Abstract
A genetic basis for tetracycline resistance in cutaneous propionibacteria was suggested by comparing the nucleotide sequences of the 16S rRNA genes from 16 susceptible and 21 resistant clinical isolates and 6 laboratory-selected tetracycline-resistant mutants of a susceptible strain. Fifteen clinical isolates resistant to tetracycline were found to have cytosine instead of guanine at a position cognate with Escherichia coli 16S rRNA base 1058 in a region important for peptide chain termination and translational accuracy known as helix 34. Cytosine at base 1058 was not detected in the laboratory mutants or the tetracycline-susceptible strains. The apparent mutation was recreated by site-directed mutagenesis in the cloned E. coli ribosomal operon, rrnB, encoded by pKK3535. E. coli strains carrying the mutant plasmid were more resistant to tetracycline than those carrying the wild-type plasmid both in MIC determinations and when grown in tetracycline-containing liquid medium. These data are consistent with a role for the single 16S rRNA base mutation in clinical tetracycline resistance in cutaneous propionibacteria.
Tetracyclines are broad-spectrum protein synthesis inhibitors which have been widely employed in the treatment of bacterial infections for several decades (21, 23, 24). Evidence suggests that the bacteriostatic effects of tetracyclines are caused by binding to 30S ribosomal subunits, where they interfere with the positioning of aminoacyl-tRNA molecules within the A site, although the exact inhibitory mechanism has not been satisfactorily explained (21, 23). A strong binding site on the 30S ribosomal subunit has been shown to involve 16S rRNA and several ribosomal proteins, especially S7 (18).
Extensive clinical use of tetracyclines for several decades has resulted in widespread resistance in major bacterial pathogens. This has severely restricted the therapeutic usefulness of the tetracyclines in recent years. Several different determinants which confer resistance by a range of mechanisms have been described. The most common of these is energy-dependent drug efflux. A number of genes encode membrane-bound proteins containing 12 or 14 hydrophobic membrane-spanning regions which probably act as multimers and reduce the intercellular concentration of the drug by exchanging protons for tetracycline-cation complexes (21, 23, 24). A second mechanism of resistance involves protection of the ribosome. Genes from gram-positive and gram-negative bacteria encode proteins with N-terminal regions that are homologous to the N-terminal regions of protein synthesis elongation factors Tu and G. These resistance proteins are able to act in vitro on susceptible ribosomes and reduce the affinity of ribosomes for tetracycline when GTP is present (4). The third type of tetracycline resistance is enzymatic inactivation by the product of the tetX locus, which has so far been detected only on two related Bacteroides transposons (24). Clinical resistance to tetracyclines by mutation occurs in several gram-negative bacterial genera by overexpression of existing chromosomal genes (mexA-mexB-oprM [14] and mar [1, 19]). The product of these loci then functions as a multidrug efflux pump of broad substrate specificity or, in the case of the mar locus, as a regulator to control expression of at least one membrane efflux system, acrAB (19). No case of clinical tetracycline resistance by mutation in a gram-positive bacterium has so far been described.
Tetracyclines (including oxytetracycline, doxycycline, and minocycline) are the most widely used antibiotics in the treatment of acne vulgaris, due to their antibacterial and immunomodulatory effects. Long-term and often sequential use of the tetracyclines in individual acne patients has exerted considerable selective pressure for the development of resistant strains of cutaneous propionibacteria, including Propionibacterium acnes, the organism implicated in the pathogenesis of the disease (6). Tetracycline-resistant P. acnes strains were first reported in 1980, and such strains have been isolated from patients in the United States, the United Kingdom, Germany, and Japan (6, 7, 12, 13). Continuous monitoring of skin carriage of resistant propionibacteria by acne patients attending the Leeds General Infirmary during the period 1991 to 1996 has shown a steady yearly increase so that by 1996, 25.3% of 588 patients carried tetracycline-resistant strains (11). Recently we demonstrated that erythromycin resistance in cutaneous propionibacteria is mediated by point mutations within the rRNA binding site rather than by the acquisition of mobile resistance determinants (22), and conventional gel electrophoresis has failed to reveal any plasmids. This led to us to question whether tetracycline resistance in cutaneous propionibacteria may be mediated in a similar manner.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture media.
