Abstract
Purpose
Quantitative images of metabolic activity can be derived through dynamic PET. However, the conventional approach necessitates invasive blood sampling to acquire the input function, thus limiting its noninvasive nature. The aim of this study was to devise a system based on convolutional neural network (CNN) capable of estimating the time-radioactivity curve of arterial plasma and accurately quantify the cerebral metabolic rate of glucose (CMRGlc) directly from PET data, thereby eliminating the requirement for invasive sampling.
Methods
This retrospective investigation analyzed 29 patients with neurological disorders who underwent comprehensive whole-body 18F-FDG-PET/CT examinations. Each patient received an intravenous infusion of 185 MBq of 18F-FDG, followed by dynamic PET data acquisition and arterial blood sampling. A CNN architecture was developed to accurately estimate the time-radioactivity curve of arterial plasma.
Results
The CNN estimated the time-radioactivity curve using the leave-one-out technique. In all cases, there was at least one frame with a prediction error within 10% in at least one frame. Furthermore, the correlation coefficient between CMRGlc obtained from the sampled blood and CNN yielded a highly significant value of 0.99.
Conclusion
The time-radioactivity curve of arterial plasma and CMRGlc was determined from 18F-FDG dynamic brain PET data using a CNN. The utilization of CNN has facilitated noninvasive measurements of input functions from dynamic PET data. This method can be applied to various forms of quantitative analysis of dynamic medical image data.
Keywords: CNN, FDG dynamic brain PET, radioactivity curve
Introduction
PET has been used in the realm of brain functional imaging. Dynamic brain PET imaging using Fluorine-18 fluorodeoxyglucose (18F-FDG) permits the examination of the cerebral metabolic rate of glucose (CMRGlc) [1]. Nevertheless, the conventional and widely accepted technique for analyzing the CMRGlc involves the extraction of arterial blood samples and the derivation of the time-radioactivity curve from arterial plasma, which necessitates an invasive procedure for patients and exposes medical personnel to radiation [2]. Consequently, the search for noninvasive alternatives to generate input functions has been underway [3,4]. Nonetheless, existing methods suffer from limitations, such as insufficient sample size and diminished accuracy, thus failing to supplant blood sampling. Therefore, we propose a novel approach that employs a convolutional neural network (CNN)-based system to estimate the time-radioactivity curve of arterial plasma.
AI studies in the domain of PET have documented findings, including a study that automated the classification of 18F-FDG whole-body PET images into three categories: benign, malignant, or equivocal [5]. FDG-PET/computed tomography (CT) images of patients with lung cancer and lymphoma have demonstrated exceptional diagnostic performance in fully automating the localization of FDG uptake patterns and identifying foci that are either suspicious or nonsuspicious for cancer classification using a CNN [6]. Furthermore, a study reported the generation of a 511-keV photon attenuation map by inputting whole-body 18F-FDG-PET/CT images from 100 patients with cancer into a CNN, which exhibited greater reliability than the four-fraction method employed in current whole-body PET/MRI scans [7]. Moreover, a study detailed the classification of lung cancer foci on FDG-PET/CT images as either tumor stages T1-2 or T3-4 by training a CNN on images obtained from patients who underwent staging using FDG-PET/CT images [8]. Hence, most reports in nuclear medicine, particularly in the realm of PET, pertained to the classification of images and the extraction of lesions, with some demonstrating efficacy in disease classification and attenuation correction. Nevertheless, regression analyses of data derived from images have seldom been documented, despite the existence of studies that have reported regression prediction of age and weight based on whole-body PET images [9].
This study aimed to devise a CNN-based framework capable of estimating the time- radioactivity curve of arterial plasma and CMRGlc from PET data. Our hypothesis posited that such an approach would facilitate a noninvasive and quantitative investigation of cerebral energy metabolism.
Methods
Subjects
This study comprised a cohort of 29 patients diagnosed with neurological disorders (mean age: 37.3 ± 13.2 years; 11 males and 18 females). The Ethics Committee of Hokkaido University Hospital Medical Information Network Clinical Trials Registry (UMIN000018160) provided approval for this study. All participants provided written informed consent. Note however that this further investigative study utilized previously acquired data retrospectively so the need for further ethical approval was waived.
