Abstract
The phenotype of a strain of Saccharomyces cerevisiae containing a disruption of the gene encoding NADPH cytochrome P-450 oxidoreductase (CPR) was quantified biochemically and microbiologically, as were those of various transformants of this strain after expression of native CPR, cytochrome P-45051 (CYP51), and a fusion protein of CYP51-CPR (FUS). Only a 4-fold decrease in ergosterol biosynthesis was observed for the cpr strain, but ketoconazole sensitivity increased 200-fold, indicating hypersensitivity to the alternative electron donor system in cpr strains. Both phenotypes could be reversed in transformants expressing the CPR and FUS, indicating the availability of the CPR in FUS as well as the expressed native CPR for monoxygenase-associated reactions. The