenced by PFPeS, suggests that an additional source of PFPeS related to consumer product use may be present. In 2003, 3M began using perfluorobutane sulfonic acid (PFBS) as a substitute for PFOS in their Scotchgard formulation which could result in PFPeS impurities in this newer mixture.63 Indeed, in a 2020 exposure assessment conducted at a Michigan site contaminated by leachate from a landfill consisting of tannery waste contaminated with PFAS-containing Scotchgard, PFPeS was found in the serum of 86% of participants.64 This finding is supportive of the hypothesis that the third source produced by UNMIX and PMF may be linked to a consumer product exposure source.
3.3. NC Cohort: Results
Results from the NC cohort are presented in Figure 3. UNMIX and PMF both identified four-source solutions and PCA-MLR identified a two-source solution.
The UNMIX solution included all compounds. While excluding PFOA created a slightly better model solution based on a minimum S/N of 5.03, it was included in the model due to its importance as a contaminant in the NC cohort; PFOA serum concentrations in Wilmington were significantly higher than the national average.65 This solution had a minimum R2 value of 0.92, a minimum S/N of 2.47, and met all diagnostic criteria (Table 2). Source 1 (NC-UNMIX-1) accounted for 3% of the total mass and contained a majority of PFHpA. Source 2 (NC-UNMIX-2) accounted for 48% of the total mass and contained the highest loadings of PFOS and PFNA, as well as high percentages of PFHxS and PFOA. Source 3 (NC-UNMIX-3) accounted for 22% of the total mass and contained a high percentage of PFO4DA. Source 4 (NC-UNMIX-4) made up 28% of the total mass and consisted of the highest levels of Nafion byproduct 2, PFO5DoA, PFHxS, and PFOA.
For the PMF analysis, multiple models with two to six sources were fit. PFHpA was flagged as weak due to a low S/N. The model with the highest contained all compounds. The chosen model had the lowest (4.24), a relatively low (3478) and met all selection criteria in Table 2. This model included all PFASs that were measured in ≥50% of the participants. Source 1 (NC-PMF-1) made up 17% of the mass and contained high loadings of PFHpA and PFO4DA. Source 2 (NC-PMF-2) made up 36% of the total mass and contained high loading of PFOS and PFNA. Source 3 (NC-PMF-3) was 22% of the total mass and contained the highest loadings of Nafion byproduct 2 and PFO5DoA. Source 4 (NC-PMF-4) was 25% of the total and had high contributions of PFHxS and PFOA.
The PCA-MLR analysis was initially run with all chemicals. Because PCA-MLR does not have a way to control for negative concentration estimates, compounds with the most negative concentrations in each source were removed, until a solution that did not contain negative values was found. There were two eigenvalues greater than one and the final solution explained 86% of the variance. The MLR had an of 0.95. The final solution contained two sources and did not include PFO5DoA and Nafion byproduct 2. Source 1 (NC-PCAMLR-1) comprised 34% of the total mass and had the most PFHpA and PFO4DA. Source 2 (NC-PCAMLR-2) contributed 66% of the total mass and had a majority of the PFHxS, PFNA, PFOA, and PFOS.
3.4. NC Cohort: Potential Exposure Sources
Several exposure sources (NC-UNMIX-3,4; NC-PMF-1,3; NC-PCAMLR-1) contained high percent-contributions of novel fluoroethers, specifically PFO4DA for NC-UNMIX-3 and NC-PMF-1, and Nafion byproduct 2 and PFO5DoA for NC-UNMIX-4 and NC-PMF-3. The model results also all share sources that contain high percentages of PFAAs (NC-UNMIX-1,2; NC-PMF-2,4; NC-PCAMLR-2), specifically PFHpA for NC-UNMIX-1; PFNA and PFOS for NC-UNMIX-2; and PFOA, and PFHxS for NC-PMF-4.