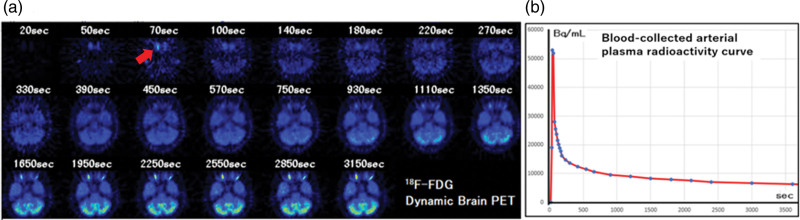
For each patient, an intravenous infusion of 185 MBq of 18F-FDG was administered, followed by dynamic PET data acquisition and arterial blood sampling. Dynamic PET images of the brain were acquired over a period of 60 min, with 22 frames. The time intervals for frame collection were as follows: 20, 50, 70, 100, 140, 180, 220, 270, 330, 390, 450, 570, 750, 930, 1110, 1350, 1650, 1950, 2250, 2550, 2850, and 3150 s (Fig. 1). Concurrently, 1 mL of arterial blood was collected from the forearm artery via an indwelling needle. Immediately after blood collection, centrifugation was performed to separate the plasma and cellular components.
Fig. 1.
(a) A typical example of slice images in which the internal carotid is most clearly depicted in each frame of 18F-FDG dynamic brain PET. (b) A typical example of the time-radioactivity curve of arterial plasma.
18F-FDG dynamic imaging and blood sampling
Arterial plasma radioactivity counts were measured using a counter (ARC-380CL manufactured by Aloka), and the arterial plasma time-radioactivity curve was obtained. The arterial blood time-radioactivity curve is not an accurate input function for compartmental model analysis of 18F-FDG accumulation in the brain, because activity is also taken up by erythrocytes. Therefore, we obtained an accurate arterial plasma time-radioactivity curve by removing erythrocyte radioactivity that did not enter brain tissue.
Scanner configuration
PET investigations were performed employing an ECAT EXACT HR+ scanner manufactured by CTI/Siemens, which exhibited in-plane and axial resolutions of 4.8 and 5.6 mm, respectively. The field of view possessed a diameter of 23.4 cm. Through continuous axial motion of the gantry, the PET scanner generated 63 tomographic images at intervals of 2.425 mm. These image slices were oriented parallel to the orbitomeatal line. To correct tissue attenuation, a transmission scan employing a 68-Ge line source was performed before emission scanning. PET images were reconstructed using the filtered back projection method employing a ramp filter. The resultant PET image from this device had a half width (FWHM) of 4.8 mm, and each pixel corresponded to an actual length of 2.5 × 2.5 × 6.3 mm. The 22 frames of the dynamic PET images were reconstructed using filtered back projection with a similar matrix size of 128 × 128 in the axial plane and a voxel size of 2.5 × 2.5 × 6.3 mm.
Convolutional neural network
A neural network represents a computational paradigm that emulates the structure and functionality of neurons in the brain. It comprises three fundamental layers: input, hidden, and output layers. Each layer consists of interconnected nodes connected by edges. A ‘deep neural network’ (DNN) refers to a neural network that incorporates multiple hidden layers. The use of DNNs in machine learning is commonly known as ‘deep learning’. A CNN stands as a specialized DNN variant that demonstrates remarkable efficacy in tasks involving image recognition. Unlike traditional approaches for image recognition, CNNs do not rely on predefined image features. In this study, we proposed adopting a CNN for classifying FDG-PET brain images.
Architectures
In this study, we used a network model based on the Xception architecture [10]. Xception has exhibited remarkable effectiveness in various medical imaging domains, encompassing clinical images of basal cell carcinoma and pigmented nevi, v, and non-contrast-enhanced CT images for identifying the ultra-high-density middle cerebral artery sign in patients with acute ischemic stroke [11–13]. Xception is a model that optimizes parameter count and computational complexity by implementing depth-wise separable convolutions.
Model training and testing
During the training phase of the model, we implemented an ‘early stopping’ technique to decrease overfitting. Early stopping serves to monitor the loss function of both training and validation processes, allowing us to stop the learning process before it becomes excessively focused. This approach has found applications in diverse machine learning methodologies [14]. The CNN used in this study was fed a complete set of image frames as inputs. Specifically, we used 22-frame PET images in a single axial plane, that captured on capturing the internal carotid most prominently. Given the use of regression models in this study, we supplemented the image inputs with arterial plasma time- radioactivity curve data. Furthermore, the output layer of the model was configured to consist of a single unit. In the model testing phase, a regression model was created using a CNN to predict the arterial plasma time-radioactivity curve. For training, validation, and evaluation, we employed the leave-one-out cross-validation (LOOCV) methodology [15]. Within the LOOCV framework, one case was chosen from 29 cases as the test case, while the remaining 28 cases were used for training and validation. Among the 28 cases, 21 were allocated for training and seven were used for validation. This process was repeated for all test cases, resulting in 29 iterations.
