Abstract
Sequential Candida glabrata isolates were obtained from the mouth of a patient infected with human immunodeficiency virus type 1 who was receiving high doses of fluconazole for oropharyngeal thrush. Fluconazole-susceptible colonies were replaced by resistant colonies that exhibited both increased fluconazole efflux and increased transcripts of a gene which codes for a protein with 72.5% identity to Pdr5p, an ABC multidrug transporter in Saccharomyces cerevisiae. The deduced protein had a molecular mass of 175 kDa and was composed of two homologous halves, each with six putative transmembrane domains and highly conserved sequences of ATP-binding domains. When the earliest and most azole-susceptible isolate of C. glabrata from this patient was exposed to fluconazole, increased transcripts of the PDR5 homolog appeared, linking azole exposure to regulation of this gene.
Failure of oropharyngeal candidiasis (OPC) to respond to fluconazole has been noted in many institutions, usually in patients with far advanced human immunodeficiency virus (HIV) infection. The majority of HIV-infected patients with fluconazole-unresponsive OPC have CD4 counts below 50/mm3 and have received many months of azole therapy (24, 33). Resistance has arisen gradually, the infection has required higher and higher doses to respond, relapses have occurred more rapidly, and response to therapy has been progressively less complete. The gradual progression of resistance had suggested a series of mutations, and molecular analysis of sequential Candida albicans strains from such patients has supported this hypothesis (27, 29, 34, 39).
Several findings indicate that increased azole efflux is a major mechanism of resistance in C. albicans, Candida glabrata, and Saccharomyces cerevisiae. Studies of azole-resistant isolates have shown increased energy-dependent azole efflux in C. glabrata (11, 17, 22), decreased intracellular azole concentrations in C. albicans and C. glabrata (10, 11, 25), and increased expression of the C. albicans multidrug transporter genes CDR1, CDR2, and MDR1 (1, 27, 30, 38). Inactivation of the CDR1 gene in C. albicans (28) and PDR5 in S. cerevisiae (31) leads to increased fluconazole susceptibility and, in C. albicans, to increased intracellular fluconazole concentrations. Another mechanism of azole resistance has been postulated to be mutation of ERG3. The resultant defect in Δ5,6 desaturation is thought to prevent the cell from accumulating a toxic 14-α-methylergosta-8,24(28)-dien-3β,6α-diol in the presence of azole (14).
There is also evidence supporting the hypothesis that azole resistance may arise from chromosome duplications leading to increased expression of ERG11 (ERG16), the gene which codes for the azole target enzyme, lanosterol 14-α demethylase (CYP51A1) (17, 35). Mutations in ERG11 that apparently alter substrate specificity may also cause azole resistance (12, 15, 29, 34). Identification of the key mutations which permit retention of lanosterol demethylation and yet block the effects of azoles are being defined (15, 29, 39).
Secondary resistance can arise during azole therapy by acquisition of a drug-resistant strain of the same or different species. C. glabrata is inherently more resistant to fluconazole than C. albicans and is found more commonly in patients receiving azoles (26). Fluconazole resistance can increase further in C. glabrata if the patient continues to receive fluconazole (37).
We describe a patient with advanced HIV infection whose oral candidiasis responded poorly to increasing doses of fluconazole. Oral cultures contained a C. glabrata strain that persisted and showed increased fluconazole resistance and increased fluconazole efflux. Using homology with the PDR5 and CDR1 genes, we cloned and sequenced a gene which appears to code for a multidrug transporter and showed increased transcription in the presence of fluconazole. The deduced amino acid sequence has the highest identity to the S. cerevisiae ATP-binding cassette transporter Pdr5p (Sts1p and Ydr1p) (4). Because we have not proven that the gene will confer the same phenotype as PDR5, we have chosen to designate the gene PDH1 (for pleomorphic drug resistance homolog) rather than PDR5. Many of the deduced amino acid sequences in Pdh1p are highly conserved in other fungal ABC transporters, including Yor1p and Snq2p (6) in S. cerevisiae, Cdr1p (23), Cdr2p (30) and Cdr3p (3) in C. albicans, bfr1+p in Schizosaccharomyces pombe (20), and Atrbp in Aspergillus nidulans (7). To date, the gene family coding for these proteins has not been studied in C. glabrata.
MATERIALS AND METHODS
Clinical isolates.
A 45-year-old male infected with HIV type 1 with a CD4 count of 60/mm3 and a 4-year history of recurrent OPC was referred because of symptomatic thrush not responding to 200 mg of fluconazole given daily. Discrete and confluent plaques of thrush were evident on the hard palate and buccal mucosa. Microscopic examination of Gram-stained smears from his mouth showed yeast and pseudohyphae. Mouth culture grew C. albicans and C. glabrata. The patient was first treated with 400 mg of fluconazole given daily and seen every 2 weeks. After 4 weeks, he had achieved only a partial response, with at least 20% of the lesions remaining. Fluconazole was increased to 800 mg for 2 additional weeks, after which only a small patch remained on his buccal mucosa and oral symptoms had resolved. Therapy with 800 mg was continued an additional 2 weeks, after which no oropharyngeal lesions were detected. Esophagoscopy was not done. Prophylaxis with fluconazole (200 mg daily) was reinitiated, according to the study protocol. After 2 weeks, his OPC had returned.
Oral specimens.
On each clinic visit, 2 weeks apart, the patient had a culture from the mouth placed on CHROMagar Candida (Hardy Diagnostics) (21). Five Candida colonies from each primary culture were subcultured on YEPD medium (1% yeast extract, 2% peptone, 2% glucose) at 30°C. Cells were suspended in 50% glycerol in water and stored at −80°C. Colonies were identified by germ tube formation in RPMI 1640 and then by use of the API 20C kit (BioMerieux Vitek).
RAPD.