Based on what is known about the NC cohort, NC-UNMIX-3,4; NC-PMF-1,3; NC-PCAMLR-1, representing 22% and 28%, 17% and 22%, and 34% exposure contribution, respectively, likely represent contributions from the Fayetteville Works Facility.27–29 This is consistent with the knowledge that Fayetteville Works is the only known source of the novel fluoroethers in the area.27–29 A high percentage of PFHpA–a PFAA –is also present in NC-PMF-1 and NC-PCAMLR-1. PFHpA was the dominant legacy PFAA measured in a 2006 wastewater discharge sample from Fayetteville Works (before methods for fluoroethers existed), and it was the second-most prevalent PFAA contributed by Fayetteville Works to the Cape Fear River based on samples collected in 2014 upstream and downstream of the facility.30–32
NC-UNMIX-1,2; NC-PMF-2,4; and NC-PCAMLR-2, representing 3% and 48%, 36% and 25%, and 66% exposure contribution, respectively, all represent contributions from PFAAs. These PFAAs have been identified in the Cape Fear River and as contaminants of New Hanover County drinking water.27,28,34,35 These model sources represent unidentified drinking water sources of PFAAs that may be separated spatially or temporally from the exposures stemming from Fayetteville Works. PFAAs have been identified upstream of the Fayetteville Works facility, and analyses over time show these shifting from longer to shorter chain PFASs.30–32
PFHxS and PFNA were spread evenly across multiple exposure sources–NC-UNMIX-2,4 and NC-PMF-2,3,4–respectively. PFHxS had the lowest in the UNMIX results (0.72) which indicates a larger problem in the model’s ability to characterize PFHxS exposure. PFNA, while primarily apportioned into NC-PMF-2,3,4, only contributed a small percentage to those sources–6%, 5%, and 6% respectively–and the total contamination. PFNA is known to be present in food (such as fish) as discussed earlier,57–61 and these models may have trouble disentangling background sources of contamination (i.e sources that are not specific to those in highly exposed communities, such as non-local diet, that many people are likely exposed to at low levels.), as evidenced by the lack of convergent results from the NHANES data. While Colorado has a very distinct and strong contamination source in the drinking water (AFFF), North Carolina may have multiple sources of water contamination, including several industrial wastewater discharges, AFFF, and runoff from fields, to which impacted biosolids are land-applied. Therefore, it is easier to determine potential additional sources beyond drinking water in the Colorado cohort.
3.5. Strengths
The major strength of this analysis is that all three receptor models found feasible solutions for PFAS sources in serum from two unique PFAS-exposed populations, the CO and NC cohorts, both of which have relatively small sample sizes (n<350). Further, UNMIX and PMF produced fairly similar results within each cohort improving confidence in conclusions. The sources that were ascertained in each cohort are supported by the known contamination in each community and are corroborated by samples taken by other researchers of the contaminated water sources.27–32
This analysis allows for a more refined understanding than simple evaluation of inter-PFAS correlations. Because these models rely in part on the correlation between the concentrations of the chemicals, we explored how the receptor models improved on the interpretation of a Spearman’s correlation (Supplemental Figures 2–4). Spearman’s correlations indicate general groupings of PFASs, but do not estimate the magnitude of individual or multiple PFAS contributions to potential sources. For example, in the CO dataset, it would be difficult to discern from Spearman’s correlations anything beyond the already suspected fact that many of the sulfonates are associated with a common source. The analysis does not give an indication that PFPeS may be behaving differently or the degree to which PFOS and PFOA may be contributing to other sources beyond AFFF-contaminated drinking water. These results provide confidence that these models could be used in the future to help investigate sources of exposure using biological as well as environmental samples.
3.6. Limitations
Along with the strengths defined above these models also have limitations. Though successful at determining solutions in the CO and NC cohorts, the models did not result in informative or feasible solutions with a larger sample size (n=1,993) in the NHANES 2015 to 2016 dataset. The CO and NC datasets both included larger arrays of PFASs (11 and 8, respectively) than the NHANES dataset with only 5 PFASs. In this case, these receptor models may be limited by the number of chemicals, and they may only be effective for populations in specific regions that are highly exposed to distinct (and common) exposure sources, which the NHANES cohort were not (supplementary figure 1). This was tested by running these models with the same 5 PFASs using the samples from the CO and NC cohorts. These tests did not return feasible or informative results and resembled the results from the NHANES data (supplementary data 1). Because these models rely on the separation of compounds using different algorithms, a larger number of compounds may help in clustering the samples into different sources. This is especially true for unmix, which identifies sources by finding “edges” wherein a compound is not present in a specific source. Having more compounds increases the opportunities for edges in the model to arise.
In addition, the models had trouble effectively partitioning compounds that result from many exposure sources (such as PFOA and PFOS). Although PFOA was found in higher concentrations compared to the general population for both CO and NC (Supplemental Figure 1), it likely originates from several sources. PFOA has been used in a variety of products for many years, making it difficult to identify its sources, especially in comparison to the novel fluoroethers observed in NC and the sulfonates associated with AFFF in CO.
A minor limitation worth noting is that in both cohorts, PCA-MLR found fewer sources than the other two models. Other studies that evaluated these three receptor methods have found that PCA-MLR is not always able to disentangle as many sources as the other two methods and may not be as effective at pulling out correlated sources.20,21,66 This may be due to the lack of negative constraints, diagnostic criteria for model improvement, and/or bootstrapping. Conversely, UNMIX and PMF appear to be very sensitive to exposure contributions and may separate single sources into multiple sources.
