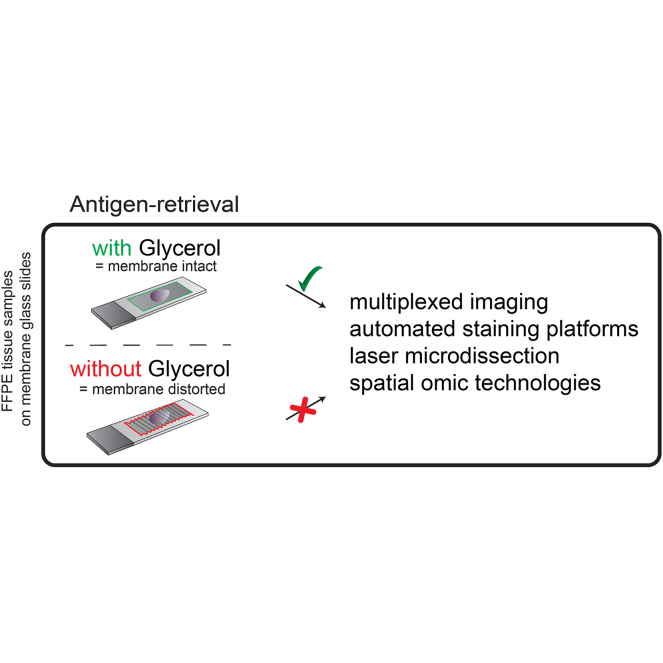
Abstract
Defining the molecular phenotype of single cells in situ is key for understanding tissue architecture in health and disease. Advanced imaging platforms have recently been joined by spatial omics technologies, promising unparalleled insights into the molecular landscape of biological samples. Furthermore, high-precision laser microdissection (LMD) of tissue on membrane glass slides is a powerful method for spatial omics technologies and single-cell type spatial proteomics in particular. However, current histology protocols have not been compatible with glass membrane slides and LMD for automated staining platforms and routine histology procedures. This has prevented the combination of advanced staining procedures with LMD. In this study, we describe a novel method for handling glass membrane slides that enables automated eight-color multiplexed immunofluorescence staining and high-quality imaging followed by precise laser-guided extraction of single cells. The key advance is the glycerol-based modification of heat-induced epitope retrieval protocols, termed “G-HIER.” We find that this altered antigen-retrieval solution prevents membrane distortion. Importantly, G-HIER is fully compatible with current antigen retrieval workflows and mass spectrometry–based proteomics and does not affect proteome depth or quality. To demonstrate the versatility of G-HIER for spatial proteomics, we apply the recently introduced deep visual proteomics technology to perform single-cell type analysis of adjacent suprabasal and basal keratinocytes of human skin. G-HIER overcomes previous incompatibility of standard and advanced staining protocols with membrane glass slides and enables robust integration with routine histology procedures, high-throughput multiplexed imaging, and sophisticated downstream spatial omics technologies.
Keywords: histology, glycerol, proteomics, antigen retrieval, membrane slides, laser microdissection, Deep Visual Proteomics, spatial proteomics
Graphical Abstract
Highlights
-
•
Glycerol-enhanced antigen retrieval enables membrane slide use in routine histology.
-
•
G-HIER is compatible with automated staining platforms and multiplexed imaging.
-
•
G-HIER integrates with laser microdissection and spatial omic technologies.
In Brief
Integrating spatial omics with laser microdissection has been challenging due to incompatibility between routine histology and glass membrane slides. We introduce glycerol-modified antigen retrieval. This standardizes membrane slide use in routine histology and aligns with automated staining platforms. Importantly, it enables integration of multiplexed imaging, laser microdissection, and spatial proteomic workflows, advancing spatial omic technologies towards clinical use.
Spatial omics technologies, such as genomics, transcriptomics, and proteomics (1, 2), have tremendously advanced our understanding of tissue architecture by resolving the complex space-dependent interplay of heterogenous cell types. Laser microdissection (LMD) enables accurate extraction of tissue and single cells from defined anatomical regions, thereby preserving the all-important spatial information (3). Furthermore, LMD is compatible with many downstream omics technologies, providing the opportunity for multiomics analyses from the same sample (4, 5). Glass or metal frame slides coated with a plastic membrane are essential for precise LMD tissue extraction, in particular when attempting to retrieve anatomical structures at cellular levels or even subcellular levels. There are different types of membranes including polyethylene naphthalate (PEN), polyphenylene sulfide, polyethylene terephthalate, polyester, or fluorocarbon, each with unique physical and imaging properties (www.leica-microsystems.com/science-lab/application-specific-consumables-for-laser-microdissection/#gallery-11). Despite this variety, the incompatibility of membrane slides with routine histology strategies such as standard heat-induced epitope retrieval (HIER) has been a long-standing problem in spatial omics technologies (6). Specifically, multiplex immunofluorescence (mIF) can visualize numerous cell types, adding an important layer for omics analyses in a single tissue, but is hampered by the absence of robust multiplexed staining procedures on membrane slides (7, 8). In addition, the majority of studies that use membrane slides employ manual staining techniques, forgoing the advantages of reproducible and high-throughput staining systems. We propose a solution to these unsolved challenges by introducing a novel approach for handling of glass membrane slides for routine histological procedures and mIF. Our protocol processes membrane glass slides in high throughput for LMD, without altering or affecting downstream proteomic analysis. We show that this enables highly precise microdissection at the single-cell level while preserving all spatial information, allowing us to profile cell types using our recently published Deep Visual Proteomics (DVP) (9).
Experimental Procedures
Tissue Samples
Formalin fixed and paraffin embedded (FFPE) human tissue samples were collected according to standard operating procedures. Briefly, skin specimens were stored in 5% formalin for 24 to 48 h at room temperature (RT), trimmed, and placed in embedding cassettes prior to automated processing (Tissue-TEK VIP; Sakura). Tissue specimens for the microarray block were fixed in formalin for 24 to 72 h at RT, trimmed, and manually embedded in paraffin.
Tissue Sectioning
Two micrometer PEN membrane slides (MicroDissect GmbH; MDG3P40AK) were pretreated with VECTABOND (Biozol; VEC-SP-1800) according to the manufacturer’s protocol and dried overnight at RT. FFPE tissue blocks were cooled to −17 °C at least 1 h prior to sectioning. Then, 2.5 μm sections were cut using a rotary microtome and placed onto a water bath at 37 °C. Sections were transferred to pretreated PEN membrane slides and dried overnight at 37 °C.
