Abstract
Background
Multidrug-resistant (MDR) bacteria are acknowledged as one of the main factors contributing to chronic illnesses and fatalities globally. Numerous diseases, including bloodstream infections, pneumonia, urinary tract infections, and surgical site infections, can be brought on by MDR bacteria. Therefore, a crucial topic of continuing research is the development of a novel and different treatment for MDR microbial pathogens. This work is introduce an alternative method for elimination of MDR bacterial isolates which are causative agents of urinary tract infection among people in Egypt. In our study, we need a novel strategy to combat MDR bacteria by green-synthesized metal nanoparticles (MNPs). That is due to the ability of MNPs to penetrate the cell wall and the cell membrane of gram-positive and gram-negative bacteria.
Methods
Clinical isolates of MDR bacteria had their antibiotic susceptibility assessed before being molecularly identified using 16 s rRNA, sequencing, and phylogenetic analysis. Also, genetic profiles of isolated strains were performed using ISSR and SDS-PAGE. Finally, characterized plant-mediated silver nanoparticles derived from lemon and pomegranate peel extracts were evaluated against isolated multidrug-resistant bacterial stains.
Results
In our present trial, one-hundred urine samples were collected from 71 females and 29 males complaining of UTI (urinary tract infection) symptoms. One-hundred microbial isolates were isolated, including 88-g negative and only 8-g positive bacteria in addition to four yeast isolates (Candida species). A total of 72% of the isolated bacteria showed MDR activity. The most prevalent MDR bacterial isolates (Escherichia coli, Pseudomonas aeruginosa, Acinetobacter baumannii, Enterococcus faecalis, and Klebsiella pneumoniae) were identified through 16S rDNA PCR sequencing as with accession numbers OP741103, OP741104, OP741105, OP741106, and OP741107, respectively. Lemon and pomegranate-mediated silver nanoparticles [Ag-NPs] were characterized by UV spectroscopy, FTIR, XRD, and TEM with average size 32 and 28 nm, respectively. Lemon and pomegranate-mediated silver nanoparticles [Ag-NPs] showed an inhibitory effect on the selected five MDR isolates at MIC 50 and 30 µg/mL, respectively. These common bacterial isolates were also genetically examined using ISSR PCR, and their total protein level was evaluated using SDS-PAGE, showing the presence of distinct genetic and protein bands for each bacterial species and emphasizing their general and protein composition as a crucial and essential tool in understanding and overcoming MDR behavior in UTI patients.
Conclusions
Lemon and pomegranate-mediated silver nanoparticles [Ag-NPs] were found to have an inhibitory effect on MDR isolates. Therefore, the study suggests that [Ag-NPs] could be a potential treatment for MDR UTI infections caused by the identified bacterial species.
Supplementary Information
The online version contains supplementary material available at 10.1186/s43141-023-00547-0.
Keywords: Lemon peel extract, Pomegranate peel extract, Silver nanoparticles, ISSR, SDS-PAGE, Multidrug-resistant bacteria
Background
UTI (bacteriuria) is the medical term for the presence of bacteria in the urine. Although clinical infection might start at 103 bacteria/mL, “remarkable” bacteriuria for epidemiological purposes should be at least 105 bacteria/mL in freshly voided urine [1]. UTI is the most frequent infectious illness in the world, impacting more than 10% of the world’s 150 million individuals per day. Clusters of virulence factors are a significant particular cause of multidrug resistance in the urinary tract [2]. MDR pathogens in urine make UTI harder to treat empirically and increase infection prevalence [3–5]. Worldwide, the uncommon appearance and growth of multidrug-resistant bacterial infections are a serious public health risk that is emerging in ever-expanding communities [6, 7]. Over the past few decades, the emergence of MDR uropathogens has led to an increase in hospital-acquired UTI infections worldwide [8, 9]. Many microbial species and isolates have not yet been identified or described [10]. Several identification techniques are currently available, ranging from total protein and isozyme profiles to DNA/DNA hybridization and sequence analysis of the 16S rDNA region [11–13]. The availability of molecular markers such as 16S rDNA [14], RAPD [15, 16], and ISSR [17] facilitates the development of phylogenetic relationships and the identification and characterization of bacterial species. Protein banding patterns using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) technique are used to distinguish infectious wild-type organisms from their mutants [18]. The biosynthesis of metal oxide nanoparticles utilizing biosynthetic agents derived from plants or microbes is best suited for producing nanomaterials that are already surrounded by cellular proteins, eliminating the requirement for additional cell outer layers or polymers. It is one of the potential approaches [19]. Metal oxide nanoparticles, such as silver, titanium, and zinc oxide nanoparticles, have been extensively researched for their antibacterial effectiveness against different MDR illnesses [20, 21]. The effect of metal oxide nanoparticles in reducing MDR bacteria growth is highlighted by a range of cellular damaging and molecular processes. Metal nanoparticles bind to cellular proteins, altering membrane permeability and protein structure, or to DNA structures, interfering with proper cell division and MDR growth [22]. Third- and fourth-generation cephalosporins and monobactams can be hydrolyzed by extended-spectrum beta-lactamase (ESBL) enzymes, whereas cephamycins and carbapenems cannot. Sulbactam or tazobactam inhibits them, and the genetic determinants are typically associated with plasmids but frequently result in co-resistance (aminoglycosides, quinolones). Due to the high prevalence of E. coli in the general population and the fluctuating incidence of K. pneumoniae (which is higher in hospitals), ESBLs pose a public health risk [23]. Size, shape, surface functionalization, surface charges, and the capacity to co-encapsulate drugs all affect how effective metal nanoparticles (MNPs) are at fighting multidrug-resistant bacterial infections. Recent research has focused heavily on the effectiveness of silver, gold, zinc oxide, selenium, copper, cobalt, and iron oxide nanoparticles against multidrug-resistant bacterial infections [24].
Methods
Isolation of clinical samples
Urine samples were collected from 100 patients (29 males and 71 females), showing symptoms of UTI, attending the outpatient clinics of Suez Canal Authority Hospital during over 8-month period (May 2020–March 2021). The patient’s age ranged from 15 to ≥ 60 years.
Antibiotic susceptibility
Antibiotic sensitivity test of twenty-five different antibiotics was performed biochemically through disc diffusion method on Muller-Hinton agar (according to guidelines of CLSI 2020). Then, the resistant bacterial isolates were confirmed using VITEK 2 system [25].
Molecular identification and genetic diversity studies
Genetic diversity was performed for the most prevalent bacterial isolates using intersimple sequence repeat (ISSR), 16 s rRNA, and sequencing, and phylogenetic tree was constructed. DNA extraction from bacterial isolates was performed using the (QIAamp DNA Mini Kit, Qiagen, Germany) according to manual instructions, six ISSR primers were used (1-6) as in Table 1.
Table 1.
Different six primers used in ISSR amplification of tested five MDR isolates
| ISSR1 5-ACACACACACACACACG-3 | ISSR2 5-ACACACACACACACACT-3 |
| ISSR3 5-ACACACACACACACACC-3 | ISSR4 5-AGAGAGAGAGAGAGAGC-3 |
| ISSR5 5-CTCTCTCTCTCTCTCTG-3 | ISSR6 5-CTCTCTCTCTCTCTCTCTA-3 |
Data analysis of genetic diversity
The SIMQUAL module was used to determine genetic similarity based on the Jaccard coefficient, and the NTSYS version 2.20 software package was used to tree the data using the unweighted pair-group method with the arithmetic mean (UPGMA) option of the SAHN module. A graph has been made. PAST version 3.16 was used to perform principal coordinate analysis (PCoA) to link genetic relationships between populations (MDR UTI-causing bacteria). Protein profiles were captured as binary data, that is, 1 or 0 depending on the presence or absence of bands, and gels were viewed immediately with the naked eye. A band was indicated as 1 if it was present in a genotype, but it was marked as 0 if it was totally present. The similarity and association between the protein traces of test strains were displayed in a dendrogram using the unweight pair-group method with arithmetic averages algorithm (UPGMA) of the protein patterns of whole-cell protein of species.
