Abstract
Background:
Pheochromocytomas and paragangliomas (PPGL) exhibit an up to 20% rate of metastatic disease that cannot be reliably predicted. This study prospectively assessed whether the dopamine metabolite, methoxytyramine, might predict metastatic disease, whether predictions might be improved using machine learning (ML) models that incorporate other features and how ML-based predictions compare to predictions by specialists in the field.
Methods:
Following prospective examination of the utility of methoxytyramine to predict metastatic disease in 267 patients with PPGL, a further retrospective dataset from 493 patients with PPGL was used to train and validate ML models according to selections of additional features. The best performing ML models were then externally validated using the prospective dataset. For comparison, 12 specialists provided predictions of metastatic disease using data from the training and external validation datasets.
Findings:
Prospective predictions indicated that plasma methoxytyramine could identify metastatic disease at respective sensitivities and specificities of 52% and 85%. The best performing ML model was based on an ensemble tree classifier algorithm that utilized nine features: plasma methoxytyramine, metanephrine and normetanephrine, age, sex, previous history of PPGL, location and size of primary tumors, presence of multifocal disease. This model presented with an area under the receiver-operating characteristic curve of 0·942 (CI:0·894-0·969) that was larger (P<0·0001) than that of the best performing specialist before (0·815, CI:0·778-0·853) and after provision of SDHB variant data (0·812, CI:0·781-0·854). Sensitivity for prediction of metastatic disease in the external validation cohort reached 83% at a specificity of 92%.
Interpretation:
Although methoxytyramine provides some utility for prediction of metastatic PPGL, sensitivity is limited. Predictive value is considerably enhanced with ML models that incorporate the above mentioned features. Our final model provides a preoperative approach to predict metastases in patients with PPGL, and thereby guide individualized patient management and follow-up.
Funding:
Deutsche Forschungsgemeinschaft, 314061271-TRR/CRC 205- 1/2
Keywords: pheochromocytoma, paraganglioma, metastases, machine learning models, predictors
Introduction
Pheochromocytomas and paragangliomas (PPGL) are neuroendocrine tumors with an up to 35% hereditary predisposition1 and approximately a 20% prevalence of metastatic disease.2,3 Unlike other tumors, there are no histopathological methods to identify metastatic disease and all PPGL must be considered to have variable potential to metastasize.4 Currently only presence of metastases at sites where no chromaffin tissue should be expected (e.g., bones, lymph nodes) establishes a definitive diagnosis of metastatic disease.4,5 Therefore, long-term follow-up is recommended for all patients with PPGL.6
Earlier therapeutic intervention in patients with metastatic PPGL is expected to reduce morbidity and mortality.7 Identification of features to reliably predict metastatic potential of PPGL at initial tumor diagnosis is therefore crucial. The relation of tumoral dopamine production to metastatic disease in patients with PPGL is established.2,8,9 Use of methoxytyramine, the O-methylated metabolite of dopamine, as a predictor of metastases offers promise, but has only been evaluated in a single retrospective patient series.2 Young age,10,11 large tumor size,2,11,12 and extra-adrenal location of primary tumors2,12 represent other established clinical predictors of metastases. Tumors due to pathogenic variants of the succinate dehydrogenase subunit B (SDHB) gene and somatic genomic alterations such as ATRX, TERT or MAML3 translocations are also associated with higher metastatic potential.13,14 However, such information is rarely available preoperatively when it can be useful to establish metastatic risk.
Despite the association of the aforementioned features with the development of metastases, there is no robust method to reliably predict metastatic disease in patients with PPGL. Some effort has been made to combine different features in scoring systems to predict metastatic PPGL, but most involve histopathological parameters.15,16 These are difficult to standardize in clinical practice,17 and lack accuracy.18 An attempt to establish a predictive score using routinely available clinical features similarly failed expectations according to a low positive predictive value.19
Advances in computational power have led to the introduction of multidimensional digitalized approaches that could potentially support decision-making in healthcare. Machine learning (ML) is one such approach for interrogating multidimensional data and an area of artificial intelligence that utilizes computational algorithms for different tasks; this is without the need for the explicit programming of previously established mathematical relationships.20,21 In diagnostics, these tasks principally involve classification.22,23
Taking the above into consideration, the present study had three aims: 1. prospectively validate use of methoxytyramine as a preoperative predictor of metastases in patients with PPGL; 2. establish ML models that incorporate methoxytyramine with other features to predict metastatic PPGL preoperatively; 3. and compare the performance of the selected ML models with the predictions of 12 clinical care specialists with expertise in the management of patients with PPGL.
Methods
Patients
This cross-sectional cohort study included 788 patients with and without metastatic PPGL enrolled at seven international tertiary centers (Supplemental Methods) under clinical protocols approved by local Ethics Committees. Clinical information included sex, age at initial tumor diagnosis, presence of multifocal and metastatic disease, initial tumor location and size as well as plasma concentrations of free normetanephrine, metanephrine and methoxytyramine.
Metastatic disease was defined as the presence of metastases in tissues distant from the primary tumor, where chromaffin cells are normally absent.4 Metastases were identified by conventional and functional imaging or histopathological examination of resected lymph nodes with further details about this and genetic testing provided in the Supplement. Testing for germline pathogenic variants of VHL, RET, SDHx, MAX, and TMEM127 was performed in 708 patients using Sanger sequencing and/or NGS, and multiplex ligation-dependent probe amplification or custom array CGH for deletion detection.