Visual Evaluation of Membrane Stability
PEN membrane slides were treated identically to the initial experimental procedure of the DVP staining protocol described later. After antigen retrieval, slides were washed twice with double distilled water (ddH2O) and imaged using the UV-scan mode of a gel documentation system (Axygen; GD-100) and Canon EOS 5D.
mIF Staining
Human tonsil tissue sections were mounted on regular glass slides (SuperFrost Plus) or PEN membrane glass slides. Mounted slides were heated at 56 °C for 20 min and deparaffinized (2 × 2 min xylene, 2 × 1 min 100% EtOH, 95% EtOH, 75% EtOH, 30% EtOH, and ddH20, respectively). Glycerol-supplemented heat-induced epitope-retrieval (termed “G-HIER”) was performed using preheated 1× target retrieval solution (TRS) pH 9 HIER buffer (DAKO; S2367)/10% glycerol (v/v; Sigma; G7757) in 50 ml conical tube placed in a water bath at 88 °C for 20 min. All further steps were performed according to the manufacturer’s instructions, using the commercially available 8-color immuno-oncology panel (Immuno8 FixVUE; Ultivue). This kit contained antibodies against CD3, CD4, CD8, CD68, FoxP3, PD1, PDL1, and cytokeratin (CK). Nuclear counterstain was performed with 4′,6-diamidino-2-phenylindole and used for image registration using the UltiStacker software (Ultivue). Images were acquired on a Zeiss Axioscan Z7 at 20× magnification with a 10% tile overlap.
DAKO Staining Platform
A small tissue microarray block composed of FFPE skin, appendix, cerebellum, adrenal gland, melanoma, and tonsillar tissue was sectioned and mounted on PEN slides as described previously. Next, sections were deparaffinized, rehydrated, and loaded wet on the fully automated instrument Omnis (Dako) based on dynamic gap staining technology and capillary forces. Sections were subjected to antigen retrieval using citrate buffer pH 6 (trisodium citrate dihydrate), TRS pH 9 (Dako; S2367), and TRS pH 6 (Dako; S2369) with or without 10% glycerol (Sigma–Aldrich/Merck; G7757) and heated for 60 min at 90 °C. Slides were subsequently incubated with anti-CK5 (1:200 dilution; Leica Biosystems, clone XM26, NCL-L-CK5) for 30 min at 32 °C. After washing and blocking of endogenous peroxidase activity, the reactions were detected/visualized using Envision FLEX+ High pH kit (Dako; GV800 + GV821) and Envision DAB+ Substrate Chromogen System (Dako; GV825) according to the manufacturer’s instructions. Finally, slides were rinsed in water, counterstained with Mayer’s hematoxylin, and air-dried prior to mounting.
Sample Preparation for Proteomics of Bulk Tissue Sections
Tonsil tissue that was stained as described previously was collected from corresponding regions on consecutive slides using LMD. Subsequently, samples were lysed in 300 mM Tris–HCl (pH 8.0), 12.5% acetonitrile (ACN) including 5 mM Tris(2-carboxyethyl)phosphine and 20 mM chloroacetamide for 10 min at 90 °C, followed by focused ultrasonication (Covaris; Adaptive Focused Acoustic technology) and repeated heating for 80 min at 90 °C. Samples were then further processed as described previously (10).
Deep Visual Proteomics
Immunofluorescence Staining
Mounted slides were heated at 56 °C for 20 min and deparaffinized (2 × 2 min xylene, 2× 1 min 100% EtOH, 95% EtOH, 75% EtOH, 30% EtOH, and ddH2O, respectively). G-HIER was performed using preheated 1× TRS (pH 9) HIER buffer (DAKO; S2367)/10% glycerol (v/v; Sigma; G7757) in 50 ml conical tube placed in a water bath at 88 °C for 20 min. Subsequent to sample blocking using 5% bovine serum albumin in PBS for 30 min at RT, primary antibodies targeting pan-CK (rabbit; DAKO, Z0622, 1:100 dilution) and KRT10 (mouse; Abcam, ab76318, 1:800 dilution) were incubated for 1.5 h at 37 °C in a wet staining chamber. Following two wash steps in PBS, an incubation with secondary antibodies against mouse immunoglobulin G (A647; Invitrogen, A32728, 1:400 dilution) and rabbit immunoglobulin G (A555; Invitrogen, A32732; 1:400 dilution) was performed for 1 h at 37 °C in a wet staining chamber, followed by 7 min incubation with SYTOX green Nucleic Acid Stain (Invitrogen, S7020; 1:700 dilution in ddH2O) at RT. Slides were then washed twice in ddH2O and allowed to dry briefly, before puncturing the membrane at the proximal end of the slide using a needle (30G) to eliminate the localized membrane elevation followed by subsequent adhesive sealing. Then, tissue sections were mounted with a cover glass using Slowfade Diamond Antifade Mountant (Invitrogen; S36967). After slide scanning, cover glasses were removed from membrane slides upon imaging by an incubation period of 5 to 10 min in ddH2O and air dried at RT.
Image Analysis and LMD
Artificial intelligence–guided cell recognition, classification, and extraction followed by mass spectrometry (MS)–based profiling was performed as described recently (9). In brief, fluorescence images were acquired on a Zeiss Axioscan Z7 at 20× magnification with a 10% tile overlap and analyzed using the Biology Image Analysis Software (Cell Signaling). Keratinocytes were identified with a deep neural network on the basis of pan-CK. After removal of duplicates at tile-overlapping regions, we used a supervised machine learning approach to differentially classify KRT10pos (e.g., suprabasal) from KRT10neg (e.g., basal) keratinocytes. Contour outlines were then exported along with reference points for image registration. Finally, 700 contours of each group were excised in quadruplicates on an LMD7 (Leica Microsystems). Each sample (consisting of 700 contours) was collected into a separate well of the underlying 384-well plate.
