Abstract
The human microbiota has been established as a key regulator of host health, in large part due to its constant interaction with, and impact on, host immunity. A range of environmental exposures, spanning from the prenatal period through adulthood are now known to impact the composition and molecular productivity of microbiomes across mucosal and dermal tissues with short- and long-term consequences for host immune function. Here we review the more recent findings in the field that provide insights into how microbial-immune interactions promote and sustain immune dysfunction associated with allergy and asthma. We consider both early life microbiome perturbation and the molecular underpinnings of immune dysfunction associated with subsequent allergy and asthma development in childhood, as well as microbiome features that relate to phenotypic attributes of allergy and asthma in older patients with established disease.
Keywords: Microbiota, Airway, Gut, Immunology, Immunometabolism
Introduction
The prevalence of allergy and asthma has increased significantly over the past several generations; data from national and state surveillance systems administered by the Centers for Disease Control and Prevention indicate that the burden of asthma in the United States population in 2019 was 7.8%. Recent meta-analyses in US populations showed that childhood asthma incidence rates vary by age, sex, parental asthma history, race/ethnicity, and calendar year with higher rates observed in younger children, particularly African American and Caribbean American populations (1, 2). Changes in disease incidence rates over time and with demographic factors (1) indicate that complex interactions between the human host and time-dependent variation in environmental and social factors underlie disease development and expression. Efforts to treat established disease have been met with some success but indicate that early intervention is key. For example, peanut oral immunotherapy is effective but age-dependent, with older children less likely to be successfully desensitized and tolerized (3), suggesting that very early-life priming of immunity is key to allergic disease development and that interventions for disease prevention should target this window of development.
These observations have led to increased interest in the developmental origins of allergy and asthma as well as the dynamic interactions with host immunity that promote pathogenesis in later life. Consistently, allergy and asthma-associated immune dysfunction is coincident with perturbation to airway, skin and gut microbiomes (4–8). That perturbed microbiomes are drivers of disease and not simply a bystander effect of disease processes comes from elegant germ-free mouse experiments (9). Transfer of feces from an infant with cow’s milk allergy to previously germ-free mice promoted an anaphylactic response to sensitization with cow’s milk allergen β-lactoglobulin (BLG) and increased BLG-specific IgE (9), indicating a functional contribution to allergic sensitization by the gut microbiome. In the airways, a relatively small number of distinct age-related airway microbiota colonization patterns are evident, that relate to risk of asthma development in later childhood (10–12), or to risk of exacerbation or loss of asthma control in older patients with diagnosed disease (13, 14). More recently it has become evident that the airway bacterial and viral microbiota exhibit seasonal dynamics, and that in the Fall, networks of upper airway bacteria interact with epithelial response to viral infection events to significantly increase asthma exacerbation risk (15). These findings underscore the complexity of the disease and highlights the temporal and dynamic nature of microbial-host interactions at the two largest mucosal surfaces that shape allergy and asthma development and disease expression.
Since immune function serves as the rheostat for human health, the capacity to sense, respond and clear noxious environmental or pathogenic microbial exposures is fundamental to host immunoprotection and relatively well understood (16). However, the ability to tolerate antigenic stimuli or, indeed, the burden and diversity of microbes that exist in human microbiomes is somewhat enigmatic. In humans, immune cell populations with the capacity for inflammatory cytokine production and established memory responses to microbes are evident in the fetal intestine as early as the second trimester of pregnancy (17, 18). In parallel, sparse colonies of viable bacteria with the capacity to induce memory T cell activation, expand fetal lymph node T cells or reduce inflammatory cytokine production by fetal-specific T cell populations are also detectable in the human fetal intestine by mid-gestation (17, 19). These data indicate that immune priming events relevant to allergy and asthma commence in utero and are dependent, in part, on maternal microbiomes. Evidence that prenatal microbial-immune priming events have long-lasting effects on immune function comes from mouse studies; prebiotic supplementation of pregnant dams promoted short chain fatty acid production by the maternal gut microbiome and increased the abundance of regulatory B and T cells in the fetus, tolerogenic effects that were sustained in the postnatal period (20). Independently, mouse models of prenatal infection indicate that maternal prenatal microbial-immune interactions shape fetal and early life immune function in part, via epigenetic imprinting (21). These processes are reliant on substrate availability and the activity of metabolic pathways which are intimately related to microbial activities. In post-natal life microbiome perturbation coupled with metabolic dysfunction precedes allergy and asthma development in childhood (22–25), relationships that remain evident in preschool aged children (26). The human microbiome develops across body sites and with advancing age (27, 28), and is influenced by a large number of exposures known to be risk factors for allergy and asthma development. This fact coupled with the knowledge that microbes tune immune function indicate that temporal and dynamic microbial-immune interactions are paramount to allergy and asthma development and chronicity. Here we provide an overview of the most recent developments implicating successional microbial mechanisms of immune dysfunction across life stages that promote allergy and asthma (Figure 1) and propose a framework for the developmental origins and temporal disease dynamics observed in patient populations.
Figure 1.
