Abstract
Adefovir dipivoxil [bis(pivaloyloxymethyl)-ester prodrug], an orally bioavailable prodrug of adefovir [9-(2-phosphonylmethoxyethyl)adenine], is currently in phase III clinical trials for the treatment of human immunodeficiency virus (HIV). In vitro experiments demonstrated that either a K65R or a K70E mutation in HIV reverse transcriptase (RT) was selected in the presence of adefovir, conferring a 16- or 9-fold decrease in susceptibility to adefovir, respectively. Previous data demonstrated that patients receiving adefovir dipivoxil monotherapy (125 mg daily) for 12 weeks experienced a median decrease in HIV RNA levels of 0.5 log10 copies/ml and that resistance to adefovir dipivoxil did not arise during that period. In the present investigation, a further study was undertaken to investigate whether RT mutations developed among viruses from patients who completed the 12-week study and who opted to enroll in a maintenance phase of prolonged (6- to 12-month) adefovir dipivoxil therapy (120 mg daily). Concomitant treatment with antiretroviral agents was permitted during the maintenance phase. The median decreases in HIV RNA levels for patients who completed 6 or 12 months of maintenance-phase dosing were 0.6 and 1.14 log10 copies/ml, respectively. The reductions in the HIV RNA levels were similar among patients who received adefovir dipivoxil with or without concomitant treatment with antiretroviral agents. Viruses from 8 of 29 patients dosed for up to 12 months developed RT mutations that were not present at baseline; these mutations may have been related to adefovir dipivoxil therapy. Viruses from two of the eight patients developed the K70E mutation while the patients were on therapy, but none of the viruses from patients developed the K65R RT substitution. Despite the development of RT mutations, sustained reductions (6 to 12 months) in viral load (≥0.7 log10 copies/ml decrease from baseline) were observed in all eight patients.
Current guidelines for the treatment of AIDS recommend combination therapy, preferably triple-drug therapy that includes a protease inhibitor (7a). Reverse transcriptase (RT) inhibitors (including nucleoside and nonnucleoside RT inhibitors) and protease inhibitors have been combined in numerous clinical studies and have been shown to provide potent and sustained suppression of human immunodeficiency virus (HIV) replication. However, the development of resistance to the RT and protease inhibitor components of these treatment regimens continues to plague the long-term success of anti-HIV therapy (34).
The development of RT resistance mutations in HIV strains from patients receiving anti-HIV therapy with nucleoside analogs is well documented. HIV strains in patients receiving zidovudine (AZT) develop numerous, specific resistance mutations in a stepwise manner (6, 26–28, 31, 32, 39). Resistance mutations resulting from didanosine (ddI) (20, 22, 29, 47, 51), zalcitabine (ddC) (18, 20, 22, 52), or lamivudine (3TC) therapy have been well characterized (20, 40, 42, 49, 50). Mutations arising during stavudine (d4T) therapy are less well described to date (30, 33). Interestingly, “hallmark” AZT-associated resistance mutations have also been shown to arise among viruses in patients receiving ddI or d4T monotherapy, although these mutations do not confer notable decreased levels of susceptibility to either of these drugs in vitro (16, 33, 51). Finally, combinations of nucleoside inhibitors may lead to novel resistance patterns in vivo, as evidenced by the multidrug-resistant viruses (41, 43, 46, 48).
Adefovir is a member of a new class of nucleotide antiviral agents and is active in vitro and in vivo against retroviruses, hepadnaviruses, and herpesviruses (4, 13, 14, 36). The orally bioavailable prodrug adefovir dipivoxil [bis-(pivaloyloxymethyl)-ester prodrug] is under clinical evaluation for the treatment of infections caused by these viruses. Adefovir is phosphorylated to its active metabolite, adefovir diphosphate, by ubiquitous cellular enzymes (1, 3, 5, 38) and demonstrates potent anti-HIV activity in monocytes and macrophages (2, 37) as well as in resting and activated T cells (46). Adefovir diphosphate is a competitive inhibitor of RT with regard to dATP and serves as an alternate substrate for incorporation into viral DNA, functioning as a chain terminator of viral DNA synthesis (10).
It has been reported previously that the lysine 65-to-arginine (K65R) and lysine 70-to-glutamic acid (K70E) mutations were selected for in vitro in the presence of adefovir (9, 19). A recombinant virus expressing the K65R mutation exhibited a 12- to 16-fold decreased level of susceptibility to adefovir in vitro (19, 23), while a recombinant virus expressing the K70E mutation showed a 9-fold decreased level of susceptibility to adefovir in vitro compared to that of the wild type (HXB2D or HIV IIIb) (9). AZT-resistant, 3TC-resistant, and multidrug-resistant viruses remain susceptible to adefovir in vitro (9, 21, 23, 48).
To address the potential development of resistance to adefovir dipivoxil in vivo, a study was undertaken to analyze samples from patients enrolled in a phase I/II clinical trial. Study GS-94-403 was a double-blind, placebo-controlled phase I/II trial of adefovir dipivoxil for the treatment of HIV. In the initial phase, patients were randomized to receive active drug (125 or 250 mg daily) or placebo for a period of 6 weeks; all patients received drug under an open-label design at one of two doses during weeks 6 to 12. The clinical data including HIV RT genotypic analyses for strains from patients enrolled in this initial phase of the study were reported recently (15). Briefly, viruses from 1 of the 19 patients who received 125 mg of adefovir dipivoxil monotherapy for 12 weeks demonstrated a change in RT (M184V) at week 12 compared to the genotype of the virus in the baseline sample. Importantly, this patient experienced a decrease in the plasma HIV-1 RNA level of 1.95 log10 copies/ml from that at the baseline, a response considerably more pronounced than the median decrease of 0.5 log10 copies/ml for this cohort, suggesting possible concomitant, surreptitious use of antiretroviral agents.
At various times (median time, 6.25 months) following the initial 12-week monotherapy phase of the study, patients were allowed to enter a maintenance dosing phase (≥6 months) during which concomitant antiretroviral therapy was permitted. The concomitant therapy used was at the discretion of the patient and the physician. Here we report the results of genotypic analyses of RT genes generated for viruses from plasma samples from patients enrolled in this maintenance phase of prolonged (6- to 12-month) adefovir dipivoxil therapy (120 mg daily) as well as the patients’ viral load responses during this time. Additionally, recombinant HIV strains were constructed, and viruses from patients who developed mutations in RT that were believed to be possibly associated with adefovir dipivoxil therapy were analyzed. Adefovir dipivoxil resistance did not arise readily during extended therapy, and sustained antiviral activity was observed.
MATERIALS AND METHODS
Patients.
GS-94-403 was a phase I/II randomized, double-blind, placebo-controlled study investigating the safety and efficacy of adefovir dipivoxil in HIV-infected adults. Patients were enrolled into this study during 1994 and 1995. Detailed descriptions of the initial monotherapy phase of the GS-94-403 study design, patient characteristics, and clinical results have been reported previously (15). Briefly, criteria for entry into the GS-94-403 study were a CD4 count of >200 cells/mm3 and a viral load of >10,000 copies/ml. After patients completed the initial phase of 12 weeks, they were eligible to enter the maintenance dosing phase (a period of ≥6 months) of the study in which the concomitant use of antiretroviral agents was permitted. A median time of 6.5 months separated completion of the initial phase and entry into the maintenance phase. For 29 patients who enrolled in the maintenance phase, plasma samples from the maintenance-phase baseline and at least one time point ≥6 months into the extended dosing period were available for genotypic analyses. At entry into the maintenance phase, these 29 patients had a median initial HIV RNA load of 45,320 copies/ml and a median CD4 cell count of 407 cells/mm3. A total of 79% of these patients were nucleoside inhibitor experienced, primarily with AZT or AZT plus ddI.