Statistical cluster analysis
Using unweighted pair-group method with average (UPGMA) and principal components analysis (PCA) was performed with PAST 3.18 software [26]. The polymorphic information content (PIC) value was calculated according to the given formula [27]. The percentage polymorphism was calculated according to the following formula. Polymorphism (%) = (number of polymorphic bands/total number of bands) × 100.
Synthesis of nanoparticles
Lemon peel and pomegranate fruit peel were hand pulverized after being taken from the whole fruit and got dried in an oven until totally dried. About 50-g dried fruit peel powders were suspended to make extracts. For 1 h, the resultant mixtures were sonicated with occasional vortexing. The suspensions were then filtered through Whatman No. 1 filter paper, excess methanol from the filtrate was evaporated using a rotary evaporator, residual water was removed by lyophilization, and the filtrate was stored at 4 °C until further use [28].
Silver nanoparticles [Ag-NPs] materials
Fresh lemon leaves and pomegranate peel as reducing agents have been purchased from commercial markets, and the identification was carried out through morphological characters at the Department of Botany, Faculty of Science, Al-Azhar University [28]. Silver nitrate [AgNO3] was bought from Sigma-Aldrich with ≥ 99.5% purity from Egypt.
Preparation of leaves extract
Fresh lemon leaves and pomegranate peels were obtained from the market and gathered and properly rinsed with tap water before being extensively cleansed with sterile Milli-Q water until removing all contaminants. To get the leaves extract, 5 g of leaves and peels were combined with 100 mL of distilled water and heated for 5 min at 80 °C before chilling and filtering through Whatman No. 1 filter paper. The collected filtered leaf extract was utilized immediately for further research [29, 30].
Silver nanoparticles synthesis
A 50-mL solution from 5-mM silver nitrate was produced. Separately, 0.05 g/mL of two extracts was gradually poured on silver nitrate solution at 60 °C to synthesize the colloid. The fast shift in hue to brownish yellow after 5 min indicates that the silver ions formed quickly, validating the creation of Ag-NPs, and there was no further change in color. The Ag-NPs were then moved to a clean container in a dark environment for future research [31, 32].
Silver nanoparticles characterization
Characterization of Ag-NPs was accomplished by visual observation and other procedures; in a visual observation, the change in color solution from colorless to reddish brown was noticed with the naked eye indicating the formation of [Ag-NPs]. UV–visible spectroscopy was used to identify Ag-NPs. FTIR spectroscopy was used to study biomolecules in lemon solution as well as the surface chemistry of Ag-NPs. Transmission electron microscopy (TEM) was used to confirm the surface shape and particle size of the Ag-NPs using high-resolution pictures [33, 34].
Antibacterial activity of NPs
Both Ag-NPs from two plant extracts were evaluated for their potential bactericidal activity against Escherichia coli, Pseudomonas aeruginosa, Acinetobacter baumannii, Enterococcus faecalis, and Klebsiella pneumoniae through the well agar diffusion technique on Muller-Hinton medium for 24 h at 37 °C. Fifty liters of NP solution was put within an agar well and then allowed to diffuse before being incubated.
Results and discussion
Isolation of bacterial isolates of UTI patients
Bacterial species isolated from UTI cases
One-hundred mid-stream urine samples were collected from 71 females and 29 males, attending the outpatient departments (OPDs) (64%) and in-patient departments)36%) of Suez Canal Authority Hospital during over 8-month period (May 2020–March 2021). Clinical evidence of urinary tract infection was determined by urologists. Patient’s age ranged from 10 to ≥ 60 years with average age 58 years. Patients on antibiotic therapy were excluded from the study. Age < 60 years was the most common age group in the study population (51%), followed by the age group of 30–60 years (34%) and < 30 years (15%). The recovered microbial isolates were divided into 88-g negative bacteria (88 isolates), gram positive ones (only eight isolates), and four yeast isolates (Candida sp.).
Standing on VITEK report of biochemical analysis of one-hundred isolates was defined as shown in the Table 2.
Table 2.
Frequency of bacterial isolates of UTI patients
| Microorganism | Number (%) |
|---|---|
| Gram-negative bacteria | |
| E. coli | 46 (46%) |
| Klebsiella pneumoniae | 24 (24%) |
| Pseudomonas aeruginosa | 10 (10%) |
| Enterobacter cloacae | 2 (2%) |
| Acinetobacter baumannii | 1 (1%) |
| Citrobacter koseri | 1 (1%) |
| Serratia liquefaciens | 1 (1%) |
| Serratia plymuthica | 1 (1%) |
| Sphingomonas paucimobilis | 1 (1%) |
| Morganella morganii | 1 (1%) |
| Gram-positive bacteria | |
| Staphylococcus lentus | 2 (2%) |
| Staphylococcus haemolyticus | 2 (2%) |
| Staphylococcus xylosus | 1 (1%) |
| Enterococcus faecalis | 1 (1%) |
| Staphylococcus saprophyticus | 1 (1%) |
| Enterococcus spp. | 1 (1%) |
| Yeast | |
| Candida tropicalis | 3 (3%) |
| Candida dubliniensis | 1 (1%) |
Antibiotic susceptibility
All isolates were tested against different groups antibiotics depending on type of genus and species of the tested isolates; after conducting statistical operations, the samples were selected of E. coli (n = 2), E. faecalis (n = 21), P. aeruginosa (n = 38), A. baumannii (n = 45), and K. pneumoniae (n = 87) as the most resistant isolates to antibiotics to conduct genetic and protein experiments on them for which their results were as follows in Table 3.
Table 3.
Antibiogram of selected microorganisms isolated from UTI-infected patients (n = 100) against different twenty-five antibiotics
| Species | Resistant to (n) (%) | Antibiotics symbols |
|---|---|---|
| E. coli | 15 (83.33%) | AMP, SAM, TZP, FOX, CAZ, CRO, FEP, MEM, AK, TOB, CIP, LEV, SXT, ATM, ETP |
| E. faecalis | 5(71.42%) | E, RD, CIP, LZD, TE |
| P. aeruginosa | 9 (81.81%) | CAZ, FEP, MEM, CN, TOB, CIP, LEV, ATM, IPM |
| A. baumannii | 10 (83.33%) | SAM, TZP, CAZ, CRO, FEP, MEM, CN, TOB, CIP, LEV |
| K. pneumoniae | 17 (94.44%) | AMP, SAM, TZP, FOX, CAZ, CRO, MEM, AK, CN, TOB, CIP, LEV, F, SXT, ATM, ETP, IPM |
AMP ampicillin, SAM ampicillin/sulbactam, TZP piperacillin/tazobactam, FOX cefoxitin, CAZ ceftazidime, CRO ceftriaxone, FEP cefepime, MEM, meropenem, AK amikacin, CN gentamicin, TOB tobramycin, CIP ciprofloxacin, F nitrofurantoin, SXT trimethoprim/sulfamethoxazole, ATM aztreonam, ETP ertapenem, IPM imipenem, E erythromycin, MXF moxifloxacin, RD rifampicin, LZD linezolid, DA clindamycin, TE tetracycline, TEC teicoplanin, LEV levofloxacin
Genetic diversity of selected isolates using ISSR
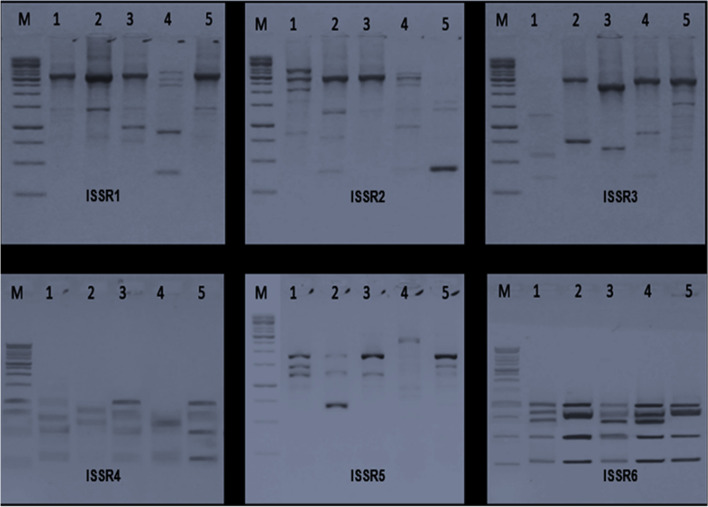
ISSR-PCR is considered to be an effective approach for phylogenetic categorization of bacterial (prokaryotic) genomes in general, as well as diagnostic genotyping of microbial pathogens. Using six ISSR primers, a DNA sample representing the five bacterial strains was submitted to PCR analysis. SSR analysis can be helpful for figuring out how genotypes relate to population information like morphology, illness susceptibility, and location [35]. The primers were produced a consistent and reproducible banding pattern for the strain tested, which is explain in Fig. 1. According to the data in Table 4, the polymorphism percentage (P%) and polymorphism information content (PIC) for the pattern formed by the ISSR marker revealed that the total DNA fragments produced were 117 fragments with molecular weights ranging from 250 to 2500 bp. The six ISSR primers used for five bacterial pathogens were found to be polymorphic, yielding a total of 36 bands, 16 of which were polymorphic. whereas 13 bands were monomorphic. The polymorphism percentages (P%) of applied six primers were 57.17, 42.85, 60.00, 20.00, 66.66, and 16.66%, respectively. Both primers (3 and 5) have the highest P% with 60.00% polymorphism.