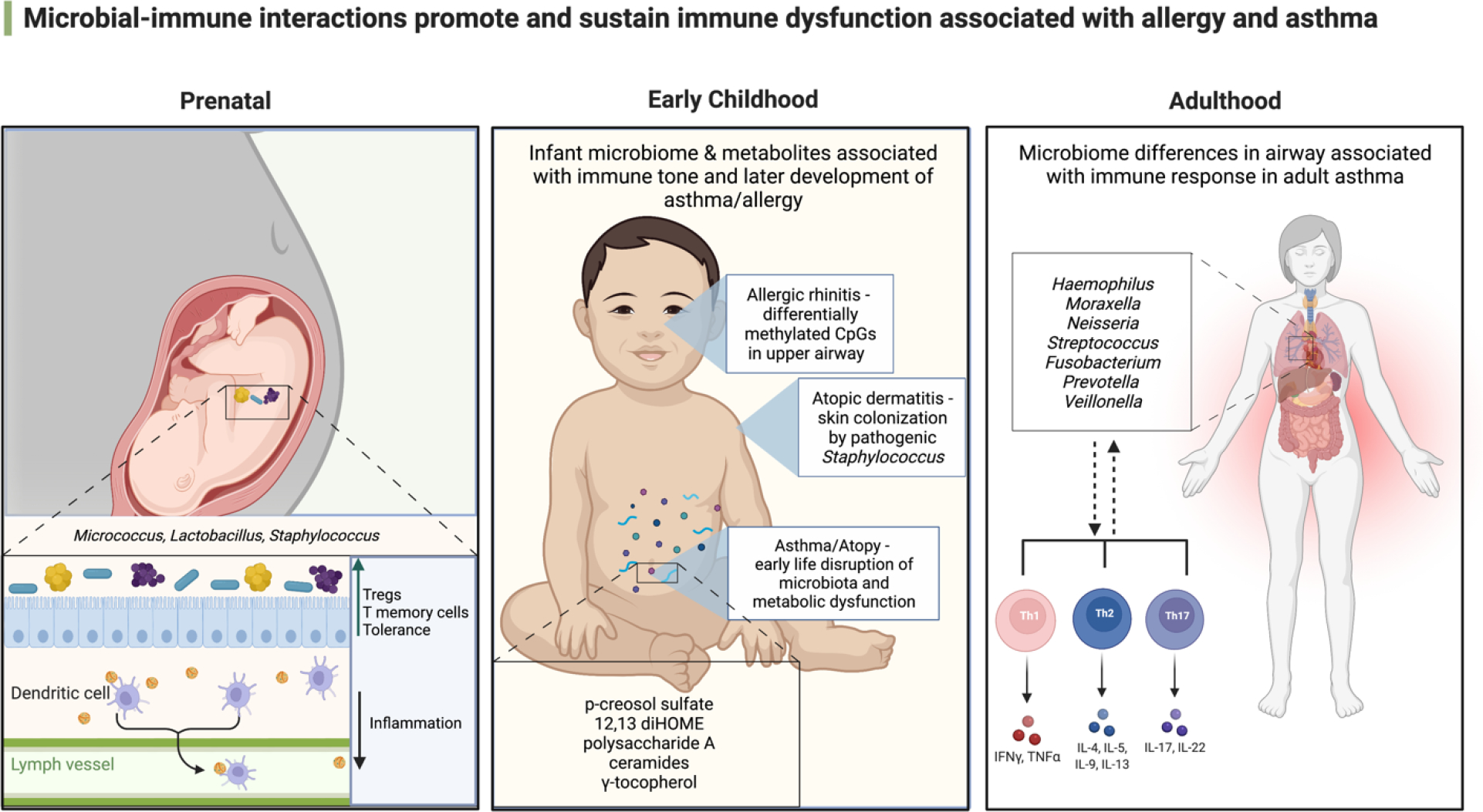
Microbial immune interactions promote and sustain immune dysfunction associated with allergy and asthma. Adapted from BioRender.com. Lipid handling in the small intestine modulates immune system homeostasis. https://app.biorender.com/biorender-templates.29
Early life development relates to childhood allergy and asthma outcomes.
Dermal, intestinal and respiratory microbiomes develop over the first several years of life, with body habitat-specific microbial colonization patterns in infancy associated with increased risk of atopy and asthma development in later childhood (10, 23, 25, 30–34). In infancy, upper airway colonization by respiratory pathogens such as Haemophilus influenzae, Moraxella catarrhalis or Streptococcus pneumoniae is associated with early life wheeze, febrile respiratory illness and asthma in later childhood (10–12). These particular species also thrive in chronically inflamed airways and are also associated with respiratory diseases like pneumonia and chronic obstructive pulmonary disease (35–37). Therefore, these microbes may be more broadly indicative of pathogenic microbial activity and dysregulation of conserved immune pathways related to these events. More recently, in the Copenhagen Prospective Studies on Asthma in Childhood (COPSAC) birth cohort, upper airway microbial composition in infants was associated with allergic rhinitis development 6 years of age, mediated in part by differentially methylated CpGs in upper airway epithelial cells (38). Studies of neonatal murine skin have determined that commensal Staphylococcus epidermidis colonization is required to promote regulatory T cell accumulation in hair follicles – a key site of bacterial colonization (39). In humans at 3 months of age, skin colonization by pathogenic Staphylococcus aureus is more prevalent in infants who develop atopic dermatitis (40). In the gut, early life microbiota perturbation and metabolic dysfunction precedes atopy and asthma development in childhood (23, 24). A number of studies to date have focused on the gut microbiota which houses the largest burden and diversity of microbes in the human body and produces a diverse repertoire of bioactive molecules. These studies provide insights into how early life microbial encounters prime immune function and offer a framework for improved understanding of the developmental origins of allergy and asthma across other body sites. Gut microbiome composition is dynamic, particularly in early life when it rapidly diversifies over the first years of life before stabilizing, with respect to dominant bacterial phylum distribution, by age 3 years (27). Despite phylum-level stability, microbial species, strains and their encoded genes continue to evolve throughout life course, in part shaped by extrinsic exposures such as pharmaceuticals, diet and antimicrobial exposures (40–43), but also by intrinsic factors such as epigenetic modifications (44). Despite these influences, recent data has indicated that bacterial strains that initially colonize the human intestine in very early life are detectable in adulthood (45), suggesting that pioneer microbial colonizers may represent sustained and contributing members to the overall function of the microbiome and its interactions with host immunity throughout an individual’s lifetime.
Early life is a pivotal time for the establishment and development of both the gut microbiome and immune system. Human fetal dendritic cells, which are crucial for effective immunity and tolerance, can migrate to lymph nodes and respond to toll-like receptor ligation (canonical receptor for microbial ligands) to induce regulatory T-cells that reduce inflammation in utero (46). Recent evidence suggests that initial intestinal encounters with viable bacteria commences in utero. Human fetal intestinal colonization by sparse colonies of a limited number of bacterial species including Lactobacillus, Staphylococcus and Micrococcus is evident as early as the second trimester of pregnancy (17, 19). The presence of these fetal bacteria corresponds with distinct programs of innate and adaptive immunity. For example, Micrococcus luteus, detected in human fetal intestinal samples, associated with increased expression of intestinal epithelial genes involved in host response to microbes, e.g. NFκB, BCL6 (modulates transcription of STAT-dependent B cell IL-4 responses), and with accumulation of fetal specific T-cells (19). Moreover, a fetal isolate of M. luteus exhibited the capacity to survive in fetal antigen presenting cells, metabolize progesterone and estradiol and reduce interferon gamma expression by T-cells, indicating strategies by which specific bacteria may survive the hostile environment of the fetal gut. These and more recent findings from independent studies suggest that the fetal intestine can support a very limited number of bacterial species, and that those species identified and isolated from this unique niche contribute to immune cell priming and development of bacterial antigen experienced memory T cell populations in utero (17, 19). These data suggest that the origins of tolerance priming can occur in utero and is, in part, dependent on translocation of maternal prenatal microbes to the developing fetus.
