Abstract
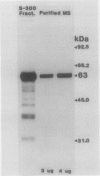
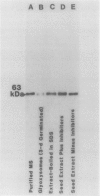
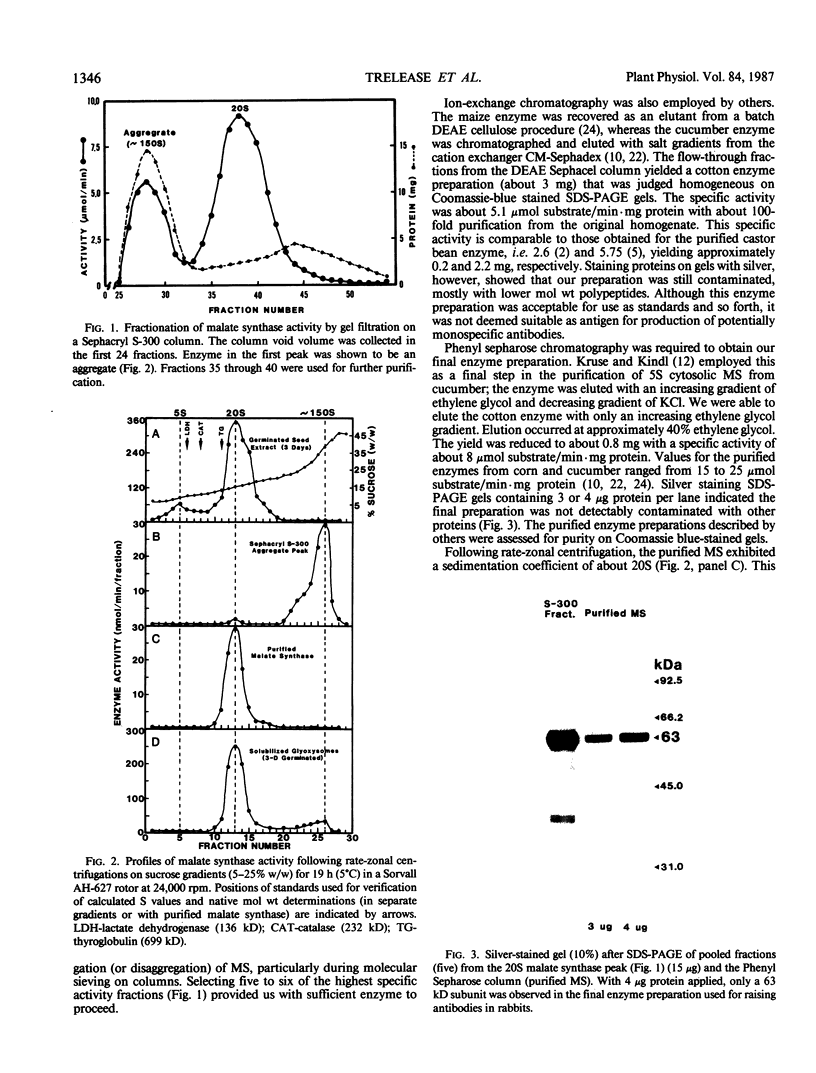
Malate synthase (EC 4.1.3.2), an enzyme unique to the glyoxylate cycle, was purified to homogeneity from cotyledons of 72-hours, darkgrown cotton (Gossypium hirsutum L.) seedlings. Homogeneity of the enzyme was assessed by silver staining SDS-PAGE gels. Purification was accomplished by using a single buffer medium through six steps involving one ammonium sulfate fractionation and chromatography on three columns (Sephacryl S-300, DEAE Sephacel, Phenyl Sepharose). Large-scale preparation of glyoxysomes, a main step in all other published procedures, was not involved. The purified enzyme and that extracted from glyoxysomes appears to be a dodecamer with a native molecular weight of 750,000 (sedimentation coefficient of >20 Svedberg units [S] on sucrose gradients) composed of identical subunits (molecular weight approximately 63,000). The monomer (5S) occurs in the cytosol. Polyclonal antibodies raised in rabbits were judged to be monospecific for malate synthase by immunotitration, double immunodiffusion, and western blotting. Double immunodiffusion experiments revealed only partial immunological identity between the 5S (cytosolic) and 20S (glyoxysomal forms, although complete identity was observed between the 5S form in immature and germinated seeds, and the 20S form in immature and germinated seeds. Cross-reactivity of the cotton antimalate synthase serum was observed with extracts from five other oilseeds. Western blot analyses showed that malate synthase protein was not present in immature seeds prior to appearance of enzyme activity, but when present, subunit molecular weight was indistinguishable in immature, desiccated, and germinated seeds.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borst P. How proteins get into microbodies (peroxisomes, glyoxysomes, glycosomes). Biochim Biophys Acta. 1986 May 5;866(4):179–203. doi: 10.1016/0167-4781(86)90044-8. [DOI] [PubMed] [Google Scholar]
- Bowden L., Lord J. M. Purification and comparative properties of microsomal and glyoxysomal malate synthase from castor bean endosperm. Plant Physiol. 1978 Feb;61(2):259–265. doi: 10.1104/pp.61.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choinski J. S., Trelease R. N. Control of Enzyme Activities in Cotton Cotyledons during Maturation and Germination: II. Glyoxysomal Enzyme Development in Embryos. Plant Physiol. 1978 Jul;62(1):141–145. doi: 10.1104/pp.62.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E. Aggregated Forms of Malate and Citrate Synthase are Localized in Endoplasmic Reticulum of Endosperm of Germinating Castor Bean. Plant Physiol. 1982 Jan;69(1):83–87. doi: 10.1104/pp.69.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse C., Kindl H. Malate synthase: aggregation, deaggregation, and binding of phospholipids. Arch Biochem Biophys. 1983 Jun;223(2):618–628. doi: 10.1016/0003-9861(83)90626-4. [DOI] [PubMed] [Google Scholar]
