Abstract
Melanoma remains the most aggressive and fatal form of skin cancer, despite several FDA-approved targeted chemotherapies and immunotherapies for use in advanced disease. Of the 100,350 new patients diagnosed with melanoma in 2020 in the U.S., more than half will develop metastatic disease leading to a 5-year survival rate <30%, with a majority of these developing drug-resistance within the first year of treatment. These statistics underscore the critical need in the field to develop more durable therapeutics as well as those that can overcome chemotherapy-induced drug resistance from currently approved agents. Fortunately, several of the drug-resistance pathways in melanoma including the proteins in those pathways rely in part on Hsp90 chaperone function. This presents a unique and novel opportunity to simultaneously target multiple proteins and drug-resistant pathways in this disease via molecular chaperone inhibition. Taken together, we hypothesize that our novel C-terminal Hsp90 inhibitor, KU758, in combination with current standard of care targeted therapies (e.g. vemurafenib and cobimetinib) can both synergize melanoma treatment efficacy in BRAF-mutant tumors, as well as target and overcome several major resistance pathways in this disease. Using in vitro proliferation and protein-based Western Blot analyses, our novel inhibitor, KU758 potently inhibited melanoma cell proliferation (without induction of the heat-shock-response) in vitro and synergized with both BRAF and MEK inhibitors in inhibition of cell migration and protein expression from resistance pathways. Overall, our work provides early support for further translation of C-terminal Hsp90 inhibitor and MAPK pathway inhibitor combinations as a novel therapeutic strategy for BRAF-mutant melanomas.
Keywords: Hsp90 inhibitors, BRAF and MEK inhibitors, combination therapy, drug efficacy, molecular drug targeting, apoptosis, migration, heat shock response
INTRODUCTION
Despite Food and Drug Administration (FDA) approval of several targeted chemotherapies and immunotherapies, melanoma remains the most aggressive and fatal skin cancer. Melanoma incidence is rising yearly with 100,350 new U.S. cases expected in 2020 (1). Of the patients diagnosed, >50% will present with metastatic disease decreasing their 5-year survival rate to ~25% (1). This indicates the critical need for novel, more-durable drugs and therapeutic approaches for these patients. Although several approved therapies – namely mitogen activated protein kinase (MAPK) pathway and immune checkpoint inhibitors – are reasonably effective early-on, the majority of patients develop drug resistance within the first year of treatment(2, 3).
A common clinical approach to mitigate resistance utilizes drug combinations that simultaneously target multiple pathways. Combination therapy using lower drug doses also has the potential to decrease toxicities while improving overall efficacy(4). In 2014, the FDA approved the combination of MAPK pathway inhibitors (MAPKi) targeting BRAF (e.g. vemurafenib) and MEK (e.g. cobimetinib) in patients with unresectable advanced BRAF-mutant melanomas. A year later, the PD-1 (programmed cell death protein-1) and CTLA-4 (cytotoxic T-lymphocyte associated protein-4) monoclonal antibodies were approved as first-line immunotherapies in metastatic BRAF-wildtype patients(5). While initial patient responses were promising, emergence of drug-resistance in both approaches highlights the need for development of more-durable therapies.
Targeting the 90-kDa heat shock protein (Hsp90) provides an attractive anticancer therapeutic approach as inhibition of chaperone function can target multiple cancer-promoting kinase pathways simultaneously. As chaperones, Hsps play a crucial role in cellular proteostasis, and more specifically the heat shock response (HSR), by maintaining protein folding, stabilization, and activation of a myriad of substrates, referred to as clients(6-8). Hsp clientele are involved in several cellular pathways/processes including steroid hormone receptors, protein kinases/ phosphatases for signal transduction, cell cycle regulation, and gene transcription regulators(9). Hsp90’s role in cancer was elucidated in the 1990s by Whitesell and colleagues after observing that oncogenic cell morphology in v-src transformed fibroblasts was reversed with the treatment of geldanamycin (GDA), a benzoquinone ansamycin antibiotic(10). Since then, numerous N-terminal Hsp90 inhibitors have been developed and tested in clinical cancer trials. This ongoing interest is best explained by Hsp90’s “central node” role in chaperoning client proteins involved in all the processes/hallmarks that contribute to and maintain the initiation and progression of cancer(11, 12). While no Hsp90 inhibitors (Hsp90i) to date are FDA approved, several inhibitors of Hsp90 (17-AAG, XL-888, AT13387 and genetispib) are investigated against melanoma in pre-clinical studies and clinical trials either alone or in combination with currently available BRAF and MEK inhibitor therapy(13-18).
Melanoma cells have active RAF-MEK-ERK signaling either due to the presence of BRAF V600E mutation or through upstream BRAF signaling. Although preclinical studies have shown delayed resistance to vemurafenib treatment or therapeutic synergy with vemurafenib when melanoma cells were treated with 17-AAG, phase II trial in patients with metastatic melanoma showed no objective clinical response and several adverse effects such as nausea, vomiting and fatigue were observed(19). Melanoma cells resistant to either vemurafenib or BRAF and MEK inhibitor combination were sensitized when treated with AT13387(17). Therefore, AT13387 was tested in Phase I clinical study in patients with advanced solid tumors. Unlike other ansamycin Hsp90 inhibitors, AT13387 was well tolerated and is currently under evaluation in phase I clinical trial in combination with dabrafenib and trametinib (20).
The majority of these Hsp90i in clinical trials target Hsp90’s N-terminal domain and induce the HSR with up-regulation of Hsp70. This leads to needing higher Hsp90 concentrations to maintain cellular inhibition a potential contributor to dose-limiting toxicities observed in these trials(21). To address this, we developed novel compounds that target the C-terminal as a unique approach for Hsp90 inhibition that does not stimulate the HSR as robustly. Our group has recently shown that C-terminal inhibitors (CT-Hsp90i) are effective in breast cancers, adrenocortical cancers, and head and neck squamous cell carcinomas and targeting cancer stem cells (CSC) by decreasing CSC markers and downregulating invasion/migration and epithelial-to-mesenchymal transition both in vitro and in vivo(22-24). Taken together, we hypothesize that combining a MAPKi (BRAF or MEK) with a new CT-Hsp90i could synergize anticancer-effect and provide a novel rational therapeutic strategy for metastatic melanoma patients. Such a combination should down-regulate major BRAF-resistance pathways and dampen the effect(s) of several cancer hallmarks. We evaluate here our novel CT-Hsp90i (KU758) alone and in combination with a BRAFi/MEKi for its ability to inhibit melanoma cell proliferation, migration, and target melanoma cellular processes critical in the maintenance of resistance.