Gradient-weighted class activation mapping
To address the challenge of explaining the decision-making process in artificial intelligence (AI) models developed through deep learning, we have incorporated a visualization technique known as Gradient-weighted Class Activation Mapping (Grad-CAM) [16]. Grad-CAM enables the identification of image regions that contribute to the activation of the neural network, thereby allowing for a visual understanding of the CNN’s estimation basis. In this study, we employed Grad-CAM to predict each test case and performed an additional experiment to visualize the basis for the estimation results. Grad-CAM assigns a continuous value to each pixel, and we applied a threshold of 90% of the maximum value to delineate the activated area.
Hardware and software environments
This experiment was performed in the following environments: operating system: Windows 10 pro 64 bit; CPU: Intel Core i9-10900K; GPU: NVIDIA GeForce RTX 3090 24GB; Framework: TensorFlow 2.5.0; Language: Python 3.8.5.
3-compartment model analysis
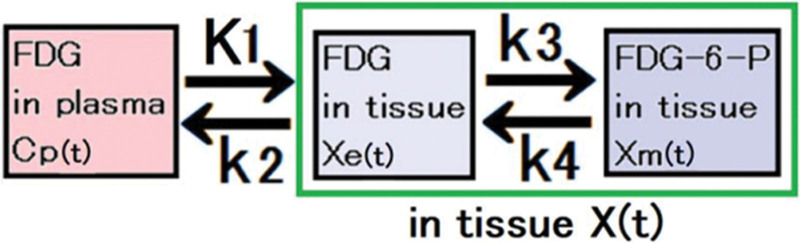
For each patient, 100 regions of interest (ROIs) were positioned on the cerebral images. The size of the ROIs was one pixel, and the length of the pixel size was 2.5 × 2.5 × 6.3 mm. The time- radioactivity curve X(t) for each ROI was calculated. The CMRGlc in each ROI was measured using the following equation in the 3-compartment model analysis:
where Xe(t) and Xm(t) are the time-radioactivity curves in the first and second tissue compartments, respectively. Cp(t) is plasma activity (Fig. 2). K1 and k2 are the rate constants of FDG uptake from the plasma into the tissue and from the tissue back into the plasma, respectively. k3 and k4 are the rate constants for the phosphorylation of FDG by hexokinase and dephosphorylation of FDG-6-PO4 to FDG, respectively (Fig. 3).
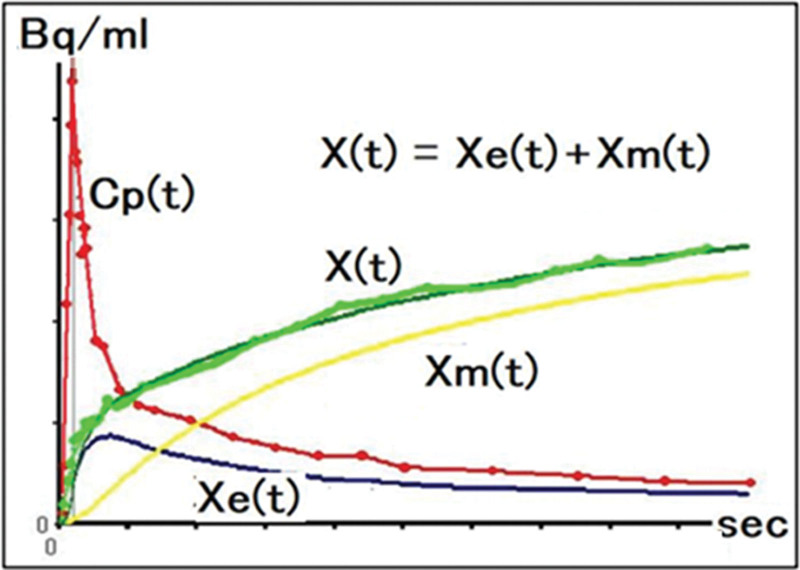
Fig. 2.
Arterial plasma time-radioactivity curve Cp(t) and tissue time-radioactivity curve X(t) at any position in the brain obtained from PET images. Where Xe(t) and Xm(t) are the time-radioactivity curves in the first and second tissue compartments, respectively. X(t) is the sum of X e(t) and Xm(t).
Fig. 3.