- Kunce C. M., Trelease R. N. Heterogeneity of catalase in maturing and germinated cotton seeds. Plant Physiol. 1986 Aug;81(4):1134–1139. doi: 10.1104/pp.81.4.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köller W., Kindl H. Glyoxylate cycle enzymes of the glyoxysomal membrane from cucumber cotyledons. Arch Biochem Biophys. 1977 May;181(1):236–248. doi: 10.1016/0003-9861(77)90502-1. [DOI] [PubMed] [Google Scholar]
- Köller W., Kindl H. Studies supporting the concept of glyoxyperoxisomes as intermediary organelles in transformation of glyoxysomes into peroxisomes. Z Naturforsch C. 1978 Nov-Dec;33(11-12):962–968. [PubMed] [Google Scholar]
- Lazarow P. B., Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Longo G. P., Bernasconi E., Longo C. P. Solubilization of enzymes from glyoxysomes of maize scutellum. Plant Physiol. 1975 Jun;55(6):1115–1119. doi: 10.1104/pp.55.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen C. R. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal Biochem. 1967 Jul;20(1):114–149. doi: 10.1016/0003-2697(67)90271-0. [DOI] [PubMed] [Google Scholar]
- Miernyk J. A., Trelease R. N., Choinski J. S. Malate synthase activity in cotton and other ungerminated oilseeds: a survey. Plant Physiol. 1979 Jun;63(6):1068–1071. doi: 10.1104/pp.63.6.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk J. A., Trelease R. N. Role of malate synthase in citric Acid synthesis by maturing cotton embryos: a proposal. Plant Physiol. 1981 May;67(5):875–881. doi: 10.1104/pp.67.5.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H., Weir E. M., Leaver C. J., Titus D. E., Becker W. M. Regulation of Glyoxysomal Enzymes during Germination of Cucumber: 3. IN VITRO TRANSLATION AND CHARACTERIZATION OF FOUR GLYOXYSOMAL ENZYMES. Plant Physiol. 1980 Jan;65(1):40–46. doi: 10.1104/pp.65.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt G. W., Mishkind M. L. The transport of proteins into chloroplasts. Annu Rev Biochem. 1986;55:879–912. doi: 10.1146/annurev.bi.55.070186.004311. [DOI] [PubMed] [Google Scholar]
- Turley R. B., Trelease R. N. Cottonseed malate synthase : biogenesis in maturing and germinated seeds. Plant Physiol. 1987 Aug;84(4):1350–1356. doi: 10.1104/pp.84.4.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]