Genotypic analyses.
The preparation of HIV RNA from patient plasma, the generation of the 1.1-kb PCR fragments carrying HIV RT, and analyses of the DNA sequences of these PCR fragments have been described previously (15, 44). Briefly, genotypic analyses were performed with RT-PCR fragments carrying HIV RT genes generated from the patients’ plasma samples taken at the maintenance-phase baseline, the 6-month time point, and when available, the 12-month time point. If a change in the RT sequence from that at the baseline was detected at the 6-month time point, the RT from viruses from the 3-month sample was also sequenced. In cases when a patient discontinued the maintenance phase between 8 and 12 months, the last available sample was analyzed. Nucleotides 1 to 900 (amino acids 1 to 300) of the HIV RT genes generated from these specified plasma samples were manually sequenced by conventional didoexy sequencing methods. A mixture of wild-type and mutant nucleotides was reliably detected by this manual sequencing method when either was present in the population at ≥20%, similar to the detection level of automated sequencing methods. Observed HIV RT mutations were interpreted in the context of each patient’s history of prior and concomitant antiretroviral therapy.
Generation of recombinant HIV.
PCR fragments corresponding to the first 1 kb of HIV type 1 (HIV-1) RT were prepared from the patients’ plasma samples obtained at baseline and after either 6 or 12 months of adefovir dipivoxil therapy. Four micrograms of the PCR fragments was cotransfected with 6 μg of the RT-deleted HIV-1 proviral molecular clone pHXB2delta2-261RT (a gift from C. Boucher) as described by Boucher et al. (7). Replication-competent viruses resulting from homologous recombination were harvested when the cultures contained notable syncytia, which was 8 to 18 days later. The RT genotypes of the recombinant viruses were confirmed by sequencing the respective RT-PCR products generated from 140 μl of DNase-treated viral supernatants.
Phenotypic analyses of recombinant HIV.
The susceptibilities of the recombinant viruses to adefovir, AZT, 3TC, and ritonavir were evaluated by a modified XTT-based assay with MT-2 cells as described previously (9). For comparison, the wild-type molecular clone HXB2D as well as RT mutants with site-directed mutations K70E (9) and T69D (a gift from J. Fitzgibbon) were evaluated. All infections were carried out at a multiplicity of infection of 0.001 and resulted in similar levels of cell death in the absence of drug.
HIV RNA quantitation.
HIV RNA loads were determined by using the Roche Amplicor technology, which has a limit of quantification of 400 copies/ml of plasma. During the maintenance phase, plasma samples were analyzed at the baseline and at months 3, 6, and 12 and/or at the time of study discontinuation for quantitation of viral load.
RESULTS
Genotypic analyses of HIV RT and viral load responses among patients enrolled in the adefovir dipivoxil maintenance phase.
For 28 patients enrolled in the maintenance phase, both baseline and 6-month plasma samples were available for HIV RT sequence analyses. For 14 of these 28 patients, a later sample was also available for analysis after a cumulative ≥8 months in the maintenance phase, including 9 patients for whom samples from the 12-month time point were available. Additionally, for one patient for whom a sample was not available at 6 months, a sample was available at 12 months and a PCR fragment carrying the RT was generated and sequenced. Therefore, sequence analysis was performed with HIV RT from all 29 patients for whom plasma samples with a detectable viral load from baseline and at least one time point ≥6 months into the maintenance phase were available. Concomitant antiretroviral agents were permitted during the maintenance phase. Among patients who added a new drug, 3TC and d4T were most commonly used.
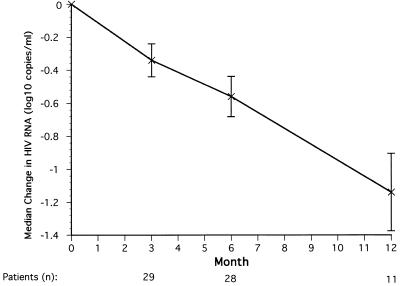
These 29 patients had a median decrease in the HIV RNA load from baseline of 0.56 log10 copies/ml after 6 months of therapy (Fig. 1). At 12 months, 18 of the 29 patients had either discontinued the study or plasma samples were not available for analyses. The remaining 11 evaluable patients had a median decrease in HIV RNA load of 1.14 log10 copies/ml at month 12 (Fig. 1). Fifteen patients received adefovir dipivoxil monotherapy for the first 6 months of the maintenance phase. These 15 patients showed median ± standard error decreases in HIV RNA load from baseline of 0.49 ± 0.17 log10 copies/ml at 3 months and 0.54 ± 0.21 log10 copies/ml at 6 months, similar to the decreases observed for all patients at those time points.
FIG. 1.
Median log change in plasma HIV RNA load (log10 copies per milliliter) for all patients after 3, 6, and 12 months of maintenance-phase therapy. Standard error bars are shown.
For 8 of 29 patients, changes in the sequence of HIV RT from that at baseline developed after ≥6 months of adefovir dipivoxil therapy; these changes may have been selected by adefovir dipivoxil (Table 1). Mutations were considered to be potentially due to adefovir dipivoxil therapy if they met one or more of the following criteria: (i) the RT mutation developed while the patient was receiving adefovir dipivoxil monotherapy, (ii) the RT mutation was previously shown to be selected for in vitro by adefovir dipivoxil, or (iii) the RT mutation developed at an amino acid associated with resistance to a different nucleoside inhibitor, although the patient was not concomitantly taking that specific drug. Many of the mutations were detected initially as mixtures of wild-type and mutant amino acids and in some cases remained so throughout the duration of the study (Table 1).
TABLE 1.
RT mutations arising during maintenance-phase therapy possibly associated with adefovir dipivoxil treatment
| Patient | Prior therapy (total no. of mo) | Nucleoside resistance mutations present at maintenance-phase baseline | Concomitant medications during maintenance phase (at the indi- cated mo of study) | RT mutations arising during maintenance phasea | Change in HIV RNA load (log10 copies/ml) from maintenance baselineb |
|---|---|---|---|---|---|
| A | AZT (<12), ddI (6) | None | None | K70E/K (3) | −0.9 (6) |
| B | AZT (22) | None | None | T215I/T (3) | −0.8 (6) |
| C | AZT (27), ddI (29) | K70R | None | M41L/M,T215N/S/Y/T (3) | −1.7 (12) |
| D | None | None | None | D67N/D,K70R,T215Y/T (12) | −1.0 (12) |
| E | AZT (3), ddI (2), 3TC (<1) | None | None (0-3), saquinavir (5-12), 3TC (7-12) | T69D (3), M184V (12) | −1.2 (12) |
| F | AZT (31) | M41L, L210W, T215Y | d4T (3-10), 3TC (8-10) | T69D/T (10), M184V (10) | −1.4 (10) |
| G | AZT (1) | None | AZT (6-8), 3TC (6-12) | K70E (12), M184V (12) | −0.9 (12) |
| H | AZT (3), d4T (6), 3TC (6) | M184V | d4T + 3TC (0-6) | K70R (6) | −0.7 (6) |
Underlining indicates RT sequences with amino acid changes from baseline that are potentially associated with adefovir dipivoxil therapy. Slashes indicate mixtures of wild-type and mutant amino acids. Values in parentheses indicate the time point (month) at which the mutation was first detected.