The diagram illustrates the velocity constants employed in the analysis of a three-compartment model. Specifically, K1 and k2 represent the rate constants governing the uptake of FDG from the plasma into the tissue and its subsequent return from the tissue to the plasma, respectively. Similarly, k3 and k4 denote the rate constants involved in the phosphorylation of FDG by hexokinase and the dephosphorylation of FDG-6-PO4 to FDG, respectively. The unit of K1 is mL/min/g, while k1, k2, and k3 are expressed in/min. These values serve as essential parameters for the calculations performed in the three-compartment model analysis.
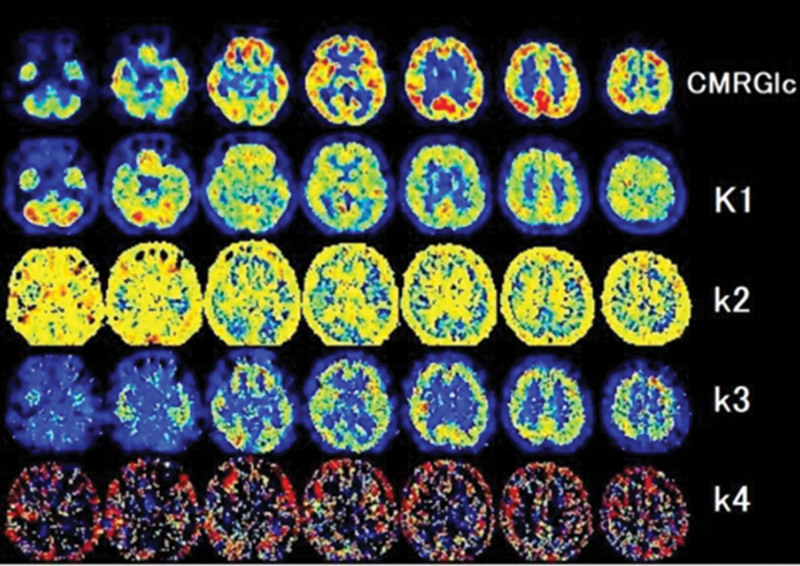
Figure 4 also shows that tissues, such as the skull and scalp, have high k2 values due to low glucose uptake and metabolism (relying predominantly on fatty acid metabolism. Xm(t) indicates the tissue that requires glucose. The CMRGlc was calculated using the following equation:
Fig. 4.
Distribution image of the rate constants K1, k2, k3, k4 in an exemplar series of 18F-FDG-PET brain images.
where LC is a lumped constant of 0.89 [17].
Results
The model underwent training for an average of 70 ± 27 epochs employing an early stopping algorithm. The training phase required an average of 560 ± 216 s, while prediction per patient took <0.1 s.
The time-radioactivity curve, estimated by the CNN employing LOOCV, was compared with the corresponding curve derived from arterial blood. In each case, the prediction error was at least 10% in at least one frame. In 21 cases, it was verified that >18 of the 22 frames exhibited a prediction error within a 10% margin. Nevertheless, the remaining eight cases showed numerous frames with prediction errors exceeding 10% (Table 1).
Table 1.
Mean error and percentage of slices within 10% error
| Mean error % | Percentage of slices within 10% error | |
|---|---|---|
| Patient_01 | 3.9 | 100% |
| Patient_02 | 6.1 | 86% |
| Patient_03 | 1.6 | 100% |
| Patient_04 | 5.0 | 86% |
| Patient_05 | 4.2 | 95% |
| Patient_06 | 1.7 | 100% |
| Patient_07 | 3.3 | 100% |
| Patient_08 | 4.3 | 95% |
| Patient_09 | 3.3 | 100% |
| Patient_10 | 17.3 | 32% |
| Patient_11 | 3.3 | 100% |
| Patient_12 | 5.5 | 91% |
| Patient_13 | 2.5 | 100% |
| Patient_14 | 17.6 | 41% |
| Patient_15 | 9.0 | 64% |
| Patient_16 | 5.1 | 91% |
| Patient_17 | 12.6 | 64% |
| Patient_18 | 3.5 | 95% |
| Patient_19 | 8.4 | 77% |
| Patient_20 | 3.4 | 100% |
| Patient_21 | 3.0 | 100% |
| Patient_22 | 2.9 | 100% |
| Patient_23 | 2.7 | 100% |
| Patient_24 | 33.8 | 18% |
| Patient_25 | 4.2 | 100% |
| Patient_26 | 47.3 | 14% |
| Patient_27 | 30.2 | 18% |
| Patient_28 | 34.5 | 9% |
| Patient_29 | 2.7 | 95% |
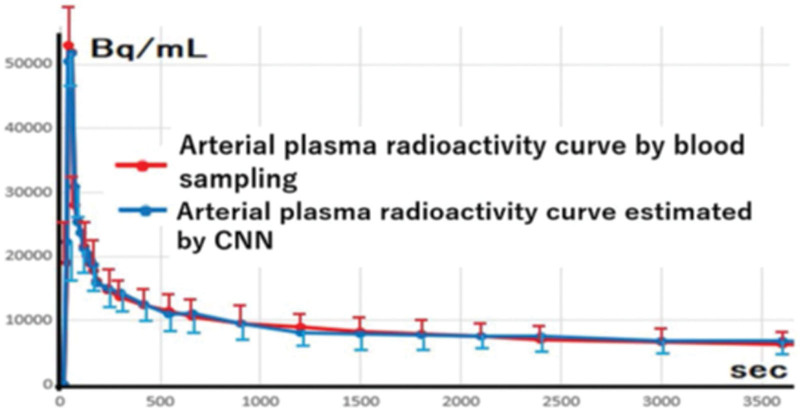
Figure 5 displays the arterial plasma time- radioactivity curves acquired through both the CNN-based method and traditional blood sampling. The accurate outcome of our proposed methodology using a CNN has demonstrated the achievement of comparable fittings, independent of the acquisition of blood samples.