Study design
Objective 1. Prospective use of plasma methoxytyramine to predict metastatic disease
The first objective of the study involved 267 patients with PPGL under the Prospective Monoamine-producing Tumor (PMT) trial (https://pmt-study.pressor.org) who presented with positive biochemical test results at initial screening. As detailed in the Supplemental Methods, one objective of the PMT-trial was to establish utility of plasma methoxytyramine to predict metastases. For this purpose, the investigator responsible for biochemical tests provided predictions of metastatic disease that were based primarily on measurements of plasma methoxytyramine, with additional consideration of the other two metabolites. These and other predictions, along with biochemical test results, were provided back to the responsible physicians at each participating center. As further detailed in the Supplemental Methods, predictions were restricted to patients with positive biochemical tests and were in the form of standardized comments that indicated strong, moderate, possible or low risk of metastases.
Objective 2. Generation of ML models to predict metastatic disease
For the second objective, we retrieved data from 493 patients with PPGL (training cohort) to generate and internally test various ML models using four different ML algorithms. The best candidate ML models were then externally validated using the dataset of 295 patients with PPGL who were enrolled in the PMT-trial (external validation cohort). These included the 267 patients with positive test results and another 28 with negative test results among whom 23 had head and neck paragangliomas (Supplemental results). After external validation, we compared ML models using multiple metrics and selected the final top performing models (Figure 1).
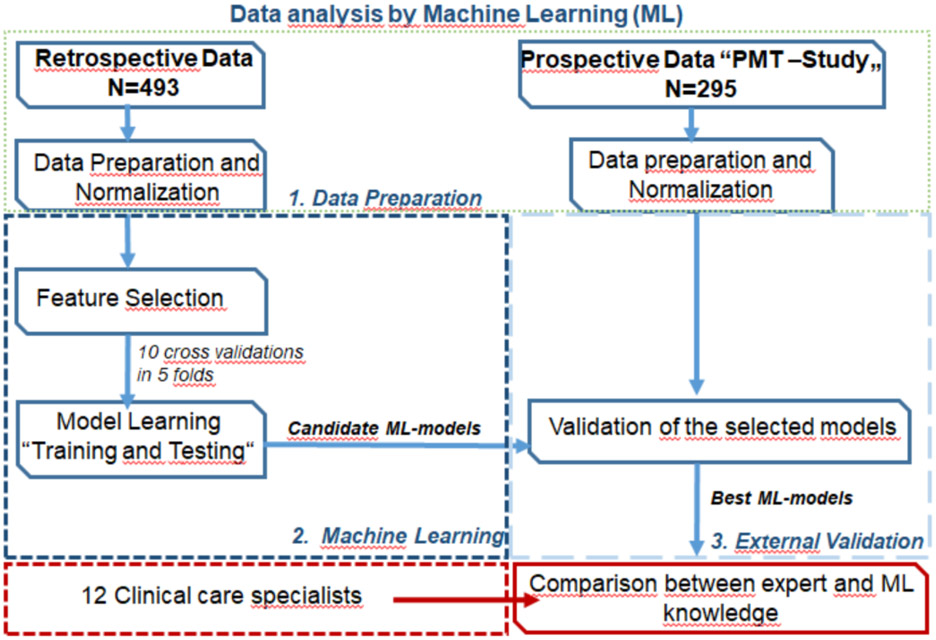
Figure 1:
Workflow for the data analysis
As detailed in the Supplemental Methods, ML models were developed after data preparation and normalization using four supervised ML algorithms with all variables included and according to ten cross-validations in five folds. The supervised ML algorithms included Decision Tree Classifier (TC), Support Vector Machine (SVM), Naive Bayes (NB), and AdaBoost Ensemble Tree Classifiers (ENS). Chi-square feature analysis for classification was carried out in the training cohort to identify invalid features containing irrelevant or redundant information.
During data preparation we performed feature analysis in the training dataset twice. The first feature analysis included nine features: 1. plasma free methoxytyramine; 2. age at initial tumor diagnosis; 3. sex; 4. previous history of PPGL (yes/no); 5.primary tumor location; 6. primary tumor size; 7. presence of multifocal disease (yes/no); 8. plasma free metanephrine; and 9. plasma free normetanephrine. The second feature analysis included the same features, supplemented by presence of SDHB pathogenic variants (positive/negative), the genetic component with the strongest anticipated metastatic predictive potential.
After feature analysis, multiple rounds of training and internal testing of ML models were performed to identify the best candidate ML models according to areas under the receiver operating characteristic (ROC) curves (AUC), and with consideration of Matthews correlation coefficient (MCC) and balanced accuracy. In order to confirm reproducibility of our results, we then externally validated the best candidate ML models in a separate cohort of patients from the PMT-trial (external validation cohort). The best ML models of the external validation were again selected by comparing their predictive performance according to AUC, with consideration of MCC and balanced accuracy. ML was performed using MATLAB MathWorks R2020a. Further details on the generation of the ML models are provided in the Supplemental Methods.
Objective 3. Predictions of metastatic disease by clinical care specialists
For the third objective, we invited 12 clinical care specialists with expertise in PPGL to provide predictions of metastatic disease for the training and external validation cohorts. Seven specialists reported experience of more than ten years, and five less than ten years. Specialists were requested to provide their own probabilities of metastatic disease using a classification score of four categories: low, possible, moderate and strong probability. Before the review process, specialists received detailed definitions for each of the four classification categories, including specific probability intervals for metastatic disease and narrative interpretations for further patient management (Supplementary Box 1).
Similar to the feature analysis and ML training, specialists were instructed to provide probabilities of metastatic disease twice. This included probabilities according to the same nine features described above for ML feature analysis. After an interval of four weeks, all specialists received a second dataset with the same features, supplemented by SDHB variant status (Supplemental Methods). Predictions of specialists were then compared with those of the top performing ML models (Figure 1).
Role of the funding source
The funder had no role in study conception, design or conduct of the study. All authors had full access to the data in the study and were involved in data interpretation and writing of the report. The corresponding author had final responsibility for the decision to submit for publication.
Statistical analysis
Details about statistical methods are outlined in the supplemental methods section.