Values in parentheses indicate the latest time point at which the HIV RNA load was available.
The viruses from five patients (patients A to E) developed mutations in RT while the patients were receiving adefovir dipivoxil monotherapy (Table 1). The viruses from patient A developed the K70E mutation, which was previously shown to be selected for in vitro in the presence of adefovir (8). The viruses from three patients (patients B to D) developed AZT-associated resistance mutations, although none of the patients reported concomitant treatment with AZT. Two of these three patients had previously received AZT. Monotherapy with ddI or d4T has been shown to select for AZT-associated resistance mutations as well (16, 33, 51). The viruses from patient E developed a T69D mutation at month 3, before the patient had started to take antiretroviral agents concomitantly. The T69D mutation has previously been shown to be associated with ddC therapy (18), although patient E did not report treatment with ddC prior to or concomitantly with adefovir dipivoxil treatment. Despite the development of RT mutations associated with adefovir dipivoxil monotherapy, sustained reductions in viral load (≥0.8 log10 copies/ml decrease from baseline) were observed in all five patients (Table 1, patients A to E).
The viruses from three additional patients (patients F to H) developed mutations in HIV RT while they were concomitantly receiving antiretroviral therapy, in addition to adefovir dipivoxil (Table 1). The viruses from patient F developed a T69D mutation, the viruses from patient G developed a K70E mutation, and the viruses from patient H developed a K70R mutation. Each of these mutations was also selected for by viruses in patients who received adefovir dipivoxil monotherapy (Table 1, patients A to E). Thus, adefovir dipivoxil may have selected the mutations in the viruses from patients F to H; however, a potential role played by concomitant therapy cannot be excluded. Similarly, while the addition of concomitant medications may have contributed to the viral RNA response in these three patients, as well as in patient E, sustained reductions in viral load were observed. In addition to developing mutations that may have been selected by adefovir dipivoxil therapy, viruses from patients E, F, and G also developed the M184V resistance mutation in the RT while the patients were concomitantly receiving 3TC treatment. In total, the viral load responses among the eight patients (patients A to H) whose viruses developed mutations associated with adefovir dipivoxil therapy were similar to the overall median decreases during the maintenance phase (month 6, −0.6 log10 copies/ml; month 12, −1.1 log10 copies/ml) (Fig. 1).
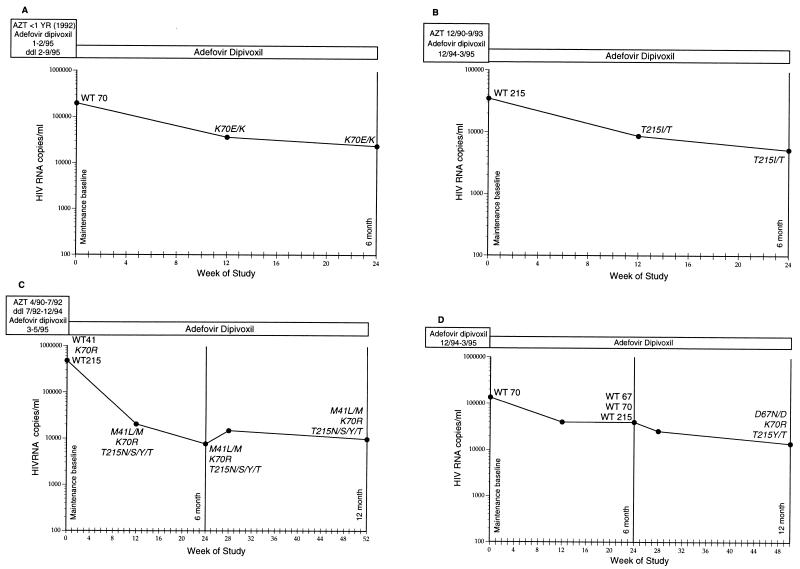
The detailed viral load responses of the four individuals (Table 1, patients A to D) who received adefovir dipivoxil monotherapy during the entire maintenance phase and whose viruses developed RT gene mutations possibly associated with adefovir dipivoxil treatment are shown in Fig. 2A to D. The data in Fig. 2A to C demonstrate that viral load suppression was maintained in the patients even 3 to 9 months after the selected mutation arose. Since the mutation which arose in the RT of the viruses from patient D developed at month 12, the effect of this sequence change on the subsequent viral RNA response could not be determined. As shown in Fig. 2, many mutations were detected initially as mixtures of wild-type and mutant amino acids and in several cases remained as mixtures through the dosing period.
FIG. 2.
Viral loads and prior anti-HIV therapies for patients who received adefovir dipivoxil monotherapy for the entire maintenance phase and whose viruses developed a sequence change from baseline that may have been associated with adefovir dipivoxil therapy. Prior and current therapies are boxed above the viral load responses. The viral load (HIV RNA copies per milliliter) is indicated on the y axis (limit of quantification, 400 copies/ml; Roche Amplicor assay). Time (in weeks) is indicated on the x axis. Each point on the graph represents the number of HIV RNA copies per milliliter in plasma measured at the identified time of treatment. Nucleotides 1 to 900 (amino acids 1 to 300) of the HIV RT gene were sequenced at all of the time points indicated. The only RT amino acids that are shown in Fig. 2 are those that were wild type at baseline and that became mutated during therapy or those that are nucleoside-associated resistance mutations present at entry into the maintenance phase. Other sequence variations between that of the RT of virus from patients and the consensus HXB2D sequence are not listed. WT, wild-type sequence; italics, a sequence change from baseline. (A) Patient A. (B) Patient B. (C) Patient C. (D) Patient D. Additional patient data are presented in Tables 1 and 2.
In addition to mutations that may have been selected by adefovir dipivoxil therapy, viruses from nine patients developed amino acid changes in RT that were associated specifically with resistance to another nucleoside analog that each patient was receiving concomitantly. Viruses from seven patients (including patients E, F, and G) developed the M184V mutation in RT while patients were taking 3TC in conjunction with adefovir dipivoxil and other anti-HIV agents. The viruses from one patient developed an M184V and an AZT-associated mutation, D67N, while the patient was concomitantly receiving AZT and 3TC, and the viruses from one patient developed an M41L mutation in the RT while the patient was concomitantly taking AZT. These nine patients had a median decrease in viral RNA load of 1.0 log10 copies/ml calculated from the maintenance-phase baseline to the time of study discontinuation (≥6 months) (data not shown).
Production and phenotypic analyses of recombinant viruses from patients who received adefovir dipivoxil monotherapy and whose viruses developed mutations in RT.