Fig. 5.
The arterial plasma time-radioactivity curve presents the calculated mean and SD from the analysis of all 29 cases. This visual representation underscores the exceptional uniformity observed in the fitting of the curves.
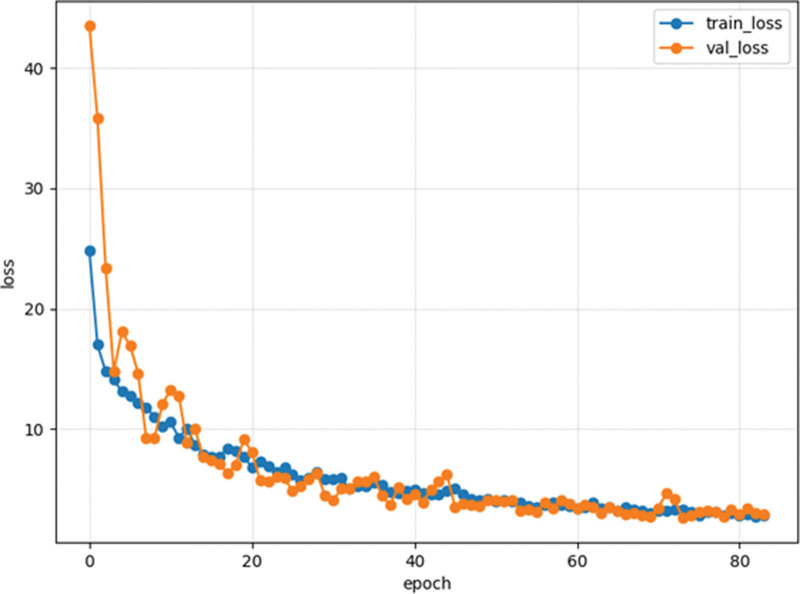
Two challenges commonly encountered in machine and deep learning are underfitting and overfitting. These phenomena can be visualized by analyzing a loss curve plotted against the number of epochs. In the case of underfitting, the loss curve continues to decrease for both the training and validation datasets. Conversely, overfitting is characterized by a training loss curve approaching 100%, while the validation loss curve diverges from it. Figure 6 illustrates the loss curve for one of the 29 trials performed in this study. In this particular instance, the training and validation loss curves did not deviate significantly, indicating that the training process achieved an optimal balance without underfitting or overfitting. Similar trends were observed across the remaining 28 trials, further supporting the absence of underfitting or overfitting.
Fig. 6.
An example of a training and verification loss curve is in this study. The vertical axis represents the value of the loss function, the horizontal axis represents the number of epochs, and the blue and orange colors depict the training and validation loss curves, respectively. The training was completed at 70 ± 27 epochs due to early stopping, and both the training and evaluation gradually declined.
