Abstract
Purpose
Machine learning models based on radiomic feature extraction from clinical imaging data provide effective and interpretable means for clinical decision making. This pilot study evaluated whether radiomics features in baseline optical coherence tomography (OCT) images of eyes with pigment epithelial detachment (PED) associated with neovascular age-related macular degeneration (nAMD) can predict treatment response to as-needed anti-vascular endothelial growth factor (VEGF) therapy.
Methods
Thirty-nine eyes of patients with PED undergoing anti-VEGF therapy were included. All eyes underwent a loading dose followed by as-needed therapy. OCT images at baseline, month 3, and month 6 were analyzed. Images were manually separated into non-responding, recurring, and responding eyes based on the presence or absence of subretinal fluid at month 6. PED radiomics features were then extracted from each image and images were classified as responding or recurring using a machine learning classifier applied to the radiomics features.
Results
Linear discriminant analysis classification of baseline features as responsive versus recurring resulted in classification performance of 64.0% (95% confidence interval [CI] = 0.63–0.65), area under the curve (AUC = 0.78, 95% CI = 0.72–0.82), sensitivity 0.79 (95% CI = 0.63–0.87), and specificity 0.58 (95% CI = 0.50–0.67). Further analysis of features in recurring eyes identified a significant shift toward non-responding mean feature values over 6 months.
Conclusions
Our results demonstrate the use of radiomics features as predictors for treatment response to as-needed anti-VEGF therapy. Our study demonstrates the potential for radiomics feature in clinical decision support for personalizing anti-VEGF therapy.
Translational Relevance
The ability to use PED texture features to predict treatment response facilitates personalized clinical decision making.
Keywords: age-related macular degeneration (AMD), artificial intelligence, image analysis
Introduction
Current recommendations regarding scheduling of anti-vascular endothelial growth factor (VEGF) therapy for patients with neovascular age-related macular degeneration (nAMD) balance clinical efficacy of treatment with costs to the healthcare system and to patients. Initial trials demonstrated efficacy of anti-VEGF therapy using scheduled monthly injections.1,2 The prohibitive costs of monthly injections motivated future studies with the goal of reducing the number of injections by increasing scheduled treatment intervals, gradual extension of treatment interval based on response (i.e. “treat-and-extend”), and only treating on an as-needed basis.3,4 The latter two of these approaches demonstrated noninferiority to scheduled monthly injections, however, one major drawback to as-needed or treat-and-extend approaches is that they are reactive in nature and only personalize treatment after a patient has experienced a recurrence of disease, exposing patients to additional complications resulting from inadequate treatment.
In recent years, there has been growing interest in prediction in the clinical setting thanks to improvements in computational power and machine learning algorithms. Applications of machine learning for prediction have shown promise for automated diagnosis of many common ophthalmic diseases, including macular degeneration and diabetic retinopathy.5–8 One drawback to many machine learning algorithms has been the inability to see into the “black box” in order to gain intuition for why a model behaves in a certain way, making nuanced decision making in a complex healthcare environment difficult.9,10 This has led to increased emphasis on the identification of biomarkers that can, in addition to improving prediction, facilitate model interpretability.11,12
One class of biomarkers that has been applied in the field of radiology and pathology is texture features, the application of which is often referred to as radiomics.13 Radiomic techniques provide summary statistics that describe features of interest in a set of images. This is useful for at least three reasons. First, it quantifies aspects of images that are of interest to clinicians. Second, radiomics features summarize image information, reducing the amount of data required to train models, improving the feasibility of prediction algorithms with fewer patients. Third, these techniques facilitate interpretability by allowing comparison of features between groups of images, thus opening the machine learning black box. Radiomics had been applied to a growing range of clinical problems (e.g. diabetic macular edema, myopia maculopathy, and age-related macular degeneration),14–16 and imaging modalities (e.g., optical coherence tomography [OCT], optical coherence tomography angiography [OCTA], and ultra-wide-field fluorescein angiography [UWFA]).17–19 Radiomic feature extraction is a promising approach to identifying biomarkers for the development of intuitive, efficient, and interpretable prediction algorithms.