After birth, mode of delivery, antimicrobial exposure and early life nutrition represent some of the key exposures that influence the pace of early life gut microbiome establishment (27, 47–50),, which has now been linked to asthma risk in later childhood (25, 32). Independent human birth cohorts in the United States and Europe have shown that delayed gut microbiota development is characteristic of infants at significantly higher risk of asthma in later childhood (25, 32). Appropriately paced infant gut microbial development with key bacteria including Lactobacillus, Bifidobacterium and Faecalibacterium, is associated with protection against atopy and asthma development in later childhood (24). These microbes thrive on human milk oligosaccharides and produce bioactive metabolites that influence brain, adipose tissue, and immune cell functional development (51–53). Breastmilk, a complex and dynamic living liquid, supports the growth and activities of lactic acid bacteria and Bifidobacteria in the gut (54), which prevent enteric pathogen invasion and inflammation (55, 56). Breastmilk also contains microbes, immune cells and their products, e.g. immunoglobulin A and anti-microbial products such as lactoferrin which further regulate early life gut microbiome species accumulation and prevent pathogenic microbial activity and associated inflammation (57–63). These data indicate that failure to regulate early-life microbial development particularly in the gastrointestinal tract, contributes to risk of atopic disease and asthma development in later childhood. Though parallel longitudinal skin microbiome studies are currently lacking, the existing cross-sectional evidence (61) and longitudinal early life airway studies (10, 30, 31) suggest that early life microbial colonization status at these sites also contributes to local immune tuning and to childhood allergy and asthma risk.
Microbial-derived metabolites shape immune function.
Numerous clinical and preclinical studies have demonstrated that early life microbial perturbation and metabolic dysfunction associates with heightened risk of allergic and asthmatic phenotypes in later childhood (23, 24). Murine studies have shown that even transient microbiome perturbation in early life is sufficient to promote long-term effects on host metabolism (64). This suggests that microbe-host metabolic interactions in infancy may have lasting consequences for host health. Indeed, a number of birth cohort studies have now shown that early life gut microbiome metabolic dysfunction is a recurring characteristic of childhood atopy and asthma development and have highlighted suites of microbial metabolites that co-vary with risk of atopy or asthma development in later childhood (23–26). Data from multiple fields now indicates that gut microbial-derived metabolites influence remote tissues and organs (65, 66). Encouragingly, a relatively consistent metabolic signature dominated by lipid metabolites (depletion of polyunsaturated fatty acids, PUFAs, and enrichment of mono-hydroxy fatty acids), is consistently observed at various stages of early life development (1 month and 3 years) in those at heightened risk of developing childhood disease (23, 24, 26). Several studies have provided evidence for the immunomodulatory effect of microbial derived metabolites (Table 1). The anti-inflammatory effects of microbial-derived short chain fatty acids are well described. We refer readers to an excellent recent review article detailing these activities (74). More recent studies have identified bacterial-derived metabolites that protect against allergic airway inflammation, including p-cresol sulfate, an L-tyrosine derivative that reduces CCL20 (a lymphocyte chemoattractant) production by airway epithelial cells (67). Bacterial-derived inflammatory mediators have also been associated with increased risk of atopy and asthma. For example, elevated fecal concentrations of 12,13-diHOME in infancy increases risk of atopy and asthma in childhood. Mechanistically, this gut bacterial-derived oxylipin promotes allergic inflammation by decreasing the frequency and IL-10 productivity of lung Treg populations (24, 68). Gamma-Tocopherol, a vitamin E metabolite, has also been implicated in increased risk of asthma in human cohorts, and has been shown to increase the inflammatory response to aeroallergens in mouse models of asthma (26, 69). Skin associated microbes and their metabolites can contribute to the development of atopic dermatitis (75). Most notably ceramides which are important for maintaining healthy skin, decrease in concentration in the context of atopic dermatitis (70). Murine models have demonstrated that decreasing Pseudomonas ceramidase activity alleviates skin inflammation, implicating microbial metabolic activity as a driver of atopic disease in this case (71). Additionally, in the colon commensal gut bacteria such as Bacteroides fragilis, produce anti-inflammatory products like polysaccharide A which decreased IL-17 production and colonic inflammation in mouse models, further identifying bacterial products of key regulators of immune cell function (73).
Table 1.
Recently identified microbial metabolites that shape immune function in early life.
| Bacterial derived metabolite | Molecule class | Influence on immunity | Allergic disease association | Reference |
|---|---|---|---|---|
| p-cresol sulfate | Amino acid derivative | Reduces CCL20 production | Atopy/asthma | 67 |
| 12,13-diHOME | oxylipin | Decreases lung Tregs and IL-10 production | Atopy/asthma | 24, 68 |
| Gamma-Tocopherol | Vitamin E metabolite | Increases CCL11, amphiregulin, activin A, and IL-5 | Atopy/asthma | 69 |
| ceramides | lipid | Reduce mast cell numbers | Atopic dermatitis | 70,71 |
| Tryptophan metabolites | Amino acids | Aryl hydrocarbon receptor ligation | asthma | 72 |
| Polysaccharide A | carbohydrate | Decreases Th17 inflammation | Colonic inflammation | 73 |
Moreover, gut microbial manipulation resulting in reprogramming of metabolism has beneficial effects on airway inflammatory responses. Oral supplementation of the gut microbiome with Lactobacillus johnsonii protects mice from airway allergic inflammatory response to allergen exposure or viral respiratory infection (76). Introduction of this single microbial species into the gut microbiome of animals subjected to airway infection with respiratory syncytial virus resulted in significant changes in the serum metabolome 48 hours post infection, including increased concentrations of circulating PUFAs and depletion of monohydroxy fatty acids, including 12,13 DiHOME (76, 77), suggesting that gut microbiome-produced or induced metabolites that shape airway response to inhaled insults. A more recent study indicates that maternal pre-natal and pup post-natal supplementation with L. johnsonii affords the greatest protection against airway inflammatory response to viral infection (78), implicating microbial-influenced pre- and post-natal immune training and function as key to protection against early life airway insults.