Comparison with CMRGlc
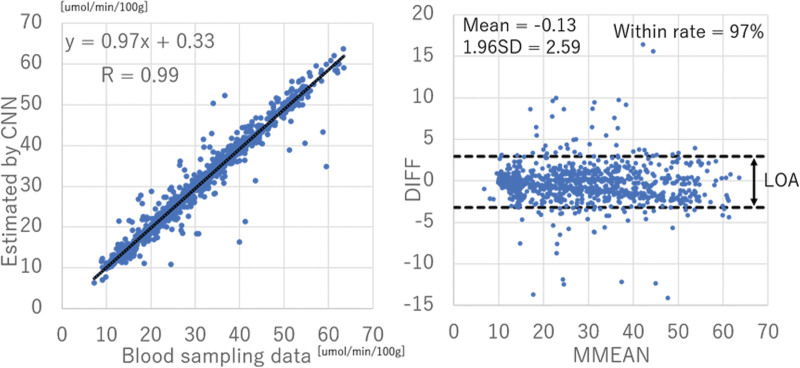
In each test patient, 100 ROIs were strategically positioned within the brain (Fig. 7). These ROIs were automatically defined as single-pixel regions every 2 cm along the vertical and horizontal axes, excluding those located within the ventricles or outside the brain. Time-radioactivity curves were obtained from brain tissue, and the CMRGlc was calculated using a 3-compartment model. The results are presented in Fig. 8a. Notably, the correlation coefficient between the CMRGlc values derived from blood sampling and those estimated by the CNN demonstrated a significantly high correlation of 0.99. According to established guidelines [18], a correlation coefficient of ≥0.5 indicates a strong correlation. Figure 8b illustrates the Brand–Altman plot, where the vertical axis represents the difference (DIFF) between the CMRGlc values obtained from blood sampling and those estimated by the CNN, and the horizontal axis represents the average (MMEAN) of these values. The dashed line represents the limits of agreement (LOA), calculated as ‘mean DIFF ± 1.96 × SD of DIFF’. If 95% of the DIFF values fall within the LOA, it indicates that the error follows a normal distribution and can be considered consistent between the CMRGlc values obtained from blood sampling and those estimated by the CNN. The findings of this study demonstrated a high level of consistency, with 97% of the DIFF values falling within the LOA.
Fig. 7.
100 regions of interest (ROI) in the brain of each case. ROIs in the image were automatically set to 1-pixel ROIs every 2 cm in the vertical and horizontal directions. However, the ROIs inside the ventricle and outside the brain were excluded.
Fig. 8.
(a) Scatter plot of the time-radioactivity curve of arterial plasma from blood sampling and by CNN. The correlation coefficient is 0.99, indicating that there is a strong correlation between the two. (b) The Brand–Altman plot (vertical axis; DIFF) (difference between CMRGlc obtained from blood sampling data and estimated by CNN), horizontal axis: MMEAN (average of CMRGlc obtained from blood sampling data and estimated by CNN). The results obtained in this study were consistent because 97% of DIFF was included in the LOA.
Grad-CAM
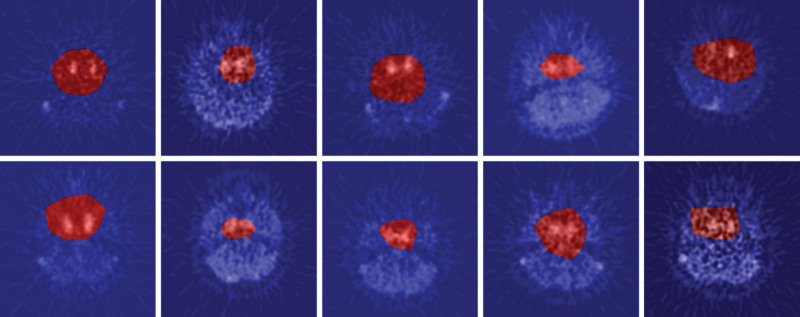
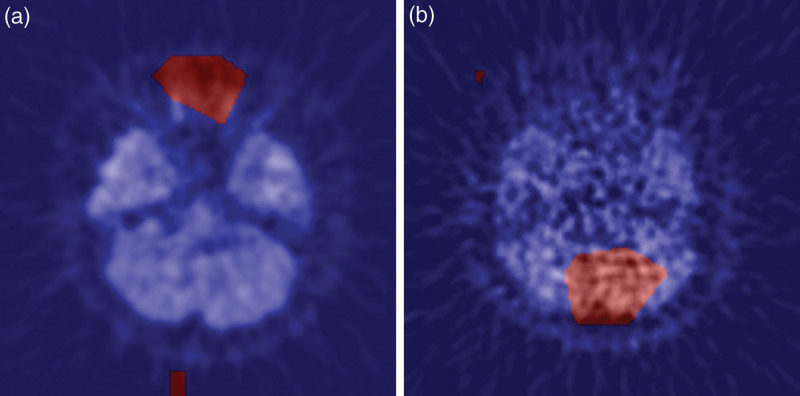
We employed Grad-CAM to discern the precise anatomical regions within the image from which the neural network extracted the most salient information. Exemplary instances of this analysis are illustrated in Fig. 9. It is noteworthy that all patients presented at least one image where an activation region encompassing the vicinity of the internal carotid artery was clearly evident. Conversely, Fig. 10 provides an example of Grad-CAM highlighting areas other than the internal carotid artery. Specifically, (A) delineates enhanced regions surrounding the optic nerve, while (B) identifies enhanced areas adjacent to the cerebellum. These particular images manifest notable prediction errors.