In this pilot study, we sought to build upon this previous work by applying radiomic feature extraction to OCT images of pigment epithelial detachment (PED) to predict treatment response to anti-VEGF therapy in nAMD. We first predicted from baseline images of responding eyes versus eyes with recurrence at 6 months. We then extracted features to track changes in PED fluid composition over time in eyes with recurrence and compared these changes to features observed in responding and nonresponding eyes.
Methods
Patient Selection and Data Acquisition
We conducted a retrospective cross-sectional study involving patients who were selected from a chart review of patients with nAMD presenting at the University of Pittsburgh Medical Center. The study was conducted in accordance with the Declaration of Helsinki and the Institutional Review Board of the University of Pittsburgh. Informed consent was obtained for all patients.
Each patient underwent a complete history and comprehensive ophthalmic examination, including baseline visual acuity, intraocular pressure, slit lamp biomicroscopy, and dilated fundus examination to confirm the diagnosis of nAMD and rule out other pathology. Inclusion criteria for this study was nAMD with PED confirmed on b-scan with a minimal height and width of 100 microns. Exclusion criteria included complicated nAMD, drusenoid PED, poor quality images, and other comorbid ocular disease. Complicated nAMD was defined as the presence of retinal pigment epithelium (RPE) rips by clinical evaluation. Both treatment naive and previously treated eyes were included in the study.
Imaging was performed using spectral domain optical coherence tomography (SD-OCT) using a Heidelberg Spectralis Device (Heidelberg Engineering, Heidelberg, Germany). Analyses were performed on a single scan passing through the fovea. B-scan resolution was 1536 × 496 with intensity range of 0 to 255.
Feature Segmentation and Feature Extraction
To facilitate image analysis and segmentation, b-scans were preprocessed using the following procedure. First, images were linearly normalized to ensure that displayed b-scan intensities had similar distributions across images. This was done by standardizing each image's pixel intensities to range between 0 and 1. Second, shadow compensation was performed to ensure proper visualization of sub-RPE tissue.20 This was necessary because signal extinction can occur secondary to shadowing from fluid and vessels in the retina.21,22 To correct for the shadowing effect, a compensation factor was calculated on a per-pixel basis by evaluating idiosyncrasies in the OCT images.21 Last, images were manually segmented by a trained annotator. This was done by manually identifying the boundary of the entire PED using ImageJ software version 1.53 using the polygon selector tool.23 The initial segmentation was then smoothed using the “fit Spline” function. Segmentation for each image was then verified by an expert clinician.
Feature Extraction
To assess the texture features of the segmented OCT images, we performed texture-based radiomic feature extraction using the Pyfeats radiomics library.24 Extracted features included first-order statistics, Gabor filter features, Law texture features, and Gray-Level Co-Occurrence Matrix (GLCM) features.25,26 First-order statistics included 10th, 25th, 75th, and 90th percentile pixel values, coefficient of variation, energy, entropy, histogram width, kurtosis, maximal gray level, mean, median, minimal gray level, mode, skewness, and variance. Gabor features were extracted by first offsetting pixel values to be between −0.5 and 0.5 and then applying Gabor filters with angles of 0, 45, 90, and 135 degrees and spatial frequencies of 0.1 and 0.4 cycles/pixel. Mean and standard deviation of Gabor filter convolution values were used as features. Law features were extracted using a mask size of 3.26,27 GLCM features included the following summary statistics of the GLCM: angular second moment, contrast, correlation sum of squares variance, inverse difference moment, sum average, sum variance, sum entropy, entropy, difference variance, difference entropy, information measure of correlation features, and maximal correlation coefficient. A total of 52 features were extracted from PED pixels in each image. Once extracted, features were standardized across images to facilitate later statistical analysis.
Feature overlay plots were obtained by measuring the above-listed features in a 20 × 20 pixel window centered on each PED segmented pixel. Extracted feature values were then normalized across images to allow comparison between images.
To identify features that explained the greatest difference between groups, a non-paired t-test was first performed for each image. This was done for comparison of baseline responding versus baseline nonresponding images, baseline recurring versus baseline responding image features, and pooled nonresponding and responding image features across all time points. The features with the smallest four P values were then plotted.
