Abbreviations
- AR
Androgen receptor
- CPGEA
Chinese Prostate Cancer Genome and Epigenome Atlas
- CHD1
Chromodomain‐Helicase DNA‐binding 1
- DMFS
Distant metastasis‐free survival
- FOXA1
Forkhead box A1
- GRID
Genomics Resource for Intelligent Discovery database
- HR
Hazard Ratio
- NCCN
National Comprehensive Cancer Network
- PAM50
Prediction analysis microarray 50
- PCa
Prostate cancer
- PSA
Prostate‐specific antigen
- PSM
Propensity score matched
- PTEN
Phosphatase and tensin homolog
- TCGA
The Cancer Genome Atlas
- TMPRSS2‐ERG
Transmembrane Protease Serine 2‐E26 transformation‐specific related gene
Dear editor,
Prostate cancer (PCa) remains a major healthcare burden in men globally [1]. Most patients present with localized disease, and treatment is recommended based on risk classification systems like the National Comprehensive Cancer Network (NCCN) [2]. However, these methods are imprecise for estimating metastasis‐free survival and prostate cancer‐specific mortality and thus biomarkers that can predict tumor aggression are needed [3, 4, 5]. Several studies have since characterized the molecular landscape of localized PCa in White [4, 5] and Black/African‐American men [6], but data is lacking in Asian men. The Chinese Prostate Cancer Genome and Epigenome Atlas (CPGEA) reported on the genomic and epigenomic landscape of 208 PCa of men from China [7]. Comparative analyses between the CPGEA cohort and data from The Cancer Genome Atlas (TCGA) revealed higher frequencies of Forkhead box A1 (FOXA1) and chromodomain‐helicase DNA‐binding 1 (CHD1) mutations, and lower frequencies of phosphatase and tensin homolog (PTEN) mutations and transmembrane protease serine 2‐E26 transformation‐specific related gene (TMPRSS2‐ERG) fusion in Chinese compared with White men [7]. These preliminary findings highlight the presence of race‐specific differences in molecular phenotypes of PCa.
Here, we conducted a study to compare the gene expression profiles across 75 signatures between an East Asian PCa cohort of 181 patients against a propensity score‐matched (PSM) cohort of 905 North American PCa patients, who were identified from a 100,529 North American PCa Genomics Resource for Intelligent Discovery database (GRID, https://decipherbio.com/grid; ClinicalTrials.gov, NCT02609269). PSM was performed at a 1:5 ratio between East Asian and North American patients using 4 factors: NCCN risk‐group, clinical‐stage, prostate‐specific antigen (PSA), and microarray quality control scores (see Supplementary Materials and Methods). Tumors were profiled using the Decipher Genomic Classifier between 2013 and 2022 (Veracyte, San Francisco, CA, USA) [8]. Treatment details of the East Asian cohort are provided in Supplementary Materials and Methods and Supplementary Table S1. Ethics approval was obtained from the SingHealth institutional review board (IRB protocol no. 2019/2177), and written informed consent was obtained from all patients.
Proportional differences of selected gene expression signatures between the East Asian and PSM‐North American cohorts are summarized in Figure 1A and Supplementary Table S2. We observed fewer PCa with ERG‐positivity (16.0% vs. 30.4%, P < 0.001), average androgen receptor (AR) activity scores (61.9% vs. 76.9%, P < 0.001), and PTEN loss (7.7% vs. 20.2%, P < 0.001) in East Asian than in North American patients. The Prediction Analysis of Microarray 50 (PAM50) classifier, which was first developed in breast cancer, bins PCa into luminal A or B, and basal subtypes, which may be predictive of sensitivity to androgen deprivation therapy [9]. We did not observe differences in the distribution of PAM50 luminal‐basal subtypes between our East Asian and North American cohorts (luminal A: 8.3% vs. 14.0%; luminal B: 39.2% vs. 39.0%; basal: 52.5% vs. 47.0%; P = 0.092). Interestingly, when we compared the distribution of luminal‐basal PCa based on the Prostate Subtyping Classifier, which revised the luminal‐basal classification of PCa into luminal differentiated, luminal proliferating, basal immune, and basal neuroendocrine [10], we observed significantly higher proportions of luminal proliferating (25.4% vs. 14.0%, P < 0.001) and basal neuroendocrine subtypes (33.7% vs. 27.9%, P < 0.001) in East Asian than in North American patients.