The mechanisms by which microbiota-derived metabolites shape immune function are well described for some classes of molecules. For example, short chain fatty acids signal through activation of G-protein coupled receptors that regulate immune function as well as by inhibition of histone deacetylase which removes acetyl groups from histones rendering DNA less accessible to transcription factors. Tryptophan metabolites which include immunomodulatory microbial-derived indoxyl metabolites, signal via the aryl hydrocarbon receptor, a transcription factor that has emerged as an important player in asthma control [Reviewed in (72)]. Beyond interactions between microbial-derived metabolites and their cognate receptors, the emerging field of immunometabolism has demonstrated that immune cell phenotypes are intimately linked to their metabolic state (79–81). Microbes, particularly those in the intestine, play a key role in defining the availability of glucose, fatty acids and amino acids which are crucial to defining the immunometabolic state of immune cells. Thus, it is unsurprising that emerging data implicates the microbiome as a regulator of immune cell function via immunometabolism (82). The importance of metabolism in the function of antigen-presenting, T and B cells that are instrumental in allergic inflammation has been demonstrated (83). Naïve T cells largely rely on OXPHOS for energy biogenesis, while activated T cells derive energy primarily through glycolysis to serve as helpers to initiate B cell activation, cytotoxic effects, and cytokine production (82, 83). Recently, studies have shown that microbial metabolites such as short-chain fatty acids and tryptophan metabolites are also essential in shaping the function of a range of immune cells including epithelial, innate, and adaptive immune cells (84).
In addition to microbial metabolites being utilized as substrates by immune cells and altering their potential function, new concepts for microbial influence over immunity are arising. One important emerging idea is that microbial metabolites can influence the long-term functioning of innate immune cells through “trained immunity”. This concept describes functional reprogramming of innate immune cells via changes to their epigenetic landscape (85). These epigenetic changes re-tune responses of innate immune cells to secondary inflammatory insults, such as bacterial lipopolysaccharide, to either decrease or increase functional responses including cytokine secretion or phagocytosis. These newer data offer insights into the molecular mechanisms by which the microbiome, particularly as it develops in early life, shape immune function. Although trained immunity in the context of allergy and asthma is a relatively new field, several potential mechanisms by which trained immunity may contribute to asthma development have recently been reviewed (86). The observations provide a framework for understanding the developmental origins of allergy and asthma in which prenatal and early life extrinsic exposures and intrinsic factors shape microbial and immune development in utero, setting the stage for post-natal microbial colonization trajectories that shape emerging immune function. With this in mind, a focus on maternal prenatal health and diet, in addition to reducing early life exposure to microbiome-perturbing factors such as stress, antimicrobials, formula feeding and processed foods, seem key and imminently implementable approaches to reduce disease burden. While the impact of reducing formula feeding and processed foods on allergy and asthma incidence has not yet been directly tested, they are both associated with perturbed microbiomes and are known to shape the early life microbiome (28, 87). However, a large population-based Canadian study demonstrated that the recent reduction in pediatric asthma incidence was attributable in part to improved antimicrobial stewardship in infancy and preservation of the gut microbiome (88), offering reason for optimism that reducing microbiome perturbing exposures, particularly in early life could significantly reduce disease burden.
Microbiome-immune interactions and asthma outcomes/endotype.
Over the last decade it has become clear from studies in adults that the airway microbiome in particular is altered in chronic asthma. Recent investigations have also shown that airway microbiota characteristics differ in relation to immune response patterns, especially level of type 2 (T2) inflammation (5, 89, 90). Far fewer studies have demonstrated clear relationships between the gut microbiome and adult asthma phenotype (89, 90), which is challenging for a number of reasons including failure to account for variance in the gut microbiome attributable to factors such as age, sex, diet and body mass index, as well as medications and age-related co-morbidities. In this section we briefly summarize and highlight key findings from studies in these areas.
First, we note that findings from most studies of the airway microbiome to date have analyzed sputum, which is easier to obtain, but analyses of samples collected directly by bronchoscopy also have been performed (5, 91–94). Secondly, many investigations have examined cohorts that focus on severe asthma, the latter representing a minority of the patient population but who experience disproportionate morbidity and healthcare utilization. With these potential caveats in mind, existing evidence from well-conducted studies has converged on the following observations. Airway microbiome characteristics differ across the spectrum of asthma severity and associate with clinical measures and immune biomarkers (5, 91–102). In addition, these associations may be further influenced by additional clinical factors such as treatments and co-morbidities (5, 92, 94).
Studies have shown that the composition of airway microbiota, in general, is shifted in severe asthma compared to milder asthma. In some severe asthma subjects, this is reflected by an enrichment in specific bacterial groups, such as members of the Proteobacteria, a large phylum that includes representative species that are potential respiratory pathogens (87, 89, 96). Far fewer studies of the microbiome have been conducted in subjects with mild asthma. However, carefully conducted comparisons have shown that even in those with mild atopic asthma, the configuration of the airway microbiome differs from both non-atopic healthy individuals and atopic individuals without asthma (5, 92, 98).
These observations, in concert with the known immunological complexity of asthma, have led investigators to further interrogate microbiome-immune relationships in chronic asthma. Studies pursuing this have generally made use of accessible biomarkers of T2 inflammation (e.g., eosinophils), with some studies further defining T2 status by airway epithelial gene expression signatures (5, 91, 92, 97). While underlying molecular mechanisms and treatment options are best understood for the T2-high endotype, this is less true for non-T2 (T2-low) asthma which represents a multitude of phenotypes. The latter may include overlapping evidence of concurrent T2 pathway activation, as seen in some severe asthma patients with increased blood eosinophils but also increased sputum neutrophils. The pathobiological drivers of such differences and variation in immune responses in chronic asthma remain incompletely understood.