It is important to be mindful that these receptor models were designed for use with environmental media, not a biological matrix like human serum. A key assumption of these models is that the contaminants do not degrade or react with one another. In the human body, some PFASs are excreted more quickly than others, may be absorbed at different rates or distributed differentially, and these models may not be distinguishing the exposure sources if toxicokinetic parameters vary drastically across individuals or different groups. These results represent a specific cross-section of time, which makes accounting for toxicokinetic differences challenging. To remedy this problem, Hu et al. separated participants by covariates that could affect toxicokinetics such as sex and age.48 When the participants in our study were separated into three groups–men, women, and children (North Carolina only)–the resultant models did not fit the selection criteria, were similar to the full models, or did not produce feasible results (Supplemental data 1). These results imply that these models, as is, may be unable to account for toxicokinetic differences between subpopulations. This could be due to a need for a larger sample size that makes up for the diminished power when separating the population into groups to account demographics that may affect toxicokinetics such as age and sex. There is also currently no way built into the model to account for individual toxicokinetic differences.2,39,40 This problem may be ameliorated by modifications to the models that allow for covariates to account for such differences. The full models on the other hand produced feasible, interpretable results that met all selection criteria. This is probably not as major a limitation to this method as it may seem. The way PFASs, like all compounds, behave in the environment, as they do in the body, is also governed by a complicated suite of interacting variables.2,67 The fate and transport of a compound after leaving a source can be affected by physical and chemical properties of the molecule itself, soil types, precipitation, etc.68 These methods are agnostic to any variable that might affect the fate of a compound, whether it is in the environment or in the human body.
3.7. Implications and Future Research
The source apportionment models evaluated performed better in areas with distinct exposure sources and may not be useful in examining broader trends in the U.S. population. This is likely due to higher measured concentrations and/or greater relative variation between PFASs in highly exposed populations. For the CO and NC cohorts, neither UNMIX nor PMF separated PFOA into separate sources effectively, possibly because PFOA exposure originates from multiple relatively small and/or overlapping sources.
This work found that UNMIX and PMF were able to extract three to four potential sources of PFAS exposure for both cohorts, while the PCA-MLR method identified two sources for each cohort. While PCA-MLR appeared to perform adequately, UNMIX and PMF were more successful at characterizing specific source groups in communities exposed to high concentrations of PFASs originating from specific contamination events. All three methods were unsuccessful in identifying specific exposure sources when the models were run with a dataset expected to be representative of background PFAS exposures in the U.S. (NHANES).
Though these three models can provide an idea of potential sources of PFAS exposure, they are limited in that they require educated judgment based on existing knowledge to identify sources. A potential preferred method of PFAS source apportionment would be CMB, however, for PFASs, CMB would require regionally specific source profiles for drinking water, dust, indoor air, diet, and consumer products. The collection and analysis of such samples would be time and resource intensive and is often not feasible, especially in situations where investigators arrive after the exposure has taken place and the profiles may have changed since the initial exposure.
The results of UNMIX and PMF in both cohorts were similar enough that it was concluded that both models worked equally well. While UNMIX and PMF produce similar results, another consideration is usability. UNMIX’s reduced requirement for user input leads to results that are easier for decision-makers to interpret. While UNMIX requires less user input, making it more usable and easier to interpret, it provides less control over the model and less methods to validate each iteration of the model than PMF. PMF contains a similar but much more robust method to compare different parameters in each models including bootstrapping and rotation of these models that isn’t present in UNMIX while letting you select the number of sources and select which species are weighted less in the model without removing them. This makes PMF more broadly useful and more adjustable to each specific situation. Overall, these models are probably most useful if used in tandem, as differences that are present in the model may reveal information about the exposure that wouldn’t be ascertained when using either separately. A more in depth look at the differences between these models can be found in table 2, in their respective manuals, and across several publications8,10–12,39,40,45.
Given the long half-life of many PFAAs53,69 and the relative lack of transformation or metabolism of PFAAs in the human body, these methods should be considered viable options for source apportionment of serum PFASs in populations that are not exposed to significant amounts of metabolically labile precursors. For PFASs with longer half-lives, serum concentration may be thought of as an integrated measure of exposure that represents both historical and current exposure, thus these methods may be able to disentangle past sources. It is important to keep in mind that PFASs that are quickly eliminated from the body (e.g., a short-chain PFAS like PFBS)70,71 often are not present in serum samples and would be excluded from source characterization. For these homologues, it is worth considering using urine and environmental media in parallel to serum for evaluation of other potential sources of PFAS exposure.
This work provides insight into the utility of applying models designed for environmental media for source apportionment of human serum. The approach could be applied in many epidemiological studies where data on environmental sources is lacking but biological samples are available. Understanding PFAS exposure contributions and how source contributions vary based on the specific exposure scenarios and profiles is essential to develop policies that are appropriately protective of public health, inform toxicity testing by identifying mixtures to test, and guide mitigation efforts by identifying the largest and most common contamination sources.
Supplementary Material
ACKNOWLEDGMENT
This publication was developed under Assistance Agreement No. R839482 awarded by the U.S. Environmental Protection Agency to Dr. Christopher P. Higgins. It has not been formally reviewed by EPA. The views expressed in this document are solely those of the authors and do not necessarily reflect those of the Agency. EPA does not endorse any products or commercial services mentioned in this publication.