Both randomly amplified polymorphic DNA (RAPD) and a contour-clamped homogeneous electric field (CHEF) were used to show that the fluconazole-resistant isolates obtained later in the course were highly similar to the original C. glabrata strain obtained from this patient. RAPD was synthesized in a 50-μl reaction volume, using 25 ng of DNA, 5 mM Mg2+, 10 nmol of deoxynucleoside triphosphate (Boehringer Mannheim), 50 ng of primer, and 5 U of Taq DNA polymerase (Boehringer Mannheim) in 1× Taq buffer (Boehringer Mannheim). PCR was performed by the method of Lehmann et al. (16), using a Perkin-Elmer Cetus DNA thermal cycler model N801050, with 45 cycles, with 1 cycle consisting of 1 min at 94°C, 1 min at 36°C, and 2 min at 72°C, with a final elongation step at 72°C for 10 min. Following thermal cycling, the amplified DNA was separated by electrophoresis in a 2% (wt/vol) agarose (SeaKem GTG; FMC BioProducts) gel slab (11 by 14 by 1 cm) containing 0.5 μg/ml of ethidium bromide. A 1-kb DNA ladder (Gibco BRL) was included in each run. The primers used for RAPD were primer S (5′-GCGATCCCCA-3′) (oligonucleotide 6 of reference 32), primer 6 (5′-AAGGATCAGA-3′ (RP-2 of reference 16), and primer 7 (5′-CACATGCTTC-3′) (RP4-2 of reference 16).
CHEF.
Pulsed-field electrophoresis was performed with 0.8% chromosomal grade agarose (Bio-Rad) in 0.5× TBE (45 mM Tris, 45 mM borate, 1 mM EDTA; buffer pH 8.0) on gels (14 by 12.5 cm) using a Bio-Rad electrophoresis chamber with a CHEF-DR 11 Drive Module (Bio-Rad). Runs were done at 150 V with 120-s switch times ramped to 240 s over 25 h, followed by 180-s switch times ramped to 360 s over 20 h.
Cloning PDH1.
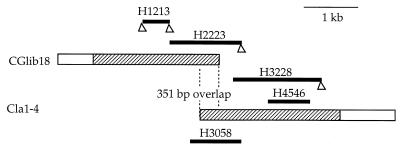
Degenerate primers H12 and H13 were designed to amplify a 0.4-kb fragment, H1213, using genomic DNA of the C. glabrata NCCLS84 as the template. Sequences of H12 and H13, shown in Table 1, were chosen on the basis of amino acid sequences conserved in PDR5 and CDR1, including a part of Walker A and B motifs. Amplification was performed with Taq DNA polymerase (Boehringer Mannheim) and a mini-thermal cycler (MJ Research) with 25 cycles, with 1 cycle consisting of 1 min at 94°C, 80 s at 48°C, and 1 min at 72°C. The amplified fragment was extracted twice with equal volumes of phenol-chloroform and once with chloroform. Fragments were blunt ended with T4 DNA polymerase (New England Biolabs) treatment and inserted in pCR-Script SK(+) vector (Stratagene). The JM109 strain of Escherichia coli (Promega) was transformed with the constructed plasmid by heat pulses as described by the supplier. Clones were sequenced by using a rhodamine terminator sequencing reaction, run on an ABI Prism 377 (Perkin-Elmer). The base sequence was analyzed with GCG (Genetics Computer Group) software. When the sequence was confirmed to have sufficient identity to PDR5, the subcloned H1213 fragment was labeled with 32P using random priming and used as a probe to screen an EcoRI-digested genomic library of the NCCLS84 strain of C. glabrata (Lambda Zap II library made by Stratagene). A 3.0-kb genomic fragment was identified, cloned into pBSK [Bluescript II SK (+); Stratagene] as CGlib18 and sequenced by the method described above. Since the sequence of the CGlib18 was shown to have high homology to the 5′ end of the PDR5 gene, a downstream fragment was obtained by PCR, using NCCLS84 genomic DNA as the template. One primer (H22) was based upon 3′ sequences from CGlib18. The other, a degenerate primer (H23) was based upon the S. cerevisiae PDR5 sequence (Table 1). A 1.3-kb fragment (H2223) was obtained, cloned into pCR-Script, and sequenced. A sense primer (H32) was derived from the 3′ sequence of H2223, and a degenerate primer (H28) was obtained from S. cerevisiae PDR5 sequence. A 1.6-kb fragment (H3228) was obtained by PCR. H4546 was amplified from H3228. Southern blot of a Cla1 (New England Biolabs) digest of NCCLS84 genomic DNA using H4546 as a probe hybridized to a single 3.6-kb fragment. A size-selected Cla1 genomic library was then made from NCCLS84, ligating the 2.5- to 4.5-kb fragments into the Cla1 site of pBSK. Using H4546 as a probe, two 3.6-kb clones, Cla1-1 and Cla1-4, were identified by colony hybridization. Cla1-4 was sequenced in its entirety. This strategy is diagrammed in Fig. 1.
TABLE 1.
Oligonucleotides used in PCR clones
| PCR clone | Template DNA | Sequence (5′→3′) (oligonucleotide name) of primer
|
|
|---|---|---|---|
| Sense | Antisense | ||
| H1213 | Genomic DNA from NCCLS84 | GTCGTTTTAGGTAG(A/G)CC[N]G (H12) | CCTTGTAGC(A/G)TT(A/G)TCCCA (H13) |
| H2223 | Genomic DNA from NCCLS84 | TAAGCAATCAAAGAGAGCTAG (H22) | (T/C)TT[N]CC[N]GC[N]CC[N]GA[N]GC (H23) |
| H3228 | Genomic DNA from NCCLS84 | TCAGAAGCAATCTTCCATTGGC (H32) | C(G/T)(A/G)TACAT(A/G)AA(A/G/T)ATCCA(A/G)AA (H28) |
| H4546 | H3228 | CAGATGCAGTTGTTGGTGTC (H45) | AGCCAATATAAATGCCTTCC (H46) |
| H3058 | Genomic DNA from NCCLS84 | CGCTGTCTTTGCCATGTC (H30) | GTTGTTTTACCTGCACCG (H58) |
FIG. 1.