To compare how changes in features in recurring eyes compared to expected feature values of nonresponding and responding eyes, the following procedure was applied. First, for each responding eye feature, average feature value was computed across all samples, including baseline, month 3, and month 6. This was repeated for nonresponding eyes and recurring eyes. Next, the values for recurring eyes were adjusted so that change toward the corresponding feature mean for responding eyes resulted in a positive value and change toward nonresponding eyes resulted in a negative value. Only features with a difference between responding and nonresponding means with paired t-test P value less than 0.2 were used. Finally, a bootstrap 95% confidence interval (CI) was computed across recurring eye mean feature changes to measure significance.
Image Classification
Standardized feature values and image labels were used to train a machine learning model to classify images as responding or recurring at 6 months. The following classification procedure was performed for all images using the sklearn python library.28 Prior to classification, image features were dimension-reduced using minimum Redundancy maximum Relevance (mRmR) to identify the most relevant 16 features within the training data.29 A linear discriminant analysis model was then used to perform classification. Model performance was assessed using three-fold hierarchical cross-validation to ensure that at least one representative of each class was included in each train and test set. Models were evaluated by measuring accuracy, receiver operator characteristic area under the curve (ROC-AUC), sensitivity, and specificity on test data. Bootstrap 95% CIs were computed across 100 iterations of the above procedure, shuffling trials with each iteration to achieve distinct cross-validation fold compositions.
Results
A total of 39 eyes from 39 patients with nAMD were included in this pilot study. Patients received scheduled monthly anti-VEGF therapy for 3 months as a loading dose followed by as-needed anti-VEGF therapy for an additional 3 months (Fig. 1). Response to anti-VEGF therapy was labeled in each eye with 23 eyes showing a consistent stable response throughout the study (referred to as “responding”), 12 eyes showing an initial response at 3 months followed by a recurrence of fluid at 6 months (referred to as “recurring”), and 4 eyes did not respond to anti-VEGF therapy at 3 months or at 6 months (referred to as “nonresponding”).
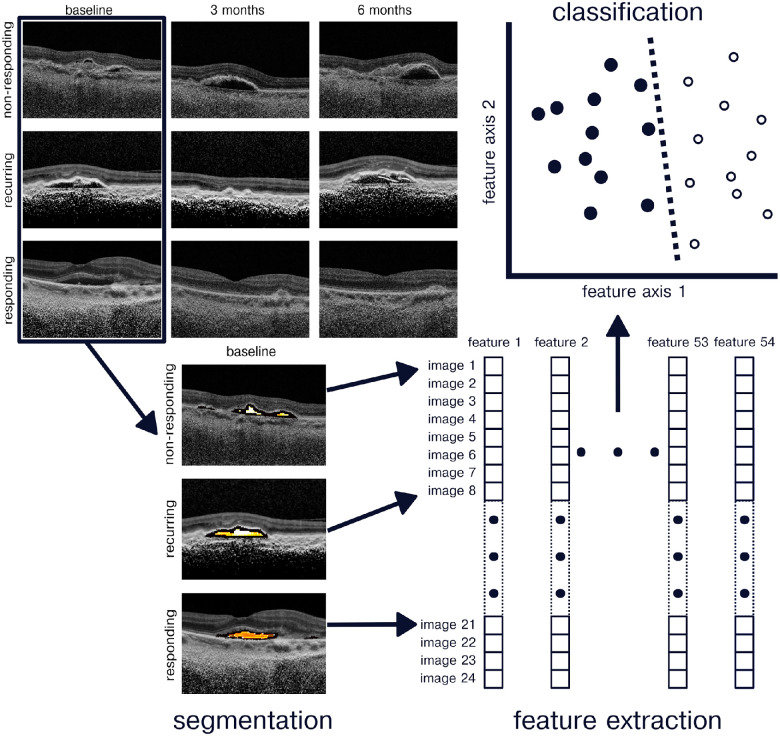
Figure 1.
Radiomics classification pipeline. Retinal OCT images were obtained at baseline, month 3, and month 6 follow-up appointments from nonresponding, recurring, and responding eyes. Images were preprocessed and then segmented to select for PED pixels. A battery of 52 texture features were then computed using the PED pixels. A linear discriminate analysis classifier was used to predict image response to as-needed anti-VEGF therapy.