FIGURE 1.
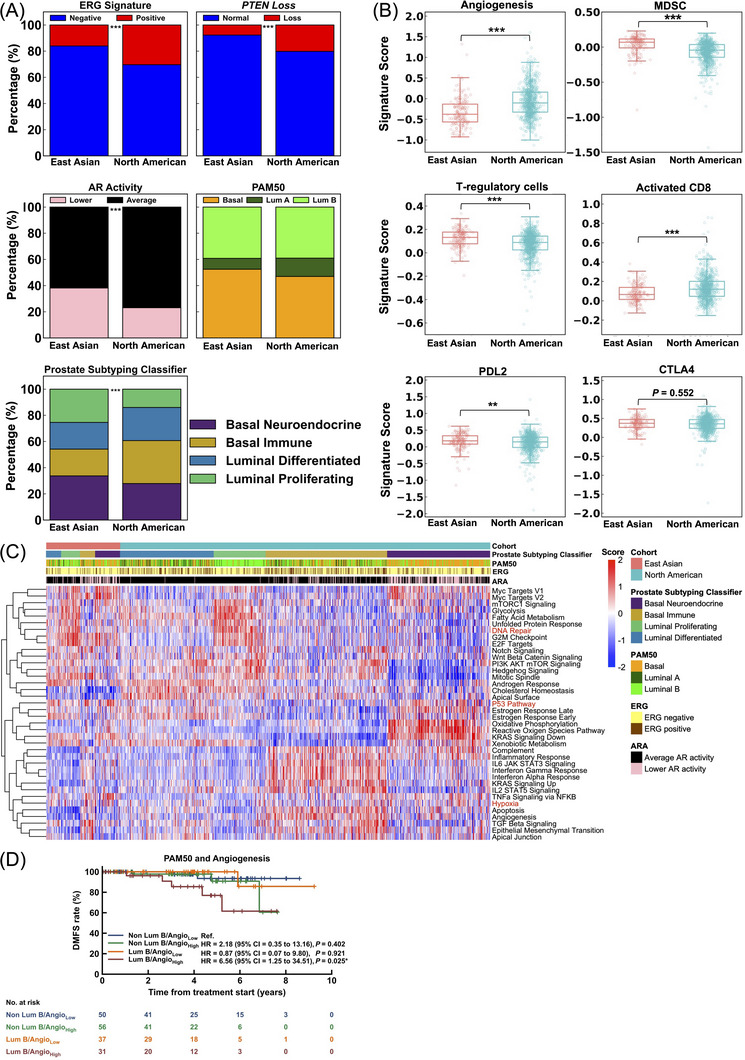
Transcriptomic profiles of 181 East Asian prostate cancer (PCa) patients and 905 North American PCa patients identified from a GRID database (NCT02609269). (A) Proportions of molecular subtypes (stacked bar charts) of tumor microenvironment‐related pathways between East Asian and North American PCa. (B) Signature scores of tumor microenvironment‐related pathways between East Asian and North American PCa. (C) Heatmap of 38 hallmarks of cancer pathways ordered by Prostate Subtyping Classifier (PSC) subtypes and East Asian or North American cohorts, including reference expression profiles for Prediction Analysis Microarray 50 (PAM50) subtypes, E26 transformation‐specific related gene (ERG) signature, and androgen receptor (AR) activity. (D) Kaplan‐Meier survival curves for distant metastasis‐free survival (DMFS) of East Asian cohort for combinatorial luminal B subtype and angiogenesis score. Hazard ratios (HR) were computed using the Cox proportional hazard model. P values were generated using the Log‐rank test and labeled as *<0.05; **<0.01; ***<0.001. Abbreviations: ERG, E26 transformation‐specific related gene; PTEN, Phosphatase and tensin homolog; AR, Androgen receptor; ARA, Androgen receptor activity; PAM50, Prediction analysis microarray 50; Lum A, Luminal A; Lum B, Luminal B; MDSC, Myeloid‐derived suppressor cells; DMFS, Distant metastasis‐free survival; Angio, Angiogenesis; HR, Hazard Ratio.