Cloned fragments of the C. glabrata PDH1 gene. Cloned genomic fragments CGlib18 and Cla1-4 are shown (the hatched areas indicate the ORFs). Five PCR subclones (H1213, H2223, H3228, H4546, and H3058) are shown, and each is aligned to the corresponding genomic sequence. Triangles at the end of PCR subclones show degenerate primers.
Probes for Southern and Northern blots.
A 0.85-kb PCR fragment, H4546, was used for both Southern and Northern analyses. An ACT1 probe was prepared as previously published (8). A 25S rRNA-encoding sequence from C. albicans (18) was 32P labeled as a probe for genes coding for rRNA (rDNA).
Southern analysis.
C. glabrata genomic DNA was extracted from strain NCCLS84 as previously published (8), digested with a restriction enzyme, and used in gel electrophoresis. The gel was denatured (1 M NaCl, 0.5 M NaOH), neutralized (1 M Tris-HCl [pH 7.5], 1 M NaCl) and transferred to a nylon membrane (Hybond-N; Amersham). Chromosomal DNA run in CHEF gel was first depurinated with 0.25 M HCl at room temperature for 20 min and then transferred by the same procedure as for the digested genomic DNA. Probes were randomly labeled with [α-32P]dCTP (Amersham) using a Prime It II kit (Stratagene). The blot was hybridized with the probe at 65°C, washed once with washing solution 1 (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS]) at room temperature, twice with washing solution 2 (0.2× SSC, 0.1% SDS) at room temperature, and then once at 65°C.
Northern analysis.
RNA was prepared from C. glabrata cells after sufficient growth in YEPD medium at 30°C to provide an optical density at 600 nm of 1 to ∼1.5. Cells were harvested and washed with RPMI 1640 medium with 300 mg of l-glutamine (Sigma) per liter, 0.165 M MOPS (morpholinepropanesulfonic acid) (Gibco BRL), and 2% glucose, pH 7.0. Cells were then rotated at 30°C for 4 h in the same RPMI 1640 medium with the addition of fluconazole (1 μg/ml). RNA was extracted from 50 mg of cells using the FastRNA Kit (Bio 101). Gel electrophoresis, blotting onto nylon membranes, and hybridization with 32P-labeled probes were done by standard methods, and the blots were washed under the same conditions as those for the Southern blots.
Quantitation of signal intensities.
Southern and Northern blots were exposed to Storage Phosphor Screens (Molecular Dynamics) for 3 h, and the screens were scanned with PhosphorImager 445 SI (Molecular Dynamics). The scanned images were quantitated with ImageQuant software (Molecular Dynamics). Quantitative volume data for the same-sized rectangular square on the blot image with each probe were used for analysis of the level of expression.
Other methods.
Intracellular fluconazole concentrations were measured after 80 min of incubation in 100 nM [3H]fluconazole (714 GBq/mmol) at 37°C with shaking at 170 rpm in phosphate-buffered saline (22). Cultures were incubated with and without 100 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP). This compound is a respiratory uncoupler which depletes intracellular stores of ATP. This concentration of CCCP did not decrease cell viability (data not shown). Specific activity of [3H]fluconazole was not sufficient to measure efflux from energy-deprived cells after glucose loading so this method was not used. MIC of fluconazole in microdilution broth was read after 48 h of incubation at 35°C. Methods of sterol analysis and antifungal susceptibility have been published elsewhere (8). Fluconazole levels in blood were performed by bioassay in the laboratory of Michael Rinaldi.
Nucleotide sequence accession number. The GenBank accession no. for the C. glabrata PDH1 gene is AF046120.
RESULTS
Studies of fluconazole resistance.
Events that occurred to the patient during study are summarized in Table 2. The concentrations of fluconazole in blood were consistent with the prescribed dose. Of the five colonies selected from mouth cultures on each of the six patient visits, two C. glabrata and three C. albicans isolates were obtained for study on all visits except those on weeks 6 and 8, when only C. glabrata colonies were detected on the CHROMagar plates. The susceptibility of the C. glabrata isolates decreased to fluconazole and itraconazole, but not to amphotericin B, during fluconazole treatment. C. glabrata colonies obtained on the same throat culture had fluconazole MICs that were within 1 dilution of one another (data not shown). The C. albicans isolates had an initial fluconazole MIC of 8 μg/ml, much higher than our reference strain of C. albicans (B311), which had an MIC of 0.25 μg/ml. The last two C. albicans strains from this patient had an MIC of 32 μg/ml. Interest focused on C. glabrata because this species was isolated on every occasion, even on week 8 when no lesions were apparent and a sharp increase in azole resistance occurred after the first visit. The relative roles of C. glabrata and C. albicans in this patient’s disease cannot be ascertained, but the absence of C. albicans during clinical response suggests this species was the major pathogen.
TABLE 2.
Course of therapy
| C. glabrata strain or isolate | Wk | Fluconazole dose (mg/day) | Oral lesions | Fluconazole level in blood (μg/ml) | MIC (μg/ml)
|
|||
|---|---|---|---|---|---|---|---|---|
|
C. glabrata
|
C. albicans, fluconazole | |||||||
| Fluconazole | Itraconazole | Amph.a | ||||||
| NCCLS84 | 32 | 2 | 0.25 | |||||
| Isolates | ||||||||
| 35a | 0 | 200b | Buccal and palate | 7 | 32 | 2 | 0.5 | 8 |
| 36a | 2 | 400 | Buccal only | 17 | 128 | >16 | 0.5 | 16 |
| 37a | 4 | 400 | Buccal only | 40 | 128 | >16 | 0.5 | 32 |
| 38a | 6 | 800 | Small patch | 55 | 128 | >16 | 0.5 | NAc |
| 40a | 8 | 800 | None | 65 | 128 | >16 | 0.5 | NA |
| 43a | 10 | 200 | Buccal and palate | 18 | 128 | >16 | 0.5 | 32 |
Amph., amphotericin B.