Randomly selected isolates of tetracycline-susceptible and -resistant propionibacteria collected from the skin surface of acne patients between 1986 and 1996 and stored under liquid nitrogen were studied. There were 12 susceptible and 17 resistant strains of P. acnes, 1 susceptible and 3 resistant strains of Propionibacterium granulosum, and 1 resistant strain of Propionibacterium avidum. Three fully susceptible type strains (P. avidum NCTC11864, P. granulosum NCTC11865, and P. acnes NCTC737) were also included. Propionibacteria were maintained on TYEG agar (2% tryptone, 1% yeast extract, 0.5% glucose) under anaerobic conditions at 34°C and identified as previously described (6). The Escherichia coli strain used in this work was JM109 (endA1 recA1 gyrA96 thi hsdR17(rK−, mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15]). E. coli cells were maintained on LB agar (tryptone, 10 g/liter; yeast extract, 5 g/liter; NaCl, 5 g/liter). Plasmid pKK3535 (2), a pBR322 derivative carrying the entire rrnB operon, was used for site-directed mutagenesis experiments. The plasmid was maintained with 100 μg of ampicillin per ml selection.
Extraction of DNA.
Actively growing propionibacteria (40-h cultures, 10-ml volumes) were exposed to 20 μg of penicillin G per ml anaerobically for 4 h to weaken the cell walls. Cells were harvested by centrifugation (4,000 × g, 5 min), resuspended to 1010 CFU/ml in TE buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA) and lysed by the addition of 100 μl of 20% sodium dodecyl sulfate (Sigma Chemical Co.). Following overnight incubation with proteinase K (100 μg/ml) at 55°C, genomic DNA was purified by phenol-chloroform extraction and ethanol precipitation. Genomic DNA was resuspended in TE buffer. Plasmid DNA was extracted from E. coli cells with Promega Wizard Plus Minipreps.
PCR and sequencing of the gene encoding 16 S rRNA.
Amplification of a 1.5-kb section of DNA encoding the 16S rRNA of propionibacteria was accomplished with primer DG74 (5′-AGGAGGTGATCCAACCGCA-3′), as described by Greisen et al. (9), and a primer (5′-GATTGGAGAGTTTGATCCTG-3′) designed from the GenBank sequences of propionibacterial 16S rRNA (accession no. M61903).
PCR amplicons were purified with the Wizard PCR purification system (Promega). Amplicons were sequenced across the entire 1.5-kb fragment length with five primers, to ensure overlap, and an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit. Sequences were determined at the Automated DNA Sequencing Facility, School of Biological Sciences, University of Leeds. Resistant isolates were sequenced in the reverse direction to confirm the presence or absence of the mutation. Five susceptible isolates were also sequenced in the reverse direction for comparison.
Site-directed mutagenesis.
A PCR method using overlap extension, as described by Ho et al. (10), was used to generate a G→C mutation at base 1058 of the 16S rRNA gene of the rrnB operon contained on pKK3535. Complementary primers encompassing the sequence to be mutated (5′-TGCTGCATGCCTGTCGTCAG-3′ and 5′-CTGACGACAGGCATGCAGCA-3′) and incorporating the required mutation (underlined) were designed. Each primer was used in conjunction with a flanking primer in two separate PCRs. This generated two PCR products, of 592 and 713 bp. These products were purified with the Wizard PCR purification system, mixed, and used as a template in a second PCR using only the flanking primers. This generated a single product of 1,286 bp. This product was digested at the unique restriction sites ApaI and XbaI to generate a 797-bp restriction fragment which was ligated to pKK3535, from which the ApaI-Xba fragment had been removed. Ampicillin-resistant transformants were sequenced across the entire PCR-generated region.