Gene expression signatures relating to angiogenesis and the immune microenvironment also differed between PCa of East Asian and North American patients (Figure 1B). We observed lower angiogenesis scores (median: ‐0.38 vs. ‐0.10, P < 0.001) and higher median scores of immune suppression signatures (myeloid‐derived suppressor cells: 0.07 vs. ‐0.04, P < 0.001; regulatory T cells: 0.13 vs. 0.09, P < 0.001; programmed death 1 ligand 2: 0.18 vs. 0.15, P = 0.004) in East Asian than in North American patients. These corresponded to a lower median activated CD8 signature score in the former than in the latter (0.06 vs. 0.12, P < 0.001).
To better understand the significance of these differences, we tested for associations between the PAM50 and Prostate Subtyping Classifier luminal‐basal status and selected gene signatures of ERG positivity, PTEN loss, and AR activity. When stratified by PAM50 status, we observed that basal tumors were strongly associated with a low rate of ERG positivity, a high rate of PTEN loss, and low AR activity in both East Asian and PSM‐North American cohorts (Supplementary Figure S1). By the Prostate Subtyping Classifier model, luminal differentiated and luminal proliferating tumors had lower rates of PTEN loss compared with basal immune and basal neuroendocrine tumors, and basal neuroendocrine PCa had the lowest AR activity in both cohorts (Supplementary Figure S2). Next, we generated a heatmap of hallmark signatures ordered by the Prostate Subtyping Classifier subtypes and cohorts (Figure 1C). Similar trends in hallmarks associated with the Prostate Subtyping Classifier subtypes were observed in both East Asian and North American cohorts, with basal tumors having higher expression of p53 and hypoxia‐related genes, while luminal proliferating tumors had higher expression of DNA repair genes.
Finally, we investigated if the different molecular subtypes were prognostic for distant metastasis‐free survival (DMFS) in the East Asian cohort. Patients with luminal A and luminal differentiated tumors had the most favorable DMFS among the luminal‐basal subtypes (Supplementary Figure S3A‐B). Among the microenvironment‐related signatures, we observed that patients with high angiogenesis signature scores had an inferior DMFS in our cohort (Hazard Ratio [HR] 3.79, 95% CI = 1.00‐14.41, P = 0.036, Supplementary Figure S3C). Interestingly, angiogenesis was able to sub‐stratify luminal B tumors for DMFS. Patients with luminal B PCa and high angiogenesis scores had the worst DMFS, compared with the other 3 subgroups (HR ref non‐luminal B + angiogenesis low = 6.56, 95% CI = 1.25‐34.51, P = 0.025, Figure 1D).
Several limitations of the present study deserve mention. First, these findings in our limited East Asian cohort, which was comprised of mostly NCCN high‐risk PCa patients, ought to be validated in other Asian cohorts, with balanced composition of the different NCCN risk groups. Second, while we recruited subjects from different geographical regions, we could not control for race effects; while 90.1% of subjects in the East Asian cohort were Chinese, we lacked curated physician‐reported race data in the North American cohort. Third, there is a need to determine the corresponding therapeutic implications of the race‐specific phenotypic and microenvironmental differences. Such analyses can only be done using well‐curated cohorts from prospective registries or clinical trials, for which archival tissues are available for molecular profiling.
To summarize, we uncovered fewer ERG‐positive, PTEN‐loss, and high AR activity tumors in East Asian than in North American PCa patients. Additionally, there was a higher proportion of luminal proliferating and basal neuroendocrine PCa in East Asian than in North American patients based on the Prostate Subtyping Classifier model. Interrogation of the tumor microenvironment revealed lower levels of angiogenesis and an overall immune suppressive state in East Asian than in North American PCa patients. Of clinical relevance, high angiogenesis and luminal B tumors had the worst DMFS in our East Asian cohort. Taken together, our results showcased race‐specific differences in gene expression profiles of PCa from East Asian and North American patients, adding to the literature on genomic and epigenomic inter‐racial heterogeneity of localized PCa. These data posit the concept that demographic host factors are highly relevant in PCa tumorigenesis, and support a larger pan‐race meta‐analysis.
DECLARATIONS
Preliminary results of this study were presented at the ASCO GU symposium 2022, San Francisco and the 2022 ESMO Congress, Paris.