Recombinant viruses were constructed from the five patients (patients A to E) whose viruses developed mutations in RT while the patients were receiving adefovir dipivoxil monotherapy. For each of these patients, a set of pre- and posttreatment recombinant viruses was prepared by homologous recombination of an RT-deleted molecular HIV clone with PCR-amplified RT genes derived from patient plasma samples (7). The RT genotypes of the resultant HIV recombinants (Table 2) corresponded to the RT genotypes of the viruses from the patients’ plasma samples in most cases (Table 1). For patient C, however, the baseline K70R mutation in recombinant virus generated from the 12-month plasma sample was not maintained, and the pre- and posttreatment recombinants expressed the T69N mutation, which was not seen in viruses from either plasma sample (although T69N was seen in the initial-phase baseline and week 12 samples). Another group has reported that the sequence of RT from recombinant viruses constructed in a similar manner matched that of the sequence of RT of viruses from plasma in 15 of 21 cases (71%) studied (51). Sequence data for the recombinant viruses also revealed that the mixtures of mutant and wild-type sequences observed in plasma samples (Table 1) often resolved into completely mutant sequences in the recombinant viruses (Table 2). Also shown in Table 2 is the marked heterogeneity of RT sequences among HIV strains from different patients.
TABLE 2.
Antiviral drug susceptibilities of recombinant HIV from patients whose viruses developed RT mutations while the patient was receiving adefovir dipivoxil monotherapy
| Patient | Time point | Recombinant virus sequencea | IC50 (μM)b
|
|||
|---|---|---|---|---|---|---|
| Adefovir | AZT | 3TC | Ritonavir | |||
| A | Baseline | A98S, E122K, I135V/I, R211K, L214F, P272A, R277K, T286A, E297A | 8.0 | 0.05 | 1.6 | 0.010 |
| 6 mo | K70E, A98S, E122K, I135V/I, R211K, L214F, R277K/R, E297A/E | 24.5 | 0.05 | 5.0 | 0.013 | |
| B | Baseline | E122K, I142V, L214F | 9.3 | 0.005 | 0.5 | 0.010 |
| 6 mo | K43R, E122K, I142V, E204G/E, R211K, L214F, T215I | 4.7 | 0.28 | 0.7 | 0.015 | |
| C | Baseline | T69N, K70R, R83K, E122K, I135T, S162C, Q207E, L214F, V245K | 18.7 | 0.57 | 1.5 | 0.013 |
| 12 mo | M41L, T69N, R83K, A98S, E122K, S162C, T200A, Q207E, L214F, T215Y | 19.0 | 1.60 | 1.5 | 0.009 | |
| D | Baseline | E122K, I135V, S162C, G196V, R211K, L214F, V276T | 10.4 | 0.14 | 1.8 | 0.028 |
| 12 mo | D67N,K70R, E122K, I135V/I, I142V, S162C, G196V, R211K, L214F, T215Y/N | 90.0 | 3.5 | 10.5 | 0.029 | |
| E | Baseline | V35I/V, A98S/A, E122K, S134R/S, G141R/G, Q207E, R211K, L214F, P272A | 7.7 | 0.15 | 1.3 | 0.014 |
| 6 mo | V35I, T69D, A98S/A, E122K, Q207E, R211K, L214F, P272A/P, L289P/L | 13.1 | 0.1 | 2.7 | 0.011 | |
| HXB2D wild-type recombinant | 15.5 | 0.25 | 2.6 | 0.014 | ||
| K70E site-directed recombinant | 76.0 | 0.20 | 7.0 | 0.018 | ||
| T69D site-directed recombinant | 45.0 | 0.50 | 35.0 | 0.020 | ||
Underlining indicates RT sequences with amino acid changes from baseline that are potentially associated with adefovir dipivoxil therapy. Slashes indicate mixtures of wild-type and mutant amino acids.
IC50s are averages for three to six independent experiments; average standard errors were <10%.
Antiviral drug susceptibility assays were performed with these matched sets of recombinant viruses from the patients as well as with wild-type (HXB2D) and two site-directed mutant RT clones of HIV (K65R and T69D). The 50% inhibitory concentrations (IC50s) of adefovir, AZT, 3TC, and ritonavir are presented in Table 2. The protease inhibitor ritonavir functioned as an internal control since the protease gene is identical among these recombinant viruses. As expected, the IC50s of ritonavir for all the virus pairs varied by less than 1.5-fold. The recombinant viruses from patients B and C did not demonstrate any significant decrease in susceptibility to adefovir, in spite of the presence of their noted mutations. The viruses from patient C did show notable decreases in AZT susceptibility, likely due to the presence of K70R and M41L-T215Y RT mutations in the pre- and posttreatment recombinants, respectively. The pretherapy recombinant from patient B, on the other hand, was unexpectedly AZT hypersensitive; the acquisition of the T215I substitution, among others, by the posttherapy virus returned the level of susceptibility to AZT to that for the wild type. In contrast to viruses from patients B and C, viruses from patient A, which developed the K70E RT mutation, demonstrated a 3.1-fold decrease in susceptibilities to both adefovir and 3TC. Interestingly, the IC50s of adefovir and 3TC for viruses with this genetic background were less than those observed for the site-directed K70E recombinant. Recombinant viruses from patient E, which developed the T69D mutation, showed very mild decreases in adefovir and 3TC susceptibility. Again, the IC50s changed less for the patient-derived recombinant than for the site-directed T69D recombinant. The final pair of viruses, derived from patient D, demonstrated decreases in susceptibility to all of the RT inhibitors tested. The data in Table 2 indicate that pre- and posttreatment recombinant viruses derived from patients who developed mutations in RT while undergoing adefovir dipivoxil monotherapy demonstrated mild (less than threefold) or undetectable changes in adefovir susceptibility for four of five patients. These results support the clinical observations of continued viral load suppression, despite the presence of these RT mutations, for 3 to 9 months in patients A to C (Table 1; Fig. 2). In the exceptional case, recombinant virus from patient D at 12 months did appear to be resistant to multiple inhibitors. While these mutations did not result in a rapid return in viral load toward baseline (Fig. 2D), they were first detected at 12 months; thus, the longer-term clinical implications of these mutations could not be determined.
Effect of baseline nucleoside-associated resistance mutations on response to adefovir dipivoxil therapy.
Among the 29 patients who were analyzed during the maintenance phase, 23 (79%) had received prior nucleoside therapy. Twenty-two patients (76%) were AZT experienced, 9 (31%) were ddI experienced, 5 (17%) were ddC experienced, 4 (14%) were 3TC experienced, and 4 (14%) were d4T experienced. None of the patients had received prior protease inhibitor therapy.
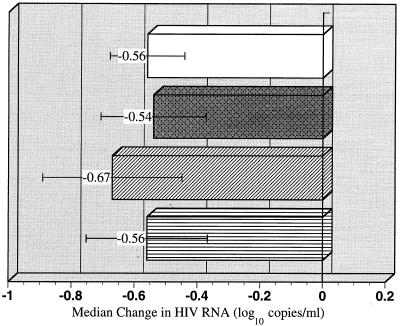
The role of baseline AZT-associated resistance mutations on the response to adefovir dipivoxil during the first 6 months of the maintenance phase was investigated. Concomitant anti-HIV therapy was used by approximately 50% of these patients during this time frame. Seven patients (25%) were AZT naive, 7 (25%) had prior AZT experience but did not have any AZT-associated resistance mutations in RT at baseline, and 14 (50%) had at least 1 AZT-associated resistance mutation (K70R, M41L, D67N, L210W, T215Y or T215F, or K219Q) at baseline. Patients in each of these three groups had similar median viral load responses (−0.6 log10 copies/ml) at month 6 of maintenance-phase dosing, and the response was equal to the overall median response (Fig. 3).