MIC determinations.
MICs of all antibiotics were determined by agar dilution, as described previously (6), with TYEG for propionibacteria and LB for E. coli. MICs were recorded after 3 days of anaerobic incubation for propionibacteria and overnight aerobic incubation for E. coli.
Growth kinetics of E. coli containing wild-type and mutated pKK3535 in tetracycline-containing medium.
For growth of E. coli containing pKK3535 in liquid medium, a 1% (vol/vol) inoculum from an overnight culture grown with ampicillin selection was added to a 50-ml volume of LB broth in a 250-ml flask with or without tetracycline at 2 μg/ml and grown at 37°C with shaking at 160 rpm. Growth of replicate cultures was assessed spectrophotometrically at 580 nm.
RESULTS
Propionibacterial 16S rRNA gene sequences.
To determine the base sequence of the 16S rRNA gene, total cellular DNA was extracted from 21 tetracycline-resistant propionibacteria, 16 susceptible strains, and 6 tetracycline-resistant laboratory mutants. To generate the resistant mutants, three tetracycline-susceptible P. acnes strains were grown in serial batch cultures with increasing concentrations of tetracycline or minocycline starting at 0.25 times the MIC, as determined by agar dilution. The 16S rRNA genes were amplified by PCR and the products were sequenced. The resulting sequences were compared to existing GenBank propionibacterial 16S rRNA sequences (accession no. M61903 and X53218) and to each other. A single apparent base change, G→C at E. coli-equivalent base 1058, was detected in 15 of the 21 resistant isolates but was not detected in any of the 16 susceptible strains. All six resistant isolates which did not have a cytosine at base 1058 were isolated before 1988. Since this time, 10 of 10 resistant isolates have had the cytosine present at base 1058. The altered base lies within helix 34 of the 16S rRNA (Fig. 1). No other base changes were detected in 16S rRNA genes. The sequence of helix 34 between E. coli and cutaneous propionibacteria is conserved except for bases C1051 and G1207, which are G and C, respectively, in the propionibacterial sequence (Fig. 1). The six laboratory-generated resistant mutants contained no base changes compared to their susceptible parent strains. There was no evidence of heterozygosity in the 16S rRNA gene sequence despite the presence of three copies of the rrn operon in P. acnes (22).
FIG. 1.
Secondary structure of helix 34 of 16S rRNA of E. coli. The location of the G→C mutation at position 1058 associated with tetracycline resistance is indicated, as is the position of the cross-link of U1052 to the 3′ base of the A-site codon (5). Tet indicates bases whose reactivities to chemical attack are enhanced by tetracycline (16). The CG base pair, C1051 and G1207, which is not conserved between E. coli and cutaneous propionibacteria is indicated.
Cross-resistance patterns and MICs of tetracyclines for sensitive and resistant isolates.
Resistant isolates with either a cytosine or a guanine at base 1058 demonstrated cross-resistance patterns similar to those of tetracyclines of differing lipophilicities (Table 1). They were most resistant to the relatively hydrophilic tetracycline and least resistant to the more lipophilic minocycline. The mean increase in MIC compared with fully susceptible strains was 64-fold for tetracycline and doxycycline and 16-fold for minocycline. The laboratory-generated resistant mutants were indistinguishable, on the basis of MICs, from resistant clinical isolates.
TABLE 1.
MICs of tetracyclines for bacterial strains used in this study
| Bacterial strain(s) | MIC (μg/ml)a
|
||
|---|---|---|---|
| TET | DOX | MIN | |
| Susceptible propionibacteria (n = 16) | 0.12–1 | 0.06–0.5 | 0.06–0.5 |
| Resistant propionibacteria (n = 21) | 2–64 | 1–32 | 0.25–4 |
| Lab-generated resistant propionibacteria (n = 6) | 2–8 | 1–4 | 1–2 |
| E. coli(pKK3535) | 2.0 | 2.0 | 1.5 |
| E. coli(pKK1058C-1) | 4.5 | 4.0 | 3.0 |
| E. coli(pKK1058C-2) | 4.5 | 4.0 | 3.0 |
| E. coli(pKK1058C-3) | 4.5 | 4.0 | 3.0 |
TET, tetracycline; DOX, doxycycline; MIN, minocycline. MICs for propionibacteria are ranges for all strains tested.