Funding Sources
This work was supported by the National Institute of Environmental Health Sciences (NIEHS) under Grants No. R21ES029394 (CO), T32ES007046, 1R21ES029353 & P42ES031009 (NC) and the Environmental Protection Agency under Grant No. G18A112656081. Any opinion, findings, conclusions, or recommendations expressed are those of the authors and do not necessarily reflect the views of the NIH or the EPA.
ABBREVIATIONS
- PFAS
per and polyfluoroalkyl substances
- NC
North Carolina
- CO
Colorado
- USEPA
United States Environmental Protection Agency
- PMF
positive matrix factorization
- PCA-MLR
principal component analysis-multiple linear regression
- AFFF
aqueous film forming foams
- PFAA
perfluoroalkyl acid
- PFHpA
Perfluoro-n-heptanoic acid
- PFOA
Perfluorooctanoic acid
- PFNA
Perfluorononanoic acid
- PFDA
Perfluorodecanoic acid
- PFUdA
Perfluoroundecanoic acid
- PFPeS
Perfluoropentanesulfonic acid
- PFHxS
Perfluorohexanesulfonic acid
- PFHpS
Perfluoroheptanesulfonic acid
- U-PFOS
Unsaturated perfluorooctanesulfonic acid
- PFOS
Perfluorooctanesulfonic acid
- MeFOSAA
2-(N-Methylperfluorooctanesulfonamido)acetic acid
- Nafionbp2
Perfluoro-2-{[perfluoro-3-(perfluoroethoxy)-2-propanyl]oxy}ethanesulfonic acid
- PFO4DA
Perfluoro-3,5,7,9-tetraoxadecanoic acid
- PFO5DoA
Perfluoro-3,5,7,9,11-pentaoxadodecanoic acid
- Nafionbp2
Nafion byproduct 2
- S/N
signal to noise ratio
Footnotes
ASSOCIATED CONTENT
Supporting Information. The following files are available free of charge.
Selected baseline characteristics of the CO and NC study populations; Model assumptions and selection criteria; Comparison of PFAS Concentration between the CO cohort, the NC cohort and the 2015–2016 NHANES cohort (U.S.); Spearman’s Correlation Structures for CO, NC and NHANES datasets (PDF)
REFERENCES
- (1).Buck RC; Franklin J; Berger U; Conder JM; Cousins IT; de Voogt P; Jensen AA; Kannan K; Mabury SA; van Leeuwen SP Perfluoroalkyl and Polyfluoroalkyl Substances in the Environment: Terminology, Classification, and Origins. Integr Environ Assess Manag 2011, 7 (4), 513–541. 10.1002/ieam.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Agency for Toxic Substances and Disease Registry (ATSDR). Toxicological Profile for Perfluoroalkyls; U.S. Department of Health and Human Services, Public Health Service: Atlanta, GA: U.S, 2021. [PubMed] [Google Scholar]
- (3).Sunderland EM; Hu XC; Dassuncao C; Tokranov AK; Wagner CC; Allen JG A Review of the Pathways of Human Exposure to Poly- and Perfluoroalkyl Substances (PFASs) and Present Understanding of Health Effects. J Expo Sci Environ Epidemiol 2019, 29 (2), 131–147. 10.1038/s41370-018-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Vestergren R; Cousins IT Tracking the Pathways of Human Exposure to Perfluorocarboxylates. Environmental science & technology 2009, 43 (15), 5565–5575. [DOI] [PubMed] [Google Scholar]
- (5).Armitage JM; MacLeod M; Cousins IT Comparative Assessment of the Global Fate and Transport Pathways of Long-Chain Perfluorocarboxylic Acids (PFCAs) and Perfluorocarboxylates (PFCs) Emitted from Direct Sources. Environmental science & technology 2009, 43 (15), 5830–5836. [DOI] [PubMed] [Google Scholar]
- (6).Mueller R; Yingling V History and Use of Per-and Polyfluoroalkyl Substances (PFAS). Interstate Technology & Regulatory Council, USA 2017. [Google Scholar]
- (7).Land M; De Wit CA; Bignert A; Cousins IT; Herzke D; Johansson JH; Martin JW What Is the Effect of Phasing out Long-Chain per-and Polyfluoroalkyl Substances on the Concentrations of Perfluoroalkyl Acids and Their Precursors in the Environment? A Systematic Review. Environmental Evidence 2018, 7 (1), 1–32. [Google Scholar]
- (8).Hopke PK Review of Receptor Modeling Methods for Source Apportionment. Journal of the Air & Waste Management Association 2016, 66 (3), 237–259. 10.1080/10962247.2016.1140693. [DOI] [PubMed] [Google Scholar]