METHODS
Cell Culture and Reagents
For cell culture, unless otherwise noted, DNA-fingerprint validated human melanoma cell lines harboring a BRAFV600E-mutation (UACC-257, UACC-62, and Skmel19) and wild type BRAF cell lines Skmel103 and Skmel173 were maintained in T-75 flasks (Thermo Fisher Scientific, Waltham, MA) in high glucose and L-glutamine Dulbecco’s Modified Eagle’s Medium (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS (Sigma-Aldrich, St. Louis, MO) and 1% penicillin/streptomycin (Thermo Fisher Scientific). Cell lines were incubated at 37°C in 5% CO2. The same protocol was followed to culture a human fetal fibroblast (FF) cell line as a control. Cells were washed with 1X phosphate buffer solution (PBS), then detached using Trypsin-EDTA (Sigma-Aldrich, St. Louis, MO). Drug compounds used included: BRAF inhibitor (BRAFi) vemurafenib (Ve) (Selleck Chemicals, Houston, TX), MEK inhibitor (MEKi) cobimetinib (Cb) (AdooQ Bioscience, Irvine, CA), novel CT-Hsp90i KU758 obtained from Dr. Brian S.J. Blagg (Univ. of Notre Dame, Notre Dame, ID), N-terminal Hsp90i (NT-Hsp90i) XL888 (AdooQ Bioscience, Irvine, CA), and natural product Hsp90i withalongolide A 4,19,27-triacetate (WGA-TA) obtained from Dr. Barbara Timmermann (Kansas Univ., Lawrence, KS).
Cell Viability Assay
Cell lines were plated in polystyrene, clear flat-bottom 96-well plates (Thermo Fisher Scientific, Waltham, MA) at 1,500 cells/well and adhered overnight. Cells were treated in serial concentrations of each inhibitor and left to incubate for 24hrs. To determine cell viability, ATP was quantified using a CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Madison, WI) following the manufacturer’s instructions. The luminescence was read using a BioTek Synergy Neo plate reader and Gen5 software (BioTek, Winooski, VT). The values collected were imported into GraphPad Prism (GraphPad Prism Software, Inc., La Jolla, CA) to obtain dose-response curves and calculate the half maximal inhibition concentration (IC50) of each drug in all cell lines. Cell viability for FF cells was determined using an MTS assay. Cells were plated in 96-well plates at 6,000 cells/well. MTS reagent (Owen’s reagent) was added to each well after 24hrs treatment period. Absorbance and IC50 values were read and calculated, respectively, as stated above.
Combination Assay
Cells were plated in 96-well plates, as aforementioned, then treated with a BRAFi or MEKi plus a Hsp90i or BRAFi plus MEKi for a total of seven combinations. The drug concentrations used were based on the IC50 values of each compound and cells were treated with each inhibitor at a range of concentrations (e.g. 1/4x, 1/2x, and 1x IC50) for 24hrs. The plating and CellTiter-Glo® protocols described above were used for combination assays. Following luminescence reading, values were analyzed using the CompuSyn software (Compusyn, Inc., Paramus, NJ), based on the Chou-Talalay theorem, to calculate the combination index (CI) of each treatment and determine synergistic combinations (e.g. CI<1). Surface maps of combination effects were plotted using Combenefit software (Cancer Research UK Cambridge Institute) to illustrate extent of biological (viability as percent of control) and synergistic/antagonistic (coloring of map) effects.
Immunoblot Analysis
Cell lines were cultured to 70% confluency, then treated for 24hrs with a drug alone or two in combination – concentrations used: 0.5μM KU758, 0.1μM WGA-TA, 0.3μM XL888, 0.125μM Cb, and 50μM Ve. Next, cells were collected, then lysed via suspension in a lysis buffer cocktail containing 2μL/mL PR protease inhibitor, 1μL/mL 100M phenylmethane sulfonyl fluoride, 2μL/mL 0.5M NaF, and 10μL/mL 100M Na3VO4 followed by sonication (Qsonica Sonicators, Newtown, CT). After centrifuging (Eppendorf, Hamburg, Germany) samples for 20mins at 4°C and 14,000rpm, protein supernatant was collected then quantified using a BSA protein standard assay (Thermo Fisher Scientific, Waltham, MA) following manufacturer’s instructions. Protein samples were prepped with 5X loading dye, loaded in equal amounts, and separated using SDS-PAGE. The Precision Plus Protein Dual Color Standards (Bio-Rad Laboratories, Hercules, CA) was used as a protein ladder. Briefly, SDS-PAGE gels were transferred to nitrocellulose blotting membranes (GE Healthcare Life Sciences, Pittsburgh, PA), washed with PBS and PBS-Tween20, and probed overnight in primary antibodies (Cell Signaling Technologies, Inc., Danvers, MA). Next blots were probed with appropriate horseradish peroxidase-linked anti-rabbit or anti-mouse secondary antibodies (Cell Signaling Technologies, Inc., Danvers, MA), washed again, and enhanced chemiluminescence horseradish peroxidase substrate [SuperSignal West Pico PLUS or Femto (Thermo Fisher Scientific, Waltham, MA)] was used to visualize nitrocellulose blots with a digital image using a ChemiDoc Imaging System (BioRad, Hercules, CA). β-actin was used as a loading control. ImageJ software (National Institutes of Health, Bethesda, MD) was used to perform densitometry of protein bands. Immunoblot quantification (densitometry) of a given protein band is described as a normalized, relative value to the corresponding β-actin expression.
Cell Cycle Analysis
UACC 257 melanoma cells grown to about 60-70% confluence in 60 mm plates were treated with the concentrations of drugs that induced synergistic effect for 24h (Ve = 50μM, 0.5μM KU758 and 0.125μM Cb) . The cells were then harvested, resuspended in 0.43 mL of ice cold 1X phosphate buffered saline (PBS) followed by 1 mL of ice-cold ethanol to get a 70% ethanol fixative solution and stored at −20°C until analysis. Finally, the cells were pelleted by centrifugation, stained with propidium iodide (PI) solution (40 μg/mL PI and 100 mg/mL RNaseA), and incubated at 37°C for 30 minutes before cell cycle analysis using ZE5 cell analyzer (Bio-Rad, Hercules, CA) at the University of Michigan Flow Cytometry Core. Each experiment was repeated thrice and only viable cells without DNA fragmentation were analyzed using FCS Express (De Novo software, Pasadena, CA).