Fig. 9.
Typical examples of Grad-CAM: CNN was found to respond to the internal carotid in slices containing the internal carotid.
Fig. 10.
There were cases in which Grad-CAM enhanced areas other than the internal carotid. (a) The area around the optic nerve is enhanced. (b) The area around the cerebellum was enhanced.
Discussions
In this study, we employed a CNN architecture known as Xception that leverages depth-wise separable convolution effectively diminishing the number of parameters and computational complexity compared with conventional approaches. Consequently, in 21 cases, we ascertained that >18 of the 22 frames manifested a prediction error within a range of 10%. Moreover, the correlation coefficient between the estimated CMRGlc derived from blood samples and those acquired from the CNN exhibited a highly significant value of 0.98. These compelling findings indicate that the CNN can accurately estimate the arterial plasma time-radioactivity curve using 18F-FDG dynamic brain PET data, thereby enabling noninvasive quantitative analysis of diverse dynamic imaging data.
We performed deep learning analysis to estimate the arterial blood time-radioactivity curve from dynamic brain 18F-FDG-PET images specifically targeting the internal carotid region. As a straightforward approach, we postulated that the arterial blood time-radioactivity curve can be derived by setting the ROI encompassing the internal carotid in the dynamic image and computing the maximum radioactivity at each timepoint. This choice is motivated by the fact that the PET device used in this study had an FWHM of 4.8 mm, while the pixel size of the PET image output was 2.5 mm. Furthermore, the inner diameter of the internal carotid in the skull ranges from approximately 2–3 mm, and considering the partial volume effect, accurate quantification of arterial blood radioactivity passing through the internal carotid lumen is unfeasible with an image possessing an FWHM of 4.8 mm. The desired parameter for analysis is not the complete time-radioactivity curve of whole blood, but rather the arterial plasma’s time-radioactivity curve after the removal of erythrocytes. Given that blood cell components are not taken up by brain tissue, the time-radioactivity curve of 18F-FDG absorbed by blood cells should be excluded from the input function during compartment model analysis. Therefore, in this study, we developed a CNN model that estimates the arterial plasma time-radioactivity curve, instead of the arterial blood time-radioactivity curve, using dynamic brain PET images of 18F-FDG.
Numerous techniques have been reported in the literature or estimating the input function in brain PET using image data. Typically, these approaches involve defining an ROI encompassing the relevant vascular structures in a PET scan to generate an input function. However, a persistent challenge lies in generating a metabolite-corrected plasma curve from a whole blood curve. Consequently, reliance on arterial or venous blood sampling remains necessary [2]. In the proposed methodology, blood sampling is used to train the CNN. Nevertheless, the findings of this study suggest that the use of a well-trained CNN might obviate the need for blood sampling during examinations.
On the contrary, the criteria employed by the CNN remain opaque, making the cause behind the heightened prediction error in the eight cases unknown. Nevertheless, it was observed that in instances where the prediction was unsuccessful, the visibility of the internal carotid was diminished compared with other cases. Furthermore, as depicted in Fig. 9, Grad-CAM exhibited a proclivity to focus on regions beyond the internal carotid. Based on these observations, it is plausible to speculate that the challenge in visualizing the internal carotid may have contributed to the diminished prediction accuracy, despite our efforts to identify frames where the internal carotid was most prominently discernible. To address this, we propose that augmenting the number of images showcasing characteristics similar to those of the training data would prove useful.
CNNs are renowned for their ability to classify images based on distinctive features present within them. Grad-CAM, a visualization technique, can illuminate the ‘region of AI focus’ and holds promise in promoting the development of explainable AI, in contrast to the conventional ‘black box’ paradigm, thereby fostering increased user confidence. The findings of this investigation substantiate that in numerous instances, CNNs exhibit discernible responses to the internal carotid, as exemplified in Fig. 9. Nonetheless, there were certain cases where Grad-CAM accentuated regions distinct from the internal carotid, as illustrated in Fig. 10. Consequently, this study validates the potential for visualizing the AI’s zone of interest, enabling further exploration of the foundations underpinning its predictions.