Fig. 1.
Patterns of ISSR electrophoresis of selected strains. Lane 1: DNA ladder (PageRuler™ Plus Prestained). Lanes 2–6: represent isolates from (1–5) which represent as E. coli, P. aeruginosa, A. baumannii, E. faecalis, and K. pneumoniae
Table 4.
Polymorphism generated by ISSR of the selected strains
| Primer | MW | E. coli | P. aeruginosa | A. baumannii | E. faecalis | K. pneumoniae | Frequency | Polymorphism | PB | MB | Unique | TB | P% | PIC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ISSR1 | 4000 | 0 | 0 | 0 | 1 | 0 | 20 | Unique | ||||||
| 3000 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | 4 | 1 | 2 | 7 | 57.17 | 0.320 | |
| 2500 | 1 | 1 | 0 | 1 | 0 | 60 | Polymorphic | |||||||
| 1500 | 1 | 1 | 1 | 0 | 1 | 80 | Polymorphic | |||||||
| 750 | 0 | 0 | 1 | 1 | 0 | 40 | Polymorphic | |||||||
| 500 | 0 | 0 | 0 | 1 | 1 | 40 | Polymorphic | |||||||
| 250 | 0 | 0 | 0 | 1 | 0 | 20 | Unique | |||||||
| ISSR2 | 3500 | 1 | 0 | 0 | 0 | 1 | 40 | Polymorphic | 4 | 1 | 2 | 7 | 57.14 | 0.297 |
| 3100 | 1 | 0 | 0 | 0 | 1 | 40 | Polymorphic | |||||||
| 3000 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | |||||||
| 1500 | 1 | 0 | 0 | 0 | 0 | 20 | Unique | |||||||
| 1000 | 1 | 0 | 0 | 0 | 0 | 20 | Unique | |||||||
| 750 | 0 | 1 | 1 | 1 | 1 | 80 | Polymorphic | |||||||
| 400 | 0 | 1 | 0 | 1 | 1 | 60 | Polymorphic | |||||||
| ISSR3 | 3000 | 0 | 1 | 1 | 1 | 1 | 80 | Polymorphic | 3 | 1 | 1 | 5 | 60.00 | 0.320 |
| 1500 | 0 | 0 | 0 | 0 | 1 | 20 | Unique | |||||||
| 1000 | 1 | 0 | 0 | 0 | 1 | 40 | Polymorphic | |||||||
| 750 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | |||||||
| 500 | 1 | 0 | 0 | 1 | 0 | 40 | Polymorphic | |||||||
| ISSR4 | 1000 | 1 | 0 | 1 | 0 | 1 | 60 | Polymorphic | 1 | 4 | 0 | 5 | 20.00 | 0.0961 |
| 750 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | |||||||
| 650 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | |||||||
| 500 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | |||||||
| 250 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | |||||||
| ISSR5 | 2500 | 0 | 0 | 0 | 1 | 0 | 20 | Unique | 4 | 0 | 3 | 7 | 57.14 | 0.3733 |
| 2200 | 1 | 1 | 1 | 0 | 1 | 80 | Polymorphic | |||||||
| 2000 | 1 | 0 | 0 | 0 | 1 | 40 | Polymorphic | |||||||
| 1500 | 1 | 1 | 1 | 0 | 1 | 80 | Polymorphic | |||||||
| 750 | 1 | 0 | 1 | 1 | 1 | 40 | Polymorphic | |||||||
| 700 | 0 | 0 | 0 | 1 | 0 | 20 | Unique | |||||||
| 600 | 0 | 1 | 0 | 0 | 0 | 20 | Unique | |||||||
| ISSR6 | 1000 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | 1 | 5 | 0 | 6 | 16.66 | 0.081 |
| 950 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | |||||||
| 850 | 1 | 0 | 1 | 1 | 0 | 60 | Polymorphic | |||||||
| 750 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | |||||||
| 500 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | |||||||
| 250 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | |||||||
| No. of fragments | 27 | 20 | 21 | 21 | 28 | 117 fragments | 16 | 13 | 7 | |||||
PB number of polymorphic bands, MB number of monomorphic bands, TB number of total bands, %P percent polymorphism, PIC polymorphism information content
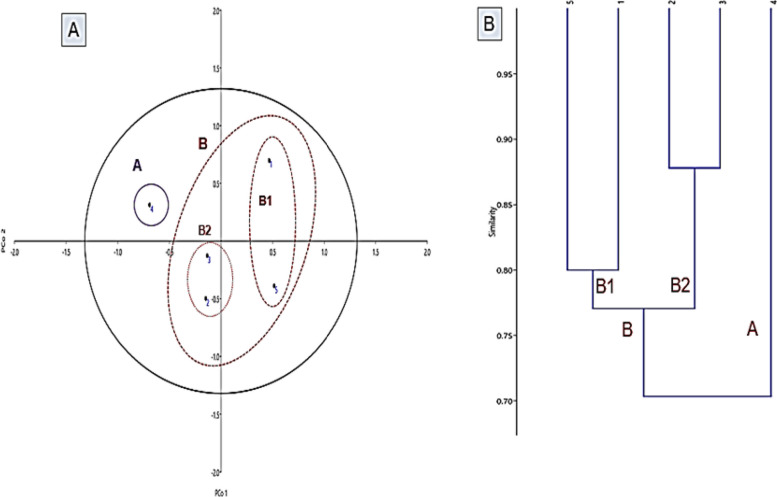
Two unique bands produced by three primers (1, 2, and 5) for pathogens are as follows: E. coli and K. pneumoniae, respectively. Also, polymorphism information contents (PIC) for ISSR primers (1, 2, 3, 4, 5, and 6) were calculated as 0.320, 0.297, 0.320, 0.0961, 0.3733, and 0.081 generated from ISSR pattern. As shown in Fig. 2, principal correlation analysis (PCoA) of isolated bacterial strains using ISSR pattern showed three clouds: A, B1, and B2. These results were similar and agree with [36] who used PCR-based SSR amplification followed by amplicon size determination to analyze the spread of the pathogens (Haemophilus influenzae). Furthermore, the utility of inter-simple sequence repeat PCR (ISSR-PCR) assay in the characterization and elucidation of the phylogenetic relationship between the pathogenic isolates of Vibrio cholerae [37]. The application of principal component analysis (PcoA) is the most basic multivariate data reduction statistical technique. The application revealed that two out of five principal components were significant (eigenvalue > 1) and contributed 58.07% of the total variation. Pco1 accounted for 32.33%, and Pco2 accounted for 25.71% of the total variation; also, the same grouping pattern was found in the cluster analysis, indicating that significant variation exists in this study. The separation of Pco1 and Pco2 showed that the five bacterial isolates were dispersed in all quarters, indicating a high level of genotypic variation among the bacterial isolates. These results were parallel to [38] who performed molecular identification and genetic diversity among Photorhabdus and Xenorhabdus microbes.