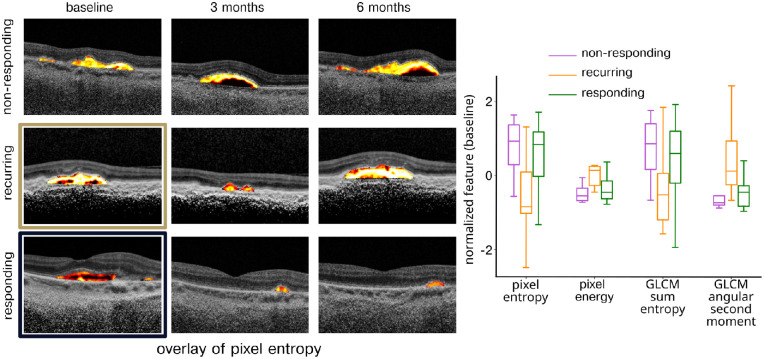
We were first interested in assessing whether it was possible to identify eyes that could transition to as-needed therapy after observing an initial response to treatment between baseline and 3-month follow-up. To do this, image features were compared at baseline between responding and recurring eyes (Fig. 2). The four features with the most significant differences between the two groups were pixel entropy (P = 0.0012), pixel energy (P = 0.019), GLCM sum entropy (P = 0.028), and GLCM angular second moment (P = 0.035). The first two of these features represent “first-order” pixel statistics that describe the average information across pixels and the average pixel intensity within the PED. The GLCM features reflect higher-order texture captured by measurement of how often specific pixel values occur near each other in space. Classification was then performed using an LDA model with a classification accuracy of 0.64 (95% CI = 0.63–0.65), ROC-AUC 0.78 (95% CI = 0.72–0.82), sensitivity 0.79 (95% CI = 0.63–0.87), and specificity 0.58 (95% CI = 0.50–0.67).
Figure 2.
Features distinguishing responding and recurring eyes. Features distinguishing responding and recurring eyes were identified based on lowest P value on a non-paired t-test. Left: Overlay image showing the most distinguishing feature, pixel entropy, for nonresponding, recurring, and responding eyes at baseline, month 3, and month 6. Right: Distribution of four features most distinguishing responding and recurring eyes separated into nonresponding, recurring, and responding feature values.
Next, to facilitate comparison of recurring eyes to responding eyes and nonresponding eyes over time, features most distinguishing responding and nonresponding eyes were identified. This was done by pooling responding and nonresponding eye images from all time points (Fig. 3, left column). The four features with the most significant differences between the two groups were mean Gabor angle 0 degrees frequency 0.1 cycles/pixel (P = 4.6 × 10−4), mean Gabor angle 90 degrees frequency 0.1 cycles/pixel (P = 6.2 × 10−4), standard deviation of Gabor angle 90 degrees frequency 0.1 cycles/pixel (P = 2.6 × 10−3), and GLCM correlation mean (P = 2.7 × 10−3). The first of two these features capture the amounts of horizontal (angle 0 degrees) and vertical (angle 90 degrees) edges and the third feature captures variation in vertical edges within the PED.
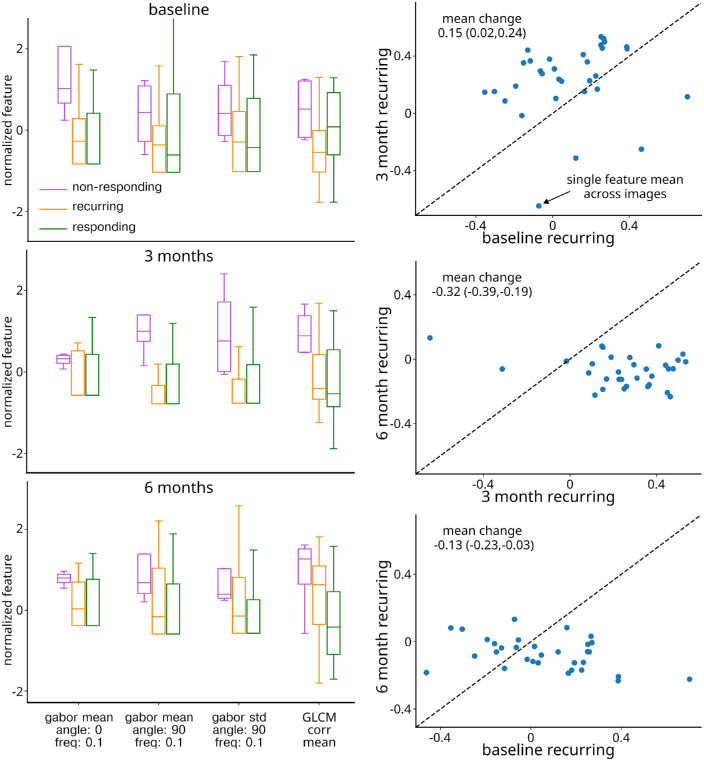
Figure 3.