See text for prior therapy. The patient was receiving 200 mg of fluconazole daily upon entry into the study.
NA, not applicable (no isolate).
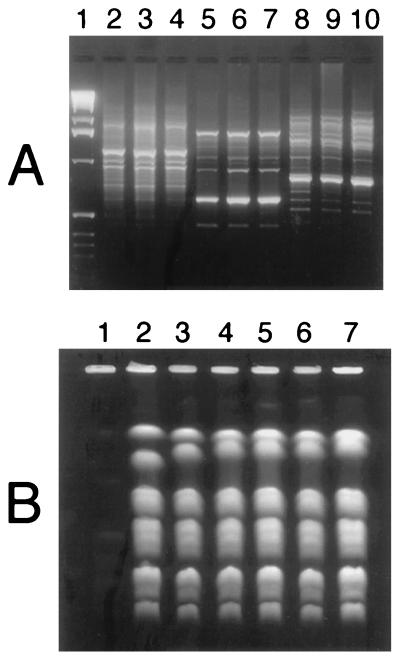
First, an attempt was made to establish whether the same strain had persisted throughout the study period. RAPDs of the C. glabrata isolates with three different sets of primers remained the same throughout treatment (Fig. 2A). CHEF analysis showed that in the earliest isolate the second largest chromosome was smaller than in subsequent isolates (Fig. 2B). The second isolate obtained from this patient also had a slightly smaller second band. A 32P-labeled rDNA probe hybridized with the two largest bands on a CHEF blot of isolates 35a and 40a (data not shown), indicating that changes in the rDNA copy number may have accounted for the size difference. The other bands, with sizes below 1.6 Mb, were unchanged. When conditions were changed to separate the smaller bands more clearly on the CHEF, no differences in the smaller bands were seen (data not shown). It was concluded that the earliest two isolates differed slightly from the other isolates, which appeared identical.
FIG. 2.
(A) RAPD gel analysis of C. glabrata DNA. Isolates were obtained at the start of the study (isolate 35a [lanes 2, 5, and 8]), at 2 weeks (isolate 36a [lanes 3, 6, and 9]), and at 4 weeks (isolate 37a [lanes 4, 7, and 10]). Primer S (lanes 2 to 4), primer 6 (lanes 5 to 7), and primer 7 (lanes 8 to 10) were used. A molecular ladder is shown in lane 1. (B) CHEF gel using isolates from weeks 0, 2, 4, 6, 8, and 10 (isolates 35a, 36a, 37a, 38a, 40a, and 43a, respectively) are shown in lanes 2 to 7, respectively. The Saccharomyces control is in lane 1.
Nonesterified sterol analysis detected only ergosterol in the six C. glabrata isolates (isolates 35a, 36a, 37a, 38a, 40a, and 43a), indicating that altered membrane sterols could not account for azole resistance. Increased C. glabrata resistance to fluconazole has recently been reported to be associated with increased energy-dependent fluconazole efflux (22). For this reason, fluconazole intracellular concentration was measured in cultures that had been incubated with or without 100 μM CCCP, a respiratory uncoupler (Fig. 3). In the absence of CCCP, C. glabrata isolates after the first visit showed markedly reduced intracellular fluconazole concentrations. CCCP increased intracellular fluconazole concentrations in all isolates, showing that energy-dependent drug efflux made a major contribution to the observed differences in intracellular fluconazole concentration. The initial isolate, isolate 35a, accumulated more fluconazole both with and without CCCP, consistent with lesser fluconazole efflux than the later isolates.
FIG. 3.
Intracellular fluconazole concentrations. Isolates Y33.90 and Y33.91 were azole-susceptible and -resistant C. glabrata isolates from the previous report (22). Isolates 35a, 36a, 37a, 38a, 40a, and 43a were obtained at 0, 2, 4, 6, 8, and 10 weeks, respectively. Fluconazole accumulation without CCCP (white bars) and with CCCP (black bars) is shown. The standard errors are indicated by the error bars. OD600, optical density at 600 nm.
Sequence of PDH1.