Fig. 2.
A Principal correlation analysis (PcoA) selected strains. B Dendrogram of ISSR based on unweighted pair-group method with arithmetic averages algorithm (UPGMA) for selected strains, isolates code (1–5) was represented as E. coli, P. aeruginosa, A. baumannii, E. faecalis, and K. pneumoniae
Genetic diversity of selected isolates using SDS-PAGE
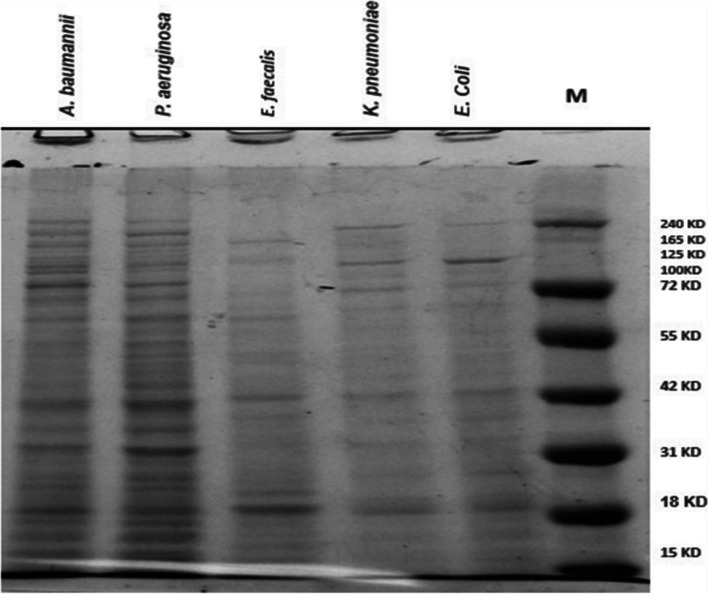
SDS-PAGE as an important simple, powerful, and rapid molecular technique for microbial species identification for comparing microbial pathogens [39]. SDS-PAGE analysis showed a total of 66 bands with molecular weight ranging from 10 to 250 KD. Eight polymorphic protein bands at the molecular size of 250, 130.0, 70.0, 35.6, 30.00, 28.00, 18.55, and 10.0 KD. Also, eight monomorphic protein bands as fragment sizes of 100, 75.0, 55.5, 25.0, 20.0, 15.0, 14.0, and 12.0 KD for all tested bacterial isolates (Fig. 3).
Fig. 3.
Patterns of SDS-PAGE electrophoretic protein of the selected strains. Lane1: protein ladder (PageRuler™ Plus Prestained). Lane (2–6): represent isolates from (1–5) which represent as E. coli, K. pneumoniae, E. faecalis, P. aeruginosa, and A. baumannii
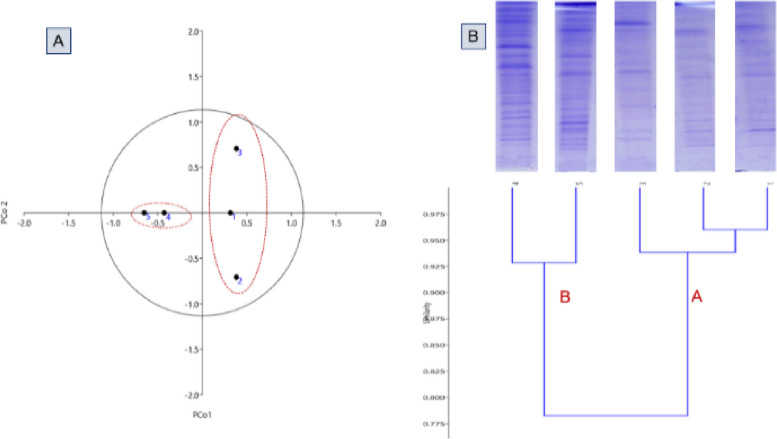
Data presented in Table 5 explained that polymorphism percentage (%) was 50.0% for all polymorphic bands. Total fragments of all isolates were 66 fragments, and bacterial isolate E. coli, K. pneumoniae, E. faecalis, P. aeruginosa, and A. baumannii were produced 13, 12, 12, 15, and 13 fragments, respectively. Also, the polymorphism information content (PIC) was calculated as 0.231 generated from the SDS-PAGE pattern. Therefore, our results was similar with [40] reporting that the genetic similarity and protein profile of the isolated field German strains were compared to Mycoplasma bovis reference strain using SDS-PAGE. In addition, [39] examined the total proteins of 20 isolates that were taken from the NIH Islamabad using SDS-PAGE to determine the variability in the gene pool of E. coli, as shown in Fig. 4. Principal component analysis (PCA), a powerful tool and the fundamental multidimensional data reduction statistical approach, found that two of the five principal component analysis remained significant (eigenvalue > 1) and accounted for 85.86% of the total variance. PC1 accounted for 72.99% of the overall variance, whereas PC2 accounted for 12.87%. When PC1 and PC2 were separated, different microbial strains emerged in all quarters, showing a significant amount of genotypic variety.
Table 5.
Polymorphism generated by SDS-PAGE of the selected strains
| MW | E. coli | K. pneumoniae | E. faecalis | P. aeruginosa | A. baumannii | Frequency | Polymorphism | PB | MB | Unique | TB | P% | PIC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 250.0 | 0 | 0 | 0 | 1 | 1 | 40.0 | Polymorphic | 8 | 8 | - | 16 | 50.00 | 0.231 |
| 130.0 | 1 | 1 | 0 | 1 | 1 | 80.0 | Polymorphic | ||||||
| 100.0 | 1 | 1 | 1 | 1 | 1 | 100 | Monomorphic | ||||||
| 75.00 | 1 | 1 | 1 | 1 | 1 | 100.0 | Monomorphic | ||||||
| 70.00 | 1 | 1 | 1 | 1 | 0 | 80.0 | Polymorphic | ||||||
| 55.60 | 1 | 1 | 1 | 1 | 1 | 100.0 | Monomorphic | ||||||
| 35.04 | 1 | 1 | 1 | 1 | 0 | 80.0 | Polymorphic | ||||||
| 30.00 | 1 | 1 | 1 | 0 | 0 | 60.0 | Polymorphic | ||||||
| 28.00 | 0 | 0 | 0 | 1 | 1 | 40.0 | Polymorphic | ||||||
| 25.00 | 1 | 1 | 1 | 1 | 1 | 100.0 | Monomorphic | ||||||
| 20.00 | 1 | 1 | 1 | 1 | 1 | 100.0 | Monomorphic | ||||||
| 18.55 | 1 | 0 | 1 | 1 | 1 | 80.0 | Polymorphic | ||||||
| 15.00 | 1 | 1 | 1 | 1 | 1 | 100.0 | Monomorphic | ||||||
| 14.00 | 1 | 1 | 1 | 1 | 1 | 100.0 | Monomorphic | ||||||
| 12.00 | 1 | 1 | 1 | 1 | 1 | 100.0 | Monomorphic | ||||||
| 10.00 | 0 | 0 | 0 | 1 | 1 | 40.0 | Polymorphic | ||||||
| Total | 13 | 12 | 12 | 15 | 13 | 66 bands | |||||||
PPB number of polymorphic bands, MB number of monomorphic bands, TB number of total bands, %P percent polymorphism, PIC polymorphism information content
Fig. 4.