Migration Assays
Cells were plated in polystyrene, clear flat-bottom 6-well plates at 250,000 cells/well and adhered overnight. A pipette tip was used to scratch a clear vertical line in well. Culture media was removed, and wells were washed with PBS to remove cell debris. Images of each well were captured in triplicate using the EVOS FLc microscope and camera (Thermo Fisher Scientific, Waltham, MA). Fresh cell culture media was added to each well, then treated with each inhibitor alone and in combination with Ve or Cb using the determined synergistic drug combination concentrations (see above), for a total of 12 treatment conditions and one untreated control. Cells incubated at 37°C in 5% CO2 until untreated cells migrated and covered previously scratched area (24hrs). Images of each well were recaptured in triplicate. For each well, we calculated the percent of the scratch zone covered by migrated cells at t=24hrs (higher percentage indicates more cell migration).
Additionally, Boyden chambers with 8.0μM pore size and PET track-etched membranes were used (Falcon, Tewksbury, MA). Cells were seeded at 50,000 cells/chamber in serum-free DMEM media. Chambers were submerged into polystyrene, clear flat-bottom 24-well plate wells with 20% FBS DMEM media and left to migrate for 24hrs at 37°C in 5% CO2. After 24hrs, each chamber was removed from the well, then fixed with formalin for 15mins and stained with 1% crystal violet, 20% methanol dye for 30mins. Chambers were washed and images captured using an EVOS FLc microscope and camera at 10X magnification. To quantify cell migration, images were used to count cells after 24hrs treatment.
Statistical Analysis
All in vitro experiments were replicated in triplicate. Significance was determined using Student’s t-test (*p < 0.05 and **p < 0.001). Dose-response curves were normalized to untreated control for all inhibitors and used to calculate IC50 values with 95% confidence intervals in GraphPad Prism, as stated above. Data is presented as mean values with standard deviation error bars. SPSS version 25 (IBM, Almont NY) was used to verify all statistical calculations.
RESULTS
KU758 Selectively and Potently Inhibits Growth of BRAF-Mutant Melanocytes
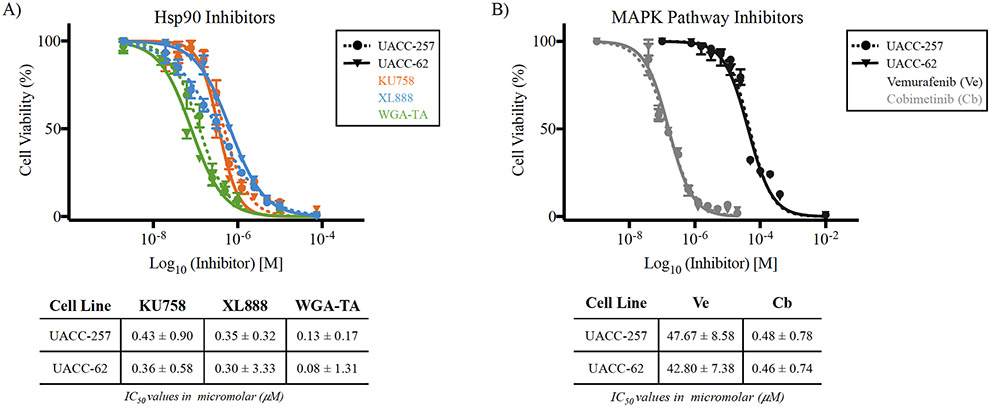
Before characterizing the combinational effect of KU758+MAPKi on resistance pathways and cancer hallmarks, it was pertinent to determine the potency and selectivity of KU758 in melanocytes. To evaluate this, we performed a proliferation assay (CellTiter-Glo®) and compared the IC50 values of 3 different Hsp90i: our novel CT-Hsp90i (KU758), the NT-Hsp90i (XL888), and the natural product inhibitor (WGA-TA). All in vitro cell viability assays were performed in UACC-257, UACC-62 and Skmel19 human BRAF-mutant melanocytes, wild type BRAF cell lines Skmel103 and Skmel173 and FF cell line controls. The IC50 of KU758 was consistent across BRAF-mutant cell lines (360-430nM; Figure 1A (UACC-257 and UACC-62), and Supplemental Figure 3 (Skmel19) and this was comparable to XL888 and WGA-TA. In BRAF-wild type cell lines, the IC50 for KU758 was 1.34 μM and 1.9 μM respectively for Skmel103 and Skmel173 respectively (Supplemental Figure 3). The IC50 of the BRAFi (Ve) and MEKi (Cb) are also listed in Figure 1B. As a non-malignant control, fetal fibroblasts (FF cells) were treated with all three Hsp90i. Each inhibitor demonstrated a cancer-specific selectivity over normal cells from 10-fold (KU758) to 300-fold (XL888) (Supplemental Figure 1).
Figure 1 – Melanocyte viability and IC50 after Hsp90i or MAPKi treatment –
All inhibitors exhibited efficacy in BRAF-mutant melanocyte (UACC-257 and UACC-62) death after 24hrs treatment with a range of potencies (IC50 value). A) The dose-response curves of all three Hsp90i show similar potencies in the sub-micromolar range (~0.1-0.4μM) – ascending IC50: WGA-TA (green) < XL888 (blue) < KU758 (orange). B) Melanocytes were treated with the BRAFi Ve (dark grey) and MEKi Cb (light grey) at several concentrations and the dose-response curves indicate higher potency of Cb over Ve. All inhibitor IC50 concentrations are listed below dose-response curves in micromolar with 95% confidence interval.