Results
Objective 1. Prospective use of plasma methoxytyramine to predict metastases
As outlined in Table 1, there were 295 patients with PPGL from the PMT-trial. Among these there were 267 patients with positive biochemical test results, in whom predictions of metastatic disease were possible according to the prospective study design (Supplemental Results). The majority of patients (79%) were correctly classified by specialist-based predictions. Specifically, predictions were correct for 186 patients without [specificity:85%, (186/219)] and 23 patients with metastases [sensitivity:52%, (23/48)] (Supplementary Table 1). Low sensitivity largely reflected patients with normal or mildly elevated plasma concentrations of methoxytyramine (Supplementary Figure 1A). Among 13 cases classified with a strong risk for metastases, 11 (85%) were correctly classified. The higher sensitivity in this particular category, reflected high (>678 pg/mL) plasma concentrations of methoxytyramine in all 11 cases.
Table 1.
Characteristics of patients with PPGL according to presence versus absence of metastatic disease for the training and external validation (PMT-trial) cohorts.
| Training cohort | External validation cohort | |||||
|---|---|---|---|---|---|---|
| Without metastases |
With Metastases |
P Value | Without metastases |
With metastases |
P Value | |
| Number | 327 | 166 | 238 | 57 | ||
| Sex (males) | 48% (156/327) | 57% (95/166) | 0·0450 | 39% (93/238) | 58% (33/57) | 0·0100 |
| Age (years) # | 39·6 (37·9-41·3) | 31·8 (31.9-35.4) | <0·0001 | 44·7 (43·2-46·2) | 40·6 (39.2-42.1) | 0·0250 |
| Tumor size (cm) * | 2·7 (2·4-2·9) | 4·4 (4.3-4.5) | <0·0001 | 2·8 (2·6-2·9) | 5·4 (5.3-5.5) | <0.0001 |
| Location (extra-adrenal) | 17% (55/327) | 71% (118/166) | <0·0001 | 22% (53/238) | 58% (33/57) | <0·0001 |
| Multifocal | 21% (67/327) | 20% (33/166) | 0·0870 | 17% (41/238) | 23% (13/57) | 0·3180 |
| Presence ofSDHB mutation € | 6% (16/267) | 50% (74/149) | <0·0001 | 3% (7/236) | 27% (15/56) | <0·0001 |
| Previous history of PPGL $ | 10% (31/327) | 70% (116/166) | <0·0001 | 14% (34/238) | 70% (40/57) | <0·0001 |
| Biochemistry (pg/mL) | ||||||
| Normetanephrine | 598·3 (594-602) | 832·9 (827-838) | 0·0260 | 549·5 (523.5-531) | 526·1 (521-531) | 0·8130 |
| Metanephrine | 144·8 (139-150) | 42·1 (38-45) | <0·0001 | 124·2 (118-129) | 52·5 (48-56) | <0·001 |
| Methoxytyramine | 13·6 (10-16) | 46·2 (44-47) | <0·0001 | 15·1 (12-17) | 49·5 (40-58) | <0·0001 |
| Follow-up (months) | 82·8 (79-85) | 95·6 (92-98) | 0·1430 | 49·3 (46-51) | 99·7 (96-103) | <0·0001 |
Continuous parameters are shown as geometric means with confidence intervals, #: age at initial tumor diagnosis; *: initial tumor size; €: for 60 patients without and for 17 with metastases in the “training” cohort and for two patients without and one with metastases in the “external validation” cohort, genetic testing was not available; $: local recurrence and/or new tumor
Objective 2. Generation of ML models to predict metastases
Patient characteristics
As outlined in Supplementary Table 2, the 493 patients with PPGL in the training dataset showed some differences from the 295 patients in the PMT-trial used for external validation. Nevertheless, in both datasets, patients with metastases were more often males and younger than those without metastases (Table 1). Patients with metastases presented more often with larger, extra-adrenal tumors than those without. There was also a higher prevalence of SDHB variants and recurrent disease in the former than the latter group. Finally, patients with metastases presented with lower metanephrine, but higher methoxytyramine concentrations than those without metastases. All above differences were highly significant (P<0·0001).
Training and testing of ML models in the learning phase
Feature and ML analyses were performed in the training cohort twice. The first analysis included nine clinical and biochemical features, whereas the second was supplemented with SDHB variant status (Supplementary Figure 2). Among the nine features in the first analysis, the top five that predicted metastases included previous history of PPGL, extra-adrenal primary tumor location, large primary tumor size, high plasma methoxytyramine and low plasma metanephrine concentrations (Supplementary Figure 2A). For the ten-feature analysis, the five most important features were previous history of PPGL, extra-adrenal primary tumor location, presence of SDHB variants, large primary tumor size and high plasma methoxytyramine concentrations (Supplementary Figure 2B).