AUTHORS CONTRIBUTION
Study conception and design: MLKC, ED
Patient recruitment and data acquisition: All authors
Data analysis and interpretation: All authors
Statistical analyses: MLKC, AH, EHWO, BHH, YL, ED
Obtained funding: MLKC
Administrative, technical, or material support: MLKC, EHWO, ED, LYK
Study supervision: MLKC, ED
Drafting of manuscript: MLKC, AH, EHWO, BHH, JJM, YL, ED, LYK
Approval of final manuscript: All authors
CONFLICT OF INTEREST STATEMENT
Melvin L.K. Chua reports personal fees from Astellas, Bayer, Pfizer, MSD, AstraZeneca, Varian, Janssen, IQVIA, Telix Pharmaceuticals; non‐financial support from AstraZeneca; non‐financial support from Veracyte Inc; grants from Ferring; consults for immunoSCAPE Inc.; and is a co‐inventor of the patent of a High Sensitivity Lateral Flow Immunoassay For Detection of Analyte in Sample (10202107837T), Singapore, and serves on the Board of Directors of Digital Life Line Pte Ltd that owns the licensing agreement of the patent, outside the submitted work.
Ravindran Kanesvaran received personal fees from Astella, BMS, Ipsen, J&J, Merck, Novartis, Amgen, and Eisai.
Alexander Hakansson, Julian Ho, Xin Zhao, Elai Davicioni and Yang Liu are employees of Veracyte, Inc.
All other authors do not declare any conflicts of interests.
CONSENT FOR PUBLICATION
The article does not contain any person's identifiable data.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the SingHealth Institutional Review Board (IRB protocol no. 2019/2177), and all patients provided written informed consent.
FUNDING INFORMATION
Melvin L.K. Chua is supported by the National Medical Research Council Singapore Clinician Scientist Award (NMRC/CSA‐INV/0027/2018, CSAINV20nov‐0021), the Duke‐NUS Oncology Academic Program Goh Foundation Proton Research Program, NCCS Cancer Fund, and the Kua Hong Pak Head and Neck Cancer Research Program.
The funding organizations had no role in the design and conduct of this trial; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Supporting information
Supporting Information
ACKNOWLEDGEMENTS
The authors thank the patients and their families for their participation in this study.
Melvin L.K. Chua, Alexander K. Hakansson, Enya H.W. Ong, Boon Hao Hong and Jing Jing Miao contributed to this work equally.
DATA AVAILABILITY STATEMENT
Data can be made available for bona fide researchers who request it from the authors. The data used in this manuscript will be deposited in the institutional repository at the National Cancer Centre Singapore.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 2. Carroll PH, Mohler JL. NCCN Guidelines Updates: Prostate Cancer and Prostate Cancer Early Detection. J Natl Compr Canc Netw. 2018;16(5s):620–3. [DOI] [PubMed] [Google Scholar]
- 3. Epstein JI, Amin MB, Reuter VE, Humphrey PA. Contemporary Gleason Grading of Prostatic Carcinoma: An Update With Discussion on Practical Issues to Implement the 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol. 2017;41(4):e1–e7. [DOI] [PubMed] [Google Scholar]
- 4. Cancer Genome Atlas Research Network . The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163(4):1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, et al. Genomic hallmarks of localized, non‐indolent prostate cancer. Nature. 2017;541(7637):359–64. [DOI] [PubMed] [Google Scholar]
- 6. Yadav S, Anbalagan M, Baddoo M, Chellamuthu VK, Mukhopadhyay S, Woods C, et al. Somatic Mutations in the DNA Repairome in Prostate Cancers in African Americans and Caucasians. Oncogene. 2020, 39(21), 4299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li J, Xu C, Lee HJ, Ren S, Zi X, Zhang Z, et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature. 2020;580(7801):93–9. [DOI] [PubMed] [Google Scholar]
- 8. Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One. 2013;8(6):e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao SG, Chang SL, Erho N, Yu M, Lehrer J, Alshalalfa M, et al. Associations of Luminal and Basal Subtyping of Prostate Cancer With Prognosis and Response to Androgen Deprivation Therapy. JAMA Oncol. 2017;3(12):1663–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weiner AB, Liu Y, Hakansson A, Zhao X, Proudfoot JA, Ho J, et al. A novel prostate cancer subtyping classifier based on luminal and basal phenotypes. Cancer. 2023. manuscript in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
Data can be made available for bona fide researchers who request it from the authors. The data used in this manuscript will be deposited in the institutional repository at the National Cancer Centre Singapore.