Sample Processing and MS
Sample replicates were processed following our recently published workflow (9). We used a liquid handling platform (Agilent Technologies; Bravo) to ensure reproducibility and high-throughput processing of samples. In brief, single-cell shapes of the same type were collected at the bottom of each well using centrifugation and vacuum evaporation upon the addition of ACN. Subsequently, cells were lysed in 4 μl 60 mM triethylamine bicarbonate for 60 min at 95 °C, followed by further 60 min at 75 °C in 12% (v/v) ACN. Protein digest was performed overnight using 4 ng LysC and 6 ng trypsin in a total sample volume of 7.5 μl, respectively. The enzymatic reaction was quenched in a final concentration of 1% (v/v) TFA. Subsequently, samples were loaded on Evotips Pure tips following the manufacturer’s instructions.
LC–MS/MS Analysis of Bulk and Ultra-High Sensitivity Data
Bulk samples were reconstituted in buffer A∗ (2% ACN/0.1% formic acid [FA] in LC–MS grade water). MS data were acquired by an EASY nanoLC 1200 (Thermo Fisher Scientific) coupled to a timsTOF Pro2 mass spectrometer (Bruker Daltonics) with a nano-electrospray ion source (CaptiveSpray; Bruker Daltonics). Peptide amounts were determined using a Nanodrop VIS-spectrophotometer (Thermo Fisher Scientific), and 200 ng of peptides were loaded on a 50 cm in-house packed HPLC column (75 μm inner diameter packed with 1.9 μm ReproSil-Pur C18-AQ silica beads; Dr Maisch GmbH). The column temperature was kept at 60 °C by an in-house manufactured oven. Sample analytes were separated using a linear 120 min gradient from 3 to 30% buffer B in 95 min, followed by an increase to 60% for 5 min and to 95% buffer B for 5 min, as well as a 5 min wash at 95% buffer B and re-equilibration for 5 min at 5% buffer B (buffer A: 0.1% FA/ddH2O; buffer B: 0.1% FA, 80% ACN, and 19.9% ddH2O). The flow rate was kept constant at 300 nl/min. The mass spectrometer was operated in data-dependent acquisition-parallel accumulation serial fragmentation (PASEF) mode as previously described (11). Briefly, one MS1 scan was followed by ten PASEF MS/MS scans per acquisition cycle. The ion accumulation and ramp time in the dual TIMS analyzer was 100 ms, and ion mobility (IM) range was set from 1/K0 = 1.6 V cm−2 to 0.6 V cm−2. Single charged precursor ions were excluded with a polygon filter (timsControl, version 3.0.20.0; Bruker Daltonics), and precursors for MS/MS were picked at an intensity threshold of 2500 arbitrary units and resequenced until reaching a target value of 20,000 arbitrary units considering a dynamic exclusion of 40 s elution.
For the high-sensitivity DVP samples, samples were loaded onto Evotips Pure and measured with the Evosep One LC system (Evosep) coupled to a timsTOF SCP mass spectrometer (Bruker Daltonics) employing a nano-electrospray ion source (Bruker Daltonics). The Whisper 20 samples per day method was used with the Aurora Elite CSI third generation column with 15 cm and 75 μm ID (AUR3-15075C18-CSI, IonOpticks) at 50 °C. The mobile phases comprised 0.1% FA in LC–MS grade water as buffer A and 99.9% ACN/0.1% FA as buffer B. The timsTOF was operated in data-independent acquisition (DIA)-PASEF mode with variable window widths. Optimal DIA-PASEF methods cover the precursor cloud highly efficient in the m/z–IM plane while providing deep proteome coverage. For method generation with py_diAID, the precursor density distribution in m/z and IM was estimated based on a tryptic 48 high-pH fraction library (12). We calculated the optimal cycle time based on the chromatographic peak width of 5 ng HeLa single runs. The optimal DIA-PASEF method consisted of one MS1 scan followed by 12 DIA-PASEF scans with two IM ramps per DIA-PASEF scan, covering an m/z range from 300 to 1200 and IM of 0.7 to 1.3 V cm−2. All other settings were as described previously.
Data Processing
Data acquired in data-dependent acquisition-PASEF mode were analyzed with MaxQuant (version 2.0.1.0) using standard settings in reference to UniProt human databases (UP000005640_9606.fasta containing 21,010 proteins/20,243 genes and UP000005640_9606_additional.fasta containing 58,856 proteins/15,630 genes) (13). Oxidation (M) and acetyl (protein-N-term) was set as variable and carbamidomethyl (C) as fixed modification. The maximum number of modifications per peptide was set to 3. Trypsin/P was selected as protease, and the maximum number of missed cleavages was set to 2. MS data acquired in DIA-PASEF mode were processed in the DIA-NN software (version 1.8.1, (14)) using the same fasta files and standard settings. Search parameters deviated from the standard settings as follows: peptide length range is 7 to 55, precursor m/z range is 100 to 1700, fragment ion m/z range is 100 to 1700, quantification strategy “any LC (high accuracy),” cross-run normalization “global,” with and without MBR (“match between runs”) enabled. Mass tolerance for precursor ions was determined by DIA-NN (15.049 ppm, MBR) and 15.0349 ppm (no MBR). Mass tolerance for fragment ions was determined by DIA-NN (20.1046 ppm, MBR and 20.1046 ppm, no MBR). Precursor false discovery rate (FDR) was set to 1%. The report.pg_matrix output file of DIA-NN was used for further data analysis.
Bioinformatics Data Analysis
Data analysis was performed in R, version 4.2.2. Statistical analysis of bulk data was performed with limma, version 3.52.4. For the comparisons shown, the number of significant hits (FDR <5%) was corrected for multiple testing using a Benjamini–Hochberg correction. The theoretical isoelectric point of a protein was calculated with the computePI function of seqinr v4.2-23 for every protein within one protein group. Hydrophobicity was estimated with the hydrophobicity function (scale = “KyteDoolittle”) in the package peptides, version 2.4.4. Amino acid sequences for UniProt identifiers were retrieved with the UniProt.ws package v2.36.5. For protein groups (sequences that cannot be distinguished by the underlying peptide identifications), the mean value of individual proteins is presented.
