Abstract
Fifteen isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamases (ESBLs) isolated during a nosocomial outbreak were studied. The strains belonged to the same clonal type, as shown by pulsed-field gel electrophoretic analysis of chromosomal DNA. All the isolates were resistant to extended-spectrum cephalosporins, aztreonam, gentamicin, and fluoroquinolones and were susceptible to carbapenems, tobramycin, netilmicin, and amikacin. None of the isolates expressed the OmpK36 porin. Eight isolates, for which the MICs of cefoxitin were ≥64 μg/ml, showed a diminished level or no expression of a 35-kDa porin. The MICs of meropenem, cefotaxime, and cefpirome were three to eight times higher for porin-deficient isolates than for isolates expressing the 35-kDa porin, but the MICs of imipenem increased two times for porin-deficient isolates compared to those for isolates expressing the porin. This MIC increase reverted to a level similar to that for the parental strain when porin-deficient isolates were transformed with the gene coding for the K. pneumoniae porin OmpK36. It is concluded that the high level of resistance to cefoxitin and the increase in the MICs of meropenem, cefotaxime, and cefpirome for the ESBL-producing K. pneumoniae isolates studied are associated with porin deficiency.
Klebsiella pneumoniae is an important human pathogen that has been associated in recent decades with nosocomial outbreaks. After the use of extended-spectrum cephalosporins, extended-spectrum β-lactamase (ESBL)-producing K. pneumoniae has become an increasingly serious problem worldwide (3, 11, 12, 25). This class of β-lactamases consists of plasmid-mediated enzymes that are able to hydrolyze expanded-spectrum cephalosporins and monobactams. In K. pneumoniae cefoxitin resistance may be due to β-lactamase production (7, 24) or the loss of porins (15, 23, 30).
Porins are outer membrane proteins (OMPs) that allow the nonspecific diffusion of small molecules into the bacterial cell. Most of the studies about OMPs have been carried out with Escherichia coli, in which two major porins (OmpC and OmpF) have been characterized. Loss of either of them has been related to antibiotic resistance (21). Decreased permeability can produce significant levels of resistance that may be increased when it is combined with enzymatic inactivation (21). In K. pneumoniae, two main porins have been characterized: OmpK35 (the homolog of OmpF) and OmpK36 (the homolog of OmpC) (1, 10). Recently, loss of the OmpK36 porin has been associated with both cefoxitin resistance and increases in cephalosporin and quinolone MICs (15). The association between the loss of porins and increased MICs of carbapenems has recently been described for K. pneumoniae producing a plasmid-mediated AmpC-like β-lactamase (2, 16). Expression of OmpK36 and/or inactivation of AmpC abolished carbapenem resistance in this particular type of strain (16).
From May 1993 to June 1995, a nosocomial outbreak due to K. pneumoniae producing ESBL involved 150 patients in our hospital (25). During the outbreak, 4% of the ESBL-producing K. pneumoniae isolates showed high levels of resistance to cefoxitin (MIC, >64 μg/ml). The aim of this study was to analyze the mechanism of cefoxitin resistance among these strains.
MATERIALS AND METHODS
Bacterial isolates.
Fifteen strains of ESBL-producing K. pneumoniae isolated from 12 colonized or infected patients during the outbreak period were studied. Eight of them were highly cefoxitin resistant (MICs, ≥64 μg/ml), for four strains cefoxitin MICs were between 16 and 32 μg/ml, and the remaining three strains were cefoxitin susceptible (MICs, between 2 and 4 μg/ml). These last three isolates were recovered together with highly resistant isolates from a pharyngeal swab, catheter, and blood of three patients, respectively.
Susceptibility studies.
MICs were determined by the microdilution method (19) and the E-test (AB Biodisk, Solna, Sweden). The following antibiotics were tested: amikacin, amoxicillin, amoxicillin-clavulanic acid (2:1), aztreonam, cefotaxime, cefoxitin, cefoxitin-clavulanic acid (2:1), cefpirome, ceftazidime, ceftazidime-clavulanic acid (2:1), ciprofloxacin, gentamicin, imipenem, meropenem, netilmicin, ofloxacin, piperacillin, piperacillin-tazobactam (2:1), sparfloxacin, and tobramycin.