FIG. 3.
Median change in HIV RNA loads (log10 copies per milliliter) after 6 months of adefovir dipivoxil therapy among different groups of patients on the basis of prior AZT experience and presence of AZT resistance mutations at initiation of maintenance-phase adefovir dipivoxil dosing. Other antiretroviral agents were concomitantly used by 50% of the patients. The four patient groups include all patients (n = 28), patients who were AZT naive (n = 7), patients who were AZT experienced but whose viruses had no AZT-associated resistance mutations (n = 7), and patients who were AZT experienced and whose viruses had one or more AZT-associated resistance mutations (n = 14), as indicated by the bars from top to bottom, respectively. Standard error bars are indicated.
The effects of other baseline RT mutations on the response to adefovir dipivoxil therapy were also investigated. The I135V or I135T RT mutation has been associated with AZT or ddI therapy (24, 35). Three patients (11%) had an I135V or I135T RT mutation at baseline that was not in the presence of other nucleoside-associated resistance mutations. These three patients received adefovir dipivoxil monotherapy during this phase and had a median decrease in HIV RNA load of 0.9 log10 copies/ml at month 6.
Viruses from one patient (4%) had a 3TC-associated M184V RT mutation at baseline that was not in the presence of other nucleoside-associated resistance mutations. This patient was receiving concomitant medications and had a decrease in HIV RNA load of 0.7 log10 copies/ml at month 6. Two additional patients entered the maintenance phase with viruses expressing an M184V mutation in RT in the presence of AZT-associated resistance mutations. Both patients were receiving concomitant medications (including 3TC) and had viral RNA load responses better than the median response at 6 months. No maintenance-phase baseline samples contained the nucleoside-associated K65R, L74V, T69D, or Q151M mutations.
DISCUSSION
Adefovir dipivoxil sustained viral load reductions in HIV-1-infected patients receiving extended therapy for ≥6 months with or without concomitant treatment with antiretroviral agents (Fig. 1). This pattern of viral load suppression is in contrast to that achieved by many other nucleoside inhibitors such as AZT, 3TC, or ddI (11, 17, 25, 29, 31, 32, 35, 42, 43, 50), with which a gradual return toward baseline is usually seen, often coincident with the development of drug-resistant viruses. Since adefovir dipivoxil is known to have activity in a variety of cell types (2, 37, 45), it is possible that the prolonged decrease in viral load observed during adefovir dipivoxil therapy is due to its activity in the longer-lived cells of the monocyte/macrophage lineage as well as the lack of significant resistance development.
The K70E and K65R mutations were selected for in vitro and exhibited approximately 9- to 16-fold decreases in sensitivity to adefovir (9, 19, 23). Notably, viruses from two patients developed the K70E mutation during maintenance therapy, and both patients responded with better than median decreases in viral load. One patient (Table 1, patient A; Fig. 2A) was receiving monotherapy, and the viruses from that patient developed a mixture of wild-type and mutant codons at amino acid 70 (K70E/K) after 3 months of maintenance dosing. Even in the presence of this mutation the patient had a decrease in viral RNA load of 0.9 log10 copies/ml after 6 months of maintenance-phase dosing. Interestingly, the HIV in this patient’s plasma remained a mixture of mutant K70E and wild-type virus between months 3 and 6, despite continued treatment with adefovir dipivoxil monotherapy. It has previously been shown that the K70E recombinant virus demonstrated modestly reduced growth kinetics in vitro (9). The K70E virus may also have a reduced replication capacity in vivo, perhaps contributing to the sustained activity of adefovir dipivoxil in this patient. The moderate decrease in susceptibility to adefovir in recombinants generated from patient A is also consistent with the lack of a viral load rebound in this patient. Viruses from the second patient (Table 1, patient G) developed the K70E mutation in RT after 12 months of adefovir dipivoxil therapy while the patient was concomitantly receiving AZT and 3TC. Since this patient was receiving concomitant therapy and also developed the M184V mutation during the maintenance phase, it is not clear what role the K70E mutation played in the virological response (decrease in viral load of 0.9 log10 copies/ml from maintenance-phase baseline at month 12) of this patient. No patient entered the maintenance phase with a virus with a K65R mutation in RT or was infected with a virus that developed that mutation during therapy, so the effect of this mutation on the virological response to adefovir dipivoxil is as yet unknown.
Viruses from two patients (Table 1, patients E and F) developed the T69D mutation while the patients were receiving adefovir dipivoxil therapy. While this mutation has previously been associated with ddC therapy (18), it has not been associated directly with treatment with other nucleosides and was not selected for in vitro by adefovir. Neither of the patients whose viruses developed the T69D mutation reported prior ddC use. Both patients had sustained decreases in viral load (−1.4 and −1.2 log10 copies/ml) subsequent to the development of this mutation. However, since both patients were concomitantly receiving other medications during the maintenance phase, the effects of the T69D mutation on the virus responses to adefovir dipivoxil treatment are not clear. The minor decrease in adefovir susceptibility measured for the posttherapy recombinant virus from patient E may suggest that adefovir dipivoxil is still able to effectively suppress replication of the T69D virus.
Interestingly, during the maintenance phase viruses from four patients (Table 1, patients B, C, D, and H) developed mutations that are characteristically associated with AZT resistance, although the patients did not report receiving concomitant AZT therapy. It is curious that among the viruses from these patients, the majority of the viruses with AZT-associated resistance mutations, once identified, maintained mixtures of mutations but did not develop into full mutants. This finding suggests that the selective pressure exerted by adefovir dipivoxil that allows the outgrowth of viruses harboring the AZT-associated resistance mutations may be quite moderate. Previous phenotypic assays performed with clinical isolates containing different combinations of AZT-resistant mutations exhibited little to no decrease in susceptibility to adefovir in vitro (8, 21, 23). Clearly, recombinant viruses from patients B and C showed no measurable decrease in susceptibility to adefovir in vitro (Table 2), and the viral loads in these patients did not rebound in the presence of viruses with these mutations (Fig. 2). Patient D is the only patient in this study whose recombinant viruses showed a greater than threefold decrease in adefovir susceptibility, yet the patient still had a notable viral RNA decrease over the course of therapy.
Three of these four patients (Table 1, patients B, C, and H) had received AZT therapy prior to the maintenance phase, and although AZT-associated mutations were not detectable at baseline, viruses with these mutations may have existed as a minority viral population in these patients. These viruses expressing AZT-associated mutations may have a slight selective advantage over wild-type viruses in the presence of adefovir dipivoxil and may therefore be enriched for during adefovir dipivoxil therapy, thus becoming a larger percentage of the viral population after the significant decrease in HIV load experienced by all three patients (−0.7, −0.8, and −1.8 log10 copies/ml, respectively; the last two patients were on monotherapy). The fourth patient (Table 1, patient D) was reportedly AZT naive, yet viruses from this patient still developed a complex mixture of AZT-associated resistance mutations after 12 months of monotherapy. Even so, this patient demonstrated a decrease in HIV RNA load of 1.0 log10 copies/ml after 12 months of adefovir dipivoxil monotherapy, making the clinical significance of these genotypic findings unclear. Alternatively, it is possible that patient D was originally infected with an AZT-resistant virus.