- (9).Miller MS; Friedlander SK; Hidy GM A Chemical Element Balance for the Pasadena Aerosol. Journal of Colloid and Interface Science 1972, 39 (1), 165–176. 10.1016/0021-9797(72)90152-X. [DOI] [Google Scholar]
- (10).Henry RC Multivariate Receptor Models—Current Practice and Future Trends. Chemometrics and Intelligent Laboratory Systems 2002, 60 (1), 43–48. 10.1016/S0169-7439(01)00184-8. [DOI] [Google Scholar]
- (11).Paatero P; Tapper U Positive Matrix Factorization: A Non-Negative Factor Model with Optimal Utilization of Error Estimates of Data Values. Environmetrics 1994, 5 (2), 111–126. 10.1002/env.3170050203. [DOI] [Google Scholar]
- (12).Henry RC Multivariate Receptor Modeling by N-Dimensional Edge Detection. Chemometrics and Intelligent Laboratory Systems 2003, 65 (2), 179–189. 10.1016/S0169-7439(02)00108-9. [DOI] [Google Scholar]
- (13).Harrison RM; Smith D; Luhana L Source Apportionment of Atmospheric Polycyclic Aromatic Hydrocarbons Collected from an Urban Location in Birmingham, UK. Environmental science & technology 1996, 30 (3), 825–832. [Google Scholar]
- (14).Larsen RK; Baker JE Source Apportionment of Polycyclic Aromatic Hydrocarbons in the Urban Atmosphere: A Comparison of Three Methods. Environ. Sci. Technol 2003, 37 (9), 1873–1881. 10.1021/es0206184. [DOI] [PubMed] [Google Scholar]
- (15).Reff A; Eberly SI; Bhave PV Receptor Modeling of Ambient Particulate Matter Data Using Positive Matrix Factorization: Review of Existing Methods. Journal of the Air & Waste Management Association 2007, 57 (2), 146–154. 10.1080/10473289.2007.10465319. [DOI] [PubMed] [Google Scholar]
- (16).Thurston GD; Spengler JD A Quantitative Assessment of Source Contributions to Inhalable Particulate Matter Pollution in Metropolitan Boston. Atmospheric Environment (1967) 1985, 19 (1), 9–25. [Google Scholar]
- (17).Li Y; Feng X; Zhou J; Zhu L Occurrence and Source Apportionment of Novel and Legacy Poly/Perfluoroalkyl Substances in Hai River Basin in China Using Receptor Models and Isomeric Fingerprints. Water Research 2020, 168, 115145. 10.1016/j.watres.2019.115145. [DOI] [PubMed] [Google Scholar]
- (18).Kuroda K; Murakami M; Oguma K; Takada H; Takizawa S Investigating Sources and Pathways of Perfluoroalkyl Acids (PFAAs) in Aquifers in Tokyo Using Multiple Tracers. Science of The Total Environment 2014, 488–489, 51–60. 10.1016/j.scitotenv.2014.04.066. [DOI] [PubMed] [Google Scholar]
- (19).Liu Y; Zhang Y; Li J; Wu N; Li W; Niu Z Distribution, Partitioning Behavior and Positive Matrix Factorization-Based Source Analysis of Legacy and Emerging Polyfluorinated Alkyl Substances in the Dissolved Phase, Surface Sediment and Suspended Particulate Matter around Coastal Areas of Bohai Bay, China. Environmental Pollution 2019, 246, 34–44. 10.1016/j.envpol.2018.11.113. [DOI] [PubMed] [Google Scholar]
- (20).Qi Y; He Z; Huo S; Zhang J; Xi B; Hu S Source Apportionment of Perfluoroalkyl Substances in Surface Sediments from Lakes in Jiangsu Province, China: Comparison of Three Receptor Models. Journal of Environmental Sciences 2017, 57, 321–328. 10.1016/j.jes.2016.12.007. [DOI] [PubMed] [Google Scholar]
- (21).Qi Y; Huo S; Xi B; Hu S; Zhang J; He Z Spatial Distribution and Source Apportionment of PFASs in Surface Sediments from Five Lake Regions, China. Scientific reports 2016, 6 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Xu J; Shi G; Guo C-S; Wang H-T; Tian Y-Z; Huangfu Y-Q; Zhang Y; Feng Y-C; Xu J A New Method to Quantify the Health Risks from Sources of Perfluoroalkyl Substances, Combined with Positive Matrix Factorization and Risk Assessment Models. Environmental Toxicology and Chemistry 2018, 37 (1), 107–115. 10.1002/etc.3955. [DOI] [PubMed] [Google Scholar]
- (23).Xu J; Tian Y-Z; Zhang Y; Guo C-S; Shi G-L; Zhang C-Y; Feng Y-C Source Apportionment of Perfluorinated Compounds (PFCs) in Sediments: Using Three Multivariate Factor Analysis Receptor Models. Journal of Hazardous Materials 2013, 260, 483–488. 10.1016/j.jhazmat.2013.06.001. [DOI] [PubMed] [Google Scholar]