Recent studies have offered clues into how airway microbiome-immune interactions may impact asthma phenotype. Investigations in severe asthma cohorts have reported differences in sputum bacterial diversity associated with airway inflammatory phenotypes, indicative that microbiome alterations in severe asthma are not uniform across patients. Taylor et al. (91), found that neutrophilic severe asthma patients harbored a significantly less diverse sputum bacterial community than those with eosinophilic airway inflammation. The difference was reflected by greater prevalence in the neutrophilic group of potentially pathogenic organisms (e.g. Haemophilus) coupled with a reduction in Streptococcus, Gemella, and other bacteria traditionally viewed as respiratory tract commensals (Table 2). Similar findings were noted from another severe asthma cohort in which two sputum microbiome clusters were identified (93). The smaller cluster of subjects had neutrophil-predominant sputum, comparatively reduced diversity, and increased representation of potential pathogens. Of note subjects in this cluster also had elevated sputum or blood eosinophils, suggestive of concurrently activated T2 pathways. Lastly, an earlier study examined microbiome-immune relationships in severe asthma using bronchial epithelial brushings to characterize and compare microbial relationships to gene expression signatures (97). No specific bacteria from the brushings associated with the T2-high signature, and in congruence, bronchial biopsy eosinophil numbers inversely correlated with brush bacterial burden. In contrast, a Th17 gene expression signature correlated with multiple Proteobacteria members, whose increased relative abundance also correlated with less stable asthma control.
Table 2.
Main respiratory bacteria (genus) and reported associations with asthma. Species-level differences likely exist but are not discernible by the 16S rRNA sequencing methods used in most studies.
| Bacterial genus | Compartment | Outcome association(s) | References |
|---|---|---|---|
| Haemophilus | nasopharyngeal, lower respir tract | (+) asthma (T2-low in adults) (+) exacerbations in children |
5,10–15,90,91,96 |
| Moraxella | nasopharyngeal, lower respir tract | (+) asthma (T2-low in adults) (+) exacerbations in children |
5,10–15,90,91,96 |
| Streptococcus | nasopharyngeal, lower respir tract | (+) asthma (+) exacerbations in children |
5,10–15,90,91 |
| Corynebacterium Dolosigranulum, | nasopharyngeal | (—) asthma, (—) exacerbation in children |
13,30 |
| Neisseria | lower respir tract | (+) asthma status (T2-low in adults) | 5,91 |
| Fusobacterium | lower respir tract | (+) asthma status (T2-high in adults) | 5,91 |
| Veillonella | lower respir tract | (+) asthma in adults | 5,91 |
Airway microbiome differences by T2 status have also been observed in mild asthma. Measures of bronchial bacterial burden are higher in T2-low compared to T2-high mild asthma (5, 92). However, airway bacteria differentially associated with T2-low mild asthma differ from those noted above to be associated with neutrophilic severe asthma. While criteria used to define T2 status have varied between studies (e.g., airway vs. blood eosinophils vs. epithelial gene expression), additional factors likely contribute to differences in the airway microbiome between severe and mild asthma. One hypothesized factor is effects of cumulative exposure to inhaled corticosteroids (ICS). While difficult to adjust for this in cross-sectional studies of severe asthma due to high prevalence of ICS use, results of a recent randomized, placebo-controlled trial of inhaled fluticasone in subjects with mild asthma demonstrated changes in the airway microbiome (5, 92). In particular, the overall composition of airway bacteria shifted with fluticasone intervention, particularly in non-responders, and responsiveness to fluticasone was related to differences in the baseline (pre-intervention) microbiome. Additional possible factors are other treatments not yet studied for their impact on the airway microbiome, as well as co-morbidities such as obesity. Current evidence suggests that obesity, as defined by body-mass index, is associated with differences in the airway microbiome of severe asthma patients, in addition to differences in gut microbiota (90, 97). This implicates obese state as potentially shaping the airway microbiome via mechanisms not yet defined. Lastly, many other cell types (e.g., mast cells, ILCs) and cytokines (e.g., IL-17, IL-6, IL-1b) contribute to T2 and non-T2 mechanisms in asthma (103–105). Given asthma’s clinical heterogeneity, more detailed study of microbiome relationships, both in the airways and gut, to other components of the immune response could reveal additional insights that clarify opportunities for more precise therapeutic targeting.
A recent study of upper airway samples from school-aged children with asthma in the Preventative Omalizumab or Step-up Therapy for Severe Fall Exacerbations (PROSE) trial identified six distinct airway microbiota compositions which significantly differed with respect to asthma features (14). Nasal microbiotas dominated by Moraxella were associated with increased exacerbation risk and eosinophil activation. Children with airway microbiota dominated by Staphylococcus or Corynebacterium were associated with reduced respiratory illness and exacerbation events, whereas Streptococcus-dominated assemblages increased the risk of rhinovirus infection. Only DNA was available for study, precluding the possibility of examining microbial activities that may underlie these relationships. However, a more recent study of children with severe asthma in the Mechanisms Underlying Asthma Exacerbations Prevented and Persistent With Immune-Based Therapy trial found that interactions between discrete networks of bacteria with specific epithelial transcriptional modules increase asthma exacerbation risk following respiratory viral infection (15). Specifically, discrete networks of upper airway bacteria comprising either Streptococcus or Staphylococcus exhibited opposing interactions with an exacerbation-associated SMAD3 nasal epithelial transcriptional module to significantly increase odds of subsequent exacerbation. Of note the airway microbiota exhibited temporal, seasonal dynamics and these relationships predominated in the fall (14).
Conclusions and Future Directions
The future for asthma and allergy research depends upon deeper understanding of molecular mechanisms that underlie both the trajectory to disease development and endotypes once established. This will be facilitated through the application of high-resolution immune and microbial cellular and molecular profiling approaches to samples collected longitudinally from multiple diverse and well-characterized human cohorts. Given the numerous factors that shape microbiomes which in turn tune immune function, we envision a future where cost-effective metabolomic pre- and/or postnatal screening can be utilized both as a prognostic tool to determine disease risk and as a monitor for microbial-immune status throughout development to identify those at risk and determine efficacy of interventions to alter disease course.