For the analysis of DVP data, differential protein expression was determined as described previously. Biological pathway enrichments were performed using the WebGestaltR package, v.0.4.4 for an overrepresentation analysis in reference to the “Reactome” database and an FDR threshold of 5% in the background of all identified proteins in the dataset (organism: Homo sapiens) (15). Spatial data from xml files were plotted with the package sf v1.0-9 as described previously (16). For the principal component analysis, data were filtered for 80% valid values in each group and imputed from left-shifted normal distribution (shift = 1.8, scale = 0.3). The FactoMineR v2.6 package was used to perform the principal component analyses. The Venn diagram and UpsetR package were used to display the number of intersections and unique proteins per condition. Visual evaluation of membrane stability was performed in Python 3.9.12 using the NumPy, Matplotlib, and Scikit-image packages. Intensity values of the acquired images were normalized to the median intensity of each image and then plotted as heatmaps using the “viridis” colormap, indicating intensity values.
Ethics Approval and Patient Consent Statement
Skin tissue sections were collected following informed consent and ethical approval (EK 22-0343). All experiments were performed in accordance with the Declaration of Helsinki. Regarding the tissue microarrays, according to Danish Data Protection Act (2018), they are exempted from patient consent and permission from legal authorities for fully anonymized material.
Results
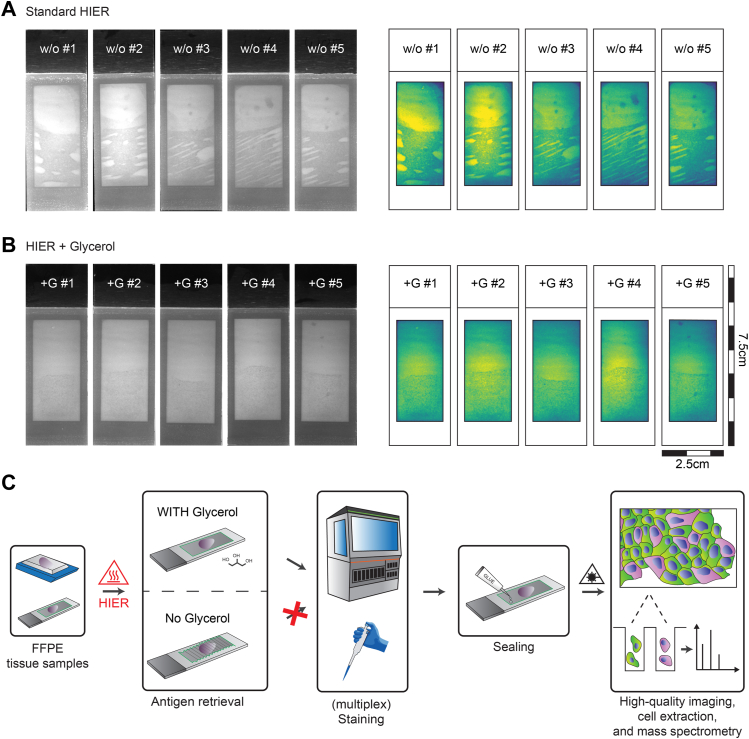
Combining digital pathology with proteomics using LMD is an area of intense interest. However, there are many practical challenges to overcome in achieving this. In our initial work with PEN membranes, we regularly observed substantial disintegration of the membrane and formation of air pockets at random locations underneath during standard HIER protocols. This renders the majority of slides incompatible with routine histological staining workflows, scanning, and potential downstream LMD procedures (Fig. 1A and supplemental Fig. S1A). To address this issue, we aimed at developing a robust membrane-compatible HIER protocol. Building on the observation that glycerol in HIER buffers enhances antigen retrieval (17), we tested the ability of glycerol in membrane stabilization. Indeed, we found that membrane integrity was preserved when using glycerol-adapted HIER, while fully eliminating the formation of random membrane distortions (Fig. 1B and supplemental Fig. S1B). Following G-HIER, tissue sections could easily be processed for staining and high-quality imaging, even in a single focal plane. For very large tissue sections, we additionally punctured the membrane at its proximal end with a syringe to remove any potentially remaining irregularities and subsequently resealed it (supplemental Fig. S1C). With this in place, G-HIER allows high-resolution and rapid whole-slide imaging in a single focal plane, minimizing the amount of data storage necessary and reducing stitching errors that are detrimental for subsequent LMD. Having optimized HIER on membrane slides for routine histology workflows, we tested its compatibility with multiplexed imaging, automated staining device compatibility (Dako Omnis; Agilent), and downstream MS-based proteomics (Fig. 1C).
Fig. 1.
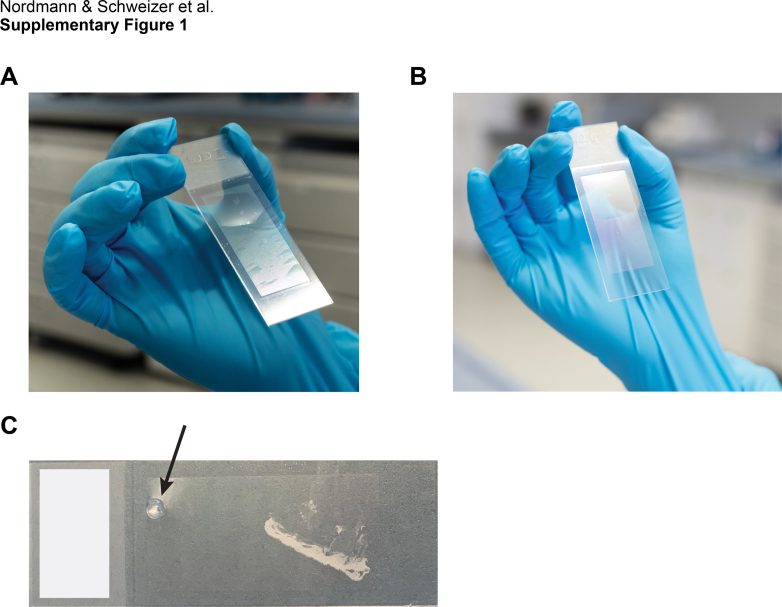
A robust workflow for heat-induced epitope retrieval (HIER)on membrane microscopy slides.A, membrane distortion resulting from a standard HIER protocol shown as UV scans (left) and heatmap of normalized median UV signal intensities across slides. B, intact membrane stability using glycerol-supplemented antigen retrieval in the same arrangement. C, workflow schematic for the preparation of LMD-compatible membrane slides. FFPE archival tissue was mounted on 2 μm PEN membrane slides as in routine pathology followed by standard HIER without and with 10% glycerol. While the slides without glycerol were unusable for further staining procedures upon heating in solution, the presence of glycerol stabilized the membrane. Minor accumulation of gas below the membrane was removed using minimal invasive methods followed by glue-based sealing. FFPE, formalin fixed and paraffin embedded; +G, HIER buffer supplemented with 10% glycerol; LMD, laser microdissection; PEN, polyethylene naphthalate; w/o, without glycerol.