KU758 Synergizes with MAPK Pathway Inhibitors
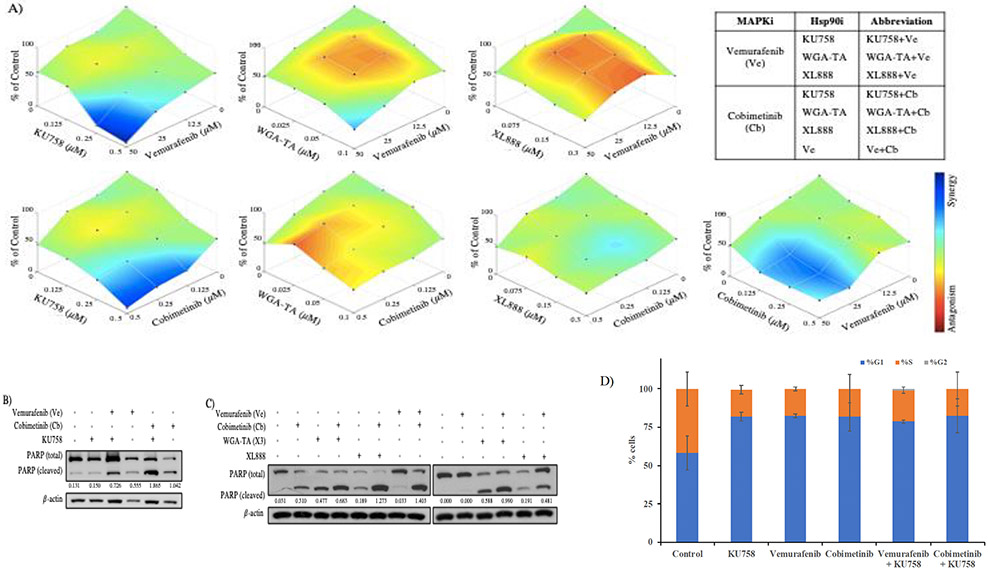
Combination therapy with targeted small molecular inhibitors, such as Ve with Cb, is one standard treatment for metastatic melanoma patients with a BRAFV600E/K-mutation status. Combination therapy success can be predicted by the extent of synergism, or lack thereof, between the two compounds of interest and is quantified by calculating the combination index (CI) of a given drug combination. A CI<1, especially <0.5 indicates synergistic combinations, while a CI=1 is an additive effect, and a CI>1 indicates an antagonistic effect (4, 25). To identify the combination effect of our novel CT-Hsp90i, KU758, and a MAPKi (Ve or Cb) on proliferation in BRAF-mutant melanocytes (UACC-257), we determined the CI of several drug combinations (Table 1). Cells were treated with Ve or Cb and KU758, XL888, or WGA-TA for seven inhibitor combinations (including Ve+Cb) at a range of concentrations based on the IC50 values. The response to combination treatment was measured as the percentage of viable cells compared to control (Figure 2A). KU758 synergized with Ve and Cb, in 7/9 combinations with six having a robust synergy (CI<0.5 for three K758+Ve combinations and CI>0.4 for three KU758+Cb combinations typically at 0.5μM KU758; dark blue and cyan coloring of the surface plots; Table 1A). The most synergistic combinations of WGA-TA+Cb and XL888+Cb were at 0.125μM Cb and 50μM Ve, so these concentrations were used for all subsequent studies. Similar to KU758, WGA-TA, synergized with BRAFi and MEKi at several drug concentrations, but with only one CI<0.5 (Figure 2A/Table 1A). Unlike the other Hsp90i, XL888+Cb synergized with relatively mid-to-low CIs, whereas CIs in XL888+Ve combinations were predominantly >1 suggesting some antagonism (Figure 2A/Table 1A). Since BRAFi and MEKi combinations are already FDA approved for use in BRAF-mutant metastatic melanoma patients, combination assays of Ve+Cb were performed and CIs were calculated for comparison. As expected, almost all (94%) Ve+Cb combinations, between the two cell lines, were synergistic (Figure 2A and Table 1B). To demonstrate replication of the synergistic combinational effect observed with KU758+MAPKi in UACC-257, experiments were repeated in UACC-62 (Supplemental Figure 2/Supplemental Table 1) and similar synergistic effects were noted. To confirm the inhibitor combinations were effective at promoting cell death, protein expression of PARP and its cleaved product were measured (Western Blot) and quantified utilizing Image-J software densitometry (compared to ß-actin). All combinations synergistically increased the expression of cleaved PARP compared to each treatment of each drug alone (p < 0.05) (Figure 2B/2C). Since, the observed changes in viability can either be due to cell cycle arrest or apoptosis, we further evaluated cell cycle using flow cytometry. Our results indicated that the percent of cells in G0/G1 phase increased from 58.32% for control cells to 81.97%, 82.55%, 81.78%, 78.8%, and 82.5% respectively after treatment, whereas the percent of cells in the S-phase decreased from 41.7% for control cells to 18%, 17.45%, 18.22%, 20.2%, and 17.5% respectively after treatment with KU758, Ve, Cb, Ve+KU758, and Cb+KU758 (Figure 2D). This indicates that KU758 either alone in combination with Ve or Cb is able to induce G0/G1 cell cycle arrest in melanoma cells. These results are consistent with previous studies in which treatment of melanoma cells with 17-AAG resulted in G0/G1 cell cycle arrest(14).
Table 1.
The CI values are listed for each inhibitor combination treatment in UACC-62 between Hsp90i and MAPKi
| KU758 |
WGA-TA |
XL888 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (μM) | 0.125 | 0.25 | 0.5 | 0.025 | 0.5 | 0.1 | 0.075 | 0.15 | 0.3 | |
| Ve | 12.5 | 1.445 | 0.428 | 0.291 | 116.814 | 0.353 | 0.070 | 8.166 | 7.199 | 2.580 |
| 25 | 1.452 | 0.340 | 0.262 | 1.636 | 0.181 | 0.045 | 5.645 | 2.136 | 1.732 | |
| 50 | 0.815 | 0.248 | 0.185 | 0.231 | 0.074 | 0.037 | 0.725 | 0.536 | 0.757 | |
| Cb | 0.125 | 1.166 | 0.735 | 0.315 | 1.198 | 1.557 | 1.701 | 0.895 | 0.236 | 0.342 |
| 0.25 | 1.014 | 0.717 | 0.235 | 0.994 | 1.341 | 1.514 | 0.362 | 0.282 | 0.410 | |
| 0.5 | 0.723 | 0.883 | 0.202 | 0.887 | 1.063 | 1.326 | 0.522 | 0.364 | 0.471 | |
Synergistic combination boxes are colored according to relative CI value: CI value <0.5 (blue), 0.5 < CI value < 1.0 (green).
Cb, cobimetinib; CI, combination index; MAPKi, mitogen-activated protein kinase inhibitor; Ve, vemurafenib; WGA-TA, withalongolide A 4,19,27-triacetate.
Figure 2 – Responses to Hsp90i+MAPKi combinations –
A) All Hsp90i, (listed left to right) KU758, WGA-TA, and XL888, showed a decrease in UACC-257 cell viability after treatment with Ve (top) or Cb (bottom). Each plot shows the percent of cell viability relative to control and illustrates the corresponding combination effect (e.g. synergistic, additive, or antagonistic) on a color spectrum. The most synergistic combinations were observed in KU758+Ve and KU758+Cb. All six Hsp90i+MAPKi combination abbreviations are tabulated. B and C) The expression of cleaved PARP increased in all inhibitor combinations compared to each drug alone. D) Cell-cycle analysis of UACC-257 by flowcytometry after drug treatments.