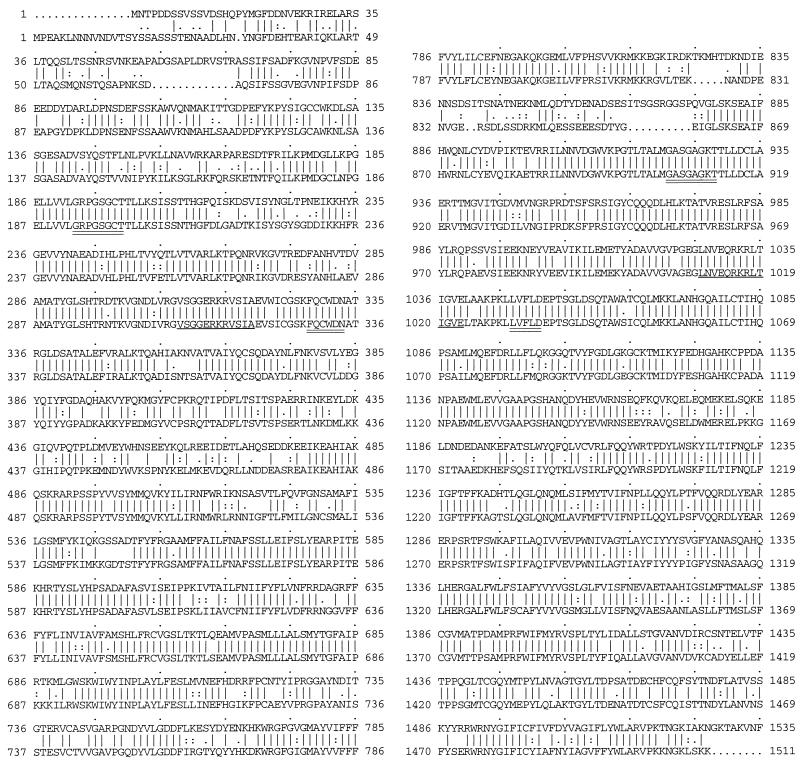
We obtained the complete PDH1 sequence from two genomic clones, CGlib18 and Cla1-4, which overlapped by 351 bp. Because CGlib18 and Cla1-4 overlapped but differed by one base, we sequenced the overlap area in another genomic clone, Cla1-1, and a PCR clone, H3058, that was directly amplified from genomic DNA with Vent DNA polymerase (New England Biolab) (Table 1 and Fig. 1). All the new clones gave the same sequence in the overlap area as Cla1-4. Analysis of the 6,225-base sequence revealed a 4,626-bp open reading frame (ORF) with 1,542 deduced amino acids. The deduced protein had a molecular mass of 175 kDa. There was no adenine at the −3 position 5′ to the initiation ATG codon, although this has been invariant in the modest number of C. glabrata genes cloned so far: ERG3, ERG11, SNF1, TRP1, HIS3, URA3, and SEC14. No intron splice sites or alternative ORF was found. In the 623-bp 5′-flanking region, there was a TATAA sequence at −105 and CAAT sequences −365 and −136 from the ATG start site. Also in the putative 5′ untranslated region was a 10-bp sequence identical to that described by Katzmann and coworkers for the S. cerevisiae PDR5 promoter (13). They described an 18-bp sequence which bound Pdr1p and Pdr3p and was critical for PDR5 expression (13) (Table 3). A palindrome that has been postulated to be a regulatory cis element in the PDR5 promoter was also found in the 5′-flanking sequence of PDH1 (19) (Table 3). The sequence TTTGCA is repeated four times between bases −468 and −502 5′ to the ATG codon. The significance of this repeat is unknown. In the 3′-flanking sequence, two putative transcription termination signals were found: the AATAAA signal of Henikoff et al. (9) at +234 from the stop codon and the TAGN28TAGTN6TTT sequence of Zaret and Sherman (40) at +175. In both the C-terminal and N-terminal homologous halves of this molecule were ABC signatures and Walker motifs (36) that are highly conserved among ABC transporter proteins in S. cerevisiae (PDR5, SNQ2, and YOR1 [GenBank accession no. L19922, X66732, and Z73066, respectively]), C. albicans (CDR1, CDR2, and CDR3 [GenBank accession no. X77589, U63812, and U89714, respectively]) (Table 4). A Kyte-Doolittle hydropathy plot (window = 7) indicated six putative transmembrane domains in each of two homologous portions, similar to those reported in the transporter proteins listed in Table 4 (data not shown). Fasta search of the amino acid sequence (Fig. 4) retrieved PDR5 as the most similar peptide with 72.5% identity in the 1,526-amino-acid overlapping sequence. CDR1, a PDR5 homolog in C. albicans, was shown to have 53.9% identity in 1,527-amino-acid sequence overlap. SNQ2, a S. cerevisiae gene closely related to PDR5, had 39.1% identity in a 1,524-amino-acid overlap.
TABLE 3.
Conserved nucleotide sequences among ABC transporters
| Gene | Positions | Nucleotide sequencec |
|---|---|---|
| PDR response elementa | ||
| PDH1 | −200 to −190 | 5′-ATCTTCCGTGGAATATCC-3′ |
| PDR5 | −205 to −188 | 5′-TGATTCCGTGGAAAGGTC-3′ |
| Palindromeb | ||
| PDH1 | −560 to −550 | TTCCG·TGGAA |
| −109 to −100 | TTCCA·CGGAA | |
| PDR5 | −536 to −526 | TTCCCACGGAA |
| −492 to −483 | TTCCG·CGGAA | |
| −376 to −367 | TTCCG·TGGAA |
PDR response element site 2 of PDR5 (13) and corresponding sequence in PDH1.
Palindrome with possible regulatory function (19).
Conserved nucleotides are shown in bold letters. A nucleotide was considered conserved if a specific base was observed at the same aligned position in more than two different genes. Gaps introduced to maximize alignment are indicated by periods.
TABLE 4.
Conserved amino acid sequences among ABC transporters
| Protein | Amino acid sequencea
|
|||||
|---|---|---|---|---|---|---|
| Walker A
|
Walker Bb
|
ABC signature
|
||||
| N-terminal | C-terminal | N-terminal | C-terminal | N-terminal | C-terminal | |
| Pdh1p | GRPGSGCTT | GASGAGKTT | FQCWDX6D | LLVFLDX6D | VSGGERKRVSIA | LNVEQRKRLTIGVE |
| Pdr5p | GRPGSGCTT | GASGAGKTT | FQCWDX6D | LLVFLDX6D | VSGGERKRVSIA | LNVEQRKRLTIGVE |
| Snq2p | GRPGAGCSS | GESGAGKTT | FQCWDX6D | LLLFLDX6D | ||
| Yor1p | GPIGTGKSS | GRTGAGKST | IYLFDX6D | KILILDX6D | ||
| Cdr1p | GRPGAGCST | GASGAGKTT | IQCWDX6D | LLLFLDX6D | VSGGERKRVSIA | LNVEQRKRLTIGVE |
| Cdr2p | GRPGAGCST | GASGAGKTT | IQCWDX6D | LLLFLDX6D | VSGGERKRVSIA | LNVEQRKRLTIGVE |
| Cdr3p | GRPGAGCST | GASGAGKTT | IQCWDX6D | LLVFLDX6D | ISGGERKRLSIA | LNVEQRKRLTIAVE |
| bfr1+p | GQPGSGCST | GESGAGKTT | IACWDX6D | LLLFLDX6D | ||
| Atrap | GRPGTGCST | GVSGAGKTT | FAAWDX6D | LLLFLDX6D | VSGGERKRVSIA | LNVEQRKLLTIGVE |
| Atrbp | GRPGSGCTT | GSSGAGKTT | VFCWDX6D | LLIFLDX6D | VSGGERKRVSIA | LSVEQRKRVTIGVE |
Conserved amino acids are shown in bold letters. An amino acid was considered conserved if a specific amino acid was observed at the same aligned position in more than two different genes. When two or more amino acids were conserved in more than two genes, no amino acid was considered conserved.