Classification performance of the ML models after external validation
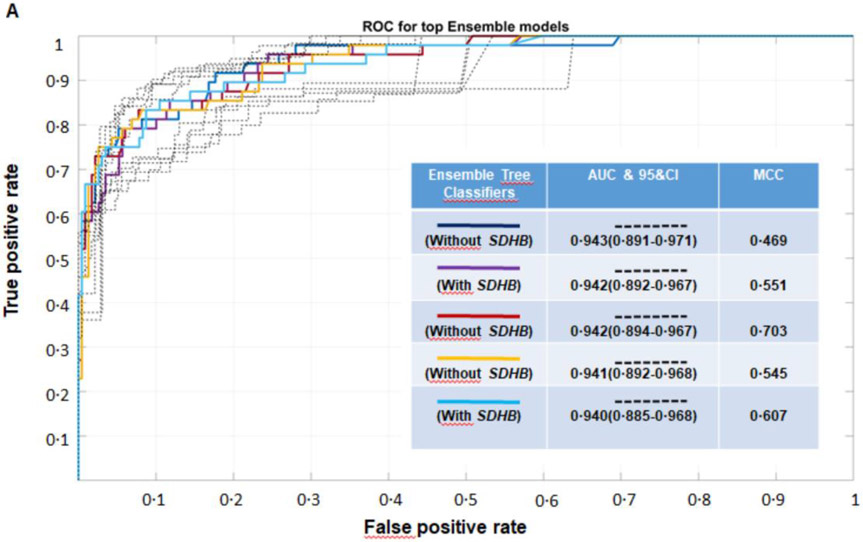
Among the 380 initial ML models evaluated in the training cohort (Supplementary Table 3), there were 40 best performing ML models that were selected for external validation. Comparisons of the AUC, with additional considerations of MCC and balanced accuracy after external validation, revealed five top performing ML models, all involving ENS algorithms (Figure 2A). All five models showed similar diagnostic performance (Supplementary Table 4). The best performing ENS model, which had an AUC of 0·942(CI:0·894-0·969), an MCC of 0·851 and a balanced accuracy of 88%, did not utilize SDHB variant status as a feature (Table 2, Figure 2A). This was followed by an ENS model that utilized SDHB variant status and displayed an AUC of 0·940(CI:0·886-0·969), an MCC 0·804 and a balanced accuracy 86%.
Figure 2:
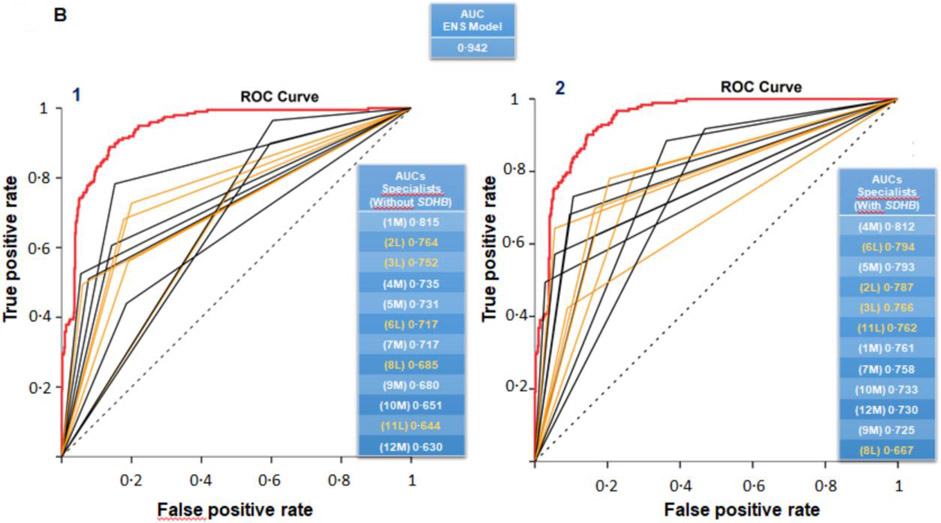
A. Predictive performance of the top five ML models after external validation according to receiver operating characteristic (ROC) curves. Areas under ROC curves (AUC) for ROC curves are shown with 95% confidence intervals (dotted curves). All five ML models presented with similar diagnostic performance in terms of AUC. The final selected ML model was a model (red) that did not utilize SDHB pathogenic variant status and presented with high MCC and balanced accuracy metrics. B. Comparison of the diagnostic performance of the selected ENS model with that of the twelve clinical care specialists according to their interpretations of likely presence or absence of metastatic disease. The classification performance of the ENS model, which was established without requirement of the SDHB pathogenic variant status, was significantly better than the performance of all specialists, both before (B1) and after provision of information about SDHB pathogenic variant status (B2).
Table 2:
Classification performance of the top performing ML models established after external validation for each of the four ML algorithms utilizing nine and ten features.
| Data set with nine features (without SDHB mutation status) | ||||
|---|---|---|---|---|
| Algorithms | TC | SVM | NB | ENS |
| AUC & CI | 0·889 (0·823-0.934) | 0·929 (0·889-0·957) | 0·839 (0·752-0·891) | 0·942 (0·894-0·969) |
| MCC | 0·863 (0·808-0·893) | 0.795 (0·735-0·840) | 0·710 (0·651-0·771) | 0·851 (0·801-0·898) |
| F1-score | 0·774 (0·699-0·863) | 0·661 (0·549-0·770) | 0·554 (0·417-0·610) | 0·755 (0·667-0·833) |
| Sensitivity | 0·854 (0·725-0·939) | 0·813 (0·687-0·909) | 0·854 (0·757-0·951) | 0·833 (0·707-0·929) |
| Specificity | 0·927 (0·894-0·957) | 0·866 (0·831-0·914) | 0·745 (0·690-0·808) | 0·922 (0·893-0·955) |
| Precision | 0·707 (0·599-0·841) | 0·557 (0·465-0·691) | 0·410 (0·308-0·506) | 0·690 (0·568-0·834) |
| Accuracy | 0·914 (0·879-0·939) | 0·857 (0·812-0·889) | 0·764 (0·723-0·805) | 0·907 (0·861-0·932) |
| Balanced Accuracy | 0·890 (0·822-0·933) | 0·839 (0·770-0·888) | 0·799 (0·758-0·840) | 0·878 (0·808-0·922) |
| Data set with ten features (with SDHB mutation status)* | ||||
| AUC & CI | 0·893 (0·823-0·936) | 0·924 (0·881-0·953) | 0.826 (0.751-0.878) | 0.940 (0.886-0.969) |
| MCC | 0·849 (0·782-0·891) | 0·795 (0·761-0·841) | 0·672 (0·617-0·719) | 0.804 (0·741-0·849) |
| F1-score | 0·750 (0·635-0·826) | 0.651 (0·533-0·726) | 0.559 (0·423-0·695) | 0·672 (0·571-0·780) |
| Sensitivity # | 0·750 (0·596-0·841) | 0·896 (0·783-0·962) | 0·791 (0·636-0·946) | 0·854 (0·777-0·939) |
| Specificity | 0·948 (0·908-0·974) | 0·821 (0·771-0·869) | 0·781 (0·726-0·836) | 0·859 (0·801-0·900) |
| Precision | 0·750 (0·578-0·834) | 0·512 (0·408-0·602) | 0·413 (0·293-0·546) | 0·554 (0·454-0·674) |
| Accuracy | 0·914 (0·877-0·942) | 0·834 (0·785-0·866) | 0·783 (0·749-0·832) | 0·856 (0·800-0·897) |
| Balanced Accuracy | 0·849 (0·793-0·910) | 0·858 (0·808-0·908) | 0·786 (0·752-0·820) | 0·855 (0.789-0·907) |
TC: Decision Tree Classifier, SVM: Support Vector Machine Classifier, NB: Naive Bayes Classifier, ENS: Ensemble Tree Classifiers, AUC: area under the Roc Curve; CI: 95% confidence intervals; MCC: Matthew`s correlation coefficient; #: sensitivity=recall rates; *Information regarding the presence or not of SDHB mutation was included as an extra feature