Major Resistance Pathway Proteins are Targeted with KU758 Combinations
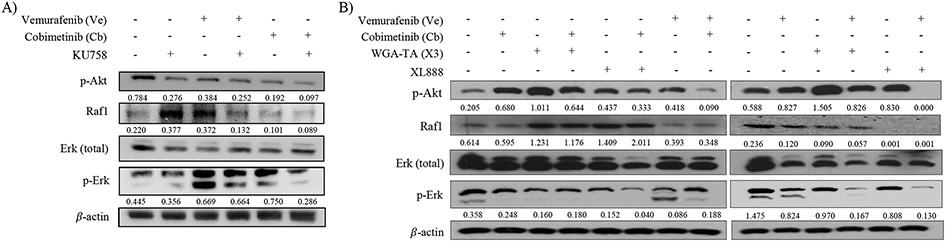
After determining the potency/selectivity of KU758 in melanocytes, and its synergy in combination with a MAPKi, we next evaluated this combination effect on two major BRAFi-resistance signaling pathways in melanoma (MAPK/Erk and PI3K/Akt pathways). We hypothesized that since Hsp90 serves as the molecular chaperone for several proteins involved in these two resistant pathways, then utilization of Hsp90i+MAPKi will lead to subsequent downregulation of these kinase drivers from the resistant pathways. To investigate this effect on resistant-pathway protein expression levels, we performed immunoblot analysis, then quantified the change in protein expression. Melanocytes treated with XL888+Cb or WGA-TA+Cb showed a 2- to 3-fold significant increase in expression of Raf1, (p<0.001). However, with XL888+Ve or WGA-TA+Ve combinations, expression of Raf1 was almost entirely ablated. In all instances, there was a significant change in protein expression compared to Cb or Ve alone (p<0.05), but not with an Hsp90i as a single agent (Figure 3B). In KU758+Ve and KU758+Cb treated cells, there was a significant decrease to Raf1 expression when compared to KU758-only treatment (p<0.05) (Figure 3A). Next, we immunoblotted for p-Erk, a kinase downstream of Raf1. In XL888+Cb and XL888+Ve combinations, the phosphorylated kinase was almost entirely knocked down (p<0.01); while it was significantly knocked down with WGA-TA+Cb, WGA-TA+Ve, and KU758+Cb (p<0.01; Figure 3A/3B). For p-Akt expression there was a 3- and 5-fold increase in cells treated with Cb or WGA-TA alone, respectively. Combinations of Hsp90i+MAPKi showed more significant knockdown than any drug alone with KU758+Cb having the highest knockdown of expression (95% vs. control), but with significant decreases also observed with the WGA-TA and XL888 combinations with Ve or Cb (all with p<0.01 vs. controls; Figure 3A/3B).
Figure 3 – Hsp90i+MAPKi combination effect on key resistance pathways –
A) In KU758 treated cells, alone or plus MAPKi, p-Akt expression decreased from untreated (control). Each combination decreased p-Akt to a greater extent than each inhibitor alone, especially KU758+Ve (p<0.001). Raf1 expression decreased in KU758+Ve, Cb, and KU758+Cb compared to control and each inhibitor alone (p<0.05). Ve, KU758+Ve, and Cb combinations increased p-Erk, while KU758+Cb effectively decreased the expression (p<0.05). B) p-Akt expression decreased in all Ve- and Cb-combinations, for WGA-TA and XL888, when compared to each inhibitor alone (p<0.05). Raf1 expression increased in WGA-TA, WGA-TA+Cb, XL888, and XL888+Cb. p-Erk expression was knocked down in WGA-TA+Ve and XL888+Ve combinations (p<0.001).
Migration is Decreased with Hsp90i+MAPKi Combinations
Since KU758+Ve and KU758+Cb combinations target key melanoma resistance pathways, we next investigated this combination effect on cell migration, since it contributes to the potential of a melanoma’s ability to move from one tissue into an adjacent one (26). Extent of cell migration in vitro is also a reasonable surrogate method to predict metastatic potential and its associated cellular processes. We hypothesized that KU758+MAPKi combinations would effectively decrease melanocyte migration since the proteins directly involved with these processes are also Hsp90 clients.
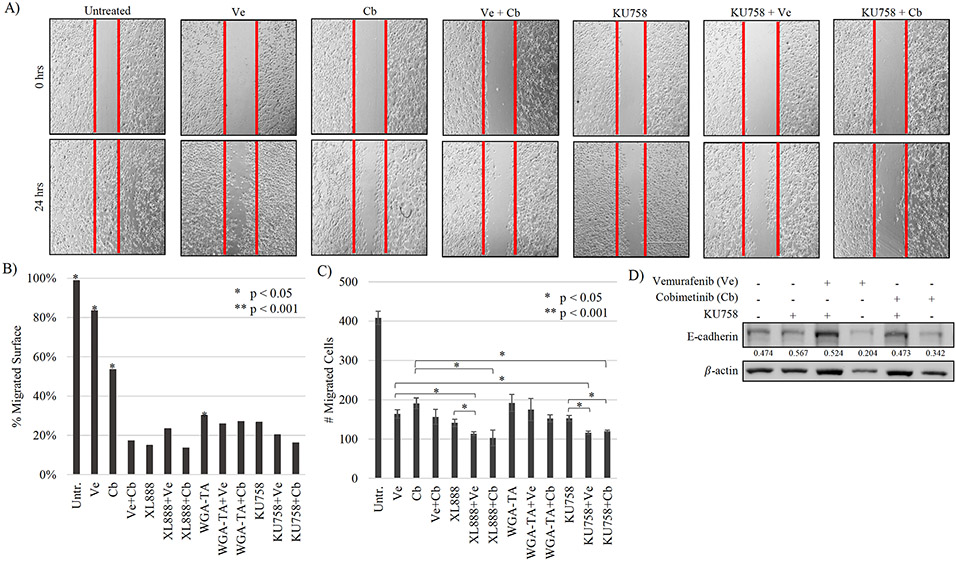
To determine the effect of Hsp90i alone and in combinations with Ve or Cb on cell migration (and indirectly on metastatic potential), we utilized scratch and Boyden chamber assays. For each treatment condition in the scratch assay, scratch distance at t=0hrs and t=24hrs was measured, then calculated as percent of cells migrated at 24hrs and significance values reflect the quantified change – lower percent migration is desired suggesting little to no migration. After 24hrs, untreated/control cells migrated to occupy 99% of the scratched area representing significant cell migration (p<0.001). Ve and Cb single-agent treatments resulted in 83% and 54% cell migration, respectively (p<0.05 vs. control), KU758-alone, KU758+Ve, and KU758+Cb resulted in 27%, 20% and 16% cell migration, respectively (all p<0.01; Figure 4A/4B). Similar decreases in migration were observed with XL888 and WGA-TA alone and in combination with a MAPKi (Figure 4B). These effects were then confirmed in our second cell line, UACC-62 (Supplemental Figure 4). Immunoblot analysis showed that expression of E-cadherin (an important determinant of tumor progression, serving as a suppressor of migration) in KU758 and MAPKi single-agent treated cells was low (p<0.05) but its protein expression was significantly increased in combination treatments indicating an increase in suppression of migration (KU758+Ve and KU758+Cb, p<0.05) (Figure 4D). Finally, we utilized Boyden chambers to quantify the number of cells that migrated from a serum-free media environment to a nutrient-rich one with 20% FBS after inhibitor treatment for 24hrs (Figure 4C). All inhibitors alone significantly interfered with the migration of cells compared to untreated/control (p<0.001). In combination, only KU758+Ve, KU758+Cb and XL888+Ve showed a significant decrease in migration compared to each drug alone (p<0.01).