Antimicrobial susceptibility tests with strains CSUB10R(pSHA19) and CSUB10R(pSHA20) (see below) were performed in Mueller-Hinton broth (Izasa, Barcelona, Spain) supplemented with 50 μg of kanamycin (Sigma, Madrid, Spain) per ml and 25 μg of chloramphenicol (Sigma) per ml.
The presence of broad extended-spectrum β-lactamase production was studied by the double-disk synergy test (14) and by the E-test method (AB Biodisk).
Conjugation experiments.
Transfer of resistance to expanded-spectrum cephalosporins and monobactams from K. pneumoniae CSUB10S and CSUB10R to E. coli J53-2 was carried out by conjugation in broth as described previously (15). Ampicillin and rifampin (100 μg/ml each) were used as selective agents.
Isoelectric focusing.
Strains were grown for 4 h in Luria broth. The growing bacteria were pelleted, resuspended in distilled water, and sonicated. Extract purifications were performed by ultracentrifugation (14). Isoelectric focusing of β-lactamase extracts was done with the PhastSystem apparatus (Pharmacia, Uppsala, Sweden) in polyacrylamide gels with a pH range of 3 to 9 (PhastGel 3-9; Pharmacia). The gels were stained with 500 μg nitrocefin (Oxoid, Hampshire, England) per ml, and pIs were determined by comparison with different β-lactamases with known pIs.
Typing methods.
Biotyping was carried out with API 20E galleries (bioMérieux, Balmes les Grottes, France) according to the manufacturer’s instructions and with MicroScan NegCombo 6I panels (DADE International, Inc., West Sacramento, Calif.).
Macrorestriction analysis of chromosomal DNA was done by pulsed-field gel electrophoresis (PFGE) by previously described procedures (8). DNA restriction was done with XbaI (New England Biolabs, Madrid, Spain) following the manufacturer’s recommendations. PFGE was performed in a CHEF-DR III apparatus (Bio-Rad, Hercules, Calif.) for 23 h at 14°C with pulse times ranging from 1 to 30 s at 6 V/cm.
OMP isolation and analysis.
For porin isolation, we used a combination of the two methods for the isolation of E. coli porins (20, 22). Cell envelopes were treated with trypsin and subjected to differential solubilization as described in detail by Alberti et al. (1). The isolated porins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and were electrophoretically transferred to Immobilon P membranes (Millipore, Barcelona, Spain) by using the buffers and conditions described by Towbin et al. (29). The membranes were stained with Ponceau red, and bands of interest were excised separately, destained, and sequenced in an Applied Biosystems 470 gas-phase sequencer (kindly done by the Servicio de Secuenciación of the Centro de Investigaciones Biológicas del Consejo Superior de Investigaciones Cientificas, Madrid, Spain).
K. pneumoniae strains were grown in Mueller-Hinton broth and sonicated, and cell envelopes were recovered by ultracentrifugation. After treatment with sodium N-lauroyl sarcosinate (Sigma, Madrid, Spain), the OMPs were collected by ultracentrifugation (15). Electrophoretic analysis of OMP by SDS-PAGE was performed in 11% acrylamide–0.35% bisacrylamide–0.1% SDS by using Laemmli’s buffers. The samples were boiled for 5 min in Laemmli’s buffer before electrophoresis. The gels were stained with Coomassie blue.
Transfer and expression of ompK36 gene.
Plasmid pSUV7, containing the gene coding for the OmpK36 porin, and plasmid pFR167, containing a truncated ompK36 gene, have been described previously (15). These plasmids include a kanamycin resistance cassette to allow their selection in the multidrug-resistant background of strain CSUB10R. Briefly, the kanamycin resistance cassette of plasmid pCSI2 (6) was obtained as an XbaI-XbaI fragment and was cloned into the unique XbaI sites of plasmids pSUV7 and pFR167. The result was plasmid pSHA19, which contains the ompK36 gene, and plasmid pSHA20, which contains a truncated ompK36 gene. The modified plasmids were introduced by electroporation into strain CSUB10R, and the transformed CSUB10R strains carrying the cloned porin genes were selected as kanamycin-resistant strains. DNA isolation, enzyme restrictions, and ligation were performed by standard procedures (28).
RESULTS
Susceptibility testing.