A Principal component analysis (PCA) of the selected strains. B Dendrogram of SDS-PAGE based on unweighted pair-group method with arithmetic averages algorithm (UPGMA) for the selected strains, 1–5 represent as E. coli, K. pneumoniae, E. faecalis, P. aeruginosa, and A. baumannii
The numerical analysis clearly revealed two distinct clusters as shown in the dendrogram; it was classified into two main clusters (A and B). Cluster A includes two isolates (P. aeruginosa and A. baumannii), and cluster B includes three isolates (E. coli, K. pneumoniae, and E. faecalis); these results were similar to [41] who reported that genetic diversity of the dominant bacteria isolated in western Iran. Also, These results were parallel to [42]. They stated that the SDS-PAGE whole cell protein patterns and 16S rRNA gene sequence analysis were used to compare the biodiversity of lactic acid-producing bacteria in garlic and green onion samples. Also Jamalzadeh et al., 2021 [43] said that the SDS-PAGE approach, in combination with computational analysis of protein profiles, is an excellent way for investigating taxonomic relationships among Bacillus species. SDS-PAGE was also used to characterize two species of bacteria, Pseudomonas aeruginosa and E. coli [44].
Genomic identification of extensive MDR isolates
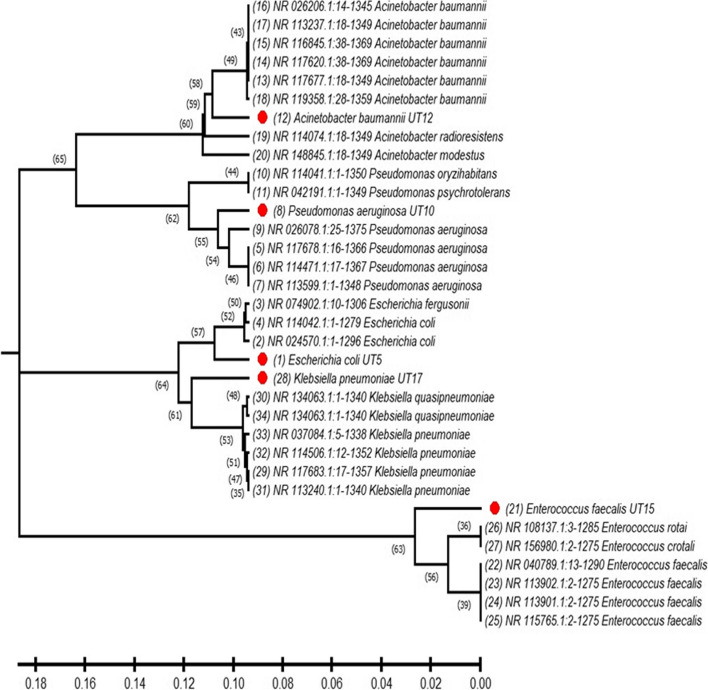
The different five extensive MDR bacterial isolates were identified by 16S rDNA gene sequencing and submitted through GenBank database. They were identified as E. coli, P. aeruginosa, A. baumannii, E. faecalis, and K. pneumoniae with accession numbers OP741103, OP741104, OP741105, OP741106, and OP741107, respectively. Also, phylogenetic trees for the five isolates were also generated in comparison with top ten hints for each one through NCBI BLASTn tool and phylogenetic trees then modified through MEGA11 software as in Fig. 5. Phylogenetic tree of multidrug-resistant bacterial isolates shows the evolutionary relationships between the five clusters of multidrug-resistant bacterial isolates. Clade A includes A. baumannii, Clade B includes P. aeruginosa, Clade C includes E. coli, Clade D includes K. pneumoniae, and Clade E includes E. faecalis. The tree suggests that these five clusters of bacteria have evolved from different common ancestors and have evolved different mechanisms of antibiotic resistance.
Fig. 5.
Phylogenetic tree of the extensive five multidrug-resistant bacterial isolates. [Clade A] include A. baumannii and other species belong to the Acinetobacter genus. [Clade B] include P. aeruginosa and other species belong to the Pseudomonas genus. [Clade C] include E. coli and other species belong to the Escherichia genus. [Clade D] include K pneumoniae and other species belong to the Klebsiella genus. [Clade E] include E. faecalis and other species belong to the Enterococcus genus
Characterization of plant-mediated NPs
UV–Vis absorption spectra
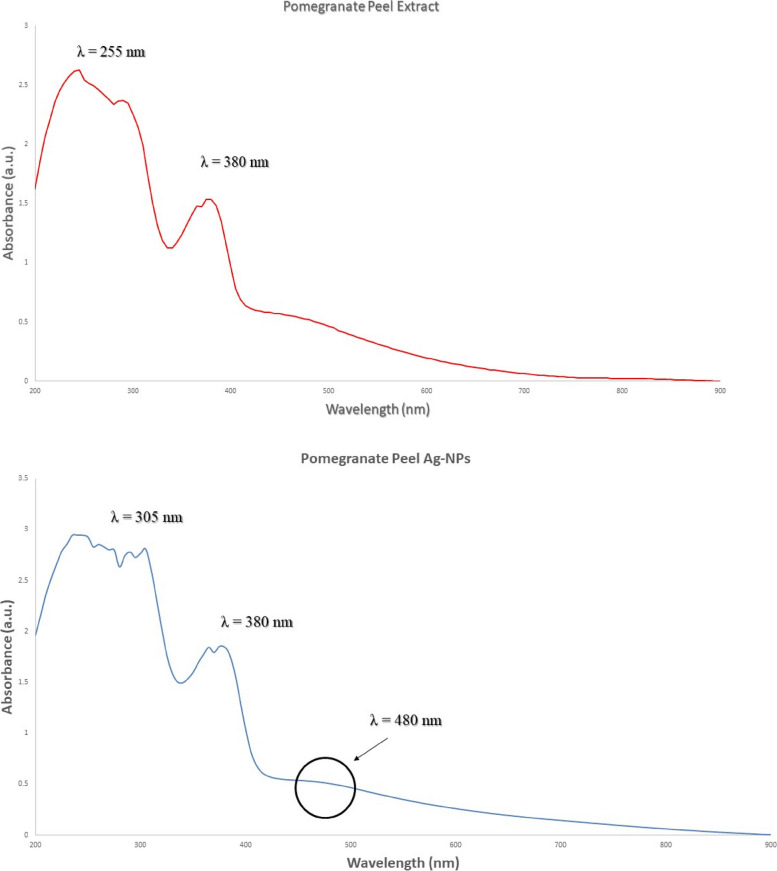
In our study, Ag-NPs were prepared via a green synthesis method. The synthesis was mediated at 32 °C under dark conditions. After the addition of plant extract supernatant, a dark brown color was obtained in the biomass after reaction with the Ag+ ions which is confirmed by UV–Vis absorption spectra after 20 min. The as-prepared Ag-NPs which synthesized in the presence of lemon extract exhibit a characteristic surface plasmon bands (SPR) at wavelength 445 nm, indicating to the formation of Ag-NPs (Fig. 6). While Ag-NPs synthesized in the presence of lemon extract exhibit a characteristic surface plasmon bands (SPR) at wavelength 480 nm, indicating to the formation of Ag-NPs (Fig. 7).
Fig. 6.
Surface plasmon absorption bands (SPR) of Ag-NPs capped with lemon extract
Fig. 7.