X6, six amino acids.
FIG. 4.
Comparison of the amino acid sequences of PDH1 and PDR5. For each pair of sequences, the top sequence is PDH1 and the bottom one is PDR5. Lines between the sequences indicate perfect matches of amino acids. Periods and colons between the sequences show similarity based on the Dayhoff table (30a) as described elsewhere (7a). There was 72.5% identity between the two sequences, with two gaps. Putative Walker A and B sites (double underlines) and putative ATP-binding sites (single underline) are indicated.
Southern and Northern analyses.
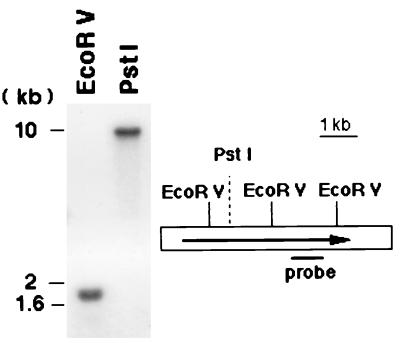
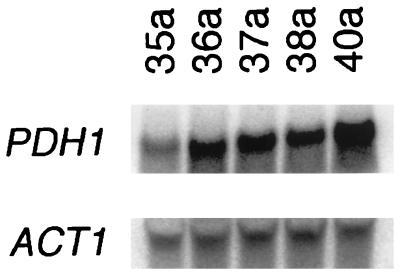
Southern analysis with the H4546 probe showed a 1.8-kb band after EcoRV digestion. A single band of about 10 kb was found on PstI digestion with the same probe. Both findings are consistent with the restriction map obtained from the sequence (Fig. 5). Considering that PDH1 appeared as a single fragment of the anticipated size in Southern analysis, it was unlikely that the PDH1 gene had a duplicate copy. By Northern analysis, using phosphorimaging quantitation, transcription of PDH1 was increased from a PDH1/ACT1 ratio of 3.5 to 32.3 when the initial, more azole-susceptible strain, strain 35a, was incubated in fluconazole. In strain 40a, transcription was not further increased by fluconazole treatment, in that the PDH1/ACT1 ratio was unchanged at 26.6 without and 24.1 with fluconazole exposure. The PDH1/ACT1 ratio of strain 40a not exposed to fluconazole was 7.5 fold higher than that in strain 35a, indicating that PDH1 was upregulated in the more azole-resistant strain. In a separate Northern analysis, sequential isolates from the patient given increasing doses of fluconazole showed a progressive increase in PDH1 transcription in the absence of fluconazole (Fig. 6). The PDH1/ACT1 ratios in these isolates, isolates 35a, 36a, 37a, 38a, and 40a, were 2.3, 2.7, 2.8, 3.4, and 4.5, respectively.
FIG. 5.
Southern analysis and restriction map for PDH1 gene. EcoRV digestion of C. glabrata genomic DNA demonstrated a single 1.8-kb fragment in the Southern analysis as expected by the restriction map. PstI digestion showed a single fragment of about 10 kb. The sequenced area of the genome (white bar) and the PDH1 ORF (arrow) are shown.
FIG. 6.
Northern blots of PDH1 transcripts in our series of C. glabrata strains. Actin transcripts were used as controls for gel loading. The amount of PDH1 transcript relative to actin (ACT1) was higher in the azole-resistant strains (strains 36a, 37a, 38a, and 40a) than in the susceptible strain (strain 35a).
Because increased ERG11 transcript in an azole-resistant C. glabrata is attributable to a roughly fourfold increase in copies of a chromosome (17), we quantitated PDH1 copy numbers using ACT1 as a control and found that the PDH1/ACT1 ratios in isolates 35a and 40a were almost equal, 1.6 and 1.7, respectively. A CHEF blot hybridized with the same PDH1 probe demonstrated a band with a size of about 1 Mb, based on the S. cerevisiae standard. The ACT1 gene in C. glabrata resides on a 1.4-Mb chromosome (17). The increased PDH1 mRNA in isolate 40a could not be attributed to increased gene copy number.
DISCUSSION
The patient when first seen had clinically apparent, symptomatic OPC despite a history of taking 200 mg of fluconazole each day and a blood level consistent with that history. Fluconazole susceptibility of the C. glabrata isolate from this patient was not different from our standard strain. It seems likely that the sudden appearance of azole resistance in serial isolates did not represent a strain change. RAPD patterns and all but the second CHEF band remained unchanged. Serial isolates of C. glabrata from the same patient often have shown variable bands with a size of >1.6 Mb, generally the top two bands. Variability has been attributed to changes in the copy number of rDNA in these bands (2). We also found rDNA in our two largest bands.
The change in azole MIC of the C. glabrata strains occurred when the fluconazole dose was increased. The more-resistant strains had a two- to threefold decrease in intracellular accumulation of fluconazole when incubated with radiolabeled drug. A metabolic inhibitor increased intracellular drug, indicative of active drug efflux. The failure of CCCP to increase intracellular concentrations to the level seen in the more-susceptible isolate may indicate that respiratory-independent mechanisms were also present.
Although decreased intracellular drug concentration may not fully explain a fourfold increase in fluconazole MIC, the change is approximately equal to the difference between the fluconazole-susceptible C. glabrata Y33.90 and the resistant strain Y33.91. The difference in intracellular concentration is also similar to that reported from five AIDS patients with susceptible and resistant C. albicans isolated from their oropharynx (27). The work reported here supports these prior observations but differs in some respects. Strains Y33.90 and Y33.91 gave different restriction fragment length polymorphism patterns and were considered different strains (11). The change in intracellular fluconazole concentrations reported here in a C. glabrata strain and in the four sets of C. albicans strains reported by Sanglard et al. (28) cannot be readily interpreted as simply strain differences because molecular typing methods detected only a modest difference. It appears more likely that increased drug efflux and resistance arose in the original strain. Cross-resistance to other azoles was found in our case, in the C. albicans-infected case reported by Hitchcock et al. (10), and in the four C. albicans-infected patients reported by Sanglard (28). Our report adds support to the concept that increased drug efflux is a major mechanism of azole resistance in C. glabrata. As noted in the introduction of this article, highly resistant isolates may have multiple mutations, particularly in the gene coding for the azole target enzyme, 14-α-demethylase. Increased azole resistance in our series of C. glabrata isolates may also reflect such mutations as well as the upregulation of ABC transporters.
It would be useful to identify the mutation which increased expression of PDH1 in the isolates obtained after isolate 35a. One possibility is increased PDH1 copy number. Marichal and coworkers found a 3.7-fold increase in the copy number of ERG11 (CYP51 and ERG16) in an azole-resistant C. glabrata, accompanied by an 8-fold increase in ERG11 mRNA (17). Almost four copies of the 740-kb chromosome were found, an aneuploid state which shifted towards euploidy during multiple subcultures. In our azole-resistant C. glabrata, there was no evidence of increased copy number on Southern blots or by probing the CHEF blot with PDH1 and ACT1. The latter gene was chosen for probing the CHEF blot because ACT1 resides on a 1.4-kb band (17), far larger than the chromosome containing PDH1. A more likely explanation for the increased expression in the azole-resistant isolates is that a mutation in a transcriptional regulator locus caused azole resistance by overexpressing ABC transporters (5). If regulation of PDH1 were analogous to regulation of PDR5, then fluconazole exposure may affect a regulatory locus which increases expression of a family of ABC transporter genes, including PDH1. The increase in PDH1 mRNA when isolate 35a was exposed to fluconazole is likely due to the same regulatory process. The 5′-flanking area of PDH1 contained a sequence similar to one of the PDR1/PDR3 binding sites which regulate PDR5 transcription in S. cerevisiae (13). PDR1/PDR3 coregulates not only PDR5 but also SNQ2 and YOR1, meaning that several genes may be upregulated by fluconazole exposure and that the fluconazole transporter may not be Pdh1p. Determining whether Pdh1p is a fluconazole transporter will require disrupting the gene or expressing the gene in a heterologous system, such as in a pdr5 S. cerevisiae mutant. Such studies are in progress. Whether or not PDH1 codes for an azole transporter, this gene provides a tool for studying the effects of azoles on regulatory loci controlling ABC transporter expression in C. glabrata.
ACKNOWLEDGMENT
We thank B. Grimberg for technical assistance.
This work was supported in part by an unrestricted grant from Pfizer, Inc.
REFERENCES
- 1.Albertson G D, Niimi M, Cannon R D, Jenkinson H F. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob Agents Chemother. 1996;40:2835–2841. doi: 10.1128/aac.40.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arif S, Barkham T, Power E G, Howell S A. Techniques for investigation of an apparent outbreak of infections with Candida glabrata. J Clin Microbiol. 1996;34:2205–2209. doi: 10.1128/jcm.34.9.2205-2209.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balan I, Alarco A-M, Raymond M. The Candida albicans CDR3 gene codes for an opaque phase ABC transporter. J Bacteriol. 1997;179:7210–7218. doi: 10.1128/jb.179.23.7210-7218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balzi E, Wang M, Leterme S, Van Dyck L, Goffeau A. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J Biol Chem. 1994;269:2206–2214. [PubMed] [Google Scholar]
- 5.Carvajal E, van den Hazel H B, Cybularz-Kolaczkowska A, Balzi E, Goffeau A. Molecular and phenotypic characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol Gen Genet. 1997;256:406–415. doi: 10.1007/s004380050584. [DOI] [PubMed] [Google Scholar]
- 6.Decottignies A, Lambert L, Catty P, Degand H, Epping E A, Moye-Rowley W S, Balzi E, Goffeau A. Identification and characterization of SNQ2, a new multidrug ATP binding cassette transporter of the yeast plasma membrane. J Biol Chem. 1995;270:18150–18157. doi: 10.1074/jbc.270.30.18150. [DOI] [PubMed] [Google Scholar]
- 7.Del Sorbo G, Andrade A C, van Nistelrooy J G M, van Kan J A L, Balzi E, De Waard M A. Multidrug resistance in Aspergillus nidulans involves novel ATP-binding cassette transporters. Mol Gen Genet. 1997;254:417–426. doi: 10.1007/s004380050434. [DOI] [PubMed] [Google Scholar]
- 7a.Dereveux J, Haeberli P, Smithies O. a comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geber A, Hitchcock C A, Swartz J E, Pullen F S, Marsden K E, Kwon-Chung L J, Bennett J E. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob Agents Chemother. 1995;39:2708–2717. doi: 10.1128/aac.39.12.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henikoff S, Kelly J D, Cohen E H. Transcription terminates in yeast distal to a control sequence. Cell. 1983;33:607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- 10.Hitchcock C A, Barrett-Bee K J, Russell N J. The lipid composition and permeability to azole of an azole- and polyene-resistant mutant of Candida albicans. J Med Vet Mycol. 1987;25:29–37. doi: 10.1080/02681218780000041. [DOI] [PubMed] [Google Scholar]
- 11.Hitchcock C A, Pye G W, Troke P F, Johnson E M, Warnock D W. Fluconazole resistance in Candida glabrata. Antimicrob Agents Chemother. 1993;37:1962–1965. doi: 10.1128/aac.37.9.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joseph-Horne T, Hollomon D W, Loeffler R S, Kelly S L. Altered P450 activity associated with direct selection for fungal azole resistance. FEBS Lett. 1995;374:174–178. doi: 10.1016/0014-5793(95)01102-k. [DOI] [PubMed] [Google Scholar]
- 13.Katzmann D J, Hallstrom T C, Mahe Y, Moye-Rowley W S. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J Biol Chem. 1996;271:23049–23054. doi: 10.1074/jbc.271.38.23049. [DOI] [PubMed] [Google Scholar]
- 14.Kelly S L, Lamb D C, Kelly D E, Maning N J, Loeffler J, Hebart H, Schumacher U, Einsele H. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol Δ5,6-desaturation. FEBS Lett. 1997;400:80–82. doi: 10.1016/s0014-5793(96)01360-9. [DOI] [PubMed] [Google Scholar]
- 15.Lamb D C, Kelly D E, Schunck W H, Shyadehi A Z, Akhtar M, Lowe D J, Baldwin B C, Kelly S L. The mutation T315A in Candida albicans sterol 14alpha-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann P F, Lin D, Lasker B A. Genotypic identification and characterization of species and strains within the genus Candida by using random amplified polymorphic DNA. J Clin Microbiol. 1992;30:3249–3254. doi: 10.1128/jcm.30.12.3249-3254.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marichal P, Vanden Bossche H, Odds F C, Nobels G, Warnock D W, Timmeran V, van Broeckhoven C, Fay S, Mose-Larsen P. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1997;41:2229–2237. doi: 10.1128/aac.41.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mercure S, Rougeau N, Montplaisir S, Lemay G. The nucleotide sequence of the 25S rRNA-encoding gene from Candida albicans. Nucleic Acids Res. 1993;21:1490. doi: 10.1093/nar/21.6.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyahara K, Hirata D, Miyakawa T. Functional analysis of the promoter of the Saccharomyces cerevisiae multidrug resistance gene YDR1, which encodes a member of the ATP Binding Cassette (ABC) superfamily. Biosci Biotechnol Biochem. 1995;59:147–149. doi: 10.1271/bbb.59.147. [DOI] [PubMed] [Google Scholar]
- 20.Nagao K, Taguchi Y, Arioka M, Kadokura H, Takatsuki A, Yoda J, Yamasaki M. bfr1+, a novel gene of Schizosaccharomyces pombe which confers brefeldin A resistance, is structurally related to the ATP-binding cassette superfamily. J Bacteriol. 1995;177:1536–1543. doi: 10.1128/jb.177.6.1536-1543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odds F C, Bernaerts R. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol. 1994;32:1923–1929. doi: 10.1128/jcm.32.8.1923-1929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkinson T, Falconer D J, Hitchcock C A. Fluconazole resistance due to energy-dependent drug efflux in Candida glabrata. Antimicrob Agents Chemother. 1995;39:1696–1699. doi: 10.1128/aac.39.8.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prasad R, Wergifosse P D, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr Genet. 1995;27:320–329. doi: 10.1007/BF00352101. [DOI] [PubMed] [Google Scholar]
- 24.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryley J F, Wilson R G, Barrett-Bee K J. Azole resistance in Candida albicans. Sabouraudia. 1984;22:53–63. [PubMed] [Google Scholar]
- 26.Sangeorzan J A, Bradley S F, Xiaogang H, Zarins L T, Ridenour G L, Tiballi R N, Kauffman C A. Epidemiology of oral candidiasis in HIV-infected patients: colonization, infection, treatment and emergence of fluconazole resistance. Am J Med. 1994;97:339–346. doi: 10.1016/0002-9343(94)90300-x. [DOI] [PubMed] [Google Scholar]
- 27.Sanglard D, Kuchler K, Ischer F, Pagani J-L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 30a.Schwartz R M, Dayhoff M O. Atlas of protein sequence and structure. Washington, D.C: National Biomedical Foundation; 1979. pp. 353–358. [Google Scholar]
- 31.Shallom J M, Golin J. The unusual inheritance of multidrug-resistance factors in Saccharomyces. Curr Genet. 1996;30:212–217. doi: 10.1007/s002940050123. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan D J, Westerneng T J, Haynes K A, Bennett D E, Coleman D C. Candida dublinensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 33.Tumbarello M, Caldarola G, Tacconelli E, Morace G, Posteraro B, Cauda R, Ortona L. Analysis of the risk factors associated with the emergence of azole resistant oral candidosis in the course of HIV infection. J Antimicrob Chemother. 1996;38:691–699. doi: 10.1093/jac/38.4.691. [DOI] [PubMed] [Google Scholar]
- 34.Vanden Bossche H, Marichal P, Gorrens J, Bellens D, Moereels H, Janssen P A. Mutation in cytochrome P450-dependent 14α-demethylase results in decreased affinity for azole antifungals. Biochem Soc Trans. 1990;18:56–59. doi: 10.1042/bst0180056. [DOI] [PubMed] [Google Scholar]
- 35.Vanden Bossche H, Marichal P, Odds F C, Le Jeune L, Coene M-C. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1992;36:2602–2610. doi: 10.1128/aac.36.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warnock D W, Burke J, Cope N J, Johnson E M, von Fraunhofer N A, Williams E W. Fluconazole resistance in Candida glabrata. Lancet. 1988;ii:1310. doi: 10.1016/s0140-6736(88)92919-4. [DOI] [PubMed] [Google Scholar]
- 38.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White T C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;41:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaret K S, Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982;28:563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]