All the strains tested were resistant to amoxicillin (MIC, >256 μg/ml) and piperacillin (MICs, 128 to >256 μg/ml). The amoxicillin-clavulanic acid MICs ranged from 4 to 16 μg/ml, whereas the piperacillin-tazobactam MIC range was from 2 to >256 μg/ml. For cefoxitin-resistant strains, the addition of clavulanic acid did not result in a reversion to cefoxitin susceptibility. Amikacin (MIC, 2 μg/ml), tobramycin (MIC range, 0.5 to 1 μg/ml), and netilmicin (MIC range, 0.5 to 2 μg/ml) were active against all K. pneumoniae isolates. All the isolates were resistant to gentamicin (MIC range, 8 to 16 μg/ml).
Table 1 presents the MICs of 11 antibiotics for cefoxitin-susceptible and -resistant isolates cultured from the same patient and the MICs of these antibiotics for CSUB10R(pSHA19) and CSUB10R(pSHA20) containing the entire and truncated ompK36 porin genes, respectively. For cefoxitin-resistant isolates, the MICs of cefotaxime (3 to 5 dilution steps), cefpirome (5 to 7 dilution steps), meropenem (5 to 6 dilution steps), imipenem (2 dilution steps), and ceftazidime-clavulanic acid (2 dilution steps) were higher than those for the cefoxitin-susceptible strains. The quinolone MICs for cefoxitin-resistant strains were always from 1 to 3 dilution steps higher than those for cefoxitin-susceptible strains. For strain CSUB10R, the cefoxitin resistance and the increased MICs of meropenem, cefotaxime, cefpirome, and ceftazidime-clavulanic acid reverted to MICs which were similar to those for strain CSUB10S after cloning of the ompK36 gene into the strain [strain CSUB10R(pSHA19)].
TABLE 1.
Characteristics of cefoxitin-resistant and -susceptible ESBL-producing K. pneumoniae pairs isolated from three patients and the transformed strains of CSUB10R containing plasmids coding for the entire (pSHA19) or truncated (pSHA20) sequence of OmpK36 porin
| Antimicrobial agent | MIC (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Patient 1
|
CSUB10R(pSHA19) | CSUB10R(pSHA20) | Patient 2
|
Patient 3
|
||||
| CSUB10R | CSUB10S | CSUB8R | CSUB8S | CSUB9R | CSUB9S | |||
| Cefoxitin | 128 | 2 | 0.6 | 128 | 128 | 4 | 128 | 4 |
| Cefotaxime | >256 | 4 | 32 | >256 | >256 | 64 | 256 | 4 |
| Cefpirome | >256 | 2 | 8 | >256 | 256 | 8 | 128 | 2 |
| Ceftazidime | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| Ceftazidime-clavulanic acid | 2 | 0.5 | 0.5 | 2 | 4 | 1 | 2 | 0.5 |
| Aztreonam | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| Imipenem | 0.5 | 0.12 | 0.06 | 0.5 | 0.5 | 0.12 | 0.5 | 0.12 |
| Meropenem | 2 | 0.06 | 0.12 | 2 | 2 | 0.03 | 2 | 0.03 |
| Ofloxacin | 4 | 2 | 0.5 | 4 | 4 | 2 | 8 | 4 |
| Ciprofloxacin | 4 | 0.5 | 0.5 | 4 | 2 | 1 | 2 | 2 |
| Sparfloxacin | 4 | 1 | 0.25 | 4 | 2 | 1 | 2 | 1 |
| Porin expression | − | + | + | − | − | + | − | + |
Table 2 presents the MICs of 11 antibiotics for K. pneumoniae isolates with different degrees of resistance to cefoxitin. For isolates for which the cefoxitin MIC was ≥64 μg/ml, the cefpirome and cefotaxime MICs increased and the meropenem MIC was 4 to 32 times higher than those for strains for which cefoxitin MICs were <64 μg/ml. The MICs of norfloxacin (MIC range, 64 to 128 μg/ml), ofloxacin (MIC range, 4 to 8 μg/ml), ciprofloxacin (MIC range, 2 to 8 μg/ml), or sparfloxacin (MIC range, 1 to 4 μg/ml) cannot be related to the degree of cefoxitin resistance.
TABLE 2.