Surface plasmon absorption bands (SPR) of Ag-NPs capped with pomegranate extract
FTIR analysis
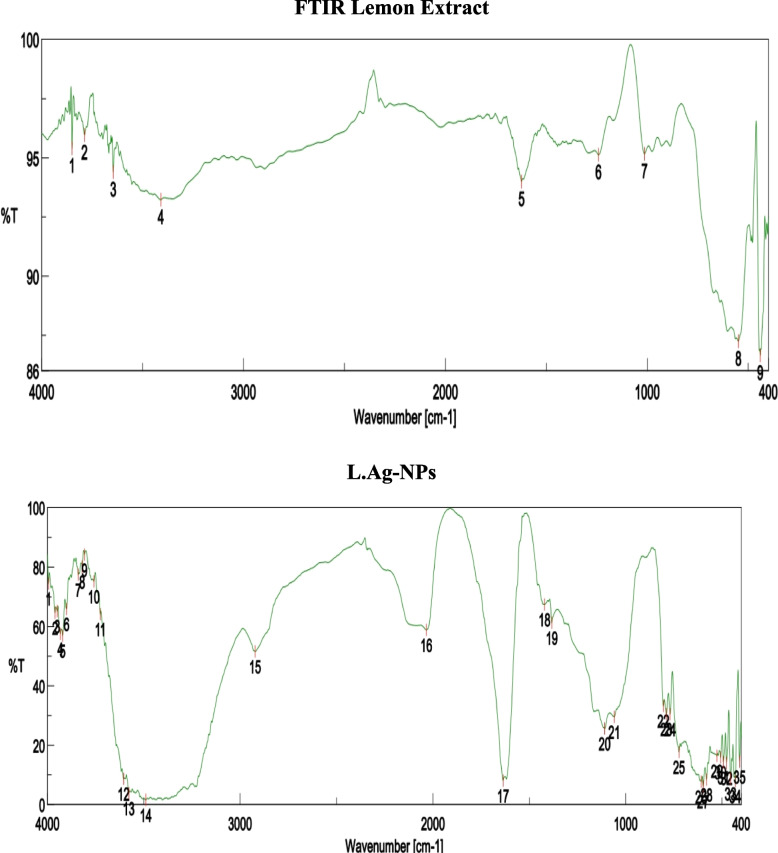
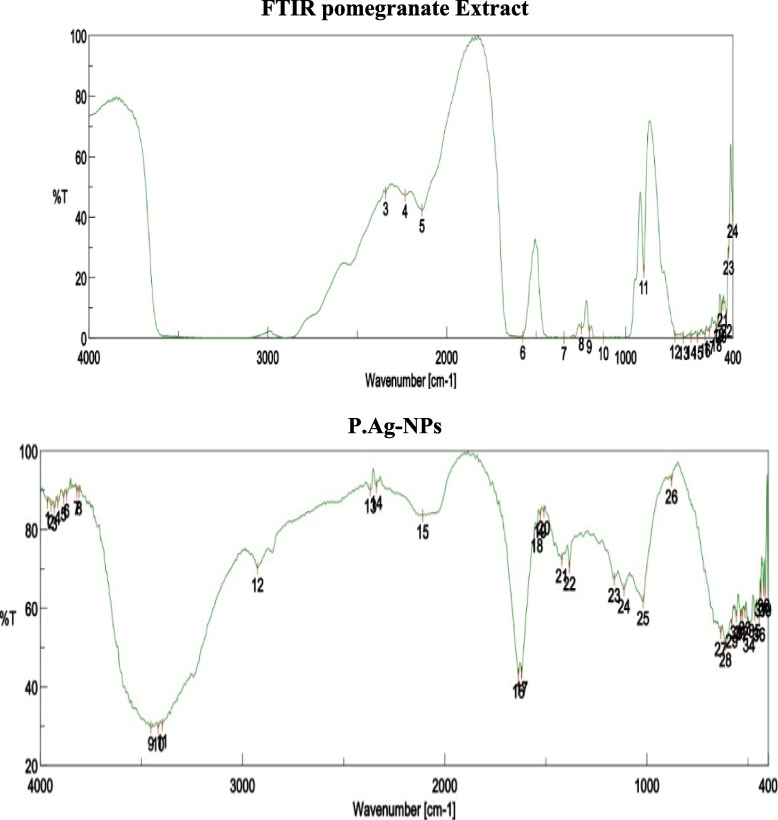
Fourier transform infrared (FTIR) spectroscopy studies were used to determine biological molecules necessary for silver nanoparticle reduction, capping, and beside stability. There were many functional groups present that may have been responsible for Ag + ion bio-reduction. The band intensities of plant extracts and silver nanoparticles in various places were investigated. The FTIR spectrum of lemon leaf extract exhibits prominent peak areas at 3409, 1623, 1241, and 548 cm−1 (Fig. 8). The presence of plant leaves extract as a capping agent with silver nanoparticles in the sample is shown by the spectra with many marginal changes in peak locations. The peaks at 400–700 cm−1 show the presence of inorganic functional groups and may suggest the existence of silver nanoparticles in the sample. The 3409 cm−1 peak corresponds to C–H stretching in alkene. The peak at 1623 cm−1 belongs to N–O stretching vibrations of the extract’s group, whereas the peak at 1241 cm−1 relates to C–N stretching vibrations of the extract’s group. Moreover, the FTIR spectra of pomegranate leaves extract exhibit large peak areas at 3396, 2926, 1383, and 400 to 700 cm–1 (Fig. 9). The spectrum from 400 to 700 cm–1 shows the presence of Ag-NPs and peaks with some marginal shifts in the region of the peaks indicating the presence of leaf extract as a coating agent in the sample. Peaks located at 400–700 cm–1 indicate the presence of inorganic functional groups and may contain silver nanoparticles in the sample. The peak located at 3396 cm–1 corresponds to the OH segment in the phenol/carboxylic group. The peak at 2926 cm–1 corresponds to the C–H stretching vibrations of the alkene group present in the extract, and a peak at 1383 cm–1 can be attributed to the group C–O stretching vibrations present in the extract. In fact, the peaks detected were due to the presence of several secondary metabolites in the leaf extract.
Fig. 8.
FTIR spectra of lemon extract and L Ag-NPs
Fig. 9.
FTIR spectra of pomegranate extract and P.Ag-NPs
XRD analysis
The XRD patterns of Ag-NPs prepared through lemon and pomegranate extracts show 20 values of 38.37°, 44.51°, 64.55°, and 77.86° and weak to moderately intense diffraction peaks at (111), (200), (220), and the (311) plane of metallic Ag with FCC crystal symmetry (Fig. 10). As a result, a higher peak intensity suggests that there are more atoms present in the crystal, which could indicate that the crystal is larger or more crystallin. A lower peak intensity, on the other hand, can indicate a smaller crystal size or a lower level of crystallinity.
Fig. 10.
Ray diffraction (XRD) pattern of Ag-NPs from both lemon and pomegranate extracts. Peaks were assigned to diffraction from the (111), (200), and (220) planes of silver. Peaks of XRD pattern can be indexed as a pattern shows the presence of the diffraction peaks corresponding to the (111), (200), (220), and 331 planes
TEM analysis
Both Ag-NPs from lemon extract and pomegranate extract were characterized through TEM which exhibits L-Ag-NPs spherical structures with size 32 nm, while P-Ag-NPs are also spherical structures with size 28 nm, with zeta potential − 20.3 and − 25.4 mV for L-Ag-NPs and P-Ag-NPs, respectively (Fig. 11). At greater concentrations, the [Ni-NPs], which have an estimated size of 50 nm, show effective anti-MRSA actions. Compared to other nosocomial bacterial infections, [Ni-NPs] exhibit more potent antibacterial activity against MRS. The UV–Vis spectroscopy, DLS, and TEM analyses of the single and probe-conjugated AuNPs were performed. Using DLS analysis, the diameter of the individual particles was 20.5 nm.
Fig. 11.
TEM image of AgNPs where A L-AgNPs while B P-AgNPs
MDR isolates growth inhibition through plant-synthesized NPs
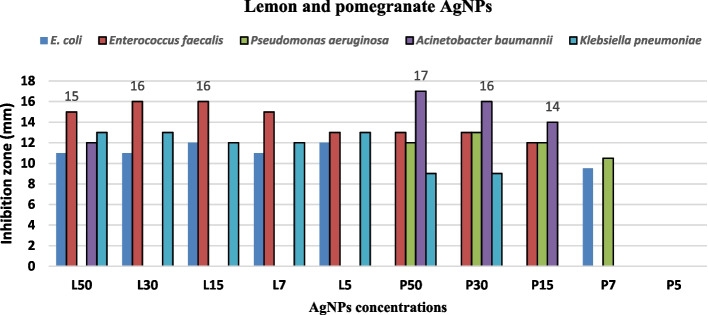
Lemon extract [L-Ag-NPs] and pomegranate extract [P-Ag-NPs] were utilized as precursors for the synthesis of silver nanoparticles, and both were measured at varied doses of 50, 30, 15, 7, and 5 ug/mL. The MIC for [L-Ag-NPs] was 50 µg/mL, and the MIC for [P-Ag-NPs] was 30 µg/mL. E. coli, E. faecalis, A. baumannii, and K. pneumoniae were the four isolates that were inhibited at [L-Ag-NPs] MIC, with inhibition zone widths of 11, 15, 12, and 13 mm, respectively. In addition, four isolates E. faecalis, P. aeruginosa, A. baumannii, and K. pneumoniae had [P-Ag-NPs] MICs that inhibited them at 13, 16, 9, and 9 mm, respectively (Figs. 12 and 13).