Three other algorithms (TC, SVM, NB) provided ML models with predictive performance that approached that of the ENS algorithm-derived models (Supplementary Tables 5-8). For the dataset that did not include SDHB variant status, the best TC model presented with an AUC of 0·889(CI:0·823-0·934), an MCC of 0·863 and a balanced accuracy of 89%. The best SVM model displayed an AUC of 0·929(CI:0·889-0·957), an MCC of 0·795 and a balanced accuracy of 84%. This was followed by the the NB model, with an AUC of 0·839(CI:0·752-0·891), an MCC of 0·710, and a balanced accuracy of 80%. (Table 2).
For the dataset supplemented with SDHB variant status, the best TC model displayed an AUC of 0·893(CI:0·823-0·936), an MCC of 0·849 and a balanced accuracy of 85%. The best SVM model displayed an AUC of 0·924(CI:0·881-0·953), an MCC of 0·795 and a balanced accuracy of 86%. This was again followed by the NB model with an AUC of 0·826(CI:0·751-0·878), an MCC of 0·672 and a balanced accuracy of 79%. (Table 2).
Objective 3. Predictions of metastatic disease by clinical care specialists
Diagnostic performance of clinical care specialists
Among the 12 specialists who provided predictions of metastatic risk, predictive performance varied widely according to the nine and ten-feature datasets without and with SDHB variant status (Table 3). The highest performance among specialists for the dataset without SDHB variant status was achieved by specialist 1M (AUC:0·815; CI:0·778-0·853), whereas the highest performance for the dataset supplemented with SDHB variant status by specialist 4M (AUC:0·812; CI:0·781-0·854).
Table 3:
Performance of twelve participants for the prediction of metastatic disease in patients with PPGL, before vs after provision of the SDHB mutation status
| AUCs (CI) | Paired comparisons |
||
|---|---|---|---|
| Dataset without SDHB status |
Dataset with SDHB status |
||
| Selected ENS model | 0·942 (0·894-0·969) | 0·940 (0·885-0·968) | - |
| Participants | |||
| 1M | 0·815 (0·778-0·853) | 0·761 (0·723-0·799) | 0·1020 |
| 2L | 0·764 (0·723-0·805) | 0·787 (0·747-0·828) | 0·2350 |
| 3L | 0·752 (0·710-0·794) | 0·766 (0·723-0·810) | 0·1090 |
| 4M | 0·735 (0·689-0·781) | 0·812 (0·781-0·854) | 0·0001 (↑) |
| 5M | 0·731 (0·687-0·776) | 0·793 (0·749-0·836) | 0·1570 |
| 6L | 0·717 (0·670-0·764) | 0·794 (0·750-0·838) | 0·0001 (↑) |
| 7M | 0·717 (0·670-0·763) | 0·758 (0·711-0·805) | 0·1420 |
| 8L | 0·685 (0·639-0·731) | 0·667 (0·617-0·717) | 0·1980 |
| 9M | 0·680 (0·642-0·719) | 0·725 (0·685-0·764) | 0·1800 |
| 10M | 0·651 (0·609-0·694) | 0·733 (0·684-0·782) | 0·1720 |
| 11L | 0·644 (0·601-0·687) | 0·762 (0·720-0·802) | 0·0001 (↑) |
| 12M | 0·630 (0·582-0·677) | 0·730 (0·684-0·776) | 0·0001 (↑) |
ENS: Ensemble Tree Classifier; M: clinical care specialists with experience on the field of PPGL more than ten years; L: clinical care specialists with experience on the field of PPGL between five to ten years; AUC: area under the Roc Curve; CI: 95% confidence intervals
The diagnostic performance of specialists did not differ among those with more than versus less than ten years experience (Table 3 & Supplementary Table 9). Specifically, for the nine-feature dataset, the specialists with more than ten years experience achieved a mean AUC of 0·708(CI:0·648-0·768), which was similar (P=0·7550) to the AUC of 0·712(CI:0·662-0·772) for those with less experience. Similarly, for the ten-feature dataset supplemented by the SDHB variant status, the specialists with more than ten years experience achieved a mean AUC of 0·758(CI:0·728-0·788), which again was similar (P=0·6390) to the AUC of 0·755(CI:0·705-0·805) for those with less experience.
Paired comparisons revealed that only four specialists (4M, 6L, 11L, 12M) improved their performance after the provision of SDHB variant status (Table 3). Overall, neither specialists with more (P=0·0630) nor less than ten years experience (P=0·1380) improved their performance after provision of SDHB variant status.