- (24).Rodenburg LA; Delistraty DA Alterations in Fingerprints of Polychlorinated Biphenyls in Benthic Biota at the Portland Harbor Superfund Site (Oregon, USA) Suggest Metabolism. Chemosphere 2019, 223, 74–82. 10.1016/j.chemosphere.2019.02.039. [DOI] [PubMed] [Google Scholar]
- (25).Rodenburg LA; Delistraty D; Meng Q Polychlorinated Biphenyl Congener Patterns in Fish near the Hanford Site (Washington State, USA). Environ Sci Technol 2015, 49 (5), 2767–2775. 10.1021/es504961a. [DOI] [PubMed] [Google Scholar]
- (26).Jovanović G; Romanić SH; Stojić A; Klinčić D; Sarić MM; Letinić JG; Popović A Introducing of Modeling Techniques in the Research of POPs in Breast Milk–A Pilot Study. Ecotoxicology and environmental safety 2019, 172, 341–347. [DOI] [PubMed] [Google Scholar]
- (27).Kotlarz N; McCord J; Collier D; Lea CS; Strynar M; Lindstrom AB; Wilkie AA; Islam JY; Matney K; Tarte P; Polera ME; Burdette K; DeWitt J; May K; Smart RC; Knappe DRU; Hoppin JA Measurement of Novel, Drinking Water-Associated PFAS in Blood from Adults and Children in Wilmington, North Carolina. Environ Health Perspect 2020, 128 (7), 077005. 10.1289/EHP6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).McCord J; Strynar M Identification of Per- and Polyfluoroalkyl Substances in the Cape Fear River by High Resolution Mass Spectrometry and Nontargeted Screening. Environ. Sci. Technol 2019, 53 (9), 4717–4727. 10.1021/acs.est.8b06017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Hopkins ZR; Sun M; DeWitt JC; Knappe DRU Recently Detected Drinking Water Contaminants: GenX and Other Per- and Polyfluoroalkyl Ether Acids: JOURNAL AWWA. Journal - American Water Works Association 2018, 110 (7), 13–28. 10.1002/awwa.1073. [DOI] [Google Scholar]
- (30).Sun M; Arevalo E; Strynar M; Lindstrom A; Richardson M; Kearns B; Pickett A; Smith C; Knappe DRU Legacy and Emerging Perfluoroalkyl Substances Are Important Drinking Water Contaminants in the Cape Fear River Watershed of North Carolina. Environ. Sci. Technol. Lett 2016, 3 (12), 415–419. 10.1021/acs.estlett.6b00398. [DOI] [Google Scholar]
- (31).Strynar M; Dagnino S; McMahen R; Liang S; Lindstrom A; Andersen E; McMillan L; Thurman M; Ferrer I; Ball C Identification of Novel Perfluoroalkyl Ether Carboxylic Acids (PFECAs) and Sulfonic Acids (PFESAs) in Natural Waters Using Accurate Mass Time-of-Flight Mass Spectrometry (TOFMS). Environ. Sci. Technol 2015, 49 (19), 11622–11630. 10.1021/acs.est.5b01215. [DOI] [PubMed] [Google Scholar]
- (32).Nakayama S; Strynar MJ; Helfant L; Egeghy P; Ye X; Lindstrom AB Perfluorinated Compounds in the Cape Fear Drainage Basin in North Carolina. Environ. Sci. Technol 2007, 41 (15), 5271–5276. 10.1021/es070792y. [DOI] [PubMed] [Google Scholar]
- (33).Barton KE; Starling AP; Higgins CP; McDonough CA; Calafat AM; Adgate JL Sociodemographic and Behavioral Determinants of Serum Concentrations of Per-and Polyfluoroalkyl Substances in a Community Highly Exposed to Aqueous Film-Forming Foam Contaminants in Drinking Water. International journal of hygiene and environmental health 2020, 223 (1), 256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Zhang C; Hopkins ZR; McCord J; Strynar MJ; Knappe DRU Fate of Per- and Polyfluoroalkyl Ether Acids in the Total Oxidizable Precursor Assay and Implications for the Analysis of Impacted Water. Environ. Sci. Technol. Lett 2019, 6 (11), 662–668. 10.1021/acs.estlett.9b00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wagner Adam; Buckland Tim. Chemours: GenX Polluting the Cape Fear since 1980, 2017. [Google Scholar]
- (36).McCord J; Newton S; Strynar M Validation of Quantitative Measurements and Semi-Quantitative Estimates of Emerging Perfluoroethercarboxylic Acids (PFECAs) and Hexfluoroprolyene Oxide Acids (HFPOAs). Journal of Chromatography A 2018, 1551, 52–58. 10.1016/j.chroma.2018.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).NHANES - About the National Health and Nutrition Examination Survey https://www.cdc.gov/nchs/nhanes/about_nhanes.htm (accessed 2021-03-23).