References
- 1.Johnson CC, Havstad SL, Ownby DR, Joseph CLM, Sitarik AR, Biagini Myers J, et al. Pediatric asthma incidence rates in the United States from 1980 to 2017. J Allergy Clin Immunol. 2021;148(5):1270–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson CC, Chandran A, Havstad S, Li X, McEvoy CT, Ownby DR, et al. US Childhood Asthma Incidence Rate Patterns From the ECHO Consortium to Identify High-risk Groups for Primary Prevention. JAMA Pediatr. 2021;175(9):919–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones SM, Kim EH, Nadeau KC, Nowak-Wegrzyn A, Wood RA, Sampson HA, et al. Efficacy and safety of oral immunotherapy in children aged 1–3 years with peanut allergy (the Immune Tolerance Network IMPACT trial): a randomised placebo-controlled study. Lancet. 2022;399(10322):359–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durack J, Huang YJ, Nariya S, Christian LS, Ansel KM, Beigelman A, et al. Bacterial biogeography of adult airways in atopic asthma. Microbiome. 2018;6(1):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durack J, Lynch SV, Nariya S, Bhakta NR, Beigelman A, Castro M, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol. 2017;140(1):63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segal LN, Clemente JC, Tsay JC, Koralov SB, Keller BC, Wu BG, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol. 2016;1:16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hufnagl K, Pali-Schöll I, Roth-Walter F, Jensen-Jarolim E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin Immunopathol. 2020;42(1):75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barcik W, Boutin RCT, Sokolowska M, Finlay BB. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity. 2020;52(2):241–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, Belda-Ferre P, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med. 2019;25(3):448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teo SM, Tang HHF, Mok D, Judd LM, Watts SC, Pham K, et al. Airway Microbiota Dynamics Uncover a Critical Window for Interplay of Pathogenic Bacteria and Allergy in Childhood Respiratory Disease. Cell Host Microbe. 2018;24(3):341–52.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17(5):704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raita Y, Pérez-Losada M, Freishtat RJ, Harmon B, Mansbach JM, Piedra PA, et al. Integrated omics endotyping of infants with respiratory syncytial virus bronchiolitis and risk of childhood asthma. Nat Commun. 2021. Jun 14;12(1):3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, Jackson D, Bacharier LB, Mauger D, Boushey H, Castro M, et al. The upper-airway microbiota and loss of asthma control among asthmatic children. Nat Commun. 2019;10(1):5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCauley K, Durack J, Valladares R, Fadrosh DW, Lin DL, Calatroni A, et al. Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J Allergy Clin Immunol. 2019;144(5):1187–97.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCauley KE, Flynn K, Calatroni A, DiMassa V, LaMere B, Fadrosh DW, et al. Seasonal airway microbiome and transcriptome interactions promote childhood asthma exacerbations. J Allergy Clin Immunol. 2022. Feb 8;S0091-6749(22)00146-4. [DOI] [PubMed] [Google Scholar]
- 16.Thompson AE. JAMA patient page. The immune system. JAMA. 2015;313(16):1686. [DOI] [PubMed] [Google Scholar]
- 17.Mishra A, Lai GC, Yao LJ, Aung TT, Shental N, Rotter-Maskowitz A, et al. Microbial exposure during early human development primes fetal immune cells. Cell. 2021;184(13):3394–409.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halkias J, Rackaityte E, Hillman SL, Aran D, Mendoza VF, Marshall LR, et al. CD161 contributes to prenatal immune suppression of IFNγ-producing PLZF+ T cells. J Clin Invest. 2019;129(9):3562–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rackaityte E, Halkias J, Fukui EM, Mendoza VF, Hayzelden C, Crawford ED, et al. Viable bacterial colonization is highly limited in the human intestine in utero. Nat Med. 2020;26(4):599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brosseau C, Selle A, Duval A, Misme-Aucouturier B, Chesneau M, Brouard S, et al. Prebiotic Supplementation During Pregnancy Modifies the Gut Microbiota and Increases Metabolites in Amniotic Fluid, Driving a Tolerogenic Environment. Front Immunol. 2021;12:712614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim AI, McFadden T, Link VM, Han SJ, Karlsson RM, Stacy A, et al. Prenatal maternal infection promotes tissue-specific immunity and inflammation in offspring. Science. 2021;373(6558). [DOI] [PubMed] [Google Scholar]
- 22.Stiemsma LT, Arrieta MC, Dimitriu PA, Cheng J, Thorson L, Lefebvre DL, et al. Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin Sci (Lond). 2016;130(23):2199–207. [DOI] [PubMed] [Google Scholar]
- 23.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. [DOI] [PubMed] [Google Scholar]
- 24.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22(10):1187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. 2018;9(1):141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee-Sarwar KA, Kelly RS, Lasky-Su J, Zeiger RS, O’Connor GT, Sandel MT, et al. Integrative analysis of the intestinal metabolome of childhood asthma. J Allergy Clin Immunol. 2019;144(2):442–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong S Factors influencing development of the infant microbiota: From prenatal period to early infancy. Clin Exp Pediatr. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.BioRender.com. Lipid handling in the small intestine modulates immune system homeostasis. Accessed May 10, 2022. https://app.biorender.com/biorender-templates
- 30.Tang HHF, Lang A, Teo SM, Judd LM, Gangnon R, Evans MD, et al. Developmental patterns in the nasopharyngeal microbiome during infancy are associated with asthma risk. J Allergy Clin Immunol. 2021;147(5):1683–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang HH, Teo SM, Belgrave DC, Evans MD, Jackson DJ, Brozynska M, et al. Trajectories of childhood immune development and respiratory health relevant to asthma and allergy. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durack J, Kimes NE, Lin DL, Rauch M, McKean M, McCauley K, et al. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat Commun. 2018;9(1):707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu T, Liu X, Kong FQ, Duan YY, Yee AL, Kim M, et al. Age and Mothers: Potent Influences of Children’s Skin Microbiota. J Invest Dermatol. 2019;139(12):2497–505.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meylan P, Lang C, Mermoud S, Johannsen A, Norrenberg S, Hohl D, et al. Skin Colonization by Staphylococcus aureus Precedes the Clinical Diagnosis of Atopic Dermatitis in Infancy. J Invest Dermatol. 2017;137(12):2497–504. [DOI] [PubMed] [Google Scholar]