Fig. 12.
Inhibitory effect of different concentrations of both lemon and pomegranate NPs on the extensive MDR isolates
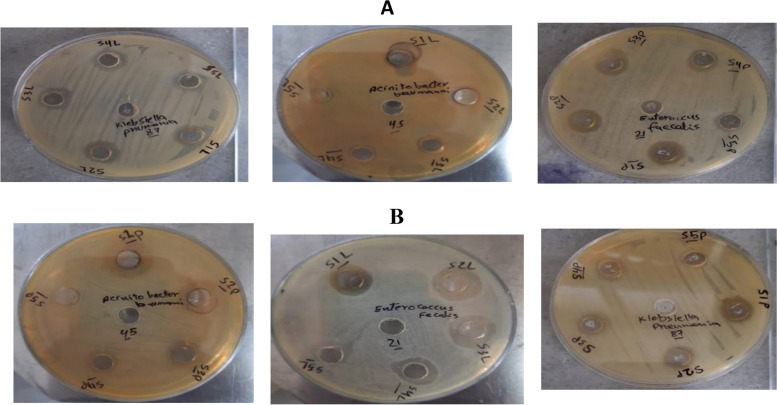
Fig. 13.
Agar well diffusion assay of plant-mediated NPs on the tested five isolates. A Lemon NPs. B pomegranate NPs
Ag-NPs prepared through Moringa oleifera plant leaves were effective as inhibitory agent at high concentrations 2 mg/mL against Enterococcus faecalis OU510, and Enterococcus faecalis ATCC29212 as a positive control, and an inhibition zone diameter of 8–10.5 mm or on agar plate. Their MIC was 0.25 mg/mL for both identified bacterial isolates [45].
Conclusion
The isolated bacterial isolates exhibited MDR activity in about 72% of cases. With the accession numbers OP741103, OP741104, OP741105, OP741106, and OP741107, respectively, the top five MDR bacterial isolates (Escherichia coli, Pseudomonas aeruginosa, Acinetobacter baumannii, Enterococcus faecalis, and Klebsiella pneumoniae) were determined through 16S rDNA PCR sequencing. ISSR PCR was used to analyze the genetic makeup of these common bacterial isolates. All of the successfully obtained genetic and protein analysis data may serve as a useful tool for understanding and combating the MDR behavior of uropathogens and improving UTI infection treatment strategies. Plant-mediated silver nanoparticles were used to inhibit four different isolates out of five. L-AgNPs, at a concentration of 50 µg/mL, inhibits E. coli, E. faecalis, A. baumannii, and K. pneumoniae, while P-AgNPs, at a concentration of 30 µg/mL, inhibits E. faecalis, P. aeruginosa, and A. baumannii. A TEM picture of two [AgNPs] revealed that L-AgNPs were 32 nm in size and P-AgNPs were 28 nm in size.
Supplementary Information
Authors’ contributions
MA, conceptualization, writing review, methodology, resources, investigation, and writing original draft. HHE, conceptualization, methodology, and resources. AY, biosynthesis and characterization of tested nanoparticles. AFES, resources, ISSR beside SDS-PAGE steps, and writing review. MK, conceptualization, writing review, methodology, resources, investigation, and writing original draft methodology. The authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
All data included in this study were presented in the form of tables and figures.
Declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Öztürk R, Murt A. Epidemiology of urological infections: a global burden. World J Urol. 2020;38:2669–2679. doi: 10.1007/s00345-019-03071-4. [DOI] [PubMed] [Google Scholar]
- 2.Murugan M, Rani KB, Wins JA, Ramachandran G, Guo F, Mothana RA. Green synthesized ZnO NPs as effective bacterial inhibitor against isolated MDRs and biofilm producing bacteria isolated from urinary tract infections. J King Saud Univ-Sci. 2022;34:101737. doi: 10.1016/j.jksus.2021.101737. [DOI] [Google Scholar]
- 3.Akram M, Shahid M, Khan AU. Etiology and antibiotic resistance patterns of community-acquired urinary tract infections in JNMC Hospital Aligarh, India. Ann Clin Microbiol Antimicrob. 2007;6:1–7. doi: 10.1186/1476-0711-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinke DT, Seaton RA, Phillips G, MacDonald TM, Davey PG. Prior trimethoprim use and trimethoprim-resistant urinary tract infection: a nested case–control study with multivariate analysis for other risk factors. J Antimicrob Chemother. 2001;47:781–787. doi: 10.1093/jac/47.6.781. [DOI] [PubMed] [Google Scholar]
- 5.Ullah A, Shah S, Almugadam B, Sadiqui S. Prevalence of symptomatic urinary tract infections and antimicrobial susceptibility patterns of isolated uropathogens in Kohat region of Pakistan. MOJ Biol Med. 2018;3:85–89. doi: 10.15406/mojbm.2018.03.00082. [DOI] [Google Scholar]
- 6.D'Andrea M, Literacka E, Zioga A, Giani T, Baraniak A, Fiett J, et al. Evolution and spread of a multidrug-resistant Proteus mirabilis clone with chromosomal AmpC-type cephalosporinases in Europe. Antimicrob Agents Chemother. 2011;55:2735–2742. doi: 10.1128/AAC.01736-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luzzaro F, Brigante G, D’Andrea MM, Pini B, Giani T, Mantengoli E, et al. Spread of multidrug-resistant Proteus mirabilis isolates producing an AmpC-type β-lactamase: epidemiology and clinical management. Int J Antimicrob Agents. 2009;33:328–333. doi: 10.1016/j.ijantimicag.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Barlow G, Nathwani D. Is antibiotic resistance a problem? A practical guide for hospital clinicians. Postgrad Med J. 2005;81:680–692. doi: 10.1136/pgmj.2005.035113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moges F, Mengistu G, Genetu A. Multiple drug resistance in urinary pathogens at Gondar College of Medical Science Hospital Ethiopia. East African Med J. 2002;79:415–420. doi: 10.4314/eamj.v79i8.8827. [DOI] [PubMed] [Google Scholar]
- 10.Drancourt M, Berger P, Raoult D. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J Clin Microbiol. 2004;42(5):2197–2202. doi: 10.1128/JCM.42.5.2197-2202.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boemare N, Akhurst R. Genera Photorhabdus and Xenorhabdus. 2002.