Recurring eye feature change over time. Left column: Each plot displays 4 features best distinguishing responding and nonresponding eyes at baseline (top), month 3 (middle), and month 6 (bottom). Right top: Comparison of baseline and month 3 feature values. Each circle represents the average value of each feature at baseline and month 3. The circles above the line represent a shift toward responding eye feature mean and circles below the line represent a shift toward nonresponding eye feature mean. Right middle: same as the right top, but for month 3 and month 6 recurring features. Right bottom: Same as right top but for baseline and month 6 recurring features.
Last, we sought to assess change in recurring features over time relative to the features distinguishing nonresponding and responding eyes identified above. Qualitatively, it was noted that recurring features tended to shift over time, with a tendency to shift toward the responding mean at 3 months and toward the nonresponding mean at 6 months. To quantify this result across a larger number of features, the features were selected from the features distinguishing responding and nonresponding eyes with P values greater than 0.2, resulting in 32 features included in the analysis. Features were further processed to ensure that recurring feature change in a positive direction indicated movement toward the responding feature means. After 3 months of scheduled anti-VEGF therapy, recurring eyes were found to have a mean normalized feature change of 0.15 (95% CI = 0.02–0.14), representing a shift toward the responding feature mean (Fig. 3, right column, top plot). In contrast, after 3 months of as-needed therapy, a recurring mean feature change of −0.32 (95% CI = −0.39 to −0.19) was observed, representing a shift toward the nonresponding eye feature mean (Fig. 3, right column, middle plot). Over the 6-month study duration, a recurring eye mean feature shift of −0.13 (95% CI = −0.23 to −0.03) occurred, indicating an overall shift in recurring features toward the nonresponding feature mean (Fig. 3, right column, bottom plot).
Discussion
This pilot study demonstrates the use of radiomic features from OCT images of nAMD eyes to predict response to scheduled and as-needed anti-VEGF therapy. The results demonstrate identification of eyes that can successfully switch to as-needed anti-VEGF therapy using a machine learning classifier applied to radiomic features. The results further leverage these features to demonstrate transition of recurring eye PED composition toward feature values associated with nonresponding eyes over the 6 months’ duration of the study. This suggests that recurring eyes may undergo a transition that makes them less responsive to anti-VEGF therapy over time.
In this study, we classified images based on whether they successfully switched to as-needed anti-VEGF therapy (i.e. responding) or had a recurrence of fluid after transitioning to as-needed anti-VEGF therapy (i.e. recurring). This is a decision frequently faced by physicians, who often rely on fixed treatment regimens based on predefined treatment intervals followed by a trial of as-needed therapy.3,4 Such an approach is unable to account for individual variability in treatment response prior to increasing treatment interval. The ability to classify individuals as likely to succeed with as-needed therapy would allow providers to individualize care to their patients. The classification approach demonstrated in this study represents a proof-of-concept decision-support tool for individualizing the anti-VEGF treatment interval based on each patient’s needs.