Multiplexed imaging enables the spatial characterization of diverse cellular population within the tissue microenvironment and has become increasingly popular in the scientific community. However, to our knowledge, it has not been performed on glass membrane slides and accordingly has not been combined with downstream LMD-based spatial omics technologies. To test the compatibility of our novel antigen retrieval method with membrane slides in combination with multiplexed imaging, we stained tonsil tissue with a commercially available 8-color immuno-oncology panel (Immuno8 FixVUE; Ultivue). Without further optimization, G-HIER resulted in high-quality images on PEN membrane slides and showed equal performance with all antibodies compared with regular glass slides (Fig. 2). Importantly, our staining unambiguously visualized the different cell types of the panel while also clearly highlighting cell boundaries required for automated cell segmentation. These results show that G-HIER enables multiplexed imaging on glass membrane slides, and both are seamlessly compatible with glass membrane slides.
Fig. 2.
Comparison of multiplex immunofluorescence imaging in human tonsil tissue. Eight-color multiplex immunofluorescence imaging (Immuno8 FixVUE; Ultivue) was performed side by side, in tissue mounted on regular (A) or PEN-membrane glass slides (B). Apart from G-HIER at 88 °C, all other steps were performed as recommended by the manufacturer. The following markers were used: CD8 (orange), PD1 (red), PDL1 (turquoise), CD68 (white), CD3 (blue), CD4 (green), FoxP3 (purple), and CK (pink). G-HIER, glycerol-supplemented heat-induced epitope retrieval; PEN, polyethylene naphthalate.
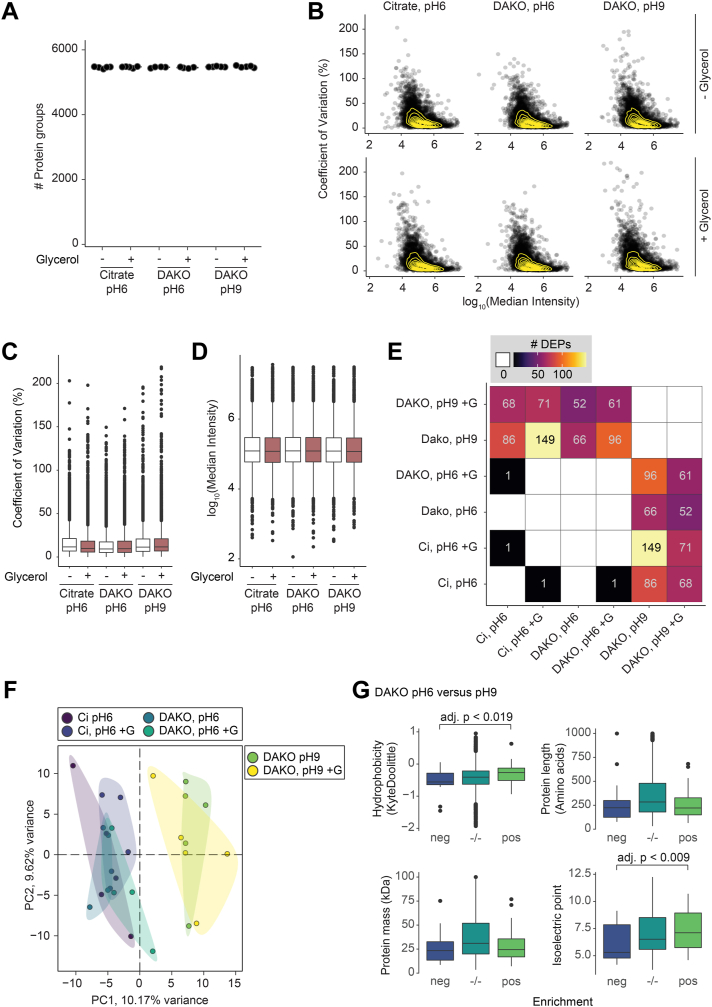
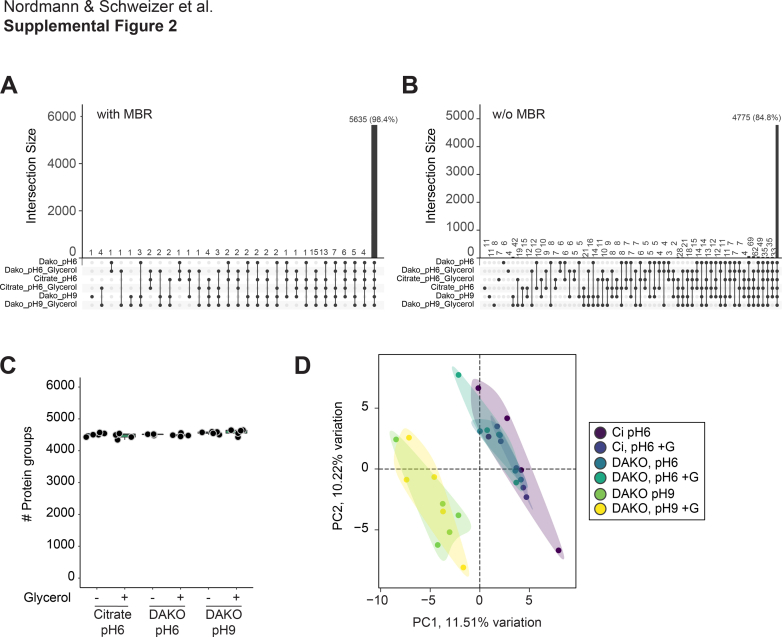
Next, we evaluated the impact of G-HIER on protein identification using MS-based proteomics. We compared the proteome depth and information content of FFPE tonsil tissue samples after antigen retrieval with six different buffer compositions: 10 mM citrate buffer at pH 6, and commercially available DAKO buffer at pH 6 and pH 9, with or without addition of 10% glycerol, respectively. To ensure a robust and efficient experimental process, we directly integrated G-HIER and CK5 staining of FFPE tissue sections mounted on PEN-membrane slides on an automated staining platform (DAKO Omnis), followed by LMD and MS. We found that membrane glass slides were fully compatible with an automated staining device when using G-HIER. Proteomic analysis revealed identical numbers of identified proteins across all buffer conditions, at 5468 ± 12 proteins (mean ± SD, Fig. 3A). In our experience, such proteome depth provides substantial coverage of most biological pathways. Likewise, distribution and median of MS intensity per protein and the coefficient of variation as a measure of variability between replicates were comparable, demonstrating that glycerol has no negative impact on proteomic data acquisition (Fig. 3, B–D). The degree of common identifications and the distinct separation between treatment groups was not affected by the “MBR” algorithm of the data processing software, despite slightly lower protein numbers (supplemental Fig. S2, A–D). Notably, in our comparison of different antigen retrieval conditions, we only found a difference because of the pH of the HIER buffer, while addition of glycerol was not a main driver of sample separation along principal component 1 and 2 (Fig. 3, E and F). We speculated that the pH could affect solubility of proteins depending on their physiochemical properties. Indeed, we found that the theoretical isoelectric point and hydrophobicity, rather than protein length or mass, affected which proteins were retained upon HIER at pH 6 compared with pH 9 (Fig. 3G). Although this does not affect G-HIER, it can be taken into consideration when establishing the staining of particular challenging protein targets. Thus, our optimized workflow efficiently combines HIER on membrane slides with downstream proteomics and automated staining techniques, without compromising reproducibility and MS data depth.