The development of AZT-associated resistance mutations (M41L, D67N, K70R, L210W, T215Y, and K219Q) in viruses from patients receiving RT inhibitors other than AZT has been documented previously. Winters et al. (51) reported that viruses from 6 of 23 patients receiving ddI monotherapy for 56 to 104 weeks developed the M41L, K70R, L210W, T215Y, and/or K219Q mutations. Two of these patients did not report receiving prior AZT therapy. Viruses from the patients who developed AZT-associated resistance mutations maintained wild-type ddI susceptibilities when phenotypic assays were performed. Demeter et al. (16) also reported that viruses from two patients receiving ddI monotherapy for >2 years developed M41L, D67N, K70R, and/or T215Y mutations. One of these patients had not received prior AZT therapy. Lin et al. (33) reported that viruses from 8 of 13 patients analyzed after ≥18 months of d4T treatment developed the AZT-associated resistance mutations M41L, D67N, L210W, T215Y, and/or K219Q or K219E. Two of these eight patients did not report receiving prior AZT therapy. Thus, as suggested for adefovir dipivoxil, both ddI and d4T may exert enough selective pressure to allow the outgrowth of viruses carrying AZT-associated resistance mutations. Infrequently, however, it appears that AZT-associated resistance mutations may be selected for in vivo in AZT-naive patients during therapy with an RT inhibitor other than AZT.
It has been shown in numerous studies that antiretroviral agent-naive patients have a greater decrease in viral load than antiretroviral agent-experienced patients. The reasons for this finding are likely numerous (stage of disease, compliance, pharmacokinetics, etc.), but at least for some patients, baseline preexisting virus mutations influence the in vivo response to a new agent. For example, patients whose viruses develop a T215Y or T215F mutation in RT during AZT therapy have been shown to have a diminished virological response to subsequent treatment with AZT-ddI, ddI alone, or AZT-ddI-delavirdine (12, 24, 25) compared to the response of patients whose viruses have the wild-type amino acid at this RT codon. The role played by other baseline nucleoside resistance mutations on the virological response to a subsequent agent is less well described to date. Interestingly, it was reported recently that patients whose viruses possess an M184V mutation at baseline and who later added AZT to their 3TC therapy and whose viruses developed multiple AZT-resistant mutations had poor virological responses to AZT-3TC combination therapy (35).
As shown in Fig. 3, the viral load responses of patients whose viruses had AZT-associated resistance mutations at baseline were similar to the overall median viral load changes among all patients in this study. However, the concomitant use of other medications by 50% of the patients, coupled with the limited number of patients in each group and the complex pattern of representation of the six individual AZT mutations present at the baseline, precludes a complete evaluation of the effects of preexisting AZT resistance mutations on the response to adefovir dipivoxil therapy. Additional studies are needed to directly address the effects of not only AZT-associated resistance mutations but also those associated with other HIV therapies on the response to adefovir dipivoxil therapy.
Few changes in the sequence of RT from baseline that could be potentially attributed to adefovir dipivoxil therapy were detected in viruses from patients who received up to 12 months of maintenance-phase dosing with or without the concomitant use of other antiretroviral agents. The lack of significant genotypic or phenotypic resistance development as well as adefovir dipivoxil’s documented activity in resting and activated lymphocytes and macrophages/monocytes (2, 37, 45) are consistent with the durable anti-HIV activity observed in this study. Ongoing blinded, controlled clinical trials will further investigate the resistance profile and antiretroviral activity of adefovir dipivoxil.
REFERENCES
- 1.Balzarini J, De Clercq E. 5-Phosphoribosyl-1-pyrophosphate synthetase converts the acyclic nucleoside phosphonates 9-(3-hydroxy-2-phosphonylmethoxypropyl)adenine and 9-(2-phosphonylmethoxyethyl)adenine directly to their antivirally active diphosphate derivatives. J Biol Chem. 1991;266:8686–8689. [PubMed] [Google Scholar]
- 2.Balzarini J, Perno C F, Schols D, De Clercq E. Activity of acyclic nucleoside phosphonate analogues against human immunodeficiency virus in monocyte/macrophages and peripheral blood lymphocytes. Biochem Biophys Res Commun. 1991;178:329–335. doi: 10.1016/0006-291x(91)91818-w. [DOI] [PubMed] [Google Scholar]
- 3.Balzarini J, Hao Z, Herdewijn P, Johns D G, DeClerq E. Intracellular metabolism and mechanism of antiretrovirus action of 9-(2-phosphonylmethoxyethyl)adenine, a potent anti-human immunodeficiency virus compound. Proc Natl Acad Sci USA. 1991;88:1499–1503. doi: 10.1073/pnas.88.4.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balzarini J, Holy A, Jindrich J, Naesens L, Snoeck R, Schols D, De Clercq E. Differential antiherpesvirus and antiretrovirus effects of the (S) and (R) enantiomers of acyclic nucleoside phosphonates: potent and selective in vitro and in vivo antiretrovirus activities of (R)-9-(2-phosphonomethoxypropyl) derivatives of adenine and 2,6-diaminopurine. Antimicrob Agents Chemother. 1993;37:332–338. doi: 10.1128/aac.37.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balzarini J, Nave J F, Becker M A, Tatiban M, DeClercq E. Kinetic properties of adenine nucleotide analogues against purified 5-phosphoribosyl-1-pyrophosphate synthetases from E. coli, rat liver, and human erythrocytes. Nucleosides Nucleotides. 1995;14:1861–1871. [Google Scholar]
- 6.Boucher C A B, O’Sullivan E, Mulder J W, Ramautarsing C, Kellam P, Darby G, Lange J M A, Goudsmit J, Larder B A. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J Infect Dis. 1992;165:105–110. doi: 10.1093/infdis/165.1.105. [DOI] [PubMed] [Google Scholar]
- 7.Boucher C A B, Kuelen W, Van Bommel T, Nijhuis M, De Jong D, De Jong M, Schipper P, Back N K T. Human immunodeficiency virus type 1 drug susceptibility determination using recombinant viruses generated from patient sera tested in a cell-killing assay. Antimicrob Agents Chemother. 1996;40:2404–2409. doi: 10.1128/aac.40.10.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Carpenter C C J, et al. Antiretroviral therapy for HIV infection in 1997. JAMA. 1997;277:1962–1969. [PubMed] [Google Scholar]
- 8.Cherrington, J. M. Unpublished data.