- (38).McDonough CA; Choyke S; Barton KE; Mass S; Starling AP; Adgate JL; Higgins CP Unsaturated PFOS and Other PFASs in Human Serum and Drinking Water from an AFFF-Impacted Community. Environmental Science & Technology 2021. 10.1021/acs.est.1c00522. [DOI] [PubMed] [Google Scholar]
- (39).Norris G; Vedantham R; Duvall R; Henry R EPA Unmix 6.0 Fundamentals & User Guide US Environmental Protection Agency, Office of Research and Development, Washington, DC: 2007, 20460. [Google Scholar]
- (40).Norris G; Duvall R; Brown S; Bai S Positive Matrix Factorization (PMF) 5.0 Fundamentals and User Guide 2014, 136. [Google Scholar]
- (41).Helsel DR Statistics for Censored Environmental Data Using Minitab and R, 2nd ed.; Wiley series in statistics in practice; Hoboken, N.J. : Wiley, [2012], 2012. [Google Scholar]
- (42).US EPA, O. Nonpoint Source Monitoring: TechNOTES https://www.epa.gov/nps/nonpoint-source-monitoring-technotes (accessed 2022-02-03).
- (43).Hekster FM; Laane RW; De Voogt P Environmental and Toxicity Effects of Perfluoroalkylated Substances. Reviews of Environmental Contamination and Toxicology 2003, 99–121. [DOI] [PubMed] [Google Scholar]
- (44).Henry RC; Christensen ER Selecting an Appropriate Multivariate Source Apportionment Model Result. Environ. Sci. Technol 2010, 44 (7), 2474–2481. 10.1021/es9018095. [DOI] [PubMed] [Google Scholar]
- (45).Hopke P A Guide to Positive Matrix Factorization, in Workshop on UNMIX and PMF as Applied to PM2. 5 Edited by Willis RD, RTP, NC; EPA 600/A-00/048, 2000. [Google Scholar]
- (46).Hopke Phillip K.. Trace Element Concentrations in Summer Aerosols at Rural Sites in New York State and Their Possible Sources and Seasonal Variations in the Composition of Ambient Sulfate-Containing Aerosols in the New York Area. Atmospheric Environment 1982, 16 (5), 1279–1280. [Google Scholar]
- (47).Pio CA; Santos IM; Anacleto TD; Nunes TV; Leal RM Particulate and Gaseous Air Pollutant Levels at the Portuguese West Coast. Atmospheric Environment. Part A. General Topics 1991, 25 (3–4), 669–680. 10.1016/0960-1686(91)90065-F. [DOI] [Google Scholar]
- (48).Hu XC; Dassuncao C; Zhang X; Grandjean P; Weihe P; Webster GM; Nielsen F; Sunderland EM Can Profiles of Poly- and Perfluoroalkyl Substances (PFASs) in Human Serum Provide Information on Major Exposure Sources? Environ Health 2018, 17 (1), 11. 10.1186/s12940-018-0355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Anderson RH; Long GC; Porter RC; Anderson JK Occurrence of Select Perfluoroalkyl Substances at US Air Force Aqueous Film-Forming Foam Release Sites Other than Fire-Training Areas: Field-Validation of Critical Fate and Transport Properties. Chemosphere 2016, 150, 678–685. [DOI] [PubMed] [Google Scholar]
- (50).Backe WJ; Day TC; Field JA Zwitterionic, Cationic, and Anionic Fluorinated Chemicals in Aqueous Film Forming Foam Formulations and Groundwater from US Military Bases by Nonaqueous Large-Volume Injection HPLC-MS/MS. Environmental science & technology 2013, 47 (10), 5226–5234. [DOI] [PubMed] [Google Scholar]
- (51).Rotander A; Toms L-ML; Aylward L; Kay M; Mueller JF Elevated Levels of PFOS and PFHxS in Firefighters Exposed to Aqueous Film Forming Foam (AFFF). Environment international 2015, 82, 28–34. [DOI] [PubMed] [Google Scholar]
- (52).Xu Y; Nielsen C; Li Y; Hammarstrand S; Andersson EM; Li H; Olsson DS; Engström K; Pineda D; Lindh CH; Fletcher T; Jakobsson K Serum Perfluoroalkyl Substances in Residents Following Long-Term Drinking Water Contamination from Firefighting Foam in Ronneby, Sweden. Environment International 2021, 147, 106333. 10.1016/j.envint.2020.106333. [DOI] [PubMed] [Google Scholar]
- (53).Li Y; Fletcher T; Mucs D; Scott K; Lindh CH; Tallving P; Jakobsson K Half-Lives of PFOS, PFHxS and PFOA after End of Exposure to Contaminated Drinking Water. Occup Environ Med 2018, 75 (1), 46–51. 10.1136/oemed-2017-104651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).McDonough CA; Choyke S; Ferguson PL; DeWitt JC; Higgins CP Bioaccumulation of Novel Per- and Polyfluoroalkyl Substances in Mice Dosed with an Aqueous Film-Forming Foam. Environ. Sci. Technol 2020, 54 (9), 5700–5709. 10.1021/acs.est.0c00234. [DOI] [PubMed] [Google Scholar]
- (55).Barzen-Hanson KA; Roberts SC; Choyke S; Oetjen K; McAlees A; Riddell N; McCrindle R; Ferguson PL; Higgins CP; Field JA Discovery of 40 Classes of Per- and Polyfluoroalkyl Substances in Historical Aqueous Film-Forming Foams (AFFFs) and AFFF-Impacted Groundwater. Environ Sci Technol 2017, 51 (4), 2047–2057. 10.1021/acs.est.6b05843. [DOI] [PubMed] [Google Scholar]
- (56).Egan Katie. FDA’s Total Diet Study: Monitoring U.S. Food Supply Safety https://www.food-safety.com/articles/4381-fdas-total-diet-study-monitoring-us-food-supply-safety (accessed 2021-10-25).