Fig. 3.
Effect of glycerol in various antigen retrieval solutions on the bulk proteome of tissue slices.A, number of proteins detected in laser-dissected tonsil tissue after LC–MS/MS. Points are individual samples, the line represents the median within one group (n > 4). B, log-transformed MS signal versus coefficient of variation (%) per solution type. Points are median values of individual proteins per group, density is estimated by yellow lines. C, coefficient of variation and (D) log10-transformed MS signal as in (B) per group (n > 4). E, number of differentially expressed proteins (multiple-testing adjusted p value <0.05) between indicated comparisons, additionally illustrated with a color gradient. White and empty indicates that no significant hits were found. F, principal component analysis of all included samples (n = 28). Colors denote groups with different retrieval solutions. The amount of variation explained per principal component is given with the axis legend. G, comparison of proteome composition after heat-induced antigen retrieval with two solutions at pH 6 or pH 9 (without glycerol). Hydrophobicity and isoelectric point are calculated values. For protein groups, the median value of each contributing protein is shown (n > 4). Boxplots represent median and the 25th and 75th percentiles, and whiskers span the 1.5-fold interquartile range. Outliers are shown, if not indicated differently. adj. p., adjusted p value; Ci, citrate; DEP, differentially expressed proteins; +G, respective buffer supplemented with 10% glycerol; MS, mass spectrometry.
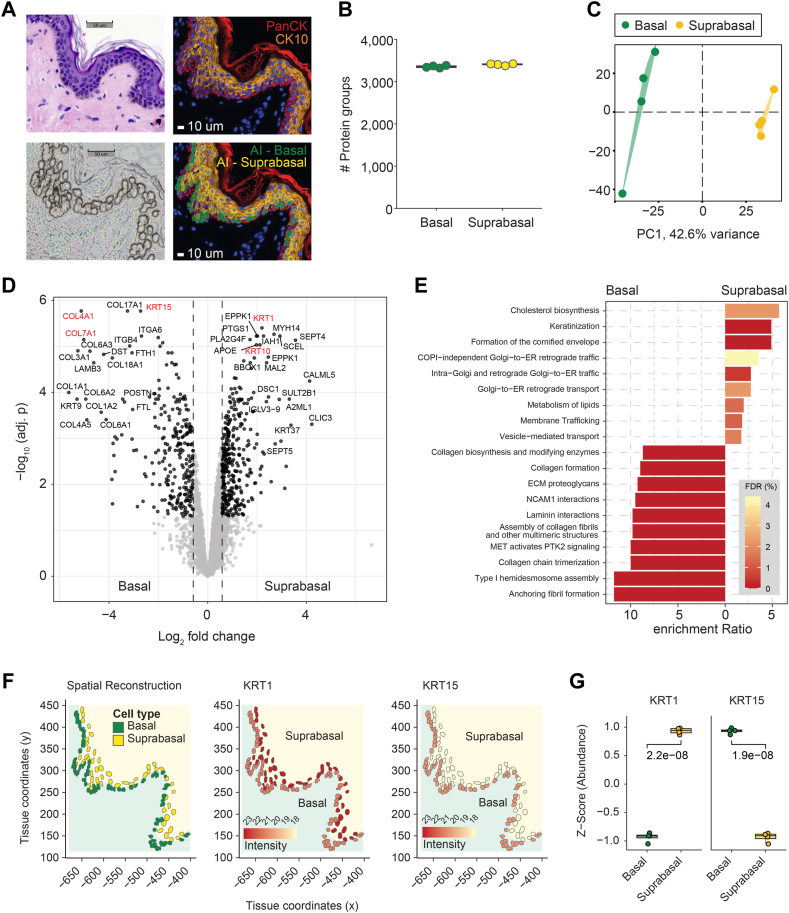
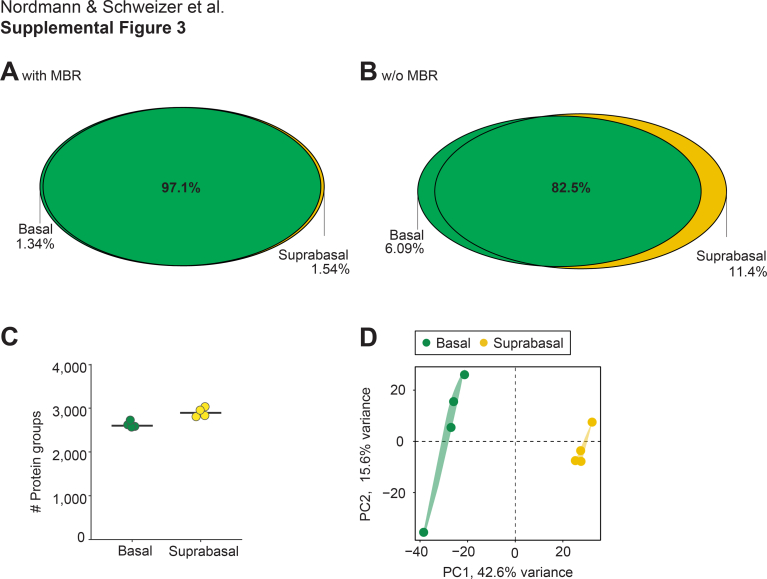
With a reliable protocol in hand for multiplexed imaging on glass membrane slides that preserves the potential of downstream proteomic analysis, we next aimed to demonstrate its applicability to the field of spatial proteomics. For this, we integrated G-HIER into the recently developed DVP workflow. Using G-HIER, we performed immunofluorescence for pan-CK and CK10 on a 3 μm thin FFPE skin section (10). Despite the inherent challenges of handling skin tissue sections during harsh histological conditions, we were able to successfully perform the entire staining procedure (including antibody titration experiments) on glass membrane slides without any signs of tissue detachment. This allowed us to profile adjacent epidermal cell types of healthy human skin, yielding spatially resolved proteomic profiles. Artificial intelligence–based algorithms segmented each cell on the basis of pan-CK and further classified it into basal and suprabasal cells by the presence or the absence of CK10 (Fig. 4A). Laser-assisted cell extraction and subsequent MS enabled clear differentiation between the cell types based on the identification and quantification of more than 3500 proteins across all samples (Fig. 4, B and C). Again, the degree of common identifications and the distinct separation between biological groups was not affected by the “MBR” algorithm (supplemental Fig. S3, A–D). Between the two cell types, 17% of the proteome was profoundly differentially expressed (minimum absolute fold change: 1.5, adjusted p value <0.05, Fig. 4D). CK10 (KRT10), the initial staining target of our study, and KRT1 were upregulated in suprabasal cells, providing positive controls. In the basal cell layer, common markers of basal keratinocytes such as CK15 (KRT15) and basal membrane anchorage fibrils COL4A1 and COL7A1 were highly enriched. Summarizing the proteomics results by a biological pathway enrichment analysis, processes such as “keratinization” and “metabolism of lipids” reflected the formation of the epidermal barrier in the suprabasal layer, whereas “type I hemidesmosome assembly” and other cell–cell interactions indicated structural anchoring of cells in the basal layer (Fig. 4E). We next took advantage of the integration of spatial and proteomic information to reconstruct the cellular architecture of the skin specimen, mapping the mean protein intensities of KRT1 and KRT15 into the cellular environment (Fig. 4, F and G). These results demonstrate the robust nature of G-HIER and DVP and how this combination enables the cell type–resolved characterization of human skin by MS-based proteomics while preserving spatial information.
Fig. 4.