Characteristics of nine K. pneumoniae strains with different degrees of resistance to cefoxitin isolated from nine patients
| Antimicrobial agent | MIC (μg/ml)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CSUB3 | CSUB7 | CSUB1 | CSUB6 | CSUB4 | CSUB5 | CSUB2 | CSUB11 | CSUB12 | |
| Cefoxitin | 16 | 16 | 32 | 32 | 64 | 64 | 128 | 128 | 128 |
| Cefotaxime | 0.25 | 4 | 2 | 2 | 128 | 64 | 4 | >256 | 32 |
| Cefpirome | 4 | 2 | 1 | 2 | 64 | 32 | 16 | >256 | 32 |
| Ceftazidime | 16 | 64 | 4 | 128 | >256 | 64 | 8 | >256 | >256 |
| Ceftazidime-clavulanic acid | 1 | 1 | 0.5 | 2 | 2 | 1 | 0.5 | 4 | 2 |
| Aztreonam | 1 | 128 | 1 | 128 | 32 | 16 | 2 | >256 | >256 |
| Imipenem | 0.12 | 0.12 | 0.06 | 0.12 | 0.25 | 0.12 | 0.12 | 0.5 | 0.5 |
| Meropenem | 0.03 | 0.03 | 0.03 | 0.03 | 0.12 | 0.12 | 0.5 | 2 | 1 |
| Ofloxacin | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 4 | 4 |
| Ciprofloxacin | 4 | 4 | 4 | 8 | 4 | 4 | 8 | 2 | 2 |
| Sparfloxacin | 4 | 2 | 4 | 4 | 4 | 2 | 4 | 1 | 1 |
| Porin expression | + | + | + | + | ±a | ± | − | − | − |
±, reduced level of porin expression.
E. coli transconjugants, with K. pneumoniae CSUB10R or CSUB10S used as donors, were resistant to expanded-spectrum cephalosporins as a result of ESBL production. Cefoxitin resistance and increased carbapenem MICs were not transferred to E. coli by conjugation of the donor.
β-Lactamase study.
The production of ESBLs was demonstrated in all the strains by the double-disk synergy test and by at least a threefold reduction in the ceftazidime MIC when clavulanic acid was added. Isoelectric focusing of β-lactamase extracts showed that all but one of the isolates produced a single enzyme: nine produced an enzyme with a pI of 8.2, and five produced an enzyme with a pI of 7.6. The remaining isolate produced two β-lactamases with pIs of 7.6 and 8.2. No relationship between pI and the degree of cefoxitin resistance was found. The cefoxitin-resistant and -susceptible strains isolated from the same patients had β-lactamases with identical pIs. Aztreonam MICs showed variations according to the β-lactamase pI. For K. pneumoniae isolates producing a β-lactamase with a pI of 8.2 the aztreonam MIC (MIC range, 128 to >256 μg/ml) was higher than that for those producing a β-lactamase with a pI of 7.6 (MIC range, 0.5 to 32 μg/ml).
Typing methods.
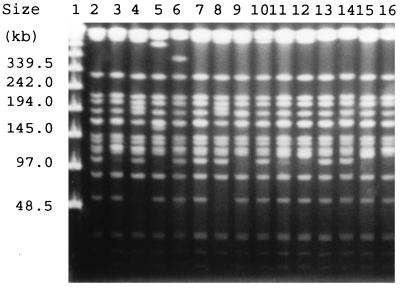
All 15 isolates exhibited the same biotype by testing both with the API 20E and the MicroScan systems. A major PFGE pattern after chromosomal DNA restriction with XbaI was found (Fig. 1). This pattern showed only five minor variations (subtypes A1 to A5), and overall, the isolates were considered to be clonally related to the dominant strain found during the outbreak. There was no relationship between the PFGE subtypes of the strains and porin expression. Pairs of cefoxitin-resistant and cefoxitin-susceptible isolates from three patients also had identical PFGE patterns.
FIG. 1.
PFGE of total DNA from K. pneumoniae cut with XbaI. Lane 1, PFGE Molecular Weight Marker (New England Biolabs); Lanes 2 and 3 and lanes 6 and 7, porin-sufficient isolates; lanes 4 and 5, strains with diminished expression of porin; lanes 8 to 10, porin-deficient isolates; lanes 11 and 12, lanes 13 and 14, and lanes 15 and 16, pairs of porin-deficient and porin-sufficient isolates (in the respective pairs of lanes) from three patients.