Our results demonstrated the trend of recurring eye features toward expressing nonresponding feature characteristics after 6 months. The cause of this transition is unclear. One possibility is that recurring eyes represent a class of eyes that are in a transition period between a treatment-responsive state and a nonresponsive state and that this transition would have occurred even if scheduled therapy had continued for a longer period of time. Another possibility is that recurring eyes simply represent eyes that would benefit from a longer period of scheduled therapy in order to achieve disease remission, after which point these eyes would be successful on as-needed therapy. Future work could address this question by tracking image features after retreatment and assessing whether features from retreated eyes return to resembling responding eyes or if they continue to shift toward nonresponding feature values.
In this study, we extracted radiomic features from images to perform image classification and to track changes in PED fluid characteristics over time. The use of radiomic features provided several key advantages during this process. First, radiomics reduced the number of input dimensions required for classification compared to performing classification using raw pixels. This reduced the number of model parameters, thereby also reducing the required number of samples for training, owing to the fact that the sample size needed to train a model scale with the number of parameters. Second, radiomic features extract meaningful and interpretable statistics from images in ways that other common dimensionality reduction methods (e.g. principal component analysis) cannot. Finally, radiomic features quantify our intuitive understanding of image texture, thus facilitating tracking of features over time. Further use of radiomic features in model development can improve the interpretability of machine learning models for nuanced clinical decision making and reduce the cost of model development through the efficient use of training data.
Several recent studies have used radiomics to classify and study retinal OCT.14,30,31 Among these, one recent study used radiomic features applied to multiple segmented fluid spaces to classify eyes as super-responders or non-super responders.31 In the current study, we build upon previous work by predicting which eyes will successfully switch to as-needed therapy without recurrence in the first 3 months. As in previous work, our study demonstrates the successful use of radiomic features to facilitate machine learning classification of OCT images with the goal of providing clinical decision support to personalize treatment to individual patient’s needs.
Limitations of this study include the small sample size, inclusion of multiple anti-VEGF treatment agents, and pooling of treatment-naive and previously treated eyes. Further work is underway to expand the study to a larger population to assess the external validity of the findings. In addition, several steps were taken to mitigate the effects of small sample size on reported classifier performance. First, we used a hierarchical cross-validation structure which ensured that test data sets included similar proportions of each class as the sampled population. Although this cannot overcome biases in sampling in the study population, it can help prevent additional bias that may arise from evaluating models on small test data sets. Second, we limited the number of parameters in our models, reducing the risk of overfitting. We did this by (1) extracting radiomic features rather than using raw pixels and (2) using relatively simple classification algorithms as opposed to more complex “deep” models which typically require larger numbers of parameters. A similar approach has been used in other studies using radiomics for prediction in retinal imaging.30,31
There are also a few areas where our radiomics-based classification could potentially be improved without increasing the sample size. For example, our study focused on single b-scans through the fovea rather than full 3-D scans consisting of multiple b-scans. Although selection of a single foveal b-scan facilitates comparison across time and limits variability that might be introduced by taking scans from different locations within each image, this approach also risks missing extrafoveal changes in PED composition. Furthermore, full 3-D scans have the benefit of potentially adding useful information with only a small increase in the number of model parameters, likely leading to better classifier performance. Single b-scan classification also requires a clinician to select the scan of interest, a step not required when using 3-D scans. A disadvantage of 3-D scans is that they are labor-intensive to manually segment. Future work will focus on using automated segmentation algorithms to increase the feasibility of studying 3-D scans.
Another potential area of improvement is in our choice of features, dimensionality reduction algorithm, and classification algorithms. We did not perform an exhaustive search of all possible combinations of these computational components in order to identify the algorithms with the best fit. This was intentional so as to avoid overfitting our model to the test data while selecting among computational components. Overfitting due to optimization of computational components could be overcome by using a separate validation data set for model selection, a step which we could include in future studies with a larger sample population.
One area of future work is to assess the significance of different features in terms of fluid composition and anatomy. For example, previous work has successfully segmented PED into serous, drusenoid, and fibrovascular fluid types.32 Understanding how such separation of fluid type relates to radiomic features would improve the interpretability of radiomics features on the one hand and possibly reveal additional subcategories of fluid that can be distinguished by radiomics features on the other hand. Further work in these areas could lead to improved clinical decision making and new understanding of the pathophysiology of PED.