Figure 4 – Changes to melanocyte migration after exposure to Hsp90i combinations –
A) KU75+MAPKi combinations decreased cell migration compared to inhibitors alone and is represented by fewer cells crossing the scratched area (red line). B) The percent of cell migration at 24hrs was calculated with the most significant cell migration (>30%) was observed in untreated, Ve, Cb, and WGA-TA treated cells (p<0.05). All other treatments mitigated cell migration into the scratched areas. C) The number of migrated cells across the membrane were counted. All treatments significantly decreased cell migration compared to untreated (p<0.001). KU758+Ve and KU758+Cb decreased cell migration to a greater extent than each inhibitor alone (p<0.05). XL888+Ve and XL888+Cb exhibited similar decrease (p<0.05).
The HSR is not Activated in KU758 Treated Cells vs. XL888
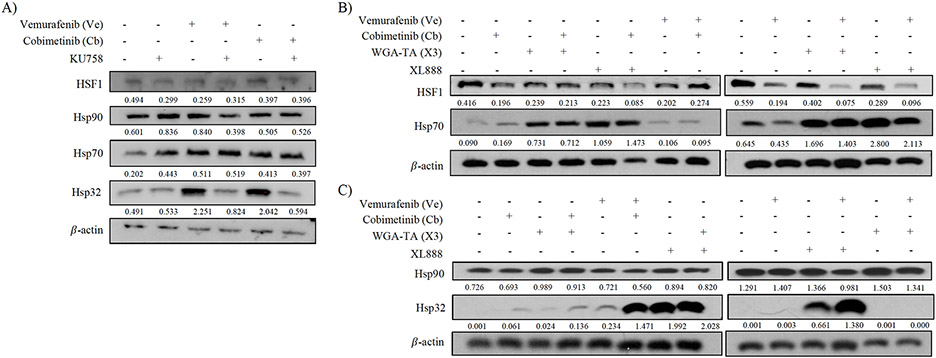
Finally, it was crucial to distinguish the effect of Hsp90i+MAPKi combinations on the HSR as 94% of Hsp90i (including XL888) target the N-terminal domain, which many experts believe stimulates a HSR with upregulation of pro-survival proteins (Hsp32 and Hsp70) requiring subsequent higher Hsp90i doses to overcome and ultimately leading to dose-limiting toxicities. As we have shown in other models [15-17], targeting Hsp90’s C-terminal domain does not upregulate the HSR so we should observe less HSR activation with KU758 and KU758+MAPKi combinations.
To test this, we quantified by Western Blot the change of expression of key Hsps – Hsp90, Hp70, Hsp32, and HSF1 in melanoma cells treated with Hsp90i alone or in combination with Ve or Cb. In all single-agent and combination treatments, Hsp90 expression was not significantly altered and remained consistent throughout (Figure 5A/5C). XL888 alone on in combinations significantly increased expression of both Hsp32 (p<0.01 vs. control) and Hsp70 (16-fold increase vs. control; p<0.01). Compared to XL888, Hsp32 and Hsp70 expression was significantly decreased in KU758 treated cells (p<0.05; Figure 5A/5B). Lastly HSF1, a transcription factor required for Hsp32/70 expression, was decreased vs controls by KU758(47% decrease), its combinations, WGA-TA+Cb, and XL888+Cb (all p<0.05; Figure 5A/5B).
Figure 5 – Effect of Hsp90i combinations on the HSR –
A) There was no significant change to HSF1 or Hsp90 expression in all KU758 and KU758+MAPKi cells. Expression of Hsp70 increased in all treatments compared to untreated (control), especially in Ve alone (p<0.05). Hsp32 increased in Ve and Cb treatments alone but decreased in KU758+Ve and KU758+Cb (p<0.05). B) Hsp70 expression increased in WGA-TA+MAPKi and XL888+MAPKi (p<0.05). No change to Hsp90 expression from control after inhibitor treatment. Hsp32 expression showed the most drastic increase in XL888 and XL88+MAPKi treated cells (p<0.05).
DISCUSSION
Since 2011, the standard treatment for metastatic melanoma patients with activating BRAF-mutations (V600E/K) has been BRAF+MEK inhibitor combination therapy, such as vemurafenib and cobimetinib (Ve+Cb). While this combination has good efficacy initially, the majority of patients acquire drug-resistance within the first year of therapy resulting in disease progression (2). This underscores the major clinical need to develop novel therapeutic strategies in melanoma with longer durability. The lack of durability of current targeted therapies is due primarily to drug resistance; two of the most prevalent resistance mechanisms resulting from BRAF-inhibition include the MAPK/Erk pathway and the PI3K/Akt pathway. In drug-resistant melanoma, these resistance pathways propagate the hallmarks of cancer and their associated processes. Since Hsp90 chaperones proteins involved in all of the hallmarks of cancer, including melanoma drug-resistance pathways, inhibiting Hsp90 function would simultaneously down-regulate multiple resistance and cancer-propagating pathways, making it an attractive therapeutic strategy (27, 28). This therapeutic approach using Hsp90 inhibition has been explored in numerous clinical trials targeting the Hsp90 N-terminal ATP binding site. N-terminal Hsp90i pan-inhibit all four Hsp90 isoforms leading to a HSR, that in turn upregulates several pro-survival processes in the cell including induction of Hsp70 levels. To overcome these pro-survival effects, higher doses of Hsp90 inhibitor are required, and as noted in clinical trials these NT-Hsp90i trials have been plagued with dose-limiting toxicities resulting in a lack of progression to FDA approval. In an attempt to overcome this Hsp90i limitation, our research team developed novel Hsp90i that target the C-terminal region of the molecular chaperone. These have been shown to be equally potent to NT-Hsp90i and selective against multiple cancers, but do not induce a significant HSR or upregulation of Hsp70 expression, and do not show any significant dose-limiting toxicity in vivo (22-24).