OMP analysis.
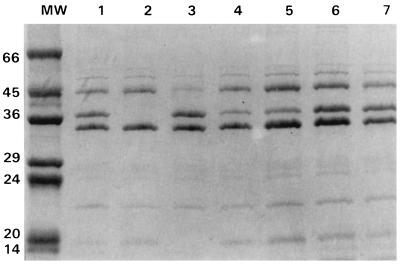
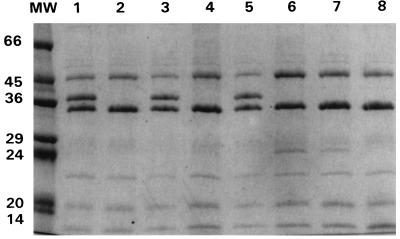
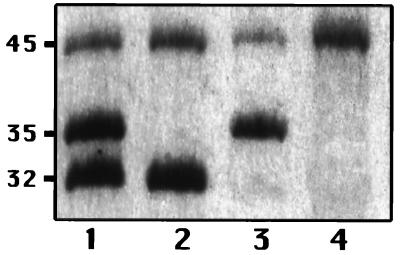
SDS-PAGE analysis of the OMPs showed that all the clinical isolates expressed two OMPs of about 32 and 45 kDa and that isolates for which cefoxitin MICs were <128 μg/ml also expressed an additional OMP of about 35 kDa (Fig. 2 and 3). The 32-kDa protein is probably the K. pneumoniae homolog of E. coli OmpA because of its increased mobility (molecular mass, about 22 kDa) in samples solubilized at 37°C (data not shown). The 45-kDa band was isolated from strain CSUB10R (Fig. 4) by a porin isolation method, and N-terminal analysis of its first 12 amino acids showed that it had complete identity with K. pneumoniae LamB. The porin isolation method, when applied to strain CSUB10S, produced the 35- and 45-kDa proteins (Fig. 4). After SDS-PAGE separation, they were transferred to a polyvinylidene difluoride membrane, and their N termini were sequenced. Sequence analysis confirmed that 45-kDa protein corresponds to LamB, while the 35-kDa sequence demonstrated that it is a nonspecific pore protein (porin). The complete identity of the first amino acids of the OmpK36 and OmpK35 porins from K. pneumoniae and other enterobacterial porins (13) prevented assignment of the 35-kDa porin to either one of the two porins of the species. This 35-kDa porin was absent from isolates for which the cefoxitin MIC was 128 μg/ml (Fig. 2 and 3). Expression of this porin, as judged by SDS-PAGE, was reduced in isolates for which cefoxitin MICs were 64 μg/ml, while sufficient expression of this porin was found in isolates for which cefoxitin MICs were 32 μg/ml (Fig. 2 and 3).
FIG. 2.
SDS-PAGE analysis of outer membrane proteins of K. pneumoniae isolates. Lane MW, molecular weight standard (in kilodaltons); lanes 1, 3, 6, and 7, porin-expressing isolates; lanes 4 and 5, isolates with diminished levels of porin expression; lane 2, porin-deficient isolate.
FIG. 3.
SDS-PAGE analysis of outer membrane proteins of K. pneumoniae isolates. Lane MW, molecular mass standard (in kilodaltons); lanes 1 and 2, lanes 3 and 4, and lanes 5 and 6, porin-sufficient and porin-deficient isolates (in the respective pairs of lanes) from the same patients; lanes 7 and 8, porin-deficient isolates.
FIG. 4.
SDS-PAGE analysis of OMPs and porins from strains CSUB10S (lanes 1 and 3) and CSUB10R (lanes 2 and 4). Lanes 1 and 2, OMP; lanes 3 and 4, porins. Numbers on the left side correspond to the approximate molecular masses of the proteins (in kilodaltons).