A second area of future work is assessment of other fluid compartments and anatomic regions. The tools discussed in this study and others are easily generalizable beyond PED and other retinal fluid compartments and could be applied to other retinal disease processes, healthy eyes with comorbid risk factors, and other types of ocular imaging.14–19 Expanded use of radiomics for prediction holds promise for improving care for an even broader patient population.
A third area of future work is to track feature changes over the course of years. The current study focused on a 3-month window of as-needed therapy. The extent to which features from recurring eyes will continue to grow to resemble features from nonresponding eyes is still unknown. Tracking PED features over a period of years will be an important step in understanding mechanisms underlying changes in treatment response over time.
In conclusion, our study evaluated the use of radiomic-based image classification on OCT images of nAMD eyes for predicting which eyes would remain clinically stable on as-needed anti-VEGF therapy. We found that our prediction pipeline achieved significant classification performance in this task. We further identified features that were likely relevant to this classification, improving model interpretability. Future work will focus on validating these results in larger study populations and expanding prediction to other anatomic compartments.
Acknowledgments
Supported by NIH CORE Grant P30 EY08098 to the Department of Ophthalmology, the Eye and Ear Foundation of Pittsburgh, and from an unrestricted grant from Research to Prevent Blindness, New York, NY.
Disclosure: R.C. Williamson, None; A. Selvam, None; V. Sant, None; M. Patel, None; S.C. Bollepalli, None; K.K. Vupparaboina, None; J.-A. Sahel, None; J. Chhablani, None
References
- 1. Rosenfeld PJ, Brown DM, Heier JS, et al.. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006; 355(14): 1419–1431. [DOI] [PubMed] [Google Scholar]
- 2. Brown DM, Kaiser PK, Michels M, et al.. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N Engl J Med. 2006; 355(14): 1432–1444. [DOI] [PubMed] [Google Scholar]
- 3. Regillo CD, Brown DM, Abraham P, et al.. Randomized, double-masked, sham-controlled trial of ranibizumab for neovascular age-related macular degeneration: PIER Study Year 1. Am J Ophthalmol. 2008; 145(2): 239–248.e5. [DOI] [PubMed] [Google Scholar]
- 4. Lalwani GA, Rosenfeld PJ, Fung AE, et al.. A variable-dosing regimen with intravitreal ranibizumab for neovascular age-related macular degeneration: year 2 of the PrONTO Study. Am J Ophthalmol. 2009; 148(1): 43–58.e1. [DOI] [PubMed] [Google Scholar]
- 5. Yim J, Chopra R, Spitz T, et al.. Predicting conversion to wet age-related macular degeneration using deep learning. Nat Med. 2020; 26(6): 892–899. [DOI] [PubMed] [Google Scholar]
- 6. Dai L, Wu L, Li H, et al.. A deep learning system for detecting diabetic retinopathy across the disease spectrum. Nat Commun. 2021; 12(1): 3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hassan B, Qin S, Ahmed R, et al.. Deep learning based joint segmentation and characterization of multi-class retinal fluid lesions on OCT scans for clinical use in anti-VEGF therapy. Comput Biol Med. 2021; 136: 104727. [DOI] [PubMed] [Google Scholar]
- 8. Bora A, Balasubramanian S, Babenko B, et al.. Predicting the risk of developing diabetic retinopathy using deep learning. Lancet Digit Health. 2021; 3(1): e10–e19. [DOI] [PubMed] [Google Scholar]
- 9. Sussillo D, Barak O.. Opening the black box: low-dimensional dynamics in high-dimensional recurrent neural networks. Neural Comput. 2013; 25(3): 626–649. [DOI] [PubMed] [Google Scholar]
- 10. Tjoa E, Guan C.. A survey on explainable artificial intelligence (XAI): toward medical XAI. IEEE Trans Neural Netw Learn Syst. 2021; 32(11): 4793–4813. [DOI] [PubMed] [Google Scholar]