- 35.Vissing NH, Chawes BL, Bisgaard H. Increased risk of pneumonia and bronchiolitis after bacterial colonization of the airways as neonates. Am J Respir Crit Care Med. 2013;188(10):1246–52. [DOI] [PubMed] [Google Scholar]
- 36.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang YJ, Erb-Downward JR, Dickson R, Curtis JL, Huffnagle GB, Han MK. Understanding the role of the microbiome in COPD: Principles, Challenges and Future Directions. Transl Res. 2017. Jan;179:71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morin A, McKennan CG, Pedersen CT, Stokholm J, Chawes BL, Malby Schoos AM, et al. Epigenetic landscape links upper airway microbiota in infancy with allergic rhinitis at 6 years of age. J Allergy Clin Immunol. 2020;146(6):1358–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scharschmidt TC, Vasquez KS, Pauli ML, Leitner EG, Chu K, Truong HA, et al. Commensal Microbes and Hair Follicle Morphogenesis Coordinately Drive Treg Migration into Neonatal Skin. Cell Host Microbe. 2017;21(4):467–77.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Kim BE, Ahn K, Leung DYM. Interactions Between Atopic Dermatitis and. Allergy Asthma Immunol Res. 2019;11(5):593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basolo A, Hohenadel M, Ang QY, Piaggi P, Heinitz S, Walter M, et al. Effects of underfeeding and oral vancomycin on gut microbiome and nutrient absorption in humans. Nat Med. 2020;26(4):589–98. [DOI] [PubMed] [Google Scholar]
- 42.Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol. 2016;14(5):273–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woo V, Alenghat T. Epigenetic regulation by gut microbiota. Gut Microbes. 2022;14(1):2022407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W, Nelson KE. Microbial Species that Initially Colonize the Human Gut at Birth or in Early Childhood Can Stay in Human Body for Lifetime. Microb Ecol. 2021;82(4):1074–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGovern N, Shin A, Low G, Low D, Duan K, Yao LJ, et al. Human fetal dendritic cells promote prenatal T-cell immune suppression through arginase-2. Nature. 2017;546(7660):662–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. [DOI] [PubMed] [Google Scholar]
- 48.Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016;65(11):1906–15. [DOI] [PubMed] [Google Scholar]
- 49.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17(5):690–703. [DOI] [PubMed] [Google Scholar]
- 50.Sugino KY, Ma T, Kerver JM, Paneth N, Comstock SS. Human Milk Feeding Patterns at 6 Months of Age are a Major Determinant of Fecal Bacterial Diversity in Infants. J Hum Lact. 2021;37(4):703–13. [DOI] [PubMed] [Google Scholar]
- 51.Tran SM, Mohajeri MH. The Role of Gut Bacterial Metabolites in Brain Development, Aging and Disease. Nutrients. 2021;13(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao H, Kang S. The Role of the Gut Microbiome in Energy Balance With a Focus on the Gut-Adipose Tissue Axis. Front Genet. 2020;11:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalbermatter C, Fernandez Trigo N, Christensen S, Ganal-Vonarburg SC. Maternal Microbiota, Early Life Colonization and Breast Milk Drive Immune Development in the Newborn. Front Immunol. 2021;12:683022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moossavi S, Miliku K, Sepehri S, Khafipour E, Azad MB. The Prebiotic and Probiotic Properties of Human Milk: Implications for Infant Immune Development and Pediatric Asthma. Front Pediatr. 2018;6:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vazquez-Gutierrez P, de Wouters T, Werder J, Chassard C, Lacroix C. High Iron-Sequestrating Bifidobacteria Inhibit Enteropathogen Growth and Adhesion to Intestinal Epithelial Cells. Front Microbiol. 2016;7:1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liévin-Le Moal V, Servin AL. Anti-infective activities of lactobacillus strains in the human intestinal microbiota: from probiotics to gastrointestinal anti-infectious biotherapeutic agents. Clin Microbiol Rev. 2014;27(2):167–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaetzel CS. Cooperativity among secretory IgA, the polymeric immunoglobulin receptor, and the gut microbiota promotes host-microbial mutualism. Immunol Lett. 2014;162(2 Pt A):10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brandtzaeg P Mucosal immunity: integration between mother and the breast-fed infant. Vaccine. 2003;21(24):3382–8. [DOI] [PubMed] [Google Scholar]
- 59.Rogier EW, Frantz AL, Bruno ME, Wedlund L, Cohen DA, Stromberg AJ, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci U S A. 2014;111(8):3074–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bode L, McGuire M, Rodriguez JM, Geddes DT, Hassiotou F, Hartmann PE, et al. It’s alive: microbes and cells in human milk and their potential benefits to mother and infant. Adv Nutr. 2014;5(5):571–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zimmermann P, Curtis N. Breast milk microbiota: A review of the factors that influence composition. J Infect. 2020;81(1):17–47. [DOI] [PubMed] [Google Scholar]
- 62.Czosnykowska-Łukacka M, Lis-Kuberka J, Królak-Olejnik B, Orczyk-Pawiłowicz M. Changes in Human Milk Immunoglobulin Profile During Prolonged Lactation. Front Pediatr. 2020;8:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakamura Y, Takahashi H, Takaya A, Inoue Y, Katayama Y, Kusuya Y, et al. Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci Transl Med. 2020;12(551). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158(4):705–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharon G, Cruz NJ, Kang DW, Gandal MJ, Wang B, Kim YM, et al. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell. 2019;177(6):1600–18.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Witkowski M, Weeks TL, Hazen SL. Gut Microbiota and Cardiovascular Disease. Circ Res. 2020;127(4):553–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wypych TP, Pattaroni C, Perdijk O, Yap C, Trompette A, Anderson D, et al. Microbial metabolism of L-tyrosine protects against allergic airway inflammation. Nat Immunol. 2021;22(3):279–86. [DOI] [PubMed] [Google Scholar]
- 68.Levan SR, Stamnes KA, Lin DL, Panzer AR, Fukui E, McCauley K, et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat Microbiol. 2019;4(11):1851–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abdala-Valencia H, Soveg F, Cook-Mills JM. γ-Tocopherol supplementation of allergic female mice augments development of CD11c+CD11b+ dendritic cells in utero and allergic inflammation in neonates. Am J Physiol Lung Cell Mol Physiol. 2016;310(8):L759–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blaess M, Deigner HP. Derailed Ceramide Metabolism in Atopic Dermatitis (AD): A Causal Starting Point for a Personalized (Basic) Therapy. Int J Mol Sci. 2019;20(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Inoue H, Someno T, Kawada M, Ikeda D. Citric acid inhibits a bacterial ceramidase and alleviates atopic dermatitis in an animal model. J Antibiot (Tokyo). 2010;63(10):611–3. [DOI] [PubMed] [Google Scholar]