- 9.Cherrington J M, Mulato A S, Fuller M D, Chen M S. Novel mutation (K70E) in human immunodeficiency virus type 1 reverse transcriptase confers decreased susceptibility to 9-[2-(phosphonomethoxy)ethyl]adenine in vitro. Antimicrob Agents Chemother. 1996;40:2212–2216. doi: 10.1128/aac.40.9.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherrington J M, Allen S J W, Bischofberger N, Chen M S. Kinetic interaction of the diphosphates of 9-(2-phosphonylmethoxyethyl)adenine and other anti-HIV active purine congeners with HIV reverse transcriptase and human DNA polymerases α, β, and γ. Antivir Chem Chemother. 1995;6:217–221. [Google Scholar]
- 11.D’Aquila R T, Johnson V A, Wells S L, Japour A J, Kuritzkes D R, DeGrutotola V, Reichelderfer P S, Coombs R W, Crumpacker C S, Kahn J O, Richman D D the AIDS Clinical Trials Group Protocol 116/117 Team and Virology Committee Resistance Working Group. Zidovudine resistance and HIV-1 disease progression during antiretroviral therapy. Ann Intern Med. 1995;122:401–408. doi: 10.7326/0003-4819-122-6-199503150-00001. [DOI] [PubMed] [Google Scholar]
- 12.Davey R T, Jr, Chaitt D G, Reed G F, Freimuth W W, Herpin B R, Metcalf J A, Eastman P S, Falloon J, Kovacs J A, Polis M A, Walker R E, Masur H, Boyle J, Coleman S, Cox S R, Wathen L, Daenzer C L, Lane H C. Randomized, controlled phase I/II trial of combination therapy with delavirdine (U-90152S) and conventional nucleosides in human immunodeficiency virus type 1-infected patients. Antimicrob Agents Chemother. 1996;40:1657–1664. doi: 10.1128/aac.40.7.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Clercq E, Holy A, Rosenberg I, Sakuma T, Balzarini J, Maudgal P C. A novel selective broad-spectrum anti-DNA virus agent. Nature (London) 1986;323:464–467. doi: 10.1038/323464a0. [DOI] [PubMed] [Google Scholar]
- 14.De Clercq E, Sakuman T, Baba M, Pauwels R, Balzarini J, Rosenberg I, Holy A. Antiviral activity of phosphonylmethoxyalkyl derivatives of purines and pyrimidines. Antivir Res. 1987;8:261–272. doi: 10.1016/s0166-3542(87)80004-9. [DOI] [PubMed] [Google Scholar]
- 15.Deeks S G, Collier A, Lalezari J, Pavia A, Rodrigue D, Drew W L, Toole J, Jaffe H S, Mulato A S, Lamy P D, Li W, Cherrington J M, Hellmann N, Kahn J. The safety and efficacy of adefovir dipivoxil, a novel anti-HIV therapy, in HIV infected adults. J Infect Dis. 1997;176:1517–1523. doi: 10.1086/514150. [DOI] [PubMed] [Google Scholar]
- 16.Demeter L M, Nawaz T, Morse G, Dolin R, Dexter A, Gerondelis P, Reichman R C. Development of zidovudine resistance mutations in patients receiving prolonged didanosine monotherapy. J Infect Dis. 1995;172:1480–1485. doi: 10.1093/infdis/172.6.1480. [DOI] [PubMed] [Google Scholar]
- 17.Eron J J, Benoit S L, Jemsek J, MacArthur R D, Santana J, Quinn J B, Kuritzkes D R, Fallon M, Rubin M for the North American HIV Working Party. Treatment with lamivudine, zidovudine, or both in HIV-positive patients with 200 to 500 CD4+ cells per cubic millimeter. N Engl J Med. 1995;333:1662–1669. doi: 10.1056/NEJM199512213332502. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgibbon J E, Howell R M, Haberzettl C A, Sperber S J, Gocke D J, Dubin D T. Human immunodeficiency virus type 1 pol gene mutations which cause decreased susceptibility to 2′,3′-dideoxycytidine. Antimicrob Agents Chemother. 1992;36:153–157. doi: 10.1128/aac.36.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foli A, Sogocio K M, Anderson B, Kavlick M, Saville M W, Wainberg M A, Gu X, Cherrington J M, Mitsuya H, Yarchoan R. In vitro selection and molecular characterization of human immunodeficiency virus type 1 with reduced sensitivity to 9-[2-(phosphonomethoxy)ethyl]adenine (PMEA) Antivir Res. 1996;32:91–98. doi: 10.1016/0166-3542(95)00985-x. [DOI] [PubMed] [Google Scholar]
- 20.Gao Q, Gu Z, Parniak M A, Cameron J, Cammack N, Boucher C, Wainberg M A. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1993;37:1390–1392. doi: 10.1128/aac.37.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gong Y-F, Marshall D R, Srinivas R V, Fridland A. Susceptibilities of zidovudine-resistant variants of human immunodeficiency virus type 1 to inhibition by acyclic nucleoside phosphonates. Antimicrob Agents Chemother. 1994;38:1683–1687. doi: 10.1128/aac.38.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu Z, Gao Q, Fang H, Salomon H, Parniak M A, Goldberg E, Cameron J, Wainberg M A. Identification of a mutation at codon 65 in the IKKK motif of reverse transcriptase that encodes human immunodeficiency virus resistance to 2′,3′-dideoxycytidine and 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1994;38:275–281. doi: 10.1128/aac.38.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu Z, Salomon H, Cherrington J M, Mulato A S, Chen M S, Yarchoan R, Foli A, Sogocio K, Wainberg M A. K65R mutation of human immunodeficiency virus type 1 reverse transcriptase encodes cross-resistance to 9-(2-phosphonylmethoxyethyl)adenine. Antimicrob Agents Chemother. 1995;39:1888–1891. doi: 10.1128/aac.39.8.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holodniy M, Mole L, Margolis D, Moss J, Dong H, Boyer E, Urdea M, Kolberg J, Eastman S. Determination of human immunodeficiency virus RNA in plasma and cellular viral DNA genotypic zidovudine resistance and viral load during zidovudine-didanosine combination therapy. J Virol. 1995;69:3510–3516. doi: 10.1128/jvi.69.6.3510-3516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holodniy M, Katzenstein D, Mole L, Winters M, Merigan T. Human immunodeficiency virus reverse transcriptase codon 215 mutations diminish virologic response to didanosine-zidovudine therapy in subjects with non-syncytium-inducing phenotype. J Infect Dis. 1996;174:854–857. doi: 10.1093/infdis/174.4.854. [DOI] [PubMed] [Google Scholar]
- 26.Hooker D J, Tachedjian G, Solomon A E, Gurusinghe A D, Land S, Birch C, Anderson J L, Roy B M, Arnold E, Deacon N J. An in vivo mutation from leucine to tryptophan at position 210 in human immunodeficiency virus type 1 reverse transcriptase contributes to high-level resistance to 3′-azido-3′-deoxythymidine. J Virol. 1996;70:8010–8018. doi: 10.1128/jvi.70.11.8010-8018.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellam P, Boucher C A B, Larder B A. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc Natl Acad Sci USA. 1992;89:1934–1938. doi: 10.1073/pnas.89.5.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellam P, Boucher C A, Tijnagel J M, Larder B A. Zidovudine treatment results in the selection of human immunodeficiency virus type 1 variants whose genotypes confer increasing levels of drug resistance. J Gen Virol. 1994;75:341–351. doi: 10.1099/0022-1317-75-2-341. [DOI] [PubMed] [Google Scholar]
- 29.Kozal M J, Kroodsma K, Winters M A, Shafer R W, Efron B, Katzenstein D A, Merigan T C. Didanosine resistance in HIV-infected patients switched from zidovudine to didanosine monotherapy. Ann Intern Med. 1994;121:263–268. doi: 10.7326/0003-4819-121-4-199408150-00005. [DOI] [PubMed] [Google Scholar]