Comparison of performance between ML models and specialists
Among the 12 specialists, none attained the diagnostic performance reached by the ENS model (Figure 2B). The average performance of specialists [AUC:0·710 (CI:0·655-0·765)] was less (P<0·0001) than the performance achieved by the ENS model [AUC:0·942 (CI:0·894-0·969)] (Figure 2B1). After provision of SDHB variant status, average performance of specialists [AUC:0·756 (CI:0·716-0·796)] also remained inferior (P<0·0001) to the performance of the ENS model (Figure 2B2 & Supplementary Table 9).
Discussion
This study introduces ML models to more accurately predict metastatic disease than previously possible. Importantly, these models allow for predictions at first diagnosis of PPGL by utilizing clinical features routinely and preoperatively available. More generally, our findings support emerging concepts that ML mathematical processes will gain traction in medicine and oncology for their potential to facilitate robust non-invasive diagnostic stratification and guide personalized patient management.
The initial prospective assessments of plasma methoxytyramine as a predictor of metastatic disease confirmed previous retrospective findings.2 However, the 52% of all patients correctly predicted by methoxtyramine with metastatic PPGL is only a little better than utility of SDHB pathogenic variants for the same purpose, according to a prevalence of up to 41% among patients with metastatic PPGL.13 This was supported in the present study by an overall 40% prevalence of SDHB variants. Nevertheless, among patients with highly increased levels of methoxytyramine, the post-test probability of metastatic disease was 85%. Overall, however, and similar to SDHB variant status, measurements of methoxytyramine alone cannot be used to accurately predict or exclude metastases.
Apart from methoxytyramine and SDHB pathogenic variants, several other features have been indicated as predictors of metastases among patients with PPGL. Our second objective was to combine routinely available clinical and biochemical features with measurements of methoxytyramine in order to develop a ML tool to predict metastases preoperatively. Large tumor size, extra-adrenal tumor location, previous history of PPGL, high plasma methoxytyramine and low metanephrine concentrations were consistently identified to predict metastases according to the feature analysis. Those risk factors likely reflect a more undifferentiated tumor phenotype associated with pseudohypoxia signaling and hypermethylation pathways.24,25 Those pathways may drive the mesenchymal transition step in metastatic progression.26,27 Findings that high plasma methoxytyramine, but low plasma metanephrine predict metastases and that both metabolites are among selected features emphasizes the importance of accurate and reliable biochemical tests carried out according to appropriate pre- and analytical procedures.
External validation of the best candidate ML models after internal testing revealed five best performing ML models with similar diagnostic performance and a mean AUC of 0·942(CI:0·891-0·968). Among those models, the ENS model provided the best MMC and balanced accuracy metrics without requirement for SDHB variant data, which is often not available at initial diagnosis. The finding that SDHB test results were not required for prediction of metastases, is explained by the key features of large tumor size, extra-adrenal location and noradrenergic/dopaminergic tumor phenotype shared between patients with SDHB-mutated metastatic PPGL and the more than twice larger group of all patients with metastatic PPGL.10,12
Apart from establishing ML models that can be easily applied preoperatively using readily available clinical information, we also validated the models in a separate cohort of patients to establish reproducibility. Furthermore, after external validation we also compared the best performing ML models with interpretations by clinical care specialists with expertise in PPGL. These comparisons established that the finally selected ENS model provided significantly improved performance over interpretations of all specialists.
Of course, one could argue that in real life clinicians incorporate more clinical information into decision making, and that the determined performance of specialists is rather artificial. In an attempt to partially eliminate this potential confounder, we investigated whether provision of SDHB variant status improved the ability of specialists to predict metastasis. Only four specialists showed improved performance; overall performance remained significantly inferior to that of the selected ENS ML model, which did not require SDHB variant status as a feature. Another argument to be considered is that clinicians focus more on “management decisions” rather than “diagnostic classifications”. In this context, we incorporated the “management decisions” in the study design and provided all specialists before the review process with narrative interpretations for further patient management for each of the four classification categories.
With the aforementioned considerations in mind, our data show that the selected ML models provide a suitable tool for prediction of metastases in patients with PPGL. Furthermore, this should be of benefit to clinicians at different levels of training and experience. The models may be implemented in digital systems or smart phone applications and used together with other routinely available data to facilitate individualized diagnostic stratification and patient management. Apart from identifying patients with low probabilities of metastases, who may then be excluded from intensive, long-term and costly follow-up programs, our ML models provide justification for preoperative functional imaging and extensive follow-up in patients with high probability of metastases. In turn, this provides opportunities of earlier disease detection and interventional strategies for improved patient outcomes.
Despite growing acceptance of the superior predictive power of ML compared to conventional statistical scores for oncological staging,28 many have considered ML a “black box” where connections between features and disease probabilities are invisible to clinicians.29 These concerns are being addressed by interfaces that integrate data with clinical decision support systems to provide automated patient-specific interpretations and narrative reports to assist clinicians towards a decision.30 Thus, ML-integrated decision support systems are expected to facilitate further the smooth and trustworthy integration of ML technologies into the clinical setting.
Our study has limitations and strengths as enlarged upon in the Supplement. The shorter duration of follow-up among patients without compared to those with metastases in the prospective PMT cohort may have impacted the importance of methoxytyramine to predict metastases by underestimating diagnostic sensitivity. We also did not develop ML models for patients with head and neck tumors separately from those with abdominal paragangliomas known to have different characteristics. Finally, the present data do not establish whether our ML models improve decision making and outcomes for patients, which requires a prospective clinical trial.
Another apparent study limitation, enlarged upon in the Supplemental Discussion, is the omission from the ML analyses of histopathological, radiological and somatic variant features that could have strengthened predictive value of ML models. Heterogeneity in radiological procedures and histopathological interpretations renders retrospective use of such features problematic. It should also be appreciated that the higher the complexity of the ML models, the lower their applicability in routine clinical practice health care setttings.