- 72.Poulain-Godefroy O, Bouté M, Carrard J, Alvarez-Simon D, Tsicopoulos A, de Nadai P. The Aryl Hydrocarbon Receptor in Asthma: Friend or Foe? Int J Mol Sci. 2020;21(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332(6032):974–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van der Hee B, Wells JM. Microbial regulation of host physiology by shortchain fatty acids. Trends Microbiol 2021;29:700–12. [DOI] [PubMed] [Google Scholar]
- 75.Fyhrquist N, Muirhead G, Prast-Nielsen S, Jeanmougin M, Olah P, Skoog T, et al. Microbe-host interplay in atopic dermatitis and psoriasis. Nat Commun 2019;10:4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A. 2014;111(2):805–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fonseca W, Lucey K, Jang S, Fujimura KE, Rasky A, Ting HA, et al. Lactobacillus johnsonii supplementation attenuates respiratory viral infection via metabolic reprogramming and immune cell modulation. Mucosal Immunol. 2017;10(6):1569–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fonseca W, Malinczak CA, Fujimura K, Li D, McCauley K, Li J, et al. Maternal gut microbiome regulates immunity to RSV infection in offspring. J Exp Med. 2021;218(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16(9):553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van den Bossche J, O’Neill LA, Menon D. Macrophage Immunometabolism: Where Are We (Going)? Trends Immunol. 2017;38(6):395–406. [DOI] [PubMed] [Google Scholar]
- 81.O’Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213(1):15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Michaudel C, Sokol H. The Gut Microbiota at the Service of Immunometabolism. Cell Metab. 2020;32(4):514–23. [DOI] [PubMed] [Google Scholar]
- 83.Shyer JA, Flavell RA, Bailis W. Metabolic signaling in T cells. Cell Res. 2020;30(8):649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Corrêa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunology. 2016;5(4):e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Netea MG, Domínguez-Andrés J, Barreiro LB, Chavakis T, Divangahi M, Fuchs E, et al. Defining trained immunity and its role in health and disease. Nat Rev Immunol. 2020;20(6):375–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lynch SV, Vercelli D. Microbiota, Epigenetics, and Trained Immunity. Convergent Drivers and Mediators of the Asthma Trajectory from Pregnancy to Childhood. Am J Respir Crit Care Med. 2021;203(7):802–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Güngör D, Nadaud P, LaPergola CC, Dreibelbis C, Wong YP, Terry N, et al. Infant milk-feeding practices and food allergies, allergic rhinitis, atopic dermatitis, and asthma throughout the life span: a systematic review. Am J Clin Nutr. 2019;109(Suppl_7):772S–99S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patrick DM, Sbihi H, Dai DLY, Al Mamun A, Rasali D, Rose C, et al. Decreasing antibiotic use, the gut microbiota, and asthma incidence in children: evidence from population-based and prospective cohort studies. Lancet Respir Med. 2020;8(11):1094–105. [DOI] [PubMed] [Google Scholar]
- 89.Barcik W, Pugin B, Westermann P, Perez NR, Ferstl R, Wawrzyniak M, et al. Histamine-secreting microbes are increased in the gut of adult asthma patients. J Allergy Clin Immunol. 2016;138(5):1491–4.e7. [DOI] [PubMed] [Google Scholar]
- 90.Michalovich D, Rodriguez-Perez N, Smolinska S, Pirozynski M, Mayhew D, Uddin S, et al. Obesity and disease severity magnify disturbed microbiome-immune interactions in asthma patients. Nat Commun. 2019;10(1):5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taylor SL, Leong LEX, Choo JM, Wesselingh S, Yang IA, Upham JW, et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J Allergy Clin Immunol. 2018;141(1):94–103.e15. [DOI] [PubMed] [Google Scholar]
- 92.Durack J, Christian LS, Nariya S, Gonzalez J, Bhakta NR, Ansel KM, et al. Distinct associations of sputum and oral microbiota with atopic, immunologic, and clinical features in mild asthma. J Allergy Clin Immunol. 2020;146(5):1016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abdel-Aziz MI, Brinkman P, Vijverberg SJH, Neerincx AH, Riley JH, Bates S, et al. Sputum microbiome profiles identify severe asthma phenotypes of relative stability at 12 to 18 months. J Allergy Clin Immunol. 2021;147(1):123–34. [DOI] [PubMed] [Google Scholar]
- 94.Denner DR, Sangwan N, Becker JB, Hogarth DK, Oldham J, Castillo J, et al. Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J Allergy Clin Immunol. 2016;137(5):1398–405.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rick EM, Woolnough KF, Seear PJ, Fairs A, Satchwell J, Richardson M, et al. The airway fungal microbiome in asthma. Clin Exp Allergy. 2020;50(12):1325–41. [DOI] [PubMed] [Google Scholar]
- 96.Huang C, Yu Y, Du W, Liu Y, Dai R, Tang W, et al. Fungal and bacterial microbiome dysbiosis and imbalance of trans-kingdom network in asthma. Clin Transl Allergy. 2020;10:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, et al. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol. 2015;136(4):874–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013;131(2):346–52.e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Abdel-Aziz MI, Vijverberg SJH, Neerincx AH, Brinkman P, Wagener AH, Riley JH, et al. A multi-omics approach to delineate sputum microbiome-associated asthma inflammatory phenotypes. Eur Respir J. 2022;59(1). [DOI] [PubMed] [Google Scholar]
- 100.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127(2):372–81 e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sharma A, Laxman B, Naureckas ET, Hogarth DK, Sperling AI, Solway J, et al. Associations between fungal and bacterial microbiota of airways and asthma endotypes. J Allergy Clin Immunol. 2019;144(5):1214–27.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Son JH, Kim JH, Chang HS, Park JS, Park CS. Relationship of Microbial Profile With Airway Immune Response in Eosinophilic or Neutrophilic Inflammation of Asthmatics. Allergy Asthma Immunol Res. 2020;12(3):412–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hudey SN, Ledford DK, Cardet JC. Mechanisms of non-type 2 asthma. Curr Opin Immunol. 2020;66:123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu W, Liu S, Verma M, Zafar I, Good JT, Rollins D, et al. Mechanism of T. J Allergy Clin Immunol. 2017;139(5):1548–58.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Choy DF, Arron JR. Beyond type 2 cytokines in asthma - new insights from old clinical trials. Expert Opin Ther Targets. 2020;24(5):463–75. [DOI] [PubMed] [Google Scholar]