Cell type–resolved spatial proteomics of the human skin.A, exemplary region of skin specimen shown in H&E (upper left), IF for pan-CK and CK10 (upper right), cell segmentation and classification by artificial intelligence (lower right), and subsequent to cell extraction by laser capture microdissection (lower left). B, number of identified protein groups for basal and suprabasal cells of the skin. C, principal component analysis separating skin cell types in the first two dimensions. D, differential protein expression of cell types. Significant proteins (minimum fold change: 1.5, adjusted p value <0.05) are highlighted in black, proof-of-principle proteins are shown in red. E, biological pathway enrichment analysis based on the “pathway Reactome” database. F, in silico spatial reconstruction of the tissue architecture showing the localization of basal and suprabasal cells in an exemplary tissue region (left panel). Enabled by DVP, mean protein intensities for the representative markers KRT1 (middle panel) and KRT15 (right panel) were mapped onto the tissue architecture. G, normalized abundances for the previously shown markers in each cell type. An unpaired Student’s t test was used to annotate the significance for each comparison. adj. p.: adjusted p value; CK, cytokeratin; DVP, Deep Visual Proteomics; IF, immunofluorescence.
Discussion
HIER workflows have not traditionally been compatible with glass membrane slides for LMD. This long-standing issue has limited the integration of LMD-based approaches with current and future technologies, such as automated staining platforms, mIF imaging, and visual omics approaches.
To address these challenges, we here establish G-HIER. This optimized technique effectively stabilizes the membrane during antigen retrieval and enables subsequent high-throughput staining and LMD. Our protocol drew inspiration from the previous finding that the addition of 10% glycerol to the antigen retrieval process greatly enhances the quality of immunostainings in FFPE tissue sections (17). This earlier study highlighted the potential benefits of glycerol in providing a universal antigen retrieval buffer that enables excellent staining properties for a large variety of epitopes. Our own findings align with these results. Beyond this advantage, we here expanded the benefits of glycerol to the favorable impact on PEN membrane slides during the antigen retrieval process. We found that our optimized workflow drastically enhances the quality of antibody-based staining results on PEN glass slides, enabling their standard use in routine pathology and even multiplexed staining procedures. In addition, fully automated staining platforms such as the Dako Omnis can now be integrated seamlessly to further increase throughput and reproducibility, while still matching the requirements for high-throughput tissue extraction via laser-capture microdissection. Furthermore, our workflow is fully compatible with downstream MS-based proteomics analysis and provides reproducible and in-depth acquisition of the proteome.
These advancements are a crucial step toward robust single-cell or cell type–specific proteomics workflows. This is especially important, when working with extremely sparse material (16). Indeed, clinical material is often precious and scarce. In this context, G-HIER provides a standardized framework for efficient and lossless use of the few sections that may be available for research purposes. We demonstrate this powerful combination by differentiating between basal and suprabasal cells of the human skin, but this can be transferred to any cell type of interest. We have now standardized on G-HIER for all applications of our novel DVP technology and are evaluating the routine combination of automated staining with G-HIER.
In summary, G-HIER now enables routine use of glass membrane slides in standard staining procedures and more complex mIF imaging, which in turn can readily be coupled to LMD-assisted tissue extraction. G-HIER broadens applications of glass membrane slides in pathology and enables their streamlined use in the growing field of spatial omics technologies.
Data Availability
MS data of this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD040281 (preliminary username: reviewer_pxd040281@ebi.ac.uk, password: NpBxbVb9).