- (57).Domingo JL; Nadal M Per-and Polyfluoroalkyl Substances (PFASs) in Food and Human Dietary Intake: A Review of the Recent Scientific Literature. Journal of agricultural and food chemistry 2017, 65 (3), 533–543. [DOI] [PubMed] [Google Scholar]
- (58).Dassuncao C; Hu XC; Nielsen F; Weihe P; Grandjean P; Sunderland EM Shifting Global Exposures to Poly- and Perfluoroalkyl Substances (PFASs) Evident in Longitudinal Birth Cohorts from a Seafood-Consuming Population. Environ. Sci. Technol 2018, 52 (6), 3738–3747. 10.1021/acs.est.7b06044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Wang Y; Liu J; Li J; Zhao Y; Wu Y Dietary Exposure of Chinese Adults to Perfluoroalkyl Acids via Animal-Origin Foods: Chinese Total Diet Study (2005–2007 and 2011–2013). Journal of agricultural and food chemistry 2019, 67 (21), 6048–6055. [DOI] [PubMed] [Google Scholar]
- (60).Macheka LR; Olowoyo JO; Mugivhisa LL; Abafe OA Determination and Assessment of Human Dietary Intake of per and Polyfluoroalkyl Substances in Retail Dairy Milk and Infant Formula from South Africa. Science of The Total Environment 2021, 755, 142697. [DOI] [PubMed] [Google Scholar]
- (61).Susmann HP; Schaider LA; Rodgers KM; Rudel RA Dietary Habits Related to Food Packaging and Population Exposure to PFASs. Environmental health perspectives 2019, 127 (10), 107003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Xu Y; Fletcher T; Pineda D; Lindh CH; Nilsson C; Glynn A; Vogs C; Norström K; Lilja K; Jakobsson K; Li Y Serum Half-Lives for Short- and Long-Chain Perfluoroalkyl Acids after Ceasing Exposure from Drinking Water Contaminated by Firefighting Foam. Environ Health Perspect 2020, 128 (7), 077004. 10.1289/EHP6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).AECOM. Perfluorobutane Sulfonic Acid (PFBS) Chemistry, Production, Uses and Environmental Fate 2019, 176. [Google Scholar]
- (64).MDHHS. Participant Demographics and Serum PFAS Summary Report Report 1 of the North Kent County Exposure Assessment; Division of Environmental Health, Translator; 2020. [Google Scholar]
- (65).Control, C. for D.; Prevention. Fourth Report on Human Exposure to Environmental Chemicals, Updated Tables Atlanta, GA: US Department of Health and Human Services, Centers for Disease Control and Prevention; 2019. [Google Scholar]
- (66).Zhang Y; Guo C-S; Xu J; Tian Y-Z; Shi G-L; Feng Y-C Potential Source Contributions and Risk Assessment of PAHs in Sediments from Taihu Lake, China: Comparison of Three Receptor Models. Water research 2012, 46 (9), 3065–3073. [DOI] [PubMed] [Google Scholar]
- (67).5 Environmental Fate and Transport Processes – PFAS — Per- and Polyfluoroalkyl Substances https://pfas-1.itrcweb.org/5-environmental-fate-and-transport-processes/ (accessed 2022-02-15).
- (68).US EPA, O. Guidance for Reporting on the Environmental Fate and Transport of the Stressors of Concern in Problem Formulations https://www.epa.gov/pesticide-science-and-assessing-pesticide-risks/guidance-reporting-environmental-fate-and-transport (accessed 2022-02-03).
- (69).U.S. Environmental Protection Agency. Third Unregulated Contaminant Monitoring Rule https://www.epa.gov/dwucmr/third-unregulated-contaminant-monitoring-rule (accessed 2020-12-20).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