Despite the aforementioned limitations, our study is the first to develop accurate ML models for the prediction of metastases in patients with PPGL using routinely available data, and without need for genetic, imaging or histopathological data. High performance of our ML models was facilitated by the availability of complete and comprehensive data in the training and external validation datasets and the long duration of follow-up in the training cohort that minimized possibilities of misclassifying patients with metastatic risk among those without evidence of metastases. Importantly, the large number of patients included in the study and its international multicentric design, supports generalizability of our ML models. Finally, the reproducibility of the selected ML models was secured not only through external validation by a different patient cohort, but also through comparisons with the performance of clinical care specialists with expertise in the care of patients with PPGL.
Conclusions
In conclusion, our study demonstrates that although plasma methoxytyramine provides some utility to predict metastases among patients with PPGL, sensitivity is limited. However, incorporation of plasma methoxytyramine in ML models along with other clinical features such as primary tumor location and size, provides a highly accurate, non-invasive approach to predict metastases in patients with PPGL, and thereby guide individualized patient management and follow-up strategies.
Supplementary Material
Research in Context.
Evidence before this study
We searched PubMed on June 10, 2022 using the search (metastatic[Title] OR malignant[Title]) AND (pheochromocytoma[Title] OR paraganglioma[Title]) AND (predictor OR predict OR diagnose OR diagnosis). We also searched abstracts of the American Society of Clinical Oncology Annual Meeting, European Society for Medical Oncology Congress and American Association for Cancer Research Annual Meeting, European Society of Endocrinology Annual Meeting, Ensat International Adrenal Cancer Symposium, within the past 3 years using the same search terms. We identified several studies on predictors of metastatic disease among patients with pheochromocytoma/paraganglioma (PPGL). In particular, retrospective observational studies have established that young age, large tumor size, extra-adrenal tumor location, presence of specific pathogenic germline (e.g. SDHB) or somatic variants (ATRX, TERT, MAML3), as well as specific long noncoding RNAs are associated with higher risk of metastatic disease among patients with PPGL. Nevertheless, none of the aforementioned features, was robust enough alone to predict metastatic disease. We also identified five studies that focused on combining features in scoring systems. Histopathological features were included in most of these scores; however, these lack reproducibility and accuracy. Similarly, a scoring system derived purely from clinical data showed inappropriately low positive predictive value. Machine learning (ML) is a new digital approach that could potentially support decision making in health care. We identified studies that established ML models to differentiate patients with PPGL from patients with other forms of hypertension, utilizing mainly metabolomics, or in the field of radiomics for the differentiation of incidental adrenal masses. However, no studies were identified that introduced ML models to predict metastatic PPGL.
Added value of this study
This clinician-designed and implemented study introduces robust noninvasive ML models to predict metastatic disease in patients with PPGL. These models utilize only routinely available features preoperatively, and can be readily applied and adapted by clinicians not only for PPGL, but also for other cancers. High performance and reproducibility of the selected ML models was secured by both external validation using a different patient cohort and also through comparisons with interpretations by an international group of clinical care specialists with expertise in the management of patients with PPGL. The latter established that the selected Ensemble Tree Classifier ML model provided significantly superior performance over interpretations of all specialists and could reliably predict metastatic disease in most patients with PPGL.
Implications of all the available evidence
We expect that clinicians will benefit from the assistance of the selected ML models, as they provide suitable tool for prediction of metastatic PPGL, and can be easily implemented in digital health care systems. Overall, our findings support emerging concepts that ML will gain traction in oncology for its potential to facilitate robust diagnostic stratification and guide personalized patient management.
Acknowledgments:
This work was supported by 314061271-TRR/CRC 205- 1/2 (to C.P., F.B., M.F., S.N., G.C., C.K., H.R., S.R.B., J.W.M.L., and G.E.), by the Free State of Saxony and TU Dresden (to A.F. and K.A.), the National Institutes of Health (to T.P., L.M., and K.P.) and by the Clinical Research Priority Program of the University of Zurich for the CRPP HYRENE (to F.B.).
Footnotes
Disclosure Statement: C.P., A.F. and G.E declare a filed German patent A5914/TUD 017, with the title “Verfahren zur Vorhersage eines Nebennierentumors sowie eines Metastaserisikos mithilfe klinisch relevanter Parameter“, relevant to this manuscript.
Conflict of Interest: The authors declare that they have no conflict of interest relevant to this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability
The data generated in this study are available in the online platform (zenodo.org), DOI 10.5281/zenodo.7749613 .