- 11. Kalra G, Kar SS, Sevgi DD, Madabhushi A, Srivastava SK, Ehlers JP.. Quantitative imaging biomarkers in age-related macular degeneration and diabetic eye disease: a step closer to precision medicine. J Pers Med. 2021; 11(11): 1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cao J, You K, Zhou J, et al.. A cascade eye diseases screening system with interpretability and expandability in ultra-wide field fundus images: A multicentre diagnostic accuracy study. eClinicalMedicine. 2022; 53: 101633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rizzo S, Botta F, Raimondi S, et al.. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018; 2(1): 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Banerjee I, de Sisternes L, Hallak JA, et al.. Prediction of age-related macular degeneration disease using a sequential deep learning approach on longitudinal SD-OCT imaging biomarkers. Sci Rep. 2020; 10(1): 15434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Du Y, Chen Q, Fan Y, et al.. Automatic identification of myopic maculopathy related imaging features in optic disc region via machine learning methods. J Transl Med. 2021; 19(1): 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kar SS, Abraham J, Wykoff CC, et al.. Computational imaging biomarker correlation with intraocular cytokine expression in diabetic macular edema: radiomics insights from the IMAGINE study. Ophthalmol Sci. 2022; 2(2): 100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prasanna P, Bobba V, Figueiredo N, et al.. Radiomics-based assessment of ultra-widefield leakage patterns and vessel network architecture in the PERMEATE study: insights into treatment durability. Br J Ophthalmol. 2021; 105(8): 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Afarid M, Mohsenipoor N, Parsaei H, et al.. Assessment of macular findings by OCT angiography in patients without clinical signs of diabetic retinopathy: radiomics features for early screening of diabetic retinopathy. BMC Ophthalmol. 2022; 22(1): 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Movahedan A, Vargas P, Moir J, et al.. Computerized texture analysis of optical coherence tomography angiography of choriocapillaris in normal eyes of young and healthy subjects. Cells. 2022; 11(12): 1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shah M, Xiao Y, Subbanna N, et al.. Evaluating intensity normalization on MRIs of human brain with multiple sclerosis. Med Image Anal. 2011; 15(2): 267–282. [DOI] [PubMed] [Google Scholar]
- 21. Girard MJA, Strouthidis NG, Ethier CR, Mari JM.. Shadow removal and contrast enhancement in optical coherence tomography images of the human optic nerve head. Invest Ophthalmol Vis Sci. 2011; 52(10): 7738–7748. [DOI] [PubMed] [Google Scholar]
- 22. Vupparaboina KK, Dansingani KK, Goud A, et al.. Quantitative shadow compensated optical coherence tomography of choroidal vasculature. Sci Rep. 2018; 8(1): 6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schneider CA, Rasband WS, Eliceiri KW.. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012; 9(7): 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Giakoumoglou N. PyFeats: open source software for image feature extraction. GitHub repository, 2021.
- 25. Haralick RM, Shanmugam K, Dinstein I.. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973; SMC-3(6): 610–621. [Google Scholar]
- 26. Laws KI. Rapid texture identification. Proc. SPIE Conf. Image Processing for Missile Guidance. 1980: 376–380. [Google Scholar]
- 27. Wu CM, Chen YC, Hsieh KS.. Texture features for classification of ultrasonic liver images. IEEE Trans Med Imaging. 1992; 11(2): 141–152. [DOI] [PubMed] [Google Scholar]
- 28. Pedregosa F, Varoquaux G, Gramfort A, et al.. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011; 12(85): 2825–2830. [Google Scholar]
- 29. Ding C, Peng H.. Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol. 2005; 03(02): 185–205. [DOI] [PubMed] [Google Scholar]
- 30. Sil Kar S, Sevgi DD, Dong V, Srivastava SK, Madabhushi A, Ehlers JP.. Multi-compartment spatially-derived radiomics from optical coherence tomography predict anti-VEGF treatment durability in macular edema secondary to retinal vascular disease: preliminary findings. IEEE J Transl Eng Health Med. 2021; 9: 1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kar SS, Cetin H, Lunasco L, et al.. OCT-derived radiomic features predict anti-VEGF response and durability in neovascular age-related macular degeneration. Ophthalmol Sci. 2022; 2(4): 100171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Selvam A, Singh SR, Arora S, et al.. Pigment epithelial detachment composition indices (PEDCI) in neovascular age-related macular degeneration. Sci Rep. 2023; 13(1): 68. [DOI] [PMC free article] [PubMed] [Google Scholar]