Conclusion
The use of a CNN has demonstrated the potential for estimating time- radioactivity curves in arterial vessels derived from 18F-FDG brain PET dynamic data. This application of CNNs offers a noninvasive method for quantifying input functions extracted from dynamic PET data, thereby presenting an effective approach for the quantitative analysis of various dynamic medical imaging datasets.
Acknowledgements
This study was supported by the Japan Society for the Promotion of Science (JSPS N0.22K0765802).
All procedures performed in studies involving human participants were by the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
The institutional review board of Hokkaido University Hospital approved the study (UMIN000018160) and waived the need for written informed consent from each patient because the study was conducted retrospectively.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Cai W, Feng D, Fulton R, Siu WC. Generalized linear least squares algorithms for modeling glucose metabolism in the human brain with corrections for vascular effects. Comput Methods Programs Biomed 2002; 68:1–14. [DOI] [PubMed] [Google Scholar]
- 2.van der Weijden CWJ, Mossel P, Bartels AL, Dierckx RAJO, Luurtsema G, Lammertsma AA, et al. Non-invasive kinetic modelling approaches for quantitative analysis of brain PET studies. Eur J Nucl Med Mol Imaging 2023; 50:1636–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett EA, Ananth M, Rossano S, Zhang M, Yang J, Lin S, et al. Quantification of positron emission tomography data using simultaneous estimation of the input function: validation with venous blood and replication of clinical studies. Mol Imaging Biol 2019; 21:926–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zanotti-Fregonara P, Chen K, Liow JS, Fujita M, Innis RB. Image-derived input function for brain PET studies: many challenges and few opportunities. J Cereb Blood Flow Metab 2011; 31:1986–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawauchi K, Furuya S, Hirata K, Katoh C, Manabe O, Kobayashi K, et al. A convolutional neural network-based system to classify patients using FDG PET/CT examinations. BMC Cancer 2020; 20:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sibille L, Seifert R, Avramovic N, Vehren T, Spottiswoode B, Zuehlsdorff S, et al. 18F-FDG PET/CT uptake classification in lymphoma and lung cancer by using deep convolutional neural networks. Radiology 2020; 294:445–452. [DOI] [PubMed] [Google Scholar]
- 7.Hwang D, Kang SK, Kim KY, Seo S, Paeng JC, Lee DS, et al. Generation of PET attenuation map for whole-body time-of-flight 18F-FDG PET/MRI using a deep neural network trained with simultaneously reconstructed activity and attenuation maps. J Nucl Med 2019; 60:1183–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirienko M, Sollini M, Silvestri G, Mognetti S, Voulaz E, Antunovic L, et al. Convolutional neural networks promising in lung cancer t-parameter assessment on baseline FDG-PET/CT. Contrast Media Mol Imaging 2018; 2018:1382309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawauchi K, Hirata K, Katoh C, Ichikawa S, Manabe O, Kobayashi K, et al. A convolutional neural network-based system to prevent patient misidentification in FDG-PET examinations. Sci Rep 2019; 9:7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chollet F. Xception: Deep learning with depthwise separable convolutions. Proceedings - 30th IEEE conference on computer vision and pattern recognition, CVPR 2017. IEEE. 2017. pp. 1800–1807. [Google Scholar]
- 11.Xie B, He X, Huang W, Shen M, Li F, Zhao S. Clinical image identification of basal cell carcinoma and pigmented nevi based on convolutional neural network. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2019; 44:1063–1070. [DOI] [PubMed] [Google Scholar]
- 12.Shinohara Y, Takahashi N, Lee Y, Ohmura T, Kinoshita T. Development of a deep learning model to identify hyperdense MCA sign in patients with acute ischemic stroke. Jpn J Radiol 2020; 38:112–117. [DOI] [PubMed] [Google Scholar]
- 13.Rahimzadeh M, Attar A. A new modified deep convolutional neural network for detecting covid-19 from x-ray images. ArXiv. arXiv; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yen C-W, Young C-N, Nagurka M. A vector quantization method for nearest neighbor classifier design. Pattern Recognition Letters. 2004; 25:725–731. [Google Scholar]
- 15.Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference, and prediction. Springer; 2009. [Google Scholar]
- 16.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. International Journal of Computer Vision. 2020; 128:336–359. [Google Scholar]
- 17.Graham MM, Muzi M, Spence AM, O’Sullivan F, Lewellen TK, Link JM, et al. The FDG lumped constant in normal human brain. J Nucl Med 2002; 43:1157–1166. [PubMed] [Google Scholar]
- 18.Cohen J. Statistical power analysis for the behavioral sciences. Second Edition. LAWRENCE ERLBAUM ASSOCIATES; 1988. [Google Scholar]