- 12.Gaugler R, Kaya HK. Entomopathogenic nematodes in biological control. Boca Raton: CRC Press; 1990. [Google Scholar]
- 13.Lee M-M, Stock SP. A multigene approach for assessing evolutionary relationships of Xenorhabdus spp. (γ-Proteobacteria), the bacterial symbionts of entomopathogenic Steinernema nematodes. J Invert Pathol. 2010;104:67–74. doi: 10.1016/j.jip.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Christensen H, Nordentoft S, Olsen JE. NOTE: phylogenetic relationships of Salmonella based on rRNA sequences. Int J Syst Evol Microbiol. 1998;48:605–610. doi: 10.1099/00207713-48-2-605. [DOI] [PubMed] [Google Scholar]
- 15.Welsh J, Petersen C, McClelland M. Polymorphisms generated by arbitrarily primed PCR in the mouse: application to strain identification and genetic mapping. Nucleic Acids Res. 1991;19:303–306. doi: 10.1093/nar/19.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zietkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics. 1994;20:176–183. doi: 10.1006/geno.1994.1151. [DOI] [PubMed] [Google Scholar]
- 18.Muhammad OI, Mahmoud UM, Fazio F, Sayed AE-DH. SDS-PAGE technique as biomarker for fish toxicological studies. Toxicol Rep. 2018;5:905–9. doi: 10.1016/j.toxrep.2018.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabiee N, Bagherzadeh M, Ghasemi A, Zare H, Ahmadi S, Fatahi Y, Dinarvand R, Rabiee M, Ramakrishna S, Shokouhimehr M. Point-of-use rapid detection of Sars-Cov-2: nanotechnology-enabled solutions for the COVID-19 pandemic. Int J Mol Sci. 2020;21:5126. doi: 10.3390/ijms21145126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao S, Zhang Y, Pan X, Zhu F, Jiang C, Liu Q, Cheng Z, Dai G, Wu G, Wang L. Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa. Int J Nanomed. 2019;14:1469–1487. doi: 10.2147/IJN.S191340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mba IE, Nweze EI. The use of nanoparticles as alternative therapeutic agents against Candida infections: an up-to-date overview and future perspectives. World J Microbiol Biotechnol. 2020;36:163. doi: 10.1007/s11274-020-02940-0. [DOI] [PubMed] [Google Scholar]
- 22.Singh R, Smitha SM, Surinder PS. The role of nanotechnology in combating multidrug resistant bacteria. J Nanosci Nanotechnol. 2014;14:1–12. doi: 10.1166/jnn.2014.9527. [DOI] [PubMed] [Google Scholar]
- 23.Mazzariol A, Bazaj A, Cornaglia G. Multi-drug-resistant gram-negative bacteria causing urinary tract infections: a review. J Chemother. 2017;29(sup1):2–9. doi: 10.1080/1120009X.2017.1380395. [DOI] [PubMed] [Google Scholar]
- 24.Mishra A, Pradhan D, Halder J, Biswasroy P, Rai VK, Dubey D et al (2022) Metal nanoparticles against multidrug-resistance bacteria. J Inorg Biochem 111938. 1–15 [DOI] [PubMed]
- 25.Schmidt TM. Encyclopedia of microbiology: Academic; 2019.
- 26.Sil S, Dey S, Mandal S. Seasonal circulation of Gulf of Khambhat, India using high frequency radars. OCEANS 2022-Chennai: IEEE; 2022. p. 1–5.
- 27.Prevost A, Wilkinson M. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet. 1999;98:107–112. doi: 10.1007/s001220051046. [DOI] [Google Scholar]
- 28.Ajitha B, Reddy YAK, Reddy PS. Green synthesis and characterization of silver nanoparticles using Lantana camara leaf extract. Mater Sci Eng C. 2015;49:373–381. doi: 10.1016/j.msec.2015.01.035. [DOI] [PubMed] [Google Scholar]
- 29.Desouky SE, Hassan SE, El-gamal MS, Ragab MAETI, Emam M. Effect of Salvia egypticae and Foeniculum vulgara extracts on quorum sensing and biofilm formation of methicillin resistant/sensitive Staphylococcus aureus isolates. World J Pharm Med Res. 2017;3:466–475. [Google Scholar]
- 30.Mohamed AA, Abu-Elghait M, Ahmed NE, Salem SS. Eco-friendly mycogenic synthesis of ZnO and CuO nanoparticles for in vitro antibacterial, antibiofilm, and antifungal applications. Biol Trace Elem Res. 2021;199:2788–2799. doi: 10.1007/s12011-020-02369-4. [DOI] [PubMed] [Google Scholar]
- 31.Abu-Elghait M, Hasanin M, Hashem AH, Salem SS. Ecofriendly novel synthesis of tertiary composite based on cellulose and myco-synthesized selenium nanoparticles: Characterization, antibiofilm and biocompatibility. Int J Biol Macromol. 2021;175:294–303. doi: 10.1016/j.ijbiomac.2021.02.040. [DOI] [PubMed] [Google Scholar]
- 32.Joseph S, Mathew B. Microwave-assisted green synthesis of silver nanoparticles and the study on catalytic activity in the degradation of dyes. J Mol Liq. 2015;204:184–191. doi: 10.1016/j.molliq.2015.01.027. [DOI] [Google Scholar]
- 33.Nahar K, Aziz S, Bashar M, Haque M, Al-Reza SM. Synthesis and characterization of silver nanoparticles from Cinnamomum tamala leaf extract and its antibacterial potential. Int J Nano Dimen. 2020;11:88–98. [Google Scholar]
- 34.Al-Dhabi NA, Mohammed Ghilan A-K, Arasu MV. Characterization of silver nanomaterials derived from marine Streptomyces sp. al-dhabi-87 and its in vitro application against multidrug resistant and extended-spectrum beta-lactamase clinical pathogens. Nanomaterials. 2018;8:279. doi: 10.3390/nano8050279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Składanowski M, et al. Silver and gold nanoparticles synthesized from Streptomyces sp. isolated from acid forest soil with special reference to its antibacterial activity against pathogens. J Cluster Sci. 2017;28:59–79. doi: 10.1007/s10876-016-1043-6. [DOI] [Google Scholar]
- 36.Dhar V, Datta S, Chaudhary RG, Upadhyay JP, Saifulla M, Mishra S, … Shamim M. Pathogenic and molecular characterisations of pigeonpea wilt pathogen Fusarium udum. Arch Phytopathol Plant Protect. 2012;45(4):423–436.
- 37.Khan AA, Alanazi AM, Nawaf A, Wani TA, Bhat MA. Pomegranate peel induced biogenic synthesis of silver nanoparticles and their multifaceted potential against intracellular pathogen and cancer. Saudi J Biol Sci. 2021;28:4191–4200. doi: 10.1016/j.sjbs.2021.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bretagne S, Costa J-M, Besmond C, Carsique R, Calderone R. Microsatellite polymorphism in the promoter sequence of the elongation factor 3 gene of Candida albicans as the basis for a typing system. J Clin Microbiol. 1997;35:1777–1780. doi: 10.1128/jcm.35.7.1777-1780.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moghaieb RE, Abdelhadi AA, El-Sadawy HA, Allam NA, Baiome BA, Soliman MH. Molecular identification and genetic diversity among Photorhabdus and Xenorhabdus isolates. 3 Biotech. 2017;7:1–9. doi: 10.1007/s13205-016-0594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravi Kumar A, Sathish V, Balakrish Nair G, Nagaraju J. Genetic characterization of Vibrio cholerae strains by inter simple sequence repeat-PCR. FEMS Microbiol Lett. 2007;272:251–258. doi: 10.1111/j.1574-6968.2007.00762.x. [DOI] [PubMed] [Google Scholar]
- 41.Durrani R, Abubakar M, Arshed MJ, Saleha S, Ullah I, Ali Q. Biological characterization and protein profiles of two model bacteria by SDS-PAGE and FT-IR. ARPN J Agric Biol Sci. 2008;3.
- 42.Salam HS, Hotzel H. Investigation of phenotypic variations amongst Mycoplasma bovis field German isolates using SDS-PAGE. Beni-Suef Univ J Basic Appl Sci. 2013;2:103–107. [Google Scholar]
- 43.Jamalzadeh A, Darvishnia M, Khodakaramian G, Bazgir E, Zafari D. Genetic diversity and plant growth-promoting activity of the dominant bacteria from canola plants in western Iran. Egypt J Biol Pest Contr. 2021;31:1–9. [Google Scholar]
- 44.Jung H-J, Hong Y, Yang H-S, Chang H-C, Kim H-Y. Distribution of lactic acid bacteria in garlic (Allium sativum) and green onion (Allium fistulosum) using SDS-PAGE whole cell protein pattern comparison and 16S rRNA gene sequence analysis. Food Sci Biotechnol. 2012;21:1457–1462. doi: 10.1007/s10068-012-0192-0. [DOI] [Google Scholar]
- 45.Yousef A, Abu-Elghait M, Barghoth MG, Elazzazy AM, Desouky SE. Fighting multidrug-resistant Enterococcus faecalis via interfering with virulence factors using green synthesized nanoparticles. Microb Pathog. 2022;173:105842. doi: 10.1016/j.micpath.2022.105842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data included in this study were presented in the form of tables and figures.