- 30.Lacey S F, Larder B A. A novel mutation (V75T) in the HIV-1 reverse transcriptase confers resistance to 2′,3′-didehydro-2′,3′-dideoxythymidine (D4T) in cell culture. Antimicrob Agents Chemother. 1994;38:1428–1432. doi: 10.1128/aac.38.6.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larder B A, Darby A G, Richman D D. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 32.Larder B A, Kemp S D. Multiple mutation in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 33.Lin P-F, Samanta H, Rose R E, Patick A K, Trimble J, Bechtold C M, Revie D R, Khan N C, Federici M E, Li H, Lee A, Anderson R E, Colonno R J. Genotypic and phenotypic analysis of human immunodeficiency virus type 1 isolates from patients on prolonged stavudine therapy. J Infect Dis. 1994;170:1157–1164. doi: 10.1093/infdis/170.5.1157. [DOI] [PubMed] [Google Scholar]
- 34.Mayers D L, Gallahan D L, Martin G J, Emmons W W, Chung R C Y, Spooner K M, Newton J A, Aronson N E, Weislow O S. Abstracts of the International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication. 1997. Drug resistance genotypes from plasma virus of HIV-infected patients failing combination drug therapy, abstr. 81. [Google Scholar]
- 35.Nijhuis M, Schuurman R, De Jong D, Van Leeuwen R, Lange J, Danner S, Keulen W, De Groot T, Boucher C A B. Lamivudine-resistant human immunodeficiency virus type 1 variants (184V) require multiple amino acid changes to become co-resistant to zidovudine in vivo. J Infect Dis. 1997;176:398–405. doi: 10.1086/514056. [DOI] [PubMed] [Google Scholar]
- 36.Pauwels R, Balzarini J, Schols D, Baba M, Desmyter J, Rosenberg I, Holy A, De Clercq E. Phosphonylmethoxyethyl purine derivatives, a new class of anti-human immunodeficiency virus agents. Antimicrob Agents Chemother. 1988;32:1025–1030. doi: 10.1128/aac.32.7.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perno C F, Balestra E, Aquaro S, Panti S, Cenci A, Lazzarino G, Tavazzi B, Di Pierro D, Balzarini J, Calio R. Potent inhibition of human immunodeficiency and herpes simplex virus type 1 by 9-(2-phosphonylmethoxyethyl)adenine in primary macrophages is determined by drug metabolism, nucleotide pools, and cytokines. Mol Pharmacol. 1996;50:359–366. [PubMed] [Google Scholar]
- 38.Robbins B L, Greenhaw J, Connelly M C, Fridland A. Metabolic pathways for activation of the antiviral agent 9-(2-phosphonylmethoxyethyl)adenine (PMEA) in human lymphoid cells. Antimicrob Agents Chemother. 1995;39:2304–2308. doi: 10.1128/aac.39.10.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rooke R, Tremblay M, Soudeyns H, DeStephano L, Yao X-J, Fanning M, Montaner J S G, O’Shaughnessy M, Gelmon K, Tsoukas C, Ruedy H, Wainberg M A. Isolation of drug-resistant variants of HIV-1 from patients on long-term zidovudine (AZT) therapy. AIDS. 1989;3:411–415. doi: 10.1097/00002030-198907000-00001. [DOI] [PubMed] [Google Scholar]
- 40.Schinazi R F, Lloyd R J, Nguyen M H, Cannon D L, McMillan A, Ilksoy N, Chu C K, Liotta D C, Bazmi H Z, Mellors J W. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother. 1993;37:875–881. doi: 10.1128/aac.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmit J-C, Cogniaux J, Hermans P, Van Vaeck C, Sprecher S, Van Remoortel B, Witvrouw M, Balzarini J, Desmyter J, De Clercq E, Vandamme A-M. Multiple drug resistance to nucleoside analogues and nonnucleoside reverse transcriptase inhibitors in an efficiently replicating human immunodeficiency virus type 1 patient strain. J Infect Dis. 1996;174:962–968. doi: 10.1093/infdis/174.5.962. [DOI] [PubMed] [Google Scholar]
- 42.Schuurman R, Nijhuis M, Van Leeuwen R, Schipper P, De Jong D, Collis P, Danner S A, Mulder J, Loveday C, Christopherson C, Kwok S, Sninsky J, Boucher C A B. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 43.Shafer R W, Iversen A K N, Winters M A, Aguiniga E, Katzenstein D A, Merigan T C the AIDS Clinical Trials Group 143 Virology Team. Drug resistance and heterogenous long-term virologic responses of human immunodeficiency virus type 1-infected subjects to zidovudine and didanosine combination therapy. J Infect Dis. 1995;172:170–178. doi: 10.1093/infdis/172.1.70. [DOI] [PubMed] [Google Scholar]
- 44.Shafer R W, Levee D J, Winters M A, Richmond K L, Huang D, Merigan T C. Comparison of QIAamp HCV kit spin columns, silica beads, and phenol-chloroform for recovering human immunodeficiency virus type 1 RNA from plasma. J Clin Microbiol. 1997;35:520–522. doi: 10.1128/jcm.35.2.520-522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirasaka T, Chokekijchai S, Yamada A, Gosselin G, Imbach J L, Mitsuya H. Comparative analysis of anti-human immunodeficiency virus type 1 activities of dideoxynucleoside analogs in resting and activated peripheral blood mononuclear cells. Antimicrob Agents Chemother. 1995;39:2555–2559. doi: 10.1128/aac.39.11.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shirasaka T, Kavlick M F, Ueno T, Gao W-Y, Kojima E, Alcaide M L, Chokekijchai S, Roy B M, Arnold E, Yarchoan R, Mitsuya H. Emergence of human immunodeficiency virus type 1 variants with resistance to multiple dideoxynucleosides in patients receiving therapy with dideoxynucleosides. Proc Natl Acad Sci USA. 1995;92:2398–2402. doi: 10.1073/pnas.92.6.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.St. Clair M H, Martin J L, Tudor-Williams G, Bach M C, Vavro C I, King D M, Kellam P, Kemp S D, Larder B A. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka M, Srinivas R V, Ueno T, Kavlick M F, Hui F K, Fridland A, Driscoll J S, Mitsuya H. In vitro induction of human immunodeficiency virus type 1 variants resistant to 2′-β-fluoro-2′,3′-dideoxyadenosine. Antimicrob Agents Chemother. 1997;41:1313–1318. doi: 10.1128/aac.41.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wainberg M A, Salomon H, Gu Z, Montaner J S G, Cooley T P, McCaffrey R, Ruedy J, Hirst H M, Cammack N, Cameron J, Nicholson W. Development of HIV-1 resistance to (−)2′-deoxy-3′-thiacytidine in patients with AIDS or advanced AIDS-related complex. AIDS. 1995;9:351–357. [PubMed] [Google Scholar]
- 50.Wainberg M A, Drosopoulos W C, Salomon H, Hsu M, Borkow G, Parniak M A, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad V R. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1995;271:1282–1284. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 51.Winters M A, Shafer R W, Jellinger R A, Mamtora G, Gingeras T, Merigan T C. Human immunodeficiency virus type 1 reverse transcriptase genotype and drug susceptibility changes in infected individuals receiving dideoxyinosine monotherapy for 1 to 2 years. Antimicrob Agents Chemother. 1997;41:757–762. doi: 10.1128/aac.41.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang D, Caliendo A M, Eron J J, DeVore K M, Kaplan J C, Hirsch M S, D’Aquila R T. Resistance to 2′,3′-dideoxyctyidine conferred by a mutation in codon 65 of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1994;38:282–287. doi: 10.1128/aac.38.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]