In this set of experiments, we demonstrate that our novel CT- Hsp90i, KU758, has similarly efficacy and potency to the NT-Hsp90i, XL888, yet is more synergistic in combination with Ve or Cb, without activating the HSR and with significantly lower Hsp70 expression modulation. In our first experiment, we demonstrate that our CT-Hsp90i, KU758, had similar efficacy, potency, and melanoma-selectively to an XL888, an NT-Hsp90i used in several clinical trials. Our 24hrs cell viability assays showed that the IC50 of KU758 in melanocytes was 0.4μM, which was comparable to the IC50 for XL888 (0.3μM) and WGA-TA (0.1μM). Additionally, these Hsp90i had 10- to 300-fold selectivity for melanocytes compared to the FF control cells. At 72hrs, these IC50 values continued to lower (data not shown), indicating increased potency when cells were exposed to each drug for a longer period of time.
After demonstrating that the three Hsp90i had similar potency and melanoma selectivity, it was crucial to identify whether they exhibited a synergistic combination effect in the presence of a MAPKi, such as vemurafenib or cobimetinib. Optimal combinations of our Hsp90i+Ve or Hsp90i+Cb had CI<1 indicating synergism, whereas additive or antagonistic combinations had CIs equal to or far greater than 1, respectively. Our compound, KU758, not only synergized with Ve and Cb, but with relatively lower CIs compared to the other two Hsp90i. It is important to note that all of the combination experiments were conducted using the highest concentration (at or near the respective compound’s IC50) of Ve, but not Cb. The reason for this is that despite KU758 synergizing with Ve at 12.5μM, 25μM, and 50μM, WGA-TA and XL888 only synergized at the highest concentration. Therefore, in order to keep as many variables consistent across all Hsp90i combinations, 50μM Ve was used for all corresponding treatments. A common observation in all of our Hsp90i+Ve or Hsp90i+Cb was that more synergism was noted in Cb-combinations compared to Ve-combinations. One potential explanation for this could be that since Cb targets MEK, the downstream effector of Ve’s target, BRAF, there is potentially a greater suppressive effect on the MAPK pathway with more distal targeting. One of the reasons MEKi were developed was to abrogate compensatory activation of the MAPK pathway resulting in the pathway’s down-regulation and this is noted especially in combination with a BRAFi (2).
After identifying that KU758 was effective, potent, and synergized with Ve and Cb, we took a mechanistic approach to understand and determine the effects of KU758+MAPKi combinations on known melanoma resistance pathways. We showed that in certain Cb-combinations there was an increase of Raf1 expression, while in Ve-combinations there was an almost complete decrease. This could in part be explained by the fact that Ve innately targets Raf1 and through this there is an overall decrease in its expression. Moreover, although Raf1 is a client of Hsp90, WGA-TA and XL888 Cb-combinations do not synergize to an extent where they result in knockdown of the protein product. On the contrary, both KU758 combinations decreased Raf1 perhaps due to a more synergistic effect. Further downstream of Raf1, there was varying p-Erk expression as the result of Hsp90i combinations. The most notable being in all XL888+Ve and WGA-TA+Ve combinations with almost complete knock down of p-Erk expression. In KU758+Ve, the lack of substantial p-Erk decrease could potentially be the effect of an alternative kinase, outside of the MAPK pathway, that activates MEK (2). Additionally, in another drug-resistant melanoma pathway (PI3K/Akt), p-Akt was almost completely knocked down in KU758+Cb treated cells. This supports our hypothesis that synergistic combinations of KU758+MAPKi help to mitigate compensatory signaling pathways in drug-resistant melanomas. All the Hsp90i tested demonstrated some synergy when combined with BRAF/MEK inhibitors and mitigated protein expression levels of key regulators from melanoma drug-resistant pathways.
Next we determined the effects of these inhibitor-combinations on cell migration, an important hallmark of cancer. While each Hsp90i showed moderate inhibition of melanocyte migration in scratch assays (with KU758 having the most inhibition), this effect was more significant when combined with a MAPKi. Evaluating cell migration results, each Hsp90i decreased cell migration significantly compared to untreated cells, and this inhibition was synergistically enhanced with KU758+MAPKi and XL888+MAPKi combinations. Compared to the current standard-of-care melanoma treatment combination (Ve+Cb), our Hsp90i+MAPKi combinations had superior effects on cell migration. This may be due to the role Hsp90i has in mitigating MAPKi drug-resistant pathways simultaneously leading to a more potent therapeutic effect on migration. E-cadherin (tumor suppressor that plays a role in the transition of stable to invasive tumors) expression is lost in melanocytes (29, 30). Down-regulation of the protein subsequently leads to a decrease in cell-adhesion molecules, and thus, increased cell motility and migration and metastatic spread. In our KU758 combinations, E-cadherin expression was significantly increased from controls, again underscoring the effectiveness and synergism of the Hsp90i+MAPKi combination on this cancer hallmark of cell migration/metastatic potential.
A distinction between each Hsp90i (and respective combinations with Ve and Cb) was its effect on the HSR. This HSR was significantly lower in the CT-Hsp90i KU758 than the NT-Hsp90i XL888. KU758 does not increase the expression of Hsp32, a major pro-survival Hsp, and rescued protein expression in KU758+Ve or KU758+Cb compared to Ve or Cb alone. This highlighted that unlike XL888, XL888+MAPKi and Ve+Cb combinations, KU758 does not activate the HSR or significantly increase Hsp32 or Hsp70 protein expression levels. This finding is significant since the HSR induction observed with NT-Hsp90i is thought by many to be a major contributing factor to their dose-limiting toxicities and failures in clinical trials. Since KU758 did not induce this HSR and its pro-survival effects, it may have clinical advantages and lower toxicity than the N-terminal inhibitors and warrants future translational studies to better evaluate this potential.
In conclusion, these studies demonstrate early proof-of-concept that the novel CT-Hsp90i KU758 is an effective therapy to target BRAF-mutated melanomas with potency similar to XL888, an NT-Hsp90i tested in clinical trials. This therapy is potent and selective against melanoma cells. It not only synergizes with BRAF and MEK inhibitors to decrease melanoma proliferation, but also induces apoptosis through PARP cleavage, inhibits cellular growth at G0/G1 phase of cell cycle, cell migration, and mitigates two important drug-resistance pathways; all without inducing the HSR, which has potential clinical advantages with lower toxicity concerns than previously tested NT-Hsp90i. Also given the advantages of KU758 in mitigating major melanoma drug-resistance pathways, this treatment approach could have more durability in the clinic than current options and would address an important treatment gap. As such, this data supports further translational testing of KU758 in metastatic melanomas in vivo to determine its future clinical potential and ideal combinational approach with MAPKi for optimal clinical synergy and benefit.
Supplementary Material
Table 2.