References
- 1.Tischler AS. Pheochromocytoma and extra-adrenal paraganglioma: updates. Arch Pathol Lab Med 2008; 132: 1272–84. [DOI] [PubMed] [Google Scholar]
- 2.Eisenhofer G, Lenders JW, Siegert G, et al. Plasma methoxytyramine: a novel biomarker of metastatic pheochromocytoma and paraganglioma in relation to established risk factors of tumour size, location and SDHB mutation status. Eur J Cancer 2012; 48: 1739–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li M, Prodanov T, Meuter L, et al. Recurrent disease in patients with sporadic pheochromocytoma and paraganglioma. J Clin Endocrinol Metab 2022: dgac563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mete O, Asa SL, Gill AJ, Kimura N, de Krijger RR, Tischler A. Overview of the 2022 WHO Classification of Paragangliomas and Pheochromocytomas. Endocr Pathol 2022; 33: 90–114. [DOI] [PubMed] [Google Scholar]
- 5.Fassnacht M, Assie G, Baudin E, et al. Adrenocortical carcinomas and malignant phaeochromocytomas: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2020; 31: 1476–1490. [DOI] [PubMed] [Google Scholar]
- 6.Lenders JWM, Kerstens MN, Amar L, P et al. 2020 Genetics, diagnosis, management and future directions of research of phaeochromocytoma and paraganglioma: a position statement and consensus of the Working Group on Endocrine Hypertension of the European Society of Hypertension. J Hypertens 2020; 38: 1443–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pamporaki C, Prodanov T, Meuter L, et al. Determinants of disease-specific survival in patients with and without metastatic pheochromocytoma and paraganglioma. Eur J Cancer 2022; 169: 32–41. [DOI] [PubMed] [Google Scholar]
- 8.Mcmillan M. Identification of hydroxytyramine in a chromaffin tumour. Lancet 1956; 271: 284. [DOI] [PubMed] [Google Scholar]
- 9.van der Harst E, de Herder WW, de Krijger RR, et al. The value of plasma markers for the clinical behaviour of phaeochromocytomas. Eur J Endocrinol 2002; 147: 85–94. [DOI] [PubMed] [Google Scholar]
- 10.Pamporaki C, Hamplova B, Peitzsch M, et al. Characteristics of Pediatric vs Adult Pheochromocytomas and Paragangliomas. J Clin Endocrinol Metab 2017; 102: 1122–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parasiliti-Caprino M, Lucatello B, Lopez C, et al. Predictors of recurrence of pheochromocytoma and paraganglioma: a multicenter study in Piedmont, Italy. Hypertens Res 2020; 43: 500–510. [DOI] [PubMed] [Google Scholar]
- 12.Ayala-Ramirez M, Feng L, Johnson MM, et al. Clinical risk factors for malignancy and overall survival in patients with pheochromocytomas and sympathetic paragangliomas: primary tumor size and primary tumor location as prognostic indicators. J Clin Endocrinol Metab 2011; 96: 717–25. [DOI] [PubMed] [Google Scholar]
- 13.Brouwers FM, Eisenhofer G, Tao JJ, et al. High frequency of SDHB germline mutations in patients with malignant catecholamine-producing paragangliomas: implications for genetic testing. J Clin Endocrinol Metab 2006; 91: 4505–9. [DOI] [PubMed] [Google Scholar]
- 14.Monteagudo M, Martínez P, Leandro-García LJ, et al. Analysis of Telomere Maintenance Related Genes Reveals NOP10 as a New Metastatic-Risk Marker in Pheochromocytoma/Paraganglioma. Cancers (Basel) 2021; 13: 4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson LD. Pheochromocytoma of the Adrenal gland Scaled Score (PASS) to separate benign from malignant neoplasms: a clinicopathologic and immunophenotypic study of 100 cases. Am J Surg Pathol 2002; 26: 551–66 [DOI] [PubMed] [Google Scholar]
- 16.Kimura N, Takayanagi R, Takizawa N, et al. Pathological grading for predicting metastasis in phaeochromocytoma and paraganglioma. Endocr Relat Cancer 2014; 21: 405–414. [DOI] [PubMed] [Google Scholar]
- 17.Wu D, Tischler AS, Lloyd RV, et al. Observer variation in the application of the Pheochromocytoma of the Adrenal Gland Scaled Score. Am J Surg Pathol 2009; 33: 599–608. [DOI] [PubMed] [Google Scholar]
- 18.Stenman A, Zedenius J, Juhlin CC. The Value of Histological Algorithms to Predict the Malignancy Potential of Pheochromocytomas and Abdominal Paragangliomas-A Meta-Analysis and Systematic Review of the Literature. Cancers (Basel) 2019; 11: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho YY, Kwak MK, Lee SE, Ahn SH, Kim H, Suh S, Kim BJ, Song KH, Koh JM, Kim JH, Lee SH. A clinical prediction model to estimate the metastatic potential of pheochromocytoma/paraganglioma: ASES score. Surgery 2018; 164: 511–517. [DOI] [PubMed] [Google Scholar]
- 20.Obermeyer Z, Lee TH. Lost in Thought - The Limits of the Human Mind and the Future of Medicine. N Engl J Med 2017; 377: 1209–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loftus TJ, Tighe PJ, Filiberto AC, et al. Artificial Intelligence and Surgical Decision-making. JAMA Surg 2020; 155: 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seymour CW, Kennedy JN, Wang S, et al. Derivation, Validation, and Potential Treatment Implications of Novel Clinical Phenotypes for Sepsis. JAMA 2019; 321: 2003–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bera K, Schalper KA, Rimm DL, et al. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 2019; 16: 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin N, de Cubas AA, Garcia-Martin R, et al. Opposing effects of HIF1α and HIF2α on chromaffin cell phenotypic features and tumor cell proliferation: Insights from MYC-associated factor X. Int J Cancer 2014; 135: 2054–64. [DOI] [PubMed] [Google Scholar]
- 25.Letouzé E, Martinelli C, Loriot C, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell 2013; 23: 739–52. [DOI] [PubMed] [Google Scholar]
- 26.Thienpont B, Steinbacher J, Zhao H, et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature 2016; 537:63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morin A, Goncalves J, Moog S, et al. TET-Mediated Hypermethylation Primes SDH-Deficient Cells for HIF2α-Driven Mesenchymal Transition. Cell Rep 2020; 30: 4551–4566.e7. [DOI] [PubMed] [Google Scholar]
- 28.Shimizu H, Nakayama KI. Artificial intelligence in oncology. Cancer Sci 2020; 111:1 452–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Wang D, Yao Z, Xin B, Wang B, Lan C, Qin Y, Xu S, He D, Liu Y. Machine Learning Models for Multiparametric Glioma Grading With Quantitative Result Interpretations. Front Neurosci 2019; 12: 1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sim I, Gorman P, Greenes RA, Haynes RB, Kaplan B, Lehmann H, Tang PC. Clinical decision support systems for the practice of evidence-based medicine. J Am Med Inform Assoc 2001; 8: 527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available in the online platform (zenodo.org), DOI 10.5281/zenodo.7749613 .