DISCUSSION
Nosocomial outbreaks due to ESBL-producing enterobacteria have become a serious problem worldwide (12, 17, 18). Treatment of infections due to these microorganisms is a difficult task because β-lactamase production inactivates most of the β-lactam antibiotics, and these microorganisms are usually resistant to other antibiotic groups such as aminoglycosides and quinolones. Cephamycins such as cefoxitin are active in vitro against these strains, but this agent can select porin-deficient mutants with increased levels of resistance to cefoxitin and other cephalosporins (15, 23, 30). Combinations of a β-lactam and a β-lactamase inhibitor are not always active against these microorganisms (27). Carbapenems also remain a good option, but the emergence of imipenem-resistant strains of Pseudomonas aeruginosa and other gram-negative bacilli could occur when imipenem is widely used (17). In addition, it has recently been described that K. pneumoniae becomes carbapenem resistant as a result of porin deficiency and plasmid-mediated AmpC-like β-lactamases (2, 16).
The loss of porins OmpC and OmpF as a cause of antibiotic resistance has been noted in several reports, especially for E. coli and Salmonella typhimurium (21). In K. pneumoniae, loss of both the OmpK35 and the OmpK36 porins has been shown to cause increased levels of resistance to cefoxitin and extended-spectrum cephalosporins and probably contributes to ciprofloxacin resistance (4, 15, 30). This resistance phenotype reverted when the strain expressed OmpK36 porin in its outer membrane after cloning of the ompK36 gene (15).
During an outbreak caused by K. pneumoniae producing ESBLs in our hospital (25), 4% of the isolates were highly resistant to cefoxitin (MICs, ≥128 μg/ml). Porin deficiency was associated with this resistance phenotype, and the diminished level of expression of this protein was related to a cefoxitin MIC of 64 μg/ml. In addition, porin deficiency was associated with increased MICs of cefotaxime and cefpirome, probably because in porin-deficient mutants the uptake of extended-spectrum cephalosporins is less effective than that in porin-sufficient strains (23). Thus, the combination of the decreased outer membrane permeation and the hydrolytic effect of ESBLs increased the MICs of expanded-spectrum cephalosporins for the resistant strains studied.
The six porin-deficient strains showed 8- to 32-fold decreased susceptibilities to meropenem but only 2-fold decreased susceptibilities to imipenem. The MICs of meropenem were four times higher than those of imipenem. When isolate CSUB10R was transformed with a gene coding for the OmpK36 K. pneumoniae porin, this resistance phenotype reverted and the MICs of carbapenems for this strain were similar to those for the CSUB10S strain that expresses the OmpK36 porin and that was isolated from the same clinical sample as strain CSUB10R. These findings suggest a main role of this porin in the decreased susceptibility of K. pneumoniae to meropenem.
The association between the loss of porins and imipenem resistance has recently been described in K. pneumoniae producing plasmid-mediated AmpC-like β-lactamase (2, 16). In other members of the family Enterobacteriaceae such as Enterobacter cloacae and Proteus rettgerii, resistance to carbapenems has been related to diminished outer membrane permeability and hydrolysis by the overproduced chromosomal β-lactamase (5, 26). It seems that, like in E. cloacae, the level of meropenem susceptibility in K. pneumoniae is more dependent on porin expression, whereas imipenem susceptibility is less affected by this resistance mechanism and is more dependent on the production of secondary β-lactamases of the AmpC type (2, 5, 16, 26). It is difficult to determine the exact mechanism by which the loss of porins results in decreased meropenem susceptibility. In independent studies, we have shown that strain CSUB10R produces an active efflux mechanism causing decreased levels of accumulation of fluoroquinolones in the cell (unpublished observation). A similar mechanism has been observed in P. aeruginosa with reduced susceptibility to meropenem when the efflux system MexAB-OprM is expressed. Experiments are in progress to determine a possible link between the efflux of meropenem and decreased susceptibility to this carbapenem.
In a comparison of the three pairs of cefoxitin-susceptible and -resistant strains isolated from the same patient, from two- to eightfold increases in the quinolone MICs were found. In addition, for strain CSUB10R from patient 1, a two- to fourfold decrease in the quinolone MICs was observed after the OmpK36 porin was introduced into this strain. This relationship has been reported previously (9, 15, 30), suggesting that loss of porin can contribute to quinolone resistance. However, this cross-resistance did not correlate with the degree of cefoxitin resistance in the other K. pneumoniae isolates studied. The relative importance of the possible mechanisms involved in quinolone resistance are under investigation; however, mutations have been detected in the quinolone resistance-determining region of gyrA but not that of parC in both CSUB10R and CSUB10S strains (unpublished observations); in addition, an active efflux mechanism causing a decreased level of accumulation of fluoroquinolones was detected in strain CSUB10R.
By PFGE there was a clonal relationship among the highly cefoxitin-resistant K. pneumoniae isolates; moreover, these isolates belonged to the same clone as the epidemic strain (which was cefoxitin susceptible) responsible for the outbreak. This suggests the in vivo selection of porin-deficient mutants from a common ancestor, i.e., the epidemic strain, as reported previously (15, 23).
ACKNOWLEDGMENTS
This work was supported in part by the grant 95/1234 from the Fondo de Investigacion Sanitaria (FIS) of the National Health Institute of Spain, by grant PB96-0197 from Comisión Interministerial de Ciencia y Tecnología (CICYT), and by a grant from Merck Sharp & Dohme de España S.A. C.A. and M.A.D. were supported by fellowships from FIS (fellowships 96/5176 and 94/1112, respectively), and S.H.-A. was supported by predoctoral fellowship from CICYT (fellowship FP94-41497233).
REFERENCES
- 1.Albertí S, Rodríguez-Quiñones F, Schirmer T, Rummel G, Tomás J M, Rosenbusch J P, Benedí V J. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional structure, and complement binding. Infect Immun. 1995;63:903–910. doi: 10.1128/iai.63.3.903-910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford P, Urban C, Mariano N, Projan S J, Rahal J J, Bush K. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob Agents Chemother. 1997;41:563–569. doi: 10.1128/aac.41.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brun-Buisson C, Legrand P, Philippon A, Montravers F, Ansquer M, Dural J. Transferable enzymatic resistance to third-generation cephalosporins during nosocomial outbreak of multiresistant Klebsiella pneumoniae. Lancet. 1987;ii:302–306. doi: 10.1016/s0140-6736(87)90891-9. [DOI] [PubMed] [Google Scholar]
- 4.Chen, H. Y., and D. M. Livermore. 1993. Activity of cefepime and other β-lactam antibiotics against permeability mutants of Escherichia coli and Klebsiella pneumoniae. J. Antimicrob. Chemother. 32(Suppl. B):63–74. [DOI] [PubMed]
- 5.Cornaglia G, Russell K, Satta G, Fontana R. Relative importance of outer membrane permeability and group 1 β-lactamase as determinants of meropenem and imipenem activities against Enterobacter cloacae. Antimicrob Agents Chemother. 1995;39:350–355. doi: 10.1128/aac.39.2.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elhai J, Wolk C P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 7.González-Leiza M, Pérez-Díaz J C, Ayala J, Casella J M, Martínez-Beltrán J, Bush K, Baquero F. Gene sequence and biochemical characterization of FOX-1 from Klebsiella pneumoniae, a new AmpC-type plasmid-mediated β-lactamase with two molecular variants. Antimicrob Agents Chemother. 1994;38:2150–2157. doi: 10.1128/aac.38.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gouby A, Neuwirth C, Bourg G, Bouziges N, Carles-Nurit M J, Despaux E, Ramuz M. Epidemiological study by pulsed-field gel electrophoresis of an outbreak of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a geriatric hospital. J Clin Microbiol. 1994;32:301–305. doi: 10.1128/jcm.32.2.301-305.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutmann L, Williamson R, Moreau N, Kitzis M-D, Collatz E, Acar J F, Goldstein F. Cross-resistance to nalidixic acid, trimethoprim, and chloramphenicol associated with alterations in outer membrane proteins of Klebsiella, Enterobacter, and Serratia. J Infect Dis. 1985;151:501–507. doi: 10.1093/infdis/151.3.501. [DOI] [PubMed] [Google Scholar]
- 10.Hernández-Allés S, Albertí S, Rubires X, Merino S, Tomás J M, Benedí V J. Isolation of Fe3-11, a bacteriophage specific for the Klebsiella pneumoniae porin OmpK36, and its use for the isolation of porin-deficient mutants. Can J Microbiol. 1995;41:399–406. [Google Scholar]