Supplemental data
This article contains supplemental data.
Conflict of interest
M. M. is an indirect investor in Evosep Biosciences. All other authors declare no competing interests.
Acknowledgments
We thank members of the department of Proteomics and Signal Transduction for help and fruitful discussions and especially Katharina Zettl for superbly performing the immunofluorescence staining and Dirk Wischnewski for meticulous sample preparations. Susanne Vondenbusch-Teetz is acknowledged for the scientific implementation of her photography skills. We thank Peter Horvath and Single Cell Technologies Ltd for their technical support and Fabian Coscia for valuable feedback. This study has further been supported by the Max-Planck Society for Advancement of Science, the Federal Ministry of Education and Research (BMBF) through project CLINSPECT-M (16LW0243K), the Chan Zuckerberg Initiative (CZF2019-002448), and by grants from the Novo Nordisk Foundation, Denmark (grant agreements: NNF14CC0001 and NNF15CC0001).
Funding and additional information
T. M. N. is supported by a Swiss National Science Foundation Early Postdoc Mobility fellowship (P 2ZHP3-199648), and F. A. R. is an EMBO postdoctoral fellow (ALTF 399-2021).
Author contributions
F. A. R. original idea; T. M. N., L. S., and F. A. R. conceptualization; T. M. N., L. S., E. R., and F. A. R. formal analysis; T. M. N., L. S., Andreas Metousis., M. T., E. R., L. M. R.-G., P.-C. S., M. B., and F. A. R. investigation; T. M. N., L. S., F. A. R., and M. M. writing–original draft; Andreas Mund and M. M. supervision.
Contributor Information
Florian A. Rosenberger, Email: rosenberger@biochem.mpg.de.
Matthias Mann, Email: mmann@biochem.mpg.de.
Supplemental Data
Supplemental Figure S1.
Supplemental Figure S2.
Supplemental Figure S3.
Comparison of unique and intersecting proteins between both groups of the DVP experiment in human skin (A) with and (B) without match between runs (MBR). C, number of identified protein groups for basal and suprabasal cells of the skin. Points are individual samples, and the line represents the median within one group (n = 4). D, principal component analysis separating skin cell types in the first two dimensions. The amount of variation explained per principal component is given with the axis legend.
References
- 1.Zhao T., Chiang Z.D., Morriss J.W., LaFave L.M., Murray E.M., Del Priore I., et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature. 2022;601:85–91. doi: 10.1038/s41586-021-04217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nirmal A.J., Maliga Z., Vallius T., Quattrochi B., Chen A.A., Jacobson C.A., et al. The spatial landscape of progression and immunoediting in primary melanoma at single-cell resolution. Cancer Discov. 2022;12:1518–1541. doi: 10.1158/2159-8290.CD-21-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Espina V., Wulfkuhle J.D., Calvert V.S., VanMeter A., Zhou W., Coukos G., et al. Laser-capture microdissection. Nat. Protoc. 2006;1:586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 4.Shen S., Li J., Huo S., Ma M., Zhu X., Rasam S., et al. Parallel, high-quality proteomic and targeted metabolomic quantification using laser capture microdissected tissues. Anal. Chem. 2021;93:8711–8718. doi: 10.1021/acs.analchem.1c01026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichterwitz S., Chen G., Aguila Benitez J., Yilmaz M., Storvall H., Cao M., et al. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat. Commun. 2016;7 doi: 10.1038/ncomms12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gjerdrum L.M., Lielpetere I., Rasmussen L.M., Bendix K., Hamilton-Dutoit S. Laser-assisted microdissection of membrane-mounted paraffin sections for polymerase chain reaction analysis: identification of cell populations using immunohistochemistry and in situ hybridization. J. Mol. Diagn. 2001;3:105–110. doi: 10.1016/S1525-1578(10)60659-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hickey J.W., Neumann E.K., Radtke A.J., Camarillo J.M., Beuschel R.T., Albanese A., et al. Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging. Nat. Methods. 2022;19:284–295. doi: 10.1038/s41592-021-01316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan W.C.C., Nerurkar S.N., Cai H.Y., Ng H.H.M., Wu D., Wee Y.T.F., et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun. (Lond.) 2020;40:135–153. doi: 10.1002/cac2.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mund A., Coscia F., Kriston A., Hollandi R., Kovacs F., Brunner A.D., et al. Deep Visual Proteomics defines single-cell identity and heterogeneity. Nat. Biotechnol. 2022;40:1231–1240. doi: 10.1038/s41587-022-01302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coscia F., Doll S., Bech J.M., Schweizer L., Mund A., Lengyel E., et al. A streamlined mass spectrometry-based proteomics workflow for large-scale FFPE tissue analysis. J. Pathol. 2020;251:100–112. doi: 10.1002/path.5420. [DOI] [PubMed] [Google Scholar]
- 11.Brunner A.D., Thielert M., Vasilopoulou C., Ammar C., Coscia F., Mund A., et al. Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation. Mol. Syst. Biol. 2022;18 doi: 10.15252/msb.202110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skowronek P., Thielert M., Voytik E., Tanzer M.C., Hansen F.M., Willems S., et al. Rapid and in-depth coverage of the (phospho-)proteome with deep libraries and optimal window design for dia-PASEF. Mol. Cell. Proteomics. 2022;21 doi: 10.1016/j.mcpro.2022.100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 14.Demichev V., Messner C.B., Vernardis S.I., Lilley K.S., Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods. 2020;17:41–44. doi: 10.1038/s41592-019-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberger F.A., Thielert M., Strauss M.T., Ammar C., Mädler S.C., Schweizer L., et al. Spatial single-cell mass spectrometry defines zonation of the hepatocyte proteome. bioRxiv. 2022 doi: 10.1101/2022.12.03.518957. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sorrelle N., Ganguly D., Dominguez A.T.A., Zhang Y., Huang H., Dahal L.N., et al. Improved multiplex immunohistochemistry for immune microenvironment evaluation of mouse formalin-fixed, paraffin-embedded tissues. J. Immunol. 2019;202:292–299. doi: 10.4049/jimmunol.1800878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS data of this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD040281 (preliminary username: reviewer_pxd040281@ebi.ac.uk, password: NpBxbVb9).