The CI values are listed for each inhibitor combination treatment in UACC-62 between Ve+Cb
| Cb | ||||
|---|---|---|---|---|
| (μM) | 0.125 | 0.25 | 0.5 | |
| Ve | 12.5 | 0.447 | 0.460 | 0.757 |
| 25 | 0.086 | 0.152 | 0.272 | |
| 50 | 0.048 | 0.098 | 0.188 | |
Synergistic combination boxes are colored according to relative CI value: CI value <0.5 (blue), 0.5 < CI value < 1.0 (green).Cb, cobimetinib; CI, combination index; Ve, vemurafenib.
ACKNOWLEDGEMENTS
Author contributions: JNS, CS, MSC participated in research design; JNS, NZ, TW, MC conducted experiments; BNT, BSJB contributed to new reagents; JNS, CS, MC and MSC performed data analysis; All authors wrote or contributed to the writing of the manuscript.
Conflicts of Interest and Acknowledgements for Sources of Funding:
No conflicts to declare. This work was funded by the National Institutes of Health - National Cancer Institute [Grants R01-CA213566, R01-CA216919 (M.S.C., B.S.J.B.); T32-CA009672 (T.W.)] and National Institute of General Medical Sciences [Grant T32-GM007544, T32-GM007767 (J.N.S.)]. Additionally, funds from the University of Michigan Departments of Surgery and Pharmacology provided lab equipment, supplies, and startup support for this work.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez JN, Wang T, Cohen MS. BRAF and MEK Inhibitors: Use and Resistance in BRAF-Mutated Cancers. Drugs. 2018;78(5):549–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell. 2017;168(4):707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayat Mokhtari R, Homayouni TS, Baluch N, Morgatskaya E, Kumar S, Das B, et al. Combination therapy in combating cancer. Oncotarget. 2017;8(23):38022–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eggermont AMM, Crittenden M, Wargo J. Combination Immunotherapy Development in Melanoma. Am Soc Clin Oncol Educ Book. 2018;38:197–207. [DOI] [PubMed] [Google Scholar]
- 6.Ritossa FM. A new puffing pattern induced by temperature shock an DNP in Drosophila. Experientia. 1962;18:571–3. [Google Scholar]
- 7.Ritossa FM. New puffs induced by temperature shock, DNP and salicilate in salivary chromosomes of D. melanogaster. Drosophilia Information Service. 1963;37:122–3. [Google Scholar]
- 8.Ritossa FM. Experimental activation of specific loci in ploytene chromosomes of drosophila. Experimental cell research. 1964;35:601–7. [DOI] [PubMed] [Google Scholar]
- 9.Csermely P, Schnaider T, Soti C, Prohaszka Z, Nardai G. The 90-kDa molecular chaperone family: structure, function, and clinical applications. A comprehensive review. Pharmacol Ther. 1998;79(2):129–68. [DOI] [PubMed] [Google Scholar]
- 10.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci U S A. 1994;91(18):8324–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. [DOI] [PubMed] [Google Scholar]
- 13.Mielczarek-Lewandowska A, Hartman ML, Czyz M. Inhibitors of HSP90 in melanoma. Apoptosis. 2020;25(1–2):12–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joshi SS, Jiang S, Unni E, Goding SR, Fan T, Antony PA, et al. 17-AAG inhibits vemurafenib-associated MAP kinase activation and is synergistic with cellular immunotherapy in a murine melanoma model. PLoS One. 2018;13(2):e0191264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eroglu Z, Chen YA, Gibney GT, Weber JS, Kudchadkar RR, Khushalani NI, et al. Combined BRAF and HSP90 Inhibition in Patients with Unresectable BRAF (V600E)-Mutant Melanoma. Clin Cancer Res. 2018;24(22):5516–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paraiso KH, Haarberg HE, Wood E, Rebecca VW, Chen YA, Xiang Y, et al. The HSP90 inhibitor XL888 overcomes BRAF inhibitor resistance mediated through diverse mechanisms. Clin Cancer Res. 2012;18(9):2502–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smyth T, Paraiso KHT, Hearn K, Rodriguez-Lopez AM, Munck JM, Haarberg HE, et al. Inhibition of HSP90 by AT13387 delays the emergence of resistance to BRAF inhibitors and overcomes resistance to dual BRAF and MEK inhibition in melanoma models. Mol Cancer Ther. 2014;13(12):2793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acquaviva J, Smith DL, Jimenez JP, Zhang C, Sequeira M, He S, et al. Overcoming acquired BRAF inhibitor resistance in melanoma via targeted inhibition of Hsp90 with ganetespib. Mol Cancer Ther. 2014;13(2):353–63. [DOI] [PubMed] [Google Scholar]
- 19.Solit DB, Osman I, Polsky D, Panageas KS, Daud A, Goydos JS, et al. Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clin Cancer Res. 2008;14(24):8302–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro GI, Kwak E, Dezube BJ, Yule M, Ayrton J, Lyons J, et al. First-in-human phase I dose escalation study of a second-generation non-ansamycin HSP90 inhibitor, AT13387, in patients with advanced solid tumors. Clin Cancer Res. 2015;21(1):87–97. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez JN, Carter TR, Cohen MS, Blagg BS. Old and New Approaches to Target the Hsp90 Chaperone. Current cancer drug targets. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian C, Kovatch KJ, Sim MW, Wang G, Prince ME, Carey TE, et al. Novel C-Terminal Heat Shock Protein 90 Inhibitors (KU711 and Ku757) Are Effective in Targeting Head and Neck Squamous Cell Carcinoma Cancer Stem cells. Neoplasia. 2017;19(12):1003–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian C, Grogan PT, Wang T, Bazzill J, Zuo A, White PT, et al. Novel C-terminal heat shock protein 90 inhibitors target breast cancer stem cells and block migration, self-renewal, and epithelial-mesenchymal transition. Molecular oncology. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T, Subramanian C, Blagg BSJ, Cohen MS. A novel heat shock protein 90 inhibitor potently targets adrenocortical carcinoma tumor suppression. Surgery. 2020;167(1):233–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–81. [DOI] [PubMed] [Google Scholar]
- 26.Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147(5):992–1009. [DOI] [PubMed] [Google Scholar]
- 27.Miyata Y, Nakamoto H, Neckers L. The therapeutic target Hsp90 and cancer hallmarks. Curr Pharm Des. 2013;19(3):347–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaeger AM, Whitesell L. Hsp90: Enabler of Cancer Adaptation. Annual Review of Cancer Biology. 2018;3:275–97. [Google Scholar]
- 29.Danen EH, de Vries TJ, Morandini R, Ghanem GG, Ruiter DJ, van Muijen GN. E-cadherin expression in human melanoma. Melanoma Res. 1996;6(2):127–31. [DOI] [PubMed] [Google Scholar]
- 30.Pecina-Slaus N. Tumor suppressor gene E-cadherin and its role in normal and malignant cells. Cancer Cell Int. 2003;3(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.