Site-directed mutagenesis of a plasmid-encoded E. coli 16S rRNA gene.
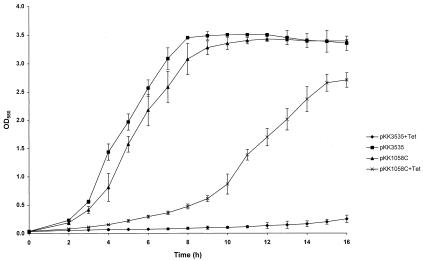
Cutaneous propionibacteria are not currently genetically manipulable organisms. In order to establish an association between the observed base change and tetracycline resistance, the mutation was recreated in the cloned ribosomal rrnB operon present on the E. coli plasmid pKK3535. A PCR method of overlap extension was used to generate a derivative of pKK3535 (pKK1058C) containing the required G→C transversion at base 1058. Three transformants containing plasmids designated pKK1058C-1, pKK1058C-2, and pKK1058C-3 were sequenced across the entire PCR-generated region to confirm that the mutation was present and to ensure that no additional mutations had been generated. All three transformants were analyzed individually to confirm that background mutations did not contribute to the phenotype. E. coli JM109 containing pKK1058C-1, -2, or -3 was twofold more resistant to tetracycline than the same strain carrying pKK3535 (Table 1) and demonstrated a similar increase in resistance to doxycycline and minocycline. To ensure that this resistance was associated with the presence of the plasmids, pKK1058C-1, -2, and -3 and pKK3535 were retransformed into E. coli JM109 and MIC determinations were repeated. The increase in tetracycline resistance was maintained for transformants carrying mutant, but not wild-type plasmids. The resistance phenotype was demonstrated by growth of pKK1058C-containing E. coli in broth culture with tetracycline at 2 μg/ml. E. coli containing pKK3535 failed to grow in this medium (Fig. 2). In the absence of selection, E. coli containing the mutant plasmid demonstrated similar growth rates and reached a similar final biomass compared to E. coli containing the wild-type plasmid but demonstrated a prolonged lag phase (Fig. 2). E. coli containing pKK1058C demonstrated no increase in MIC of rifampin or norfloxacin compared to E. coli containing pKK3535, thus ruling out activation of the mar locus.
FIG. 2.
Growth of E. coli in liquid medium with and without 2 μg of tetracycline per ml. Values are means ± 95% confidence limits. All readings were means of triplicate samples except for those of pKK1058C-containing E. coli, which were means of nine samples (three each of pKK1058C-1, -2, and -3). OD580, optical density at 580 nm.
DISCUSSION
The increased tetracycline resistance of E. coli carrying PKK1058C demonstrates a direct effect of the single base mutation on the susceptibility of cells to tetracycline. The relative increase in tetracycline resistance was greater in the propionibacterial strains than in E. coli containing pKK1058C. There are a number of possible reasons for this. Firstly, E. coli contains seven chromosomal wild-type ribosomal operons. The gene dosage from plasmids similar to PKK1058C produces approximately half of the cells’ total rRNA (8). This is likely to reduce the observable MIC below the levels which would be achievable if all ribosomes were mutant. Secondly, gram-positive and gram-negative ribosomes may differ in their responses to the introduced mutation. It is possible that the 16S rRNA mutation is only part of the story in propionibacteria and that mutations in other chromosomal loci may contribute to the resistance phenotype in an additive manner. Clearly, there must be an explanation for the similar resistance phenotypes observed in six of the resistant clinical isolates and the laboratory mutants. Mutations in ribosomal proteins are a strong possibility, especially protein S7, which has been shown to bind tetracycline by photoaffinity labelling (18).
The location of the base change at position 1058 in helix 34 may be functionally significant. Helix 34 is a conserved region which appears to be involved in peptide chain termination and translational accuracy (3, 17, 20). Base U1052 has been photo-cross-linked to the 3′ base of the A-site codon associating helix 34 with the decoding site (5). Tetracycline is thought to inhibit binding of tRNAs to the A site (21, 23). A mutation in helix 34 may weaken the binding of tetracycline to the ribosome or allow access of tRNAs in the presence of tetracycline.
Techniques such as chemical footprinting (16) and photoincorporation (18) have identified bases G693, A892, G1300, and G1338 outside helix 34 as potential sites for interaction of rRNA with tetracycline. We found no evidence of any mutations in these bases leading to tetracycline resistance, but it remains possible that such mutations do not give rise to resistance although the bases may be involved in binding the drug. Alternatively, changes at these positions may be lethal.
Point mutations within rRNA are becoming increasingly recognized as the means by which macrolide and lincosamide resistance is acquired in several clinically important genera. The mechanism seems to be restricted to organisms with low numbers of rRNA operons, and mutant generation and overgrowth usually follows prolonged exposure to the antibiotics in individual patients. Mycobacterium spp. (15, 26), Helicobacter pylori (25), and cutaneous propionibacteria (22) have all been shown to carry rRNA mutations associated with clinical resistance and to possess few copies of the rRNA operon with no evidence of heterogeneity. Thus, the effects of point mutations are not diluted or masked by the presence of multiple copies of ribosomes containing wild-type rRNA. We have previously proposed that, under selection, gene conversion may occur to copy the mutant phenotype to all rRNA operons before the phenotype becomes detectable, thus preventing detection of heterozygous strains.
At present, the clinical significance of tetracycline resistance in cutaneous propionibacteria is unclear. Although MICs for the majority of resistant isolates exceed serum tetracycline concentrations on the standard dose of 1 g per day, many resistant isolates would be inhibited by serum minocycline concentrations achievable on either a 100- or 200-mg daily dose. However, it is the drug concentration within the lumen of pilosebaceous follicles which is important, as this is where propionibacteria reside and where the changes resulting in lesion formation occur. Unfortunately, because of the technical difficulties involved such data are not available at present. Tetracycline-recalcitrant acne is relatively common but may be due to a variety of causes, including secondary infection and poor compliance, as well as overgrowth of resistant propionibacteria.
It will be interesting to determine whether similar rRNA mutations are present in other bacterial pathogens resistant to tetracyclines.
ACKNOWLEDGMENTS
We thank Stephen Douthwaite and Harry F. Noller for kind provision of strains and plasmids used in this work.
We thank Dermik Laboratories Inc. and the Leeds Foundation For Dermatological Research for financial assistance.
REFERENCES
- 1.Alekshun M N, Levy S B. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–2075. doi: 10.1128/aac.41.10.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 3.Brown C M, McCaughan K K, Tate W P. Two regions of the Escherichia coli 16S ribosomal RNA are important for decoding stop signals in polypeptide chain termination. Nucleic Acids Res. 1993;21:2109–2115. doi: 10.1093/nar/21.9.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burdett V. Tet(M)-promoted release of tetracycline from ribosomes is GTP dependent. J Bacteriol. 1996;178:3246–3251. doi: 10.1128/jb.178.11.3246-3251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dontsova O, Dokudovskaya S, Kopylov A, Bogdanov A, Rinke-Appel J, Jünke N, Brimacombe R. Three widely separated positions in the 16S RNA lie in or close to the ribosomal decoding region: a site-directed cross-linking study with mRNA analogues. EMBO J. 1992;11:3105–3116. doi: 10.1002/j.1460-2075.1992.tb05383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eady E A, Jones C E, Gardner K J, Taylor J P, Cove J H, Cunliffe W J. Tetracycline-resistant propionibacteria from acne patients are cross resistant to doxycycline, but sensitive to minocycline. Br J Dermatol. 1993;128:556–560. doi: 10.1111/j.1365-2133.1993.tb00235.x. [DOI] [PubMed] [Google Scholar]
- 7.Forssman T. Antibiotic resistance in acne patients under antibiotic treatment in comparison to an untreated control group with retrospective assessment of therapy. In: Surber C, Elsner P, Bircher A J, editors. Exogenous dermatology. Current problems in dermatology. Basel, Switzerland: Karger; 1995. pp. 91–97. [DOI] [PubMed] [Google Scholar]
- 8.Gourse R L, Stark J R, Dahlberg A E. Site-directed mutagenesis of ribosomal RNA. J Mol Biol. 1982;159:397–416. doi: 10.1016/0022-2836(82)90291-1. [DOI] [PubMed] [Google Scholar]
- 9.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 11.Jones C E, Vyakrnam S, Eady E A, Cove J H, Cunliffe W J. Antibiotic resistant propionibacteria and acne: crisis or conundrum? J Investig Dermatol. 1996;108:381. [Google Scholar]
- 12.Kurokawa I, Nishijima S, Asada Y. The antibiotic sensitivity of Propionibacterium acnes: a 15-year bacteriological study and retrospective evaluation. J Dermatol. 1988;15:149–154. doi: 10.1111/j.1346-8138.1988.tb03667.x. [DOI] [PubMed] [Google Scholar]
- 13.Leyden J J, McGinley K J, Cavalieri S, Webster G F, Mills O H, Kligman A M. Propionibacterium acnes resistance to antibiotics in acne patients. J Am Acad Dermatol. 1983;8:41–45. doi: 10.1016/s0190-9622(83)70005-8. [DOI] [PubMed] [Google Scholar]
- 14.Li X-Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meier A, Kirschner P, Springer B, Steingrube V A, Brown B A, Wallace R J, Jr, Böttger E C. Identification of mutations in 23S rRNA of clarithromycin-resistant Mycobacterium intracellulare. Antimicrob Agents Chemother. 1994;38:381–384. doi: 10.1128/aac.38.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moazed D, Noller H F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 17.Moine H, Dahlberg A E. Mutations in helix 34 of Escherichia coli 16S ribosomal RNA have multiple effects on ribosome function and synthesis. J Mol Biol. 1994;243:402–412. doi: 10.1006/jmbi.1994.1668. [DOI] [PubMed] [Google Scholar]
- 18.Oehler R, Polacek N, Steiner G, Barta A. Interaction of tetracycline with RNA: photoincorporation into ribosomal RNA of Escherichia coli. Nucleic Acids Res. 1997;25:1219–1224. doi: 10.1093/nar/25.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pagel F T, Zhao S Q, Hijazi K A, Murgola E J. Phenotypic heterogeneity of mutational changes at conserved nucleotides in 16S ribosomal RNA. J Mol Biol. 1997;267:1113–1123. doi: 10.1006/jmbi.1997.0943. [DOI] [PubMed] [Google Scholar]
- 21.Roberts M C. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 22.Ross J I, Eady E A, Cove J H, Jones C E, Ratyal A, Miller Y W, Vyakrnam S, Cunliffe W J. Clinical resistance to erythromycin and clindamycin in cutaneous propionibacteria isolated from acne patients is associated with mutations in 23S rRNA. Antimicrob Agents Chemother. 1997;41:1162–1165. doi: 10.1128/aac.41.5.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schnappinger D, Hillen W. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch Microbiol. 1996;165:359–369. doi: 10.1007/s002030050339. [DOI] [PubMed] [Google Scholar]
- 24.Speer B S, Shoemaker N B, Salyers A A. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin Microbiol Rev. 1992;5:387–399. doi: 10.1128/cmr.5.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallace R J, Jr, Meier A, Brown B A, Zhang Y, Sander P, Onyi G O, Böttger E C. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob Agents Chemother. 1996;40:1676–1681. doi: 10.1128/aac.40.7.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]