Abstract
Diabetes enhances myocardial ischemic/reperfusion (MI/R) injury via an incompletely understood mechanism. Adiponectin (APN) is a cardioprotective adipokine suppressed by diabetes. However, how hypoadiponectinemia exacerbates cardiac injury remains incompletely understood. Dysregulation of miRNAs plays a significant role in disease development. However, whether hypoadiponectinemia alters cardiac miRNA profile, contributing to diabetic heart injury, remains unclear.
Methods and Results:
Wild-type (WT) and APN knockout (APN-KO) mice were subjected to MI/R. A cardiac microRNA profile was determined. Among 23 miRNAs increased in APN-KO mice following MI/R, miR-449b was most significantly upregulated (3.98-fold over WT mice). Administrating miR-449b mimic increased apoptosis, enlarged infarct size, and impaired cardiac function in WT mice. In contrast, anti-miR-449b decreased apoptosis, reduced infarct size, and improved cardiac function in APN-KO mice. Bioinformatic analysis predicted 73 miR-449b targeting genes, and GO analysis revealed oxidative stress as the top pathway regulated by these genes. Venn analysis followed by luciferase assay identified Nrf-1 and Ucp3 as the two most important miR-449b targets. In vivo administration of anti-miR-449b in APN-KO mice attenuated MI/R-stimulated superoxide overproduction. In vitro experiments demonstrated that high glucose/high lipid and simulated ischemia/reperfusion upregulated miR-449b and inhibited Nrf-1 and Ucp3 expression. These pathological effects were attenuated by anti-miR-449b or Nrf-1 overexpression. In a final attempt to validate our finding in a clinically relevant model, high-fat diet (HFD)-induced diabetic mice were subjected to MI/R and treated with anti-miR-449b or APN. Diabetes significantly increased miR-449b expression and downregulated Nrf-1 and Ucp3 expression. Administration of anti-miR-449b or APN preserved cardiac Nrf-1 expression, reduced cardiac oxidative stress, decreased apoptosis and infarct size, and improved cardiac function.
Conclusion:
We demonstrated for the first time that hypoadiponectinemia upregulates miR-449b and suppresses Nrf-1/Ucp3 expression, promoting oxidative stress and exacerbating MI/R injury in this population. Dysregulated APN/miR-449b/oxidative stress pathway is a potential therapeutic target against diabetic MI/R injury.
Keywords: Adiponectin, microRNA, ischemia/reperfusion, diabetes, antioxidant
1. Introduction
Despite improved reperfusion strategies leading to declined mortality in the non-diabetic population, the mortality and morbidity of ischemic heart disease in diabetic patients remain high.[1] [2] Clinical studies and basic experiments demonstrate that increased oxidative stress following myocardial ischemia/reperfusion (MI/R) is a big hurdle in preventing the diabetic exacerbation of MI/R injury.[3] [4] Defining the determinants responsible for the increased oxidative stress and accelerated cardiac injury in the diabetic heart is of great clinical importance in developing novel strategies to limit cardiac damage.
Adiponectin (APN) is an adipocyte-specific cytokine critical in maintaining systematic metabolic hemostasis. Clinical and experimental results consistently demonstrate that plasma APN level is significantly reduced in type 2 diabetic individuals [5]. More importantly, hypoadiponectinemia during diabetes is correlated with increased acute myocardial infarction risk [6–8] and worse cardiac functional recovery after MI/R [9–11]. However, molecular mechanisms responsible for hypoadiponectinemia exacerbation of MI/R injury in diabetes remain incompletely understood.
MicroRNA are endogenous non-coding RNAs ranging from 18 to 24 nucleotides in length. They are highly conserved and ubiquitously expressed in all species [12, 13]. Accumulating evidence demonstrates that these small non-coding RNAs influence many cellular processes through post-transcriptional regulation of their target genes. miRNAs modulate the stability and/or the translational efficiency of target mRNAs in most biological processes, including in cellular responses to redox imbalance [14].
Given the disappointing clinical results of exogenous antioxidative supplementation treatments on the MI/R injury, enhancing endogenous antioxidative capability emerge to be of potential as a promising strategy. In addition, short of a biomarker to predict the risks of cardiovascular complication in diabetes is the bottleneck for prognosis and warning of the disorder of cardiovascular aggravation problem. Therefore, pursuing the unique signature of miRNA responsible for suppressing endogenous antioxidative molecules and promoting cardiac impairment in diabetes may hold great perspective as a novel diagnostic and therapeutic tool for diabetic cardiovascular injury.
Therefore, the aims of this study were to 1) identify cardiac miRNA(s) upregulated by hypoadiponectinemia and high-fat diet (HFD)-induced diabetes in a nonbiased fashion; 2) clarify molecular mechanism linking dysregulated mRNA(s) and enhanced cardiac injury; and 3) evaluate the possibility of identified miRNA as a potential therapeutic target against diabetic cardiac injury after MI/R.
2. Materials and Methods
2.1. Animal Procedures
Adult (8-week-old) male C57BL/6J mice were randomized to receive a high-fat diet (HFD) (60 kcal%) (Research Diets Inc. D12492i) or a 10 kcal% control normal diet (ND, D12450Bi) containing the same protein content as the HFD for 12 weeks. This is a widely accepted type 2 diabetic animal model in diabetic research filed, closely simulating the pathologic condition of Western diet-induced type 2 diabetes in human, including obesity, hyperlipidemia, hyperglycemia, hyperinsulinemia, and insulin resistance. [15–17] All animal studies were performed per the National Institutes of Health Guidelines and regulations on the Use of Laboratory Animals, and the protocols were approved by the Thomas Jefferson University Committee on Animal Care.
Adult male C57/BL6 mice, APN knockout mice (APN-KO), or HFD mice were anesthetized with 2% isoflurane. MI was induced by temporarily exteriorizing the heart via left anterior descending (LAD) coronary artery ligation with a 6–0 silk suture slipknot as previously described.[16] After 30 minutes of MI, the silk slipknot was released, and myocardial reperfusion commenced for 3 hours (for apoptosis and signaling assay) or 24 hours (for infarct size and cardiac function), as we previously reported. [18]. All assays utilized tissue from ischemia/reperfusion regions (areas at risk identified by Evans blue-negative staining). The sham-operated control group (Sham) underwent the same surgical procedures, except the suture placed under the LAD was not tied. The APN-treated group received APN (0.25 μg/g, intraperitoneal injection) immediately before coronary occlusion.
2.2. RNA isolation, miRNA array, and Target Prediction
RNA was isolated as previously described. Briefly, 100 mg of left ventricular tissue was homogenized using TRIZOL (Invitrogen) reagent, and the homogenized samples were incubated at room temperature for 5 min. Chloroform was added to the samples, vigorously mixed, and incubated at room temperature for 5 min; following incubation, the samples were centrifuged at 12,000g for 15 min at 4 °C. RNA was precipitated from the aqueous phase by addition of isopropanol to a fresh tube containing the supernatant aqueous phase. The integrity of the RNA was tested by running samples on a denaturing formaldehyde gel, and concentration was measured by Nano 3000 (Thermos Fisher).
MicroRNA array (Ohio State Comprehensive Cancer Centre, version 3.0, containing 627 probes for mature miRNA corresponding to 288 different human miRNAs) was carried out and analyzed by GenePix Pro. [19] The average values of the replicate spots for each miRNA were background subtracted, normalized, and subjected to further analysis. Global median and lowest normalization of the heart microarray data were carried out using the BRB Array Tools.
Putative miRNA target genes were identified using the microRNA databases and target prediction tools miRbase and TargetScan. Gene ontology (GO) biological process enrichment of those validated targets was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov) and metascape (http://metascape.org). Heatmap was presented by log-transformed p-value. [20]
The plasma miRNA was purified by miRNeasy Plasma kit (Qiagen). The miR-449b was detected using miScript PCR system with the miScript II RT kit (Qiagen, 218161) and PowerUp SYBR Green master Mix Kit (Thermofisher, A25742) on a QuantStudio 5 Real-Time PCR System (ABI) in a 96-well format, respectively. U6 RNA was used as a reference control.
2.3. Determination of myocardial function
In animals observed 24 and 72 hours after reperfusion, the following outcomes were measured as described previously. [21] [18] Cardiac function was determined by echocardiography (Visual Sonics Vevo 2100 system, MS550, 40-MHz frequency probe). Briefly, mice were anesthetized with 2% isoflurane. Mice were placed supine on a heated table (core temperature was maintained at 37°C), and chest hair was removed. B-mode images were acquired from a parasternal long-axis view to evaluate LV ejection fraction (EF) and fractional shortening (FS).
2.4. Determination of myocardial apoptosis and myocardial infarct size
Myocardial apoptosis was determined by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) staining, Caspase 3 activity (expressed as nmol of pNA/hour per mg of protein) and DNA fragmentation assay as described previously. [22] [23] TUNEL staining was performed via an In-Situ Cell Death Detection Kit (Roche Diagnostics, Manheim, Germany) per the manufacturer’s protocol. The percentage of apoptotic cells was calculated as the ratio of TUNEL-positive cells to the total number of cells, counted in three different random fields. The caspase-3 activity was determined using the fluorogenic substrate DEVD-7-amino-4-trifluoromethyl-coumarin (AFC). For DNA laddering determination, [24] 10ug of samples’ DNA was isolated and dissolved in DNA suspension buffer. The DNA samples were loaded and size-fractionated on a 2% agarose gel for electrophoresis. Ethidium bromide-stained DNA was visualized under UV light and analyzed.
Myocardial infarct size was determined by Evans blue and 2,3,5-triphenyl tetrazolium chloride (1%, TTC) double staining. Briefly, the LAD was re-occluded at the end of the experiment. Even’s blue dye was injected into the left ventricle to delineate the anatomic area at risk (AAR) subjected to occlusion and reperfusion (Evans blue negative) and the non-ischemic normal zone (Evans blue positive). The heart was removed and sliced into serial transverse sections. Slices were incubated at 37°C for 20 to 30 min in 1% TTC in 0.1 mol/L phosphate buffer adjusted to pH 7.4 and photographed with a digital camera. Infarcted and non-infarcted myocardium within the AAR were digitally measured using image analysis software (Image J, version 1.47, National Institutes of Health, Bethesda, MD). Infarct size was expressed as a percentage of the AAR.
2.5. Measurements of plasma Creatine kinase-MB (CK-MB)
WT, APN-KO, and HFD mice plasma were withdrawn after 30min ischemia and 3 hours of reperfusion. Plasma CK-MB levels were determined using a commercially available ELISA Kit (R&D Systems, Minneapolis, MN).
2.6. Anti-miR and miRNA mimic Administration
Anti-miR-449b and miR-449b mimic were synthesized by IDT company. The anti-miR is a full-length oligonucleotide directed against the mature sequence of mmu-microRNA-449b (5’-GCCAGCTAACAACACTGCCTTT-3’). Mimic is a full-length oligonucleotide with miR-449b (5’-AGGCAGTGTTGTTAGCTGGCTT-3’). Both molecules were modified with locked nucleic acid (LNA) technology to ensure stability. LNA are modified nucleotides in which the 2’-oxygen and 4’-carbon atoms are joined by an extra bridge. Oligonucleotides that include LNAs show enhanced specificity, sensitivity, and hybridization stability. The scrambled control sequence contained the same amount of building bases (ATCG) but in random order. Anti-miR-449b or miR-449b mimic (1 nmol/mouse) was mixed with atelocollagen (AteloGene; KOKEN, Japan) and tail vein injected into mice for 24 h before coronary occlusion.[25] [26]
2.7. Cardiomyocyte isolation, simulated ischemia-reperfusion (SI/R), and high glucose/high lipid culture
Neonatal rat cardiomyocytes (NRCMs) were isolated as described.[27] Briefly, hearts were removed from 1–2-day-old rat pups and enzymatic dissociated. Cells were plated on 6-well plates and cultured in F10 culture medium (Hyclone). When myocytes (1×106 cells per well) reached 90% confluence, the medium was changed to ischemia buffer solution (in mM: 118 NaCl, 24 NaHCO3, 1 NaH2PO4·H2O, 2.5 CaCl2·2H2O, 0.5 sodium EDTA·2H2O, 20 sodium lactates, 16 KCl, pH 6.2) in 2% oxygenation 5% CO2 incubator. [28] The cells were exposed to these conditions for 4 hours, then incubated again in F10 medium at 37 °C in 95% air and 5% CO2 (reperfusion) for 6 hours as SI/R treatment.
To mimic non-diabetic or diabetic environments in vitro, NRCMs were randomized to receive one of the following treatments: normal glucose/normal lipid culture (Control, 5.5mM D-glucose+19.5mM L-glucose); or high glucose/high lipid (HG/HL, 25mM D-glucose/250 μM palmitates, Sigma) as previously reported [29–31].
2.8. Luciferase Reporter Assay
The pLightSwitch vector was commercially purchased, and the fragment of NFE2L1 (nuclear factor erythroid 2-related factor 1, Nrf-1) full-length 3’-UTR was cloned, then co-transfected with vehicle or miR-449b into neonatal cardiomyocytes according to the instruction provided by Switchgear Genomics.[32] Forty-eight hours after transfection, LightSwitch Luciferase Assay Reagent was added to measure luciferase activity, and the signal was detected on the SpectraMax M5 microplate reader (Molecular Devices). The site-directed mutagenesis was performed using the in-fusion mutation kit. Confirmation of the mutations was done with conventional Sanger-based sequencing by GeneScript Company.
2.9. Superoxide Generation Measurement
Superoxide content was quantified by lucigenin-enhanced chemiluminescence assay, and the cellular origin of reactive oxygen species was determined by dihydroethidium staining (DHE, Molecular Probes, Carlsbad, CA) as previously described. [23] Images were acquired with Olympus DP72 System via CellSens Dimension Software Version 1.16.
2.10. Plasmid construction and transfection
The cDNA encoding Nrf-1 was subcloned into 3xflag plasmid (Sigma) with the Clontech HD In-fusion Cloning strategy (Clontech Laboratories, Inc., Mountain View). NRCMs in a 60-mm culture plate were transfected with 2μg plasmid DNA encoding Nrf-1 via TransMessenger transfection reagent (Qiagen) per manufacturer’s protocols. When cells were co-transfected with plasmid DNA, empty vector (no cDNA insert) served as control. 6 hours after transfection, the culture medium was switched to the normal culture medium. Incubation commenced 72 hours before any experiments.
2.11. Western Blot
The extraction of protein from heart tissue or cells was prepared by lysis buffer (Cell signaling), and Western Blot assay was performed as described. [33] Samples were loaded on 4–20% SDS-PAGE gels, transferred to PVDF membranes, blotted in 5% film milk, and probed with primary antibodies against Nrf-1, GAPDH, Caspase-3, Cleaved Caspase 3 (Cell Signaling). PVDF membranes were then incubated with a second antibody for 1 hour. For the detection of proteins, the Pierce ECL Substrate kit was used on a ChemiDoc MP Imager (Bo-Rad, CA).
2.12. Quantitative RT-PCR and Chip-based digital PCR (dPCR)
miRNA primers were designed by integrated DNA technology (IDT). RNA from mouse heart tissue was extracted with Ambion miRNA extraction kit according to the manufacturer’s instructions.[32] 500–1000ng RNA was reverse-transcribed to cDNA using the miScript-II RT PCR kit. Real-time PCR was performed on an ABI QuantStudio 5 system using SYBR Green. Transcript quantities were compared using the relative Ct method, where the amount of target normalized to the amount of endogenous control (U6) and relative to the control sample is given by 2−ΔΔCt.
In order to detect the absolute level of microRNA, a Chip-based dPCR assay was performed as described in the previous publication,[34–36] using the QuantStudio 3D Digital PCR System platform (Thermo Fisher Scientific) composed of the QuantStudio 3D Instrument, the Dual Flat Block GeneAmp PCR System 9700, and the QuantStudio 3D Digital PCR Chip Loader. Briefly, 1000ng of RNA was reverse-transcribed into cDNA (using the TagMan Advanced miRNA cDNA Synthesis Kit) and PCR cycling was performed with QuantStudio 3D digital PCR 20K Chip kit v2 at 96 °C for 10 min, denaturation at 98 °C for 30 s, followed by annealing/extension at 60 °C for 2 min (39 cycles), and then final extension at 60 °C for 2 min. Analysis was executed with QuantStudio 3D AnalysisSuite (Thermo Fisher Cloud, Waltham, MA, USA). The estimated copy number of miRNA-499b is expressed as the number of copies contained per 10pg heart RNA.
2.13. Patient selection
A total of 220 patients, including 110 patients with IHD (ischemic heart disease) without diabetes, 80 IHD with type 2 diabetes, and 30 healthy volunteers, were enrolled in this study. All participants were Han nationality. 110 non-diabetic IHD plasma samples including healthy volunteers were obtained from the shared online biobank center (Shanxi Medical University, China). 80 type 2 diabetes patients with IHD and 30 healthy controls were recruited in the Department of Endocrinology from August to October 2019. Exclusion criteria for this study were: type 1 diabetes, severe hepatic and renal dysfunction, active liver disease, malignancy, the inflammatory process in the past two months, pregnancy, or any factor affecting body weight such as hyperthyroidism, corticosteroids, or contraceptives. The fasting plasma samples were collected and stored at −80°C. Clinical test data were collected from hospital databases and analyzed. All subjects provided written informed consent to participate in the study.
2.14. Statistics
The statistical analyses were performed using GraphPad Prism 9.0.2 statistic software (La Jolla, CA). For continuous variables, normal distribution was evaluated by the Shapiro-Wilk test. For multiple groups, one-way ANOVA or a 2-way ANOVA was performed with post hoc Tukey multiple comparison tests with an adjusted p value of <0.05 was considered statistically significant. Normally distributed data are presented as mean±SEM of representative experiments. Spearman’s rank correlation analysis (Pearson’s correlation analysis, if normally distributed) was used to evaluate the correlation between continuous variables. The ROC curve was constructed to assess the sensitivity, specificity, and area under the curve (AUC), and the confidence interval of miR-449b was 95%. A two-sided value of P < 0.05 was considered statistically significant.
3. Results
3.1. miR-449b expression is significantly increased in APN-KO mouse heart
Since hypoadiponectinemia is a significant driver of type 2 diabetic metabolic disorder and cardiovascular complications [37], we sought to clarify whether the miRNA expression profile is altered in APN-KO mice. We performed a microarray analysis of miRNAs extracted from the WT or APN-KO hearts with or without MI/R. We focused on miRNAs that met the following criteria: the miRNAs are significantly expressed in the heart (miRNA spot intensity>100); the miRNAs are altered by MI/R (2-fold increase after MI/R); and MI/R suppressed miRNAs were rescued by APN administration. Among 23 miRNAs that met these criteria, miR-449b was identified as the most significant miRNA suppressed by APN-KO and rescued by APN administration (APN-KO MIR/APN-KO sham over WT MIR/WT Sham) after MI/R. (Figures 1A and 1B). Validation with Taqman microRNA real-time polymerase chain reaction (RT-PCR) demonstrated that APNKO without MI/R (APNKO sham vs. WT Sham) slightly increased miR-449b expression (the first bar in each group). Interestingly, MI/R (WT MI/R vs. WT sham) slightly reduced miR-449b expression in WT animals. Finally, MI/R significantly upregulated miR-449b expression in APN-KO heart (Figure 1C). To confirm the finding from APN-KO mice in a more clinically relevant animal model, miR-449b expression in HFD-induced type 2 diabetic heart was determined. Consistent with APN-KO animals, HFD modestly (but not statistically significant) increased miR-449b expression. Most importantly, HFD animals subjected to MI/R upregulated cardiac miR-449b expression to a level more remarkable than APN-KO animals after MI/R (Figure 1C). APN administration slightly (not statistically significant) reduced miR-449b in APN-KO and HFD animals without MI/R. However, APN administration significantly attenuated miR-449b expression in APN-KO as well as in HFD heart after MI/R (Figure 1C).
Figure 1. Myocardial microRNA (miRNAs) expression profiles in animals subjected to myocardial ischemia-reperfusion.
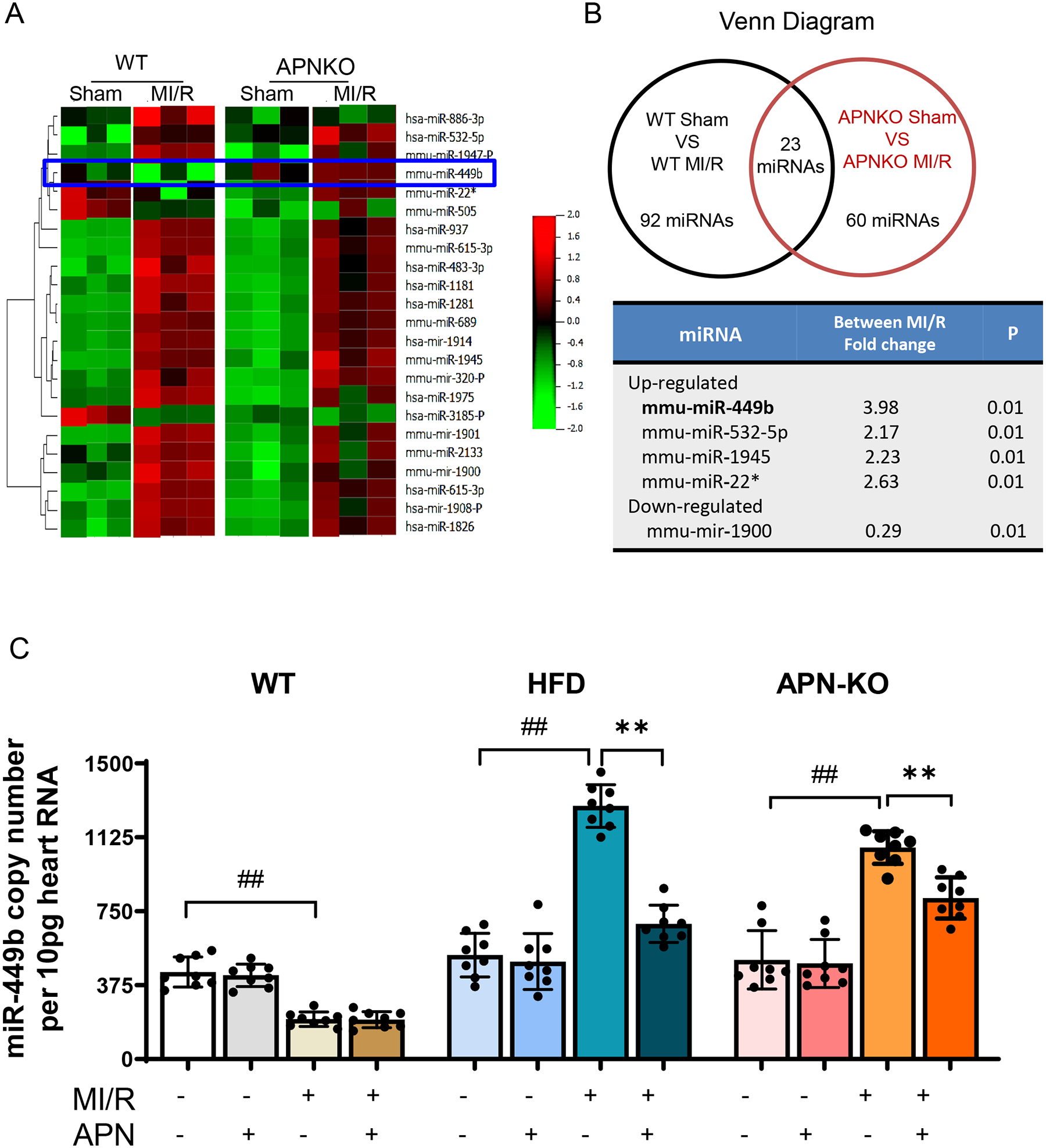
A. Hierarchal clustering heat map showing all differentially expressed microRNAs in WT and APN-KO mice hearts. B. Venn diagram showing 23 miRNAs was the same over-crossed from WT and APN-KO comparation (Up panel). miRNA expression fold change was calculated between WT and APN-KO Sham over MI/R (Down panel). C. Digital PCR validation using miRNA-specific primer assays for miR-449b in cardiac tissues. **p<0.01 versus MI/R, respectively; ##p < 0.01 versus WT-MI/R group. Statistical significance was evaluated by a two-way ANOVA. Post hoc pairwise tests for indicated group pairs were performed after Tukey correction. MI/R, myocardial ischemia/reperfusion.
To determine whether cardiomyocytes are the primary cellular source of miR-449b, additional experiments utilizing cardiomyocytes isolated from different groups of heart were performed. The evaluating miRNA-449b absolute expression level results demonstrated that miR-449 alterations observed in cardiac tissue (Figure 1C) were mirrored in isolated cardiomyocytes (Figure S4). These results indicate that cardiomyocytes-generated miR-449b is responsible for cardiac miR-449b alteration observed in disease hearts.
3.2. miR-449b regulates the cardiac apoptosis and MI/R injury
Although miR-449b family members have been reported to be involved in liver and renal ischemic injury [38, 39], little is known regarding its role in heart pathogenesis under either diabetic or I/R conditions. We, therefore, determined the role of miR-449b may contribute to MI/R injury by administration of a miR-449b mimic (gain-of-function) in WT mice (miR-449b OE) or an anti-miR-449b (loss-of-function) in APN-KO mice immediately before MI/R, respectively. Administration of miR-449b mimic significantly exacerbated cardiac dysfunction in WT mice subjected to MI/R. In contrast, cardiac function was significantly improved in anti-miR-449b treated APN-KO mice following MI/R (Figure 2A). To determine whether this protective effect retains after a longer period of reperfusion, another group of mice was subjected to 72 hours of reperfusion. Our results demonstrated a similar regulatory pattern as that observed at 24 hours (Supplemental Figure 3). Consistently, infarct size was significantly enlarged, and creatine kinase-MB (CK-MB) was further increased in miR-449b mimic treated WT mice, whereas infarct size was significantly reduced, and CK-MB was decreased in anti-miR-449b treated APN-KO mice (Figure 2B and 2C). Finally, cardiac apoptosis analysis (DNA ladder and Cleaved Caspase 3 analysis) demonstrated that administration of miR-449b mimic in WT mice exaggerated apoptosis, whereas administration of anti-miR-449b in APN-KO mice reduced apoptosis after MI/R (Figure 2D–2F).
Figure 2. Myocardial infarct size and apoptosis detection after myocardial ischemia-reperfusion in WT, APN-KO, and HFD mice.
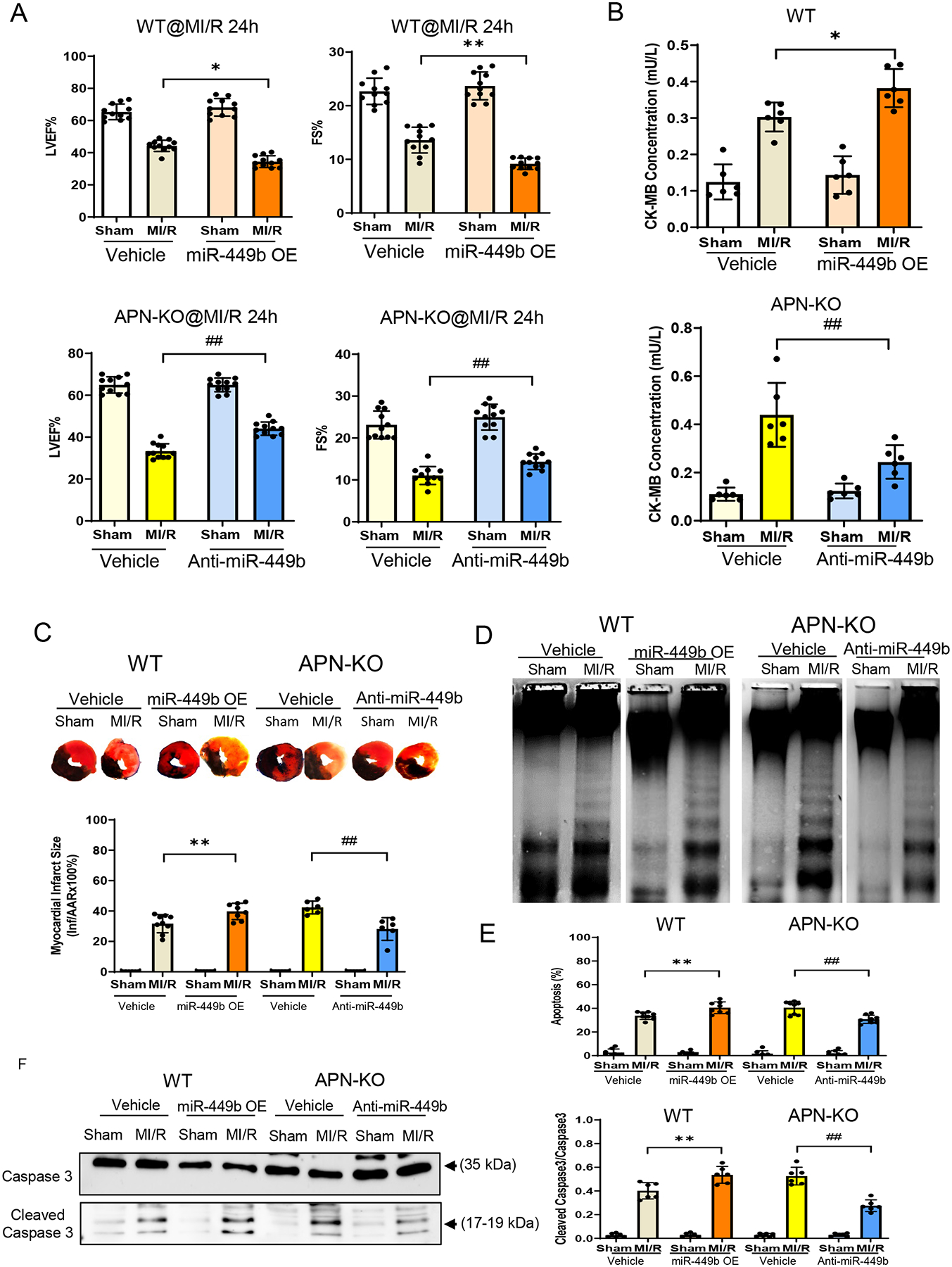
A. Left ventricular ejection fraction (LVEF%), and fractional shorting (FS%) were determined by echocardiography in WT and APN-KO mice subjected to MI/R with anti-miR-449b or miR-449b mimic (miR-449b OE) administration. **p < 0.01 vs. vehicle+MI/R group in WT mice, .##p < 0.01 vs. vehicle+MI/R group in APN-KO mice. n=10/group. B. CK-MB measurement of WT and APN-KO mice subjected to MI/R with miR-449b OE or anti-miR-449b administration. *p < 0.05 versus WT-MI/R group; **p < 0.01 versus APN-KO-MI/R group. For A and B, statistical significance was evaluated by one-way ANOVA with Tukey test for post hoc pairwise tests. C. Percentage of infarct size expressed as a percentage of the area risk (AAR). **p < 0.01 versus WT-MI/R group; ##p < 0.01 versus APN-KO-MI/R group. Statistical significance was evaluated by two-way ANOVA with Tukey test for post hoc pairwise tests. D. DNA ladder induced by MI/R on WT and APN-KO mice with miR-449b mimic or anti-miR-449b administration. E. Percentage of apoptosis expressed as a percentage of cell death. **p < 0.01 versus WT-MI/R group; ##p < 0.01 versus APN-KO-MI/R group. n=6–10/group, 3 independent experiments. Statistical significance was evaluated by two-way ANOVA with Tukey for post hoc pairwise tests. F. Cleaved Caspase 3 evaluated by Western blot in WT and APN-KO mice with miR-449b mimic or anti-miR-449b administration. **p < 0.01 versus WT-MI/R group; ##p < 0.01 versus APN-KO-MI/R group. n=6/group, 3 independent experiments. Statistical significance was evaluated by two-way ANOVA with Tukey for post hoc pairwise tests. OE, overexpression; MI/R, myocardial ischemia/reperfusion. APN-KO, Adiponectin Knockout.
3.3. miR-449b causes apoptosis by provoking oxidative stress
Next, we sought to identify the downstream signaling pathways contributing to miR-449b-induced cardiomyocyte apoptosis and cardiac dysfunction. Two databases were analyzed, including miRNA-responsive genes in human primary cells overexpressing miR-449b (GSE77251 PubMed database, Supplemental Figure 1A) and prediction of miR-449b target genes by Targetscan. Venn overlapping analysis identified 72 genes common in these two databases (Figure 3A). Among biological pathways regulated by these 72 genes (Metascape online Go analysis, Http://Metascape.org; Supplemental Fig 1B), cellular response to oxidative stress is ranked as the top pathway among that regulated by miR-449b targeting genes (Figure 3B).
Figure 3. Oxidative stress evaluation in animal model.
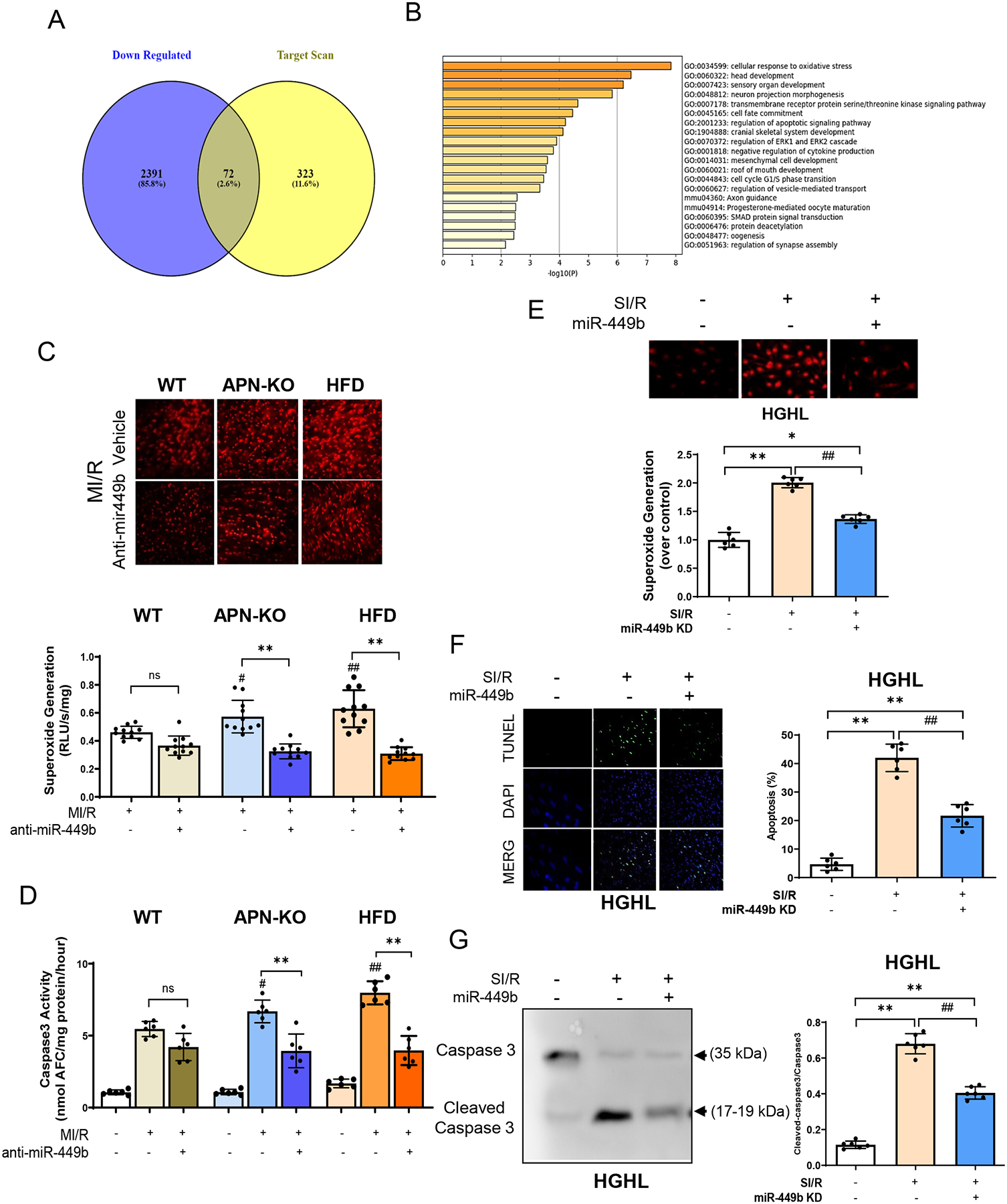
A. Venn diagram shows that 72 targets were the same over-crossed from predicted targets for miR-449b. B. Go analysis showing signaling pathways regulated by miR-449b targeting genes. C. DHE assay (up panel) and Lucigenin assay detection (down panel) for superoxide generation in WT, APN-KO, and HFD mice subjected to MI/R with anti-miR-449b administration. **p < 0.01 versus MI/R group, respectively; #p < 0.05 & ## p < 0.01 versus WT-MI/R group. D. Caspase 3 activity evaluations in generation in WT, APN-KO, and HFD mice subjected to MI/R with anti-miR-449b administration. **p < 0.01 versus MI/R group, respectively; #p < 0.05 & ## p < 0.01 versus WT-MI/R group. For C and D, statistical significance was evaluated by two-way ANOVA. Post hoc pairwise tests for indicated group pairs were performed after Tukey correction. E. Superoxide generation in cultured NRCMs subjected to SI/R with anti-miR-449b administration. *p < 0.05 & **p < 0.01 versus control group; ## p < 0.01 versus SI/R group. F. Apoptosis evaluation by TUNEL stain assay (green) in HGHL-induced diabetic NRCMs with anti-miR-449b administration. DAPI (blue). **p < 0.01 versus control group; ## p < 0.01 versus SI/R group. G. Cleaved Caspase 3 evaluated by Western blot in HGHL-induced diabetic NRCMs with anti-miR-449b administration. **p < 0.01 versus control group; ## p < 0.01 versus SI/R group. n=6–11/group, 3 independent experiments. For E-G, statistical significance was evaluated by one-way ANOVA. Tukey tests were used to correct for multiple comparisons. I/R, myocardial ischemia/reperfusion; SI/R, Simulated ischemia/reperfusion; HGHL, High glucose, and high lipids; KD, Knockdown.
Validation of miR-449b regulation of oxidative stress and apoptosis in vivo is critical to support the role of miR-449b in the diabetic exaggeration of MI/R injury. In APN-KO mice, administration of anti-miR-449b significantly decreased superoxide production after MI/R(−43.3%±14.6%, p < 0.05. Figure 3C). To confirm the oxidative/pro-apoptotic role of miR-449b in the diabetic heart after MI/R, HFD mice were treated with anti-miR-449b and subjected to MI/R. Anti-miR-449b treatment significantly reduced superoxide production and cardiac apoptosis in HFD mice (−50.9%±13.9% and −43.1%±11.1%, respectively, Figure 3C and 3D). Moreover, superoxide production and caspase-3 activity were increased in the APN-KO or HFD heart compared to WT heart after MI/R injury (Figure 3C and 3D). These results indicate diabetic hypoadiponectinemia upregulates cardiac miR-449b, promoting oxidative stress and apoptosis in the MI/R heart.
To obtain evidence that miR-449 directly regulates cardiomyocytes’ oxidative stress and apoptosis, NRCM were cultured with high glucose/high lipid (HG/HL) and subjected to simulated ischemia/reperfusion (SI/R) in the presence and absence of anti-miR-449b. Consistent with in vivo results, inhibition of miR-449b attenuated SI/R-induced cardiomyocyte superoxide production (Figure 3E) and apoptosis (TUNEL) and Cleaved caspase-3. (Figure 3F and 3G).
3.4. miR-449b promotes oxidative stress by directly targeting Nrf-1 and Ucp3
As our in vivo and in vitro experimental results demonstrate that miR-449b promotes oxidative stress and apoptosis, we next focused on miR-449b targeting genes with antioxidative properties. Among the eight identified cellular responses to oxidative stress genes, the top responded genes were Nrf-1 and Ucp3, which attracted our attention because of their previously unknown relationship with miR-449b and unidentified role in diabetic cardiac injury. Several in-silicon, in vitro, and in vivo experiments demonstrated the critical role of these two molecules in miR-449b oxidative stress and cardiac injury. First, analysis of the 3’-untranslated region (UTR) of Nrf-1 mRNA revealed two potential binding sites for miR-449b. The seed sequence in position 657–663 of Nrf-1 3’-UTR is conserved among mammals (Supplemental Figures 2A and 2B). Meanwhile, analysis of the 3’-UTR of Ucp3 mRNA identified conserved target sequences for miR-449b (Supplemental Figure 2A and 2C). Second, to examine whether miR-449b directly binds to the 3’-UTRs of Nrf-1 or Ucp3 mRNAs, wild type or two mutated miRNA binding sites of Nrf-1 (Nrf1-3’UTR1; Nrf1-3’UTR2) and Ucp3 (Ucp3-3’UTR1; Ucp3-3’UTR2) were inserted into the 3’UTR of a luciferase plasmid, and dual luciferase assays in NRCMs was performed. As shown in Figure 4A, luciferase activities were significantly decreased when miR-449b was co-overexpressed with luciferase plasmids harboring wild type Nrf-1 3’-UTR. This effect was abolished when mut 3’-UTR luciferase plasmids were co-transfected with the miR-449b overexpression plasmid. Similarly, luciferase activities analysis shown in Figure 4B demonstrated that luciferase activities were significantly decreased when miR-449b was co-overexpressed with luciferase plasmids harboring wild-type Ucp3 3’-UTR. This effect was abolished when mut 3’-UTR luciferase plasmids were co-transfected with the miR-449b overexpression plasmid. Third, NRCMs were cultured with HG/HL and subjected to SI/R in the presence and absence of miR-449b mimic (miR-449b OE) or anti-miR-449b (miR-449b KD). SI/R significantly reduced Nrf-1 and Ucp3 mRNA (Figure 5A) and protein expression (Figure 5B) in HG/HL cultured cardiomyocytes. More importantly, treatment with anti-miR-449b significantly attenuated, whereas treatment with miR-449b mimic further exacerbated SI/R downregulation of Nrf-1 and Ucp3 gene (Figure 5A) and protein expression (Figure 5B). Fourth, the effect of Nrf-1 overexpression (Nrf-1 OE) upon SI/R-stimulated superoxide production was determined. As summarized in Figures 5C and 5D, Nrf-1-OE significantly attenuated superoxide production in SI/R cardiomyocytes. Finally, the effect of APN upon Nrf-1 and Ucp3 expression in MI/R hearts in non-diabetic and diabetic animals was investigated. We also identified the Nrf-1 downstream responsors. Interestingly, MI/R had no significant effect on Nrf-1 (Figure 6A) and Ucp3 protein expression in the non-diabetic heart (Figure 6B). However, MI/R significantly suppressed both Nrf-1 and Ucp3 expression in diabetic mice (Figure 6A). Treatment with APN significantly upregulated Nrf-1 and Ucp3 expression in the non-diabetic and diabetic heart (Figures 6A and 6B). Consistently, APN treatment significantly attenuated apoptosis (determined by cleaved Caspase 3, Figure 6C) and superoxide production (Figure 6D) in non-diabetic and diabetic hearts subjected to MI/R.
Figure 4. Luciferase activity assay for 3-UTR in NFE2L1 (nuclear factor erythroid 2-related factor 1, Nrf-1).
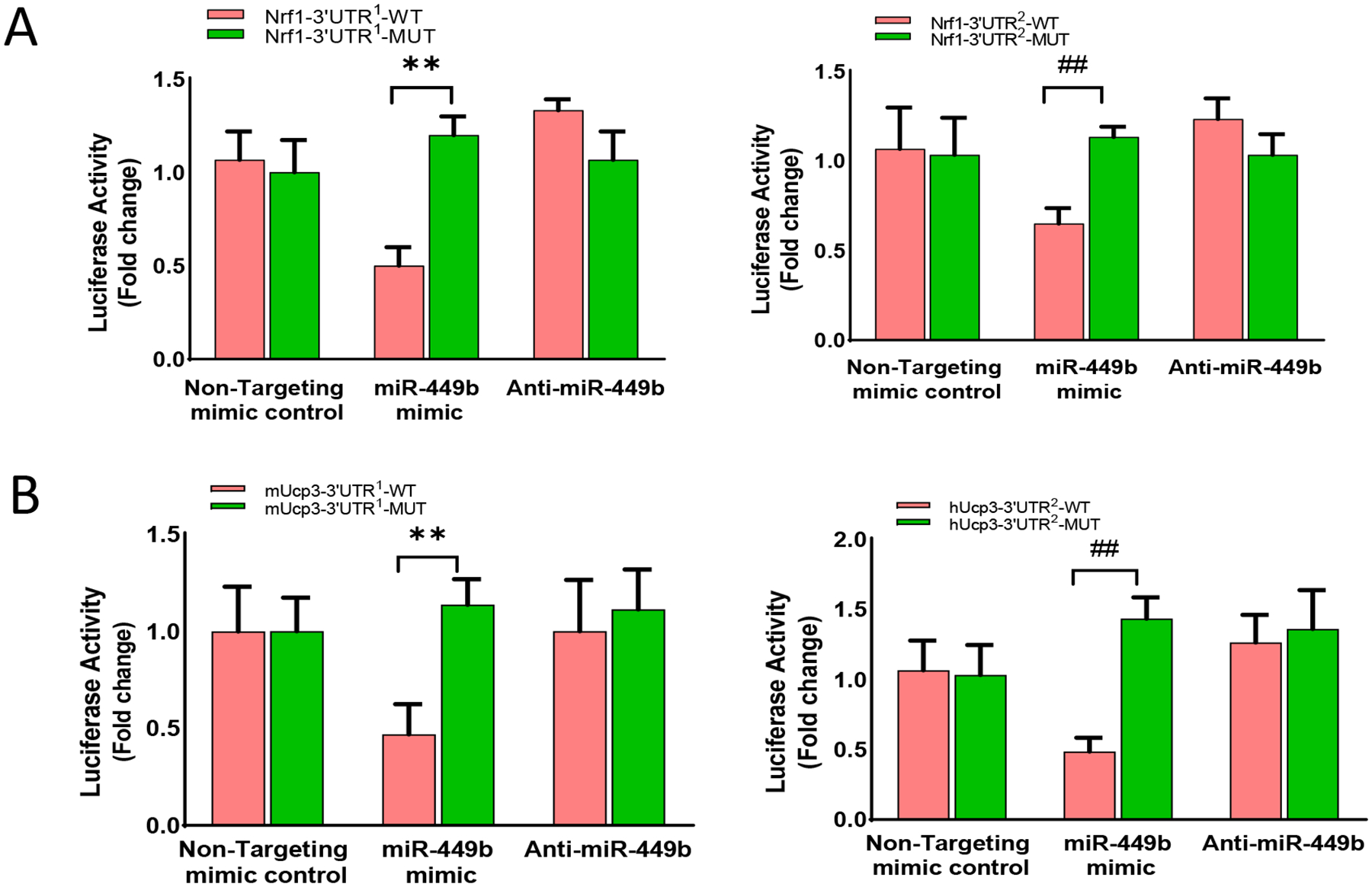
A. Luciferase assay showing co-transfected with luciferase constructs harboring 3’-UTR sequences from either wild-type (WT) or mutated (Mut) target Nrf-1 with anti-miR-449b or miR-449b mimic in NRCMs. **p < 0.01 versus Nrf1-3’UTR1-WT; ##p < 0.01 versus Nrf1-3’UTR2-WT. B. Luciferase assay showing co-transfected with luciferase constructs harboring 3’-UTR sequences from either wild-type (WT) or mutated (Mut) target Ucp3 with anti-miR-449b or miR-449b mimic in NRCMs. **p < 0.01 versus Ucp3-3’UTR1-WT; ##p < 0.01 versus Ucp3-3’UTR2-WT. n=6–9 per group, 3 independent experiments. Statistical significance was evaluated by two-way ANOVA. Post hoc pairwise tests for indicated group pairs were performed after Tukey correction.
Figure 5. Nrf-1 was regulated by anti-miR-449b and miRNA mimic administration.
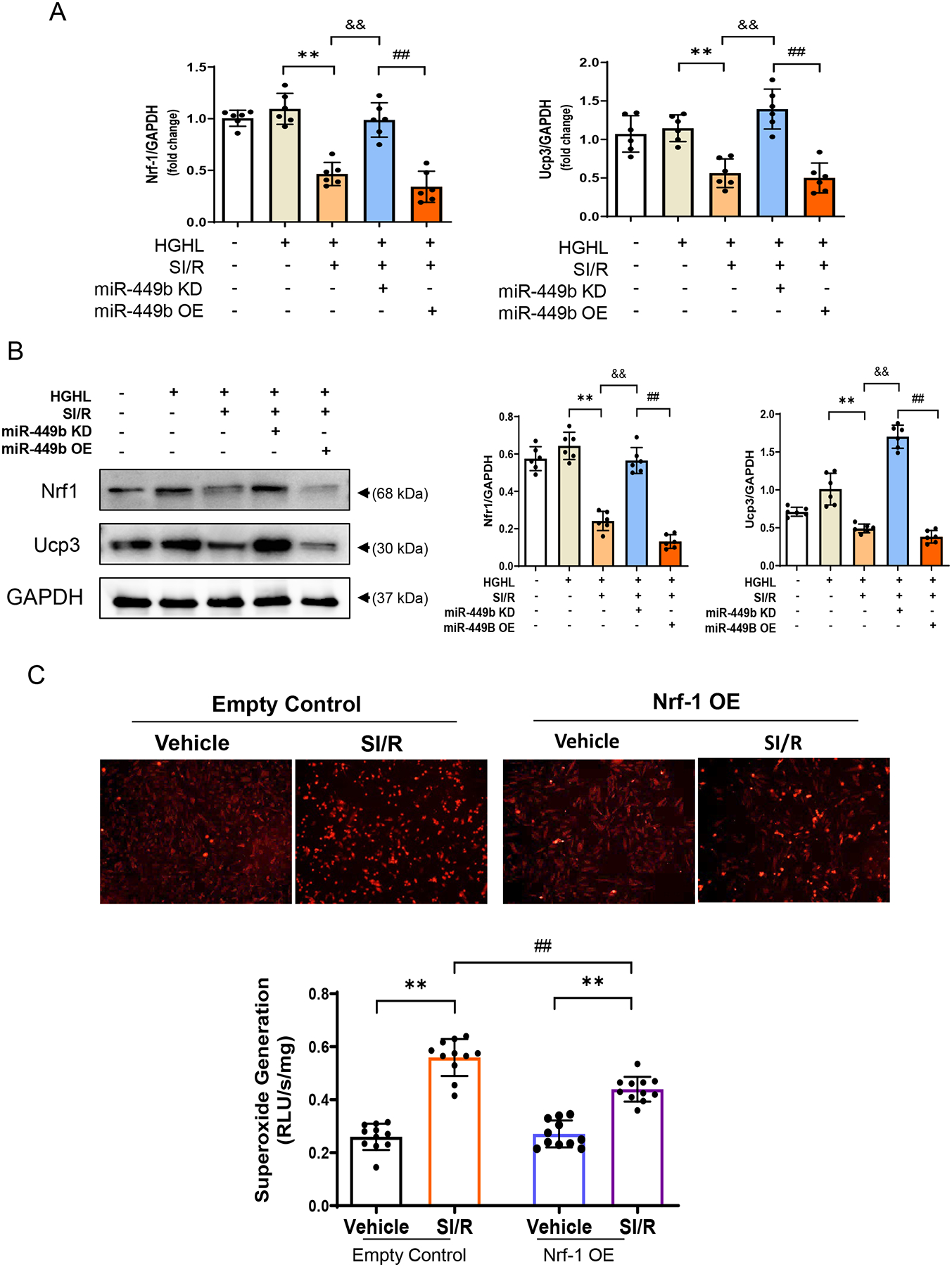
A. Real-time PCR analysis of miR-449b in NRCMs **p < 0.01 versus HG/HL group; &&p < 0.01 versus HG/HL+SI/R group; ##p < 0.01 versus HG/HL+SI/R+miR-449b KD group. B. Nrf-1 and Ucp3 expression levels were detected in NRCMs after simulated ischemia-reperfusion (SI/R) with anti-miR-449b or miR-449b mimic administration. **p < 0.01 versus HG/HL group; &&p < 0.01 versus HG/HL+SI/R group; ##p < 0.01 versus HG/HL+SI/R+miR-449b KD group. For A and B, statistical significance was evaluated by one-way ANOVA. Tukey tests were used to correct for multiple comparisons. C. DHE assay (up panel) and Lucigenin assay detection (down panel) for superoxide generation in NRCMs transfected with harboring Nrf-1 plasmid followed by SI/R challenges. **p < 0.01 versus vehicle group, respectively, ##p < 0.01 versus control+SI/R group. n=6/group, 3 independent experiments. Statistical significance was evaluated by two-way ANOVA. Post hoc pairwise tests for indicated group pairs were performed after Tukey correction. Nrf-1, Nuclear factor erythroid 2-related factor 1. HGHL, high glucose and high lipid.
Figure 6. APN administration upregulated Nrf1 and Ucp3’s level preventing HFD-induced cardiac MI/R injury.
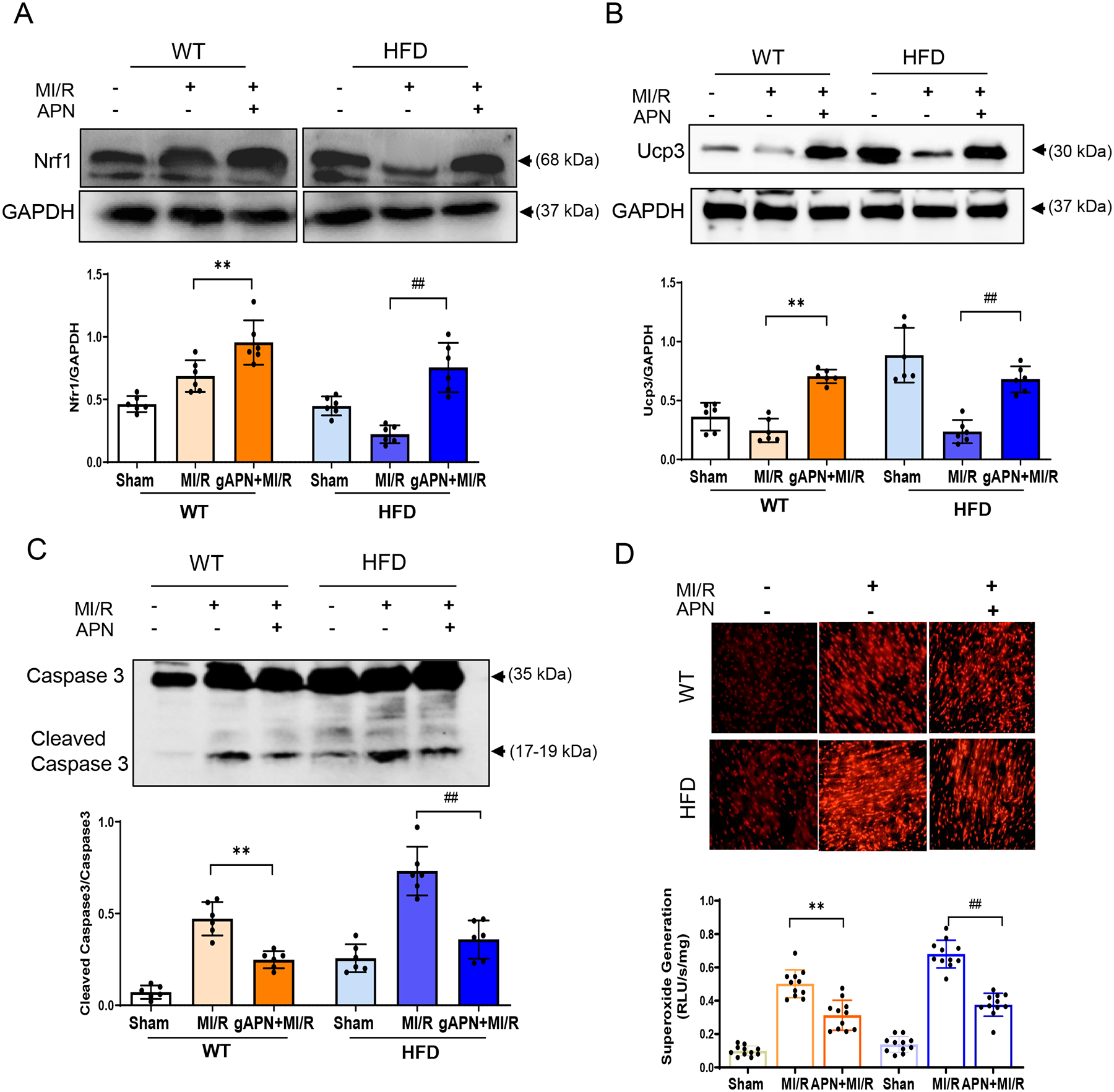
A. Nrf1 level in APN administrated WT and HFD mice subjected to MI/R. Representative Western blot, upper panel; Bar graph quantification, down panel. **p < 0.01 versus WT+MI/R group, ##p < 0.01 versus HFD+MI/R group, n = 6/group. B. Ucp3 level in APN administrated HFD mice subjected to MI/R. Representative Western blot, upper panel; Bar graph quantification, down panel. **p < 0.01 versus WT+MI/R group, ##p < 0.01 versus HFD+MI/R group, n = 6/group. C. Determination of Cleaved Caspase 3 level in HFD-induced diabetic animals with APN administration followed by MI/R. **p < 0.01 versus WT+MI/R group, ##p < 0.01 versus HFD+MI/R group, n = 6/group. D. DHE assay (up panel) and Lucigenin assay detection (down panel) for superoxide generation in HFD-induced diabetic animals with APN administration followed by MI/R challenges. **p < 0.01 versus WT+MI/R group, ##p < 0.01 versus HFD+MI/R group, n = 11/group. For A-D, statistical significance was evaluated by one-way ANOVA. Tukey tests were used to correct for multiple comparisons. HFD, high-fat diet; MI/R, myocardial ischemia/reperfusion; APN, Adiponectin.
3.5. Anti-miR-449b as a novel effective treatment against MI/R injury in diabetic mice
In a final attempt proving our concept that miR-449b upregulation plays a causative role in the diabetic exacerbation of MI/R injury, we treated HFD-fed mice with anti-miR-449b and determined its effect on MI/R injury. TTC staining of cross sections of ventricles revealed that cardiac infarct size was significantly reduced in anti-miR-449b treated diabetic MI/R mice compared with the scrambled miRNA treated MI/R mice (−34.5%±12.2%, Figure 7A and 7B). Consistent with the infarct size, anti-miR-449b treatment significantly attenuated plasma CK-MB levels (Figure 7C) and improved cardiac function (Figure 7D) in diabetic mice subjected to MI/R.
Figure 7. miR-449b regulation altered cardiomyocyte apoptosis in HFD-induced diabetic animals.
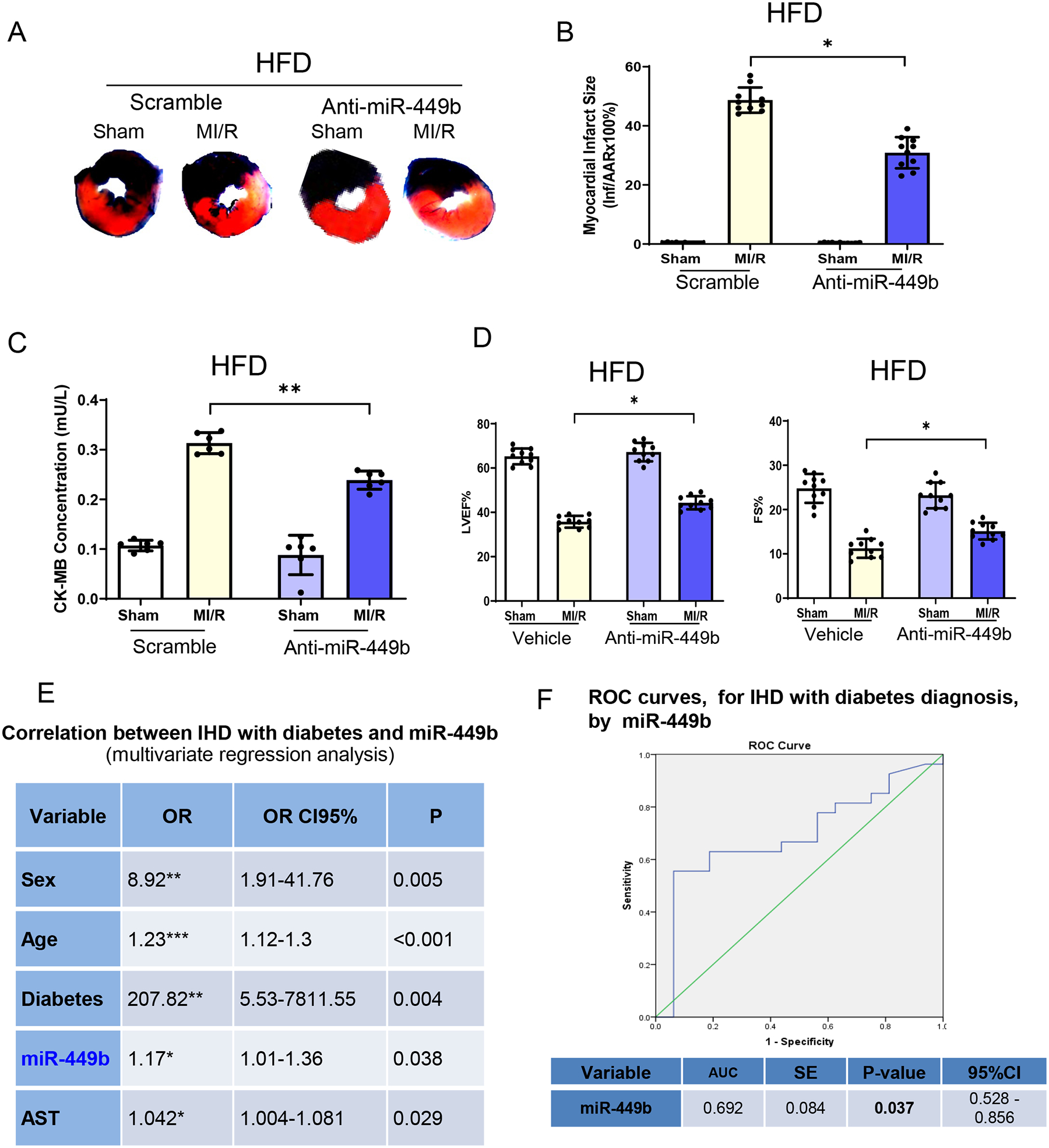
A. Anti-miR-449b administration reduced infarct size in HFD animals followed by MI/R. B. Quantification of infarct size of AAR shown in A. **p < 0.01 versus scramble+MI/R group, n = 10/group. C. CK-MB determination in anti-miR-449b administrated HFD animals followed by MI/R. **p < 0.01 versus scramble+MI/R group, n = 6/group. D. Cardiac function evaluations were determined by echocardiography in anti-miR-449b administrated HFD animals followed with MI/R. **p < 0.01 versus scramble+MI/R group, n = 10/group. For B-D, statistical significance was evaluated by one-way ANOVA. Tukey tests were used to correct for multiple comparisons. E. Correlation between IHD and miR-449b analyzed via multivariate logistics regression analysis. F. ROC curve illustrating miR-449b diagnostic ability for IHD. IHD, ischemic heart disease. ROC, Receiver operating characteristic.
To elucidate the potential roles of miR-449b in diabetic IHD patients, we determined plasma miR-449b levels in three groups of patients (110 IHD without diabetes, 80 IHD with type 2 diabetes, and 30 healthy volunteers) and analyzed their association with disease conditions with multivariate regression. Plasma miR-449b is not significantly associated with non-diabetic IHD (Supplement Table 1). However, serum miR-449b levels were significantly associated with IHD with type 2 diabetes (Figure 7E). A receiver operating characteristic (ROC) curve was constructed to evaluate the diagnostic value of miR-449b for IHD with type 2 diabetes. The AUC (area under the curve) was 0.692 (95% confidence interval 0.528–0.856, P=0.037, Figure 7F), confirming miR-449b’s potential to be a diagnostic predictor for IHD with type 2 diabetes.
4. Discussion
We demonstrated three new findings in this study. First, we identified a new cardiac miR-449b responsible for the accelerated cardiac injury, upregulated by hypoadiponectinemia and HFD-induced diabetes after MI/R. Secondly, we clarified new molecular mechanisms linking dysregulated miRNA(s) and enhanced cardiac injury. Thirdly, we evaluated the possibility of miR-449b as a potential therapeutic target against diabetic cardiac injury after MI/R.
Although clinical studies have convincingly demonstrated that the diabetic heart is more vulnerable to MI/R, multiple possibilities have been identified and reported from experimental studies. However, fewer of them could be translated into clinical. In the present study, we put efforts into identifying the key carriers which not only responsible for the link between diabetes to the exaggerated cardiac injury, but also directly targeted to cardiomyocytes injured pathways since hypoadiponectinemia correlates with the risk of CHD and is responsible for the increased sensitivity injury in the diabetic heart.[40–44]. We adopted the unbiased discovery-driven approach, microRNA deep sequencing to discover the new mediators in accelerated heart injury from adiponectin deficiency model after ischemia-reperfusion injury.
In the present studies, we found that significant enlargement of myocardial infarct size and significantly higher post-reperfusion CK-MB levels (the diagnostic hallmark for evaluating post-ischemic myocardial infarction in patients after acute myocardial ischemia [45, 46]) in APN-KO mice correlated with more severely impaired ventricular function. Through high throughput deep sequencing analysis of microRNAs, we successfully screened out miR-449b as a novel miRNA upregulated in diabetic hearts and validated its function on the deteriorated cardiac injury through the miRNA overexpression approach. In line with our current results, our studies also show significantly worse cardiac function and more severe impairment in cardiomyocytes in diabetic mice than in the control. Notably, the newly identified miR-449b was the mediator contributing to the exaggerated MI/R cardiac injury in diabetic conditions by targeting the endogenous antioxidative stress system.
Our secondary discovery reported that miR-449b upregulation in diabetes MI/R and suppression of Nrf-1 and Ucp3 antioxidants promote heart injury, indicating an adverse maladaptive response of the heart to ischemia-reperfusion in diabetic conditions. The hypoadiponectinemia increased miR-449b, while the inhibition of miR-449b prevented the development of cardiac ischemia-reperfusion injury in diabetic conditions. Accumulating studies support the concept that imbalanced oxidative stress and reduction of antioxidant capacity contribute to myocardial tissue injury secondary to ischemia-reperfusion.[47] [48, 49] More argument focus on excess oxidative stress caused by the burst of reactive oxygen species (ROS) in the presence of inadequate antioxidant defense, which has been considered a potential mechanism governing the increased sensitivity of the diabetic heart to MI/R.[50–52] However, effective strategies are short for promoting the compromised antioxidant signals. This study demonstrated that miR-449b targeted antioxidants during ischemia-reperfusion injury in diabetic conditions by inhibiting the endogenous antioxidant molecules Ucp3 and Nrf-1. Furthermore, overexpressing Nrf-1 antioxidants helps to protect against cardiac dysfunction and is insensible to the injury. The role of microRNAs in pathological stress conditions has been well established. [50] But little is known regarding miR-449b in the heart pathogenesis under either diabetic or MI/R conditions. It is worth to note that, miRNA-449b expression was measured by absolute quantitative digital PCR and confirmed by Real time quantificative PCR (Figure S5), which indicated that miRNA-449b expression differed between ischemia injuries without diabetes and those with diabetes. Despite the published data [53, 54] indicating nuance assessment of miRNA-449b, our study has developed detectable methods that have unequivocally demonstrated the superior biomarker potential of miRNA-449b compared to other miRNAs. According to our knowledge, this is the first study to report upregulation of miR-449b in cardiomyocytes under diabetic MI/R and its role in such conditions, whereas such a phenomenon wasn’t observed in those without diabetes. Antioxidant imbalance is a feature of ischemia-reperfusion injury.[55] Effective control of the overexpressed ROS may alleviate the progression of exacerbated injury in the diabetic heart. Therefore, miR-449b or Nrf-1 molecule could be promising therapeutic targets to in diabetic patients suffering MI/R. The miR-449b and the targets we identified in this study, Ucp3 and Nrf-1, provide potential therapeutic targets for a new antioxidant strengthen oriented treatment for diabetic ischemia-reperfusion injury.
It should be indicated that our study has some limitations. First, it is important to note that no cell model can fully replicate all the phenomena in an in vivo setting. To increase the translational value of in vitro experimental findings (i.e., HGHL treatment as an in vitro diabetic model), we validated in vitro findings with HFD animal model, and confirmed the targets and functional signals identified from in vitro studies. Before proceeding to clinical diagnostic application, a large scale evaluation of individuals using the developed dPCR is required although the pre-clinic verification has been achieved to some extent. Second, cell-specific anti-miR therapy is currently unavailable in a clinical setting. Systemic anti-miR treatment in diabetic patients leads to a general suppression of microRNA, which might influence inflammatory processes, leading to unwanted side effects. These concerns also apply to gene therapy for Nrf-1. Thirdly, diabetic patients with IHD is a very complex clinical condition; elevated blood glucose and lipid levels are severe risks to the internal environment. Although miR-449b is correlated with diabetic IHD, further study with a larger patient population is necessary to precisely determine the clinical role of miR-449b in diabetic IHD versus non-diabetic IHD. Fouthly, we provided evidence that miR-449b expression was dramatically increased after MI/R in APN-KO heart and HFD heart but not in WT non-diabetic heart, indicating that miR-449b is a MI/R inducible factor specifically in diabetic and hypoadiponectinemic conditions. However, how APN regulates miR-449b expression remains to be determined. Several possibilities exist. First, inflammatory and oxidative stress regulate miRNA biogenesis. [56] Previous studies demonstrate that APN protects against ischemic heart injury by decreasing inflammation. Diabetic suppression of APN may enhance inflammatory responses to upregulate miR-449b biogenesis. Second, increased superoxide production in hypoadiponectinemic and diabetic MI/R heart could activate miRNA biogenesis signals [57]; Third, hypoadiponectinemia-induced metabolic disorder could contribute to miRNA biogenesis. These possibilities will be investigated in our future study.
5. Conclusion
In summary, this study demonstrated that in high fat-induced diabetic conditions, mice displayed reduced antioxidant capacity leading to upregulated myocardial oxidative stress, which rendered the diabetic heart more vulnerable to ischemia-reperfusion injury. microRNA 449b and antioxidant molecule Nrf-1 screened out in this study exhibit strong potential to serve as the therapeutic target in treating diabetes-related cardiovascular system disease.
Supplementary Material
Acknowledgments
We appreciate Dr. Changgong Liu’s help designing the miRNA database and analyzing miRNA profiles. We also appreciate Shanxi Medical University for sharing and exploring the biobank database of IHD patients. The graphic abstract is created with BioRender.com.
Funding
This work was supported by awards from the National Institutes of Health (HL096686, Ma/ Wang, MPI; HL-123404, Ma; HL158612 and HL157495, Wang) and the American Heart Association (20TPA35490095, Wang).
Footnotes
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
References
- [1].McGovern PG, Pankow JS, Shahar E, Doliszny KM, Folsom AR, Blackburn H, Luepker RV, Recent trends in acute coronary heart disease--mortality, morbidity, medical care, and risk factors. The Minnesota Heart Survey Investigators, N Engl J Med 334(14) (1996) 884–90. [DOI] [PubMed] [Google Scholar]
- [2].Saidi O, Ben Mansour N, O’Flaherty M, Capewell S, Critchley JA, Ben Romdhane H, Analyzing recent coronary heart disease mortality trends in Tunisia between 1997 and 2009, PLoS One 8(5) (2013) e63202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Crowley A, Menon V, Lessard D, Yarzebski J, Jackson E, Gore JM, Goldberg RJ, Sex differences in survival after acute myocardial infarction in patients with diabetes mellitus (Worcester Heart Attack Study), Am Heart J 146(5) (2003) 824–31. [DOI] [PubMed] [Google Scholar]
- [4].Marso SP, Miller T, Rutherford BD, Gibbons RJ, Qureshi M, Kalynych A, Turco M, Schultheiss HP, Mehran R, Krucoff MW, Lansky AJ, Stone GW, Comparison of myocardial reperfusion in patients undergoing percutaneous coronary intervention in ST-segment elevation acute myocardial infarction with versus without diabetes mellitus (from the EMERALD Trial), Am J Cardiol 100(2) (2007) 206–10. [DOI] [PubMed] [Google Scholar]
- [5].Lau WB, Ohashi K, Wang Y, Ogawa H, Murohara T, Ma XL, Ouchi N, Role of Adipokines in Cardiovascular Disease, Circ J 81(7) (2017) 920–928. [DOI] [PubMed] [Google Scholar]
- [6].Goldstein BJ, Scalia R, Adipokines and vascular disease in diabetes, Curr. Diab. Rep 7(1) (2007) 25–33. [DOI] [PubMed] [Google Scholar]
- [7].Pischon T, Girman CJ, Hotamisligil GS, Rifai N, Hu FB, Rimm EB, Plasma adiponectin levels and risk of myocardial infarction in men, JAMA 291(14) (2004) 1730–1737. [DOI] [PubMed] [Google Scholar]
- [8].Frystyk J, Berne C, Berglund L, Jensevik K, Flyvbjerg A, Zethelius B, Serum adiponectin is a predictor of coronary heart disease: a population-based 10-year follow-up study in elderly men, J Clin. Endocrinol. Metab 92 (2007) 571–576. [DOI] [PubMed] [Google Scholar]
- [9].Kojima S, Funahashi T, Otsuka F, Maruyoshi H, Yamashita T, Kajiwara I, Shimomura H, Miyao Y, Fujimoto K, Sugiyama S, Sakamoto T, Yoshimura M, Ogawa H, Future adverse cardiac events can be predicted by persistently low plasma adiponectin concentrations in men and marked reductions of adiponectin in women after acute myocardial infarction, Atherosclerosis 194 (2006) 204–213. [DOI] [PubMed] [Google Scholar]
- [10].Shibata R, Numaguchi Y, Matsushita K, Sone T, Kubota R, Ohashi T, Ishii M, Kihara S, Walsh K, Ouchi N, Murohara T, Usefulness of adiponectin to predict myocardial salvage following successful reperfusion in patients with acute myocardial infarction, Am J Cardiol 101(12) (2008) 1712–1715. [DOI] [PubMed] [Google Scholar]
- [11].Tao L, Gao E, Jiao X, Yuan Y, Li S, Christopher TA, Lopez BL, Koch W, Chan L, Goldstein BJ, Ma XL, Adiponectin cardioprotection after myocardial ischemia/reperfusion involves the reduction of oxidative/nitrative stress, Circulation 115(11) (2007) 1408–1416. [DOI] [PubMed] [Google Scholar]
- [12].Huang Y, Zou Q, Wang SP, Tang SM, Zhang GZ, Shen XJ, The discovery approaches and detection methods of microRNAs, Mol Biol Rep 38(6) (2011) 4125–35. [DOI] [PubMed] [Google Scholar]
- [13].Song C, Wang C, Zhang C, Korir NK, Yu H, Ma Z, Fang J, Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata), BMC Genomics 11 (2010) 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Quiat D, Olson EN, MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment, J Clin Invest 123(1) (2013) 11–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yi W, Sun Y, Gao E, Wei X, Lau WB, Zheng Q, Wang Y, Yuan Y, Wang X, Tao L, Li R, Koch W, Ma XL, Reduced cardioprotective action of adiponectin in high-fat diet-induced type II diabetic mice and its underlying mechanisms, Antioxid Redox Signal 15(7) (2011) 1779–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gan L, Xie D, Liu J, Bond Lau W, Christopher TA, Lopez B, Zhang L, Gao E, Koch W, Ma XL, Wang Y, Small Extracellular Microvesicles Mediated Pathological Communications Between Dysfunctional Adipocytes and Cardiomyocytes as a Novel Mechanism Exacerbating Ischemia/Reperfusion Injury in Diabetic Mice, Circulation 141(12) (2020) 968–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu J, Meng Z, Gan L, Guo R, Gao J, Liu C, Zhu D, Liu D, Zhang L, Zhang Z, Xie D, Jiao X, Lau WB, Lopez BL, Christopher TA, Ma X, Cao J, Wang Y, C1q/TNF-related protein 5 contributes to diabetic vascular endothelium dysfunction through promoting Nox-1 signaling, Redox Biol 34 (2020) 101476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang Y, Wang X, Jasmin JF, Lau WB, Li R, Yuan Y, Yi W, Chuprun K, Lisanti MP, Koch WJ, Gao E, Ma XL, Essential role of caveolin-3 in adiponectin signalsome formation and adiponectin cardioprotection, Arterioscler Thromb Vasc Biol 32(4) (2012) 934–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu CG, Calin GA, Volinia S, Croce CM, MicroRNA expression profiling using microarrays, Nat Protoc 3(4) (2008) 563–78. [DOI] [PubMed] [Google Scholar]
- [20].Naga Prasad SV, Gupta MK, Duan ZH, Surampudi VS, Liu CG, Kotwal A, Moravec CS, Starling RC, Perez DM, Sen S, Wu Q, Plow EF, Karnik S, A unique microRNA profile in end-stage heart failure indicates alterations in specific cardiovascular signaling networks, PLoS One 12(3) (2017) e0170456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhao J, Wang F, Zhang Y, Jiao L, Lau WB, Wang L, Liu B, Gao E, Koch WJ, Ma XL, Wang Y, Sevoflurane preconditioning attenuates myocardial ischemia/reperfusion injury via caveolin-3-dependent cyclooxygenase-2 inhibition, Circulation 128(11 Suppl 1) (2013) S121–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang Y, Lau WB, Gao E, Tao L, Yuan Y, Li R, Wang X, Koch WJ, Ma XL, Cardiomyocyte-derived adiponectin is biologically active in protecting against myocardial ischemia-reperfusion injury, Am J Physiol Endocrinol Metab 298(3) (2010) E663–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang Y, Gao E, Tao L, Lau WB, Yuan Y, Goldstein BJ, Lopez BL, Christopher TA, Tian R, Koch W, Ma XL, AMP-activated protein kinase deficiency enhances myocardial ischemia/reperfusion injury but has minimal effect on the antioxidant/antinitrative protection of adiponectin, Circulation 119(6) (2009) 835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhu WZ, Wang SQ, Chakir K, Yang D, Zhang T, Brown JH, Devic E, Kobilka BK, Cheng H, Xiao RP, Linkage of beta1-adrenergic stimulation to apoptotic heart cell death through protein kinase A-independent activation of Ca2+/calmodulin kinase II, J Clin Invest 111(5) (2003) 617–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yang Y, Cheng HW, Qiu Y, Dupee D, Noonan M, Lin YD, Fisch S, Unno K, Sereti KI, Liao R, MicroRNA-34a Plays a Key Role in Cardiac Repair and Regeneration Following Myocardial Infarction, Circ Res 117(5) (2015) 450–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brennan E, Wang B, McClelland A, Mohan M, Marai M, Beuscart O, Derouiche S, Gray S, Pickering R, Tikellis C, de Gaetano M, Barry M, Belton O, Ali-Shah ST, Guiry P, Jandeleit-Dahm KAM, Cooper ME, Godson C, Kantharidis P, Protective Effect of let-7 miRNA Family in Regulating Inflammation in Diabetes-Associated Atherosclerosis, Diabetes 66(8) (2017) 2266–2277. [DOI] [PubMed] [Google Scholar]
- [27].Carbe CJ, Cheng L, Addya S, Gold JI, Gao E, Koch WJ, Riobo NA, Gi proteins mediate activation of the canonical hedgehog pathway in the myocardium, Am J Physiol Heart Circ Physiol 307(1) (2014) H66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Han L, Xu C, Jiang C, Li H, Zhang W, Zhao Y, Zhang L, Zhang Y, Zhao W, Yang B, Effects of polyamines on apoptosis induced by simulated ischemia/reperfusion injury in cultured neonatal rat cardiomyocytes, Cell Biol Int 31(11) (2007) 1345–52. [DOI] [PubMed] [Google Scholar]
- [29].Liu GZ, Liang B, Lau WB, Wang Y, Zhao J, Li R, Wang X, Yuan Y, Lopez BL, Christopher TA, Xiao C, Ma XL, Wang Y, High glucose/High Lipids impair vascular adiponectin function via inhibition of caveolin-1/AdipoR1 signalsome formation, Free Radic Biol Med 89 (2015) 473–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang J, Song Y, Elsherif L, Song Z, Zhou G, Prabhu SD, Saari JT, Cai L, Cardiac metallothionein induction plays the major role in the prevention of diabetic cardiomyopathy by zinc supplementation, Circulation 113(4) (2006) 544–54. [DOI] [PubMed] [Google Scholar]
- [31].Yao Q, Ke ZQ, Guo S, Yang XS, Zhang FX, Liu XF, Chen X, Chen HG, Ke HY, Liu C, Curcumin protects against diabetic cardiomyopathy by promoting autophagy and alleviating apoptosis, Journal of molecular and cellular cardiology 124 (2018) 26–34. [DOI] [PubMed] [Google Scholar]
- [32].Li P, Zhu N, Yi B, Wang N, Chen M, You X, Zhao X, Solomides CC, Qin Y, Sun J, MicroRNA-663 regulates human vascular smooth muscle cell phenotypic switch and vascular neointimal formation, Circ Res 113(10) (2013) 1117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang Y, Gao E, Lau WB, Wang Y, Liu G, Li JJ, Wang X, Yuan Y, Koch WJ, Ma XL, G-protein-coupled receptor kinase 2-mediated desensitization of adiponectin receptor 1 in failing heart, Circulation 131(16) (2015) 1392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bissels U, Wild S, Tomiuk S, Holste A, Hafner M, Tuschl T, Bosio A, Absolute quantification of microRNAs by using a universal reference, RNA 15(12) (2009) 2375–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].D’Alessandra Y, Valerio V, Moschetta D, Massaiu I, Bozzi M, Conte M, Parisi V, Ciccarelli M, Leosco D, Myasoedova VA, Poggio P, Extraction-Free Absolute Quantification of Circulating miRNAs by Chip-Based Digital PCR, Biomedicines 10(6) (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M, Absolute quantification by droplet digital PCR versus analog real-time PCR, Nat Methods 10(10) (2013) 1003–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tao L, Wang Y, Gao E, Zhang H, Yuan Y, Lau WB, Chan L, Koch WJ, Ma XL, Adiponectin: an indispensable molecule in rosiglitazone cardioprotection following myocardial infarction, Circ Res 106(2) (2010) 409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang Y, Lv J, Wu G, Li W, Zhang Z, Li W, Lei X, MicroRNA-449b-5p targets HMGB1 to attenuate hepatocyte injury in liver ischemia and reperfusion, J Cell Physiol (2019). [DOI] [PubMed] [Google Scholar]
- [39].Xu Y, Niu Y, Li H, Pan G, Downregulation of lncRNA TUG1 attenuates inflammation and apoptosis of renal tubular epithelial cell induced by ischemia-reperfusion by sponging miR-449b-5p via targeting HMGB1 and MMP2, Inflammation 43(4) (2020) 1362–1374. [DOI] [PubMed] [Google Scholar]
- [40].Cao C, Liu HM, Li W, Wu Y, Leng Y, Xue R, Chen R, Tang LH, Sun Q, Xia Z, Tang QZ, Shen DF, Meng QT, Role of adiponectin in diabetes myocardial ischemia-reperfusion injury and ischemic postconditioning, Acta Cir Bras 35(1) (2020) e202000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Eroglu S, Sade LE, Bozbas H, Haberal A, Ozbicer S, Demir O, Muderrisoglu H, Association of serum adiponectin levels and coronary flow reserve in women with normal coronary angiography, Eur J Cardiovasc Prev Rehabil 16(3) (2009) 290–6. [DOI] [PubMed] [Google Scholar]
- [42].Hashimoto N, Kanda J, Nakamura T, Horie A, Kurosawa H, Hashimoto T, Sato K, Kushida S, Suzuki M, Yano S, Iwai R, Takahashi H, Yoshida S, Association of hypoadiponectinemia in men with early onset of coronary heart disease and multiple coronary artery stenoses, Metabolism 55(12) (2006) 1653–7. [DOI] [PubMed] [Google Scholar]
- [43].Kawano T, Saito T, Yasu T, Saito T, Nakamura T, Namai K, Tamemoto H, Kawakami M, Saito M, Ishikawa SE, Close association of hypoadiponectinemia with arteriosclerosis obliterans and ischemic heart disease, Metabolism 54(5) (2005) 653–6. [DOI] [PubMed] [Google Scholar]
- [44].Kumada M, Kihara S, Sumitsuji S, Kawamoto T, Matsumoto S, Ouchi N, Arita Y, Okamoto Y, Shimomura I, Hiraoka H, Nakamura T, Funahashi T, Matsuzawa Y, C.A.D.S.G.C.a.d. Osaka, Association of hypoadiponectinemia with coronary artery disease in men, Arterioscler Thromb Vasc Biol 23(1) (2003) 85–9. [DOI] [PubMed] [Google Scholar]
- [45].Di Chiara A, Dall’Armellina E, Badano LP, Meduri S, Pezzutto N, Fioretti PM, Predictive value of cardiac troponin-I compared to creatine kinase-myocardial band for the assessment of infarct size as measured by cardiac magnetic resonance, J Cardiovasc Med (Hagerstown) 11(8) (2010) 587–92. [DOI] [PubMed] [Google Scholar]
- [46].Christenson RH, Vollmer RT, Ohman EM, Peck S, Thompson TD, Duh SH, Ellis SG, Newby LK, Topol EJ, Califf RM, Relation of temporal creatine kinase-MB release and outcome after thrombolytic therapy for acute myocardial infarction. TAMI Study Group, Am J Cardiol 85(5) (2000) 543–7. [DOI] [PubMed] [Google Scholar]
- [47].Galinanes M, Fowler AG, Role of clinical pathologies in myocardial injury following ischaemia and reperfusion, Cardiovasc Res 61(3) (2004) 512–21. [DOI] [PubMed] [Google Scholar]
- [48].Zhu HJ, Wang DG, Yan J, Xu J, Up-regulation of microRNA-135a protects against myocardial ischemia/reperfusion injury by decreasing TXNIP expression in diabetic mice, Am J Transl Res 7(12) (2015) 2661–71. [PMC free article] [PubMed] [Google Scholar]
- [49].Yang Z, Laubach VE, French BA, Kron IL, Acute hyperglycemia enhances oxidative stress and exacerbates myocardial infarction by activating nicotinamide adenine dinucleotide phosphate oxidase during reperfusion, J Thorac Cardiovasc Surg 137(3) (2009) 723–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kitano D, Takayama T, Nagashima K, Akabane M, Okubo K, Hiro T, Hirayama A, A comparative study of time-specific oxidative stress after acute myocardial infarction in patients with and without diabetes mellitus, BMC Cardiovasc Disord 16 (2016) 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Katakami N, Sakamoto K, Kaneto H, Matsuhisa M, Ohno K, Shimizu I, Ishibashi F, Osonoi T, Kashiwagi A, Kawamori R, Yamasaki Y, Cumulative effect of oxidative stress-related gene polymorphisms on myocardial infarction in type 2 diabetes, Diabetes Care 32(5) (2009) e55. [DOI] [PubMed] [Google Scholar]
- [52].Di Filippo C, Cuzzocrea S, Rossi F, Marfella R, D’Amico M, Oxidative stress as the leading cause of acute myocardial infarction in diabetics, Cardiovasc Drug Rev 24(2) (2006) 77–87. [DOI] [PubMed] [Google Scholar]
- [53].Huang W, Feng Y, Liang J, Yu H, Wang C, Wang B, Wang M, Jiang L, Meng W, Cai W, Medvedovic M, Chen J, Paul C, Davidson WS, Sadayappan S, Stambrook PJ, Yu XY, Wang Y, Loss of microRNA-128 promotes cardiomyocyte proliferation and heart regeneration, Nat Commun 9(1) (2018) 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Park TJ, Park JH, Lee GS, Lee JY, Shin JH, Kim MW, Kim YS, Kim JY, Oh KJ, Han BS, Kim WK, Ahn Y, Moon JH, Song J, Bae KH, Kim DH, Lee EW, Lee SC, Quantitative proteomic analyses reveal that GPX4 downregulation during myocardial infarction contributes to ferroptosis in cardiomyocytes, Cell Death Dis 10(11) (2019) 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Climent M, Viggiani G, Chen YW, Coulis G, Castaldi A, MicroRNA and ROS Crosstalk in Cardiac and Pulmonary Diseases, Int J Mol Sci 21(12) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ge Q, Gerard J, Noel L, Scroyen I, Brichard SM, MicroRNAs regulated by adiponectin as novel targets for controlling adipose tissue inflammation, Endocrinology 153(11) (2012) 5285–96. [DOI] [PubMed] [Google Scholar]
- [57].Akbari A, Majd HM, Rahnama R, Heshmati J, Morvaridzadeh M, Agah S, Amini SM, Masoodi M, Cross-talk between oxidative stress signaling and microRNA regulatory systems in carcinogenesis: Focused on gastrointestinal cancers, Biomed Pharmacother 131 (2020) 110729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