- 11.Jacoby G A, Han P. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996;34:908–911. doi: 10.1128/jcm.34.4.908-911.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacoby G A, Medeiros A A. More extended spectrum β-lactamases. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeanteur D, Lakey J H, Pattus F. The bacterial porin superfamily: sequence alignment and structure prediction. Mol Microbiol. 1991;5:2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- 14.Legrand P, Fournier G, Buré A, Jarlier V, Nicolas M H, Decré D, Durval J, Philippon A. Detection of extended broad-spectrum beta-lactamases in Enterobacteriaceae in four French hospitals. Eur J Clin Microbiol Infect Dis. 1989;8:527–529. doi: 10.1007/BF01967473. [DOI] [PubMed] [Google Scholar]
- 15.Martínez-Martínez L, Hernández-Allés S, Abertí S, Tomás J M, Benedí V J, Jacoby G A. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 1996;40:342–348. doi: 10.1128/aac.40.2.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez-Martínez L, Pascual A, Suarez A I, Hernández-Allés S, Benedí V J, Jacoby G A. Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Resistance to carbapenems in porin-deficient Klebsiella pneumoniae mediated by plasmid-encoded AmpC β-lactamases, abstr. C-94; p. 62. [Google Scholar]
- 17.Meyer K S, Urban C, Eagan J A, Berger B J, Rahal J J. Nosocomial outbreak of Klebsiella infection resistant to late generation cephalosporins. Ann Intern Med. 1993;119:353–358. doi: 10.7326/0003-4819-119-5-199309010-00001. [DOI] [PubMed] [Google Scholar]
- 18.Morosini M I, Cantón R, Martínez-Beltrán J, Negri M C, Pérez-Díaz J C, Baquero F, Blázquez J. New extended-spectrum TEM-type β-lactamase from Salmonella enterica subsp. enterica isolated in a nosocomial outbreak. Antimicrob Agents Chemother. 1995;39:458–461. doi: 10.1128/aac.39.2.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. Approved standard M7-A4. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th edition. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 20.Nikaido H, Rosenberg E Y. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J Bacteriol. 1983;153:241–252. doi: 10.1128/jb.153.1.241-252.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989;33:1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nurminem M. A mild procedure to isolate the 34K, 35K and 36K porins of the outer membrane of Salmonella typhimurium. FEMS Microbiol Lett. 1978;3:331–334. [Google Scholar]
- 23.Pangon B, Bizet C, Buré A, Pichon F, Philippon A, Regnier B, Gutman L. In vivo selection of cephamycin-resistant, porin deficient mutant of Klebsiella pneumoniae producing TEM-3 beta-lactamase. J Infect Dis. 1989;159:1005–1006. doi: 10.1093/infdis/159.5.1005. [DOI] [PubMed] [Google Scholar]
- 24.Papanicolaou G A, Medeiros A A, Jacoby G A. Novel plasmid-mediated β-lactamase (MIR-1) conferring resistance to oxymino- and α-methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1990;34:2200–2209. doi: 10.1128/aac.34.11.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peña, C., M. Pujol, C. Ardanuy, A. Ricart, R. Pallarés, J. Liñares, J. Ariza, and F. Gudiol. Epidemiology and successful control of a large outbreak due to Klebsiella pneumoniae producing extended-spectrum β-lactamases. Antimicrob. Agents Chemother. 42:53–58. [DOI] [PMC free article] [PubMed]
- 26.Raimond A, Traverso A, Nikaido H. Imipenem- and meropenem-resistant mutants of Enterobacter cloacae and Proteus rettgeri lack porins. Antimicrob Agents Chemother. 1991;35:1174–1180. doi: 10.1128/aac.35.6.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice L B, Carias L L, Bonomo R A, Shlaes D M. Molecular genetics of resistance to both ceftazidime and β-lactam–β-lactamase inhibitor combinations in Klebsiella pneumoniae and in vivo response to β-lactam therapy. J Infect Dis. 1996;173:151–158. doi: 10.1093/infdis/173.1.151. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Klundert J A M, van Gestel M H, Meerdink G, de Marie S. Emergence of bacterial resistance to cefamandole in vivo due to outer membrane protein deficiency. Eur J Clin Microbiol Infect Dis. 1988;7:776–777. doi: 10.1007/BF01975046. [DOI] [PubMed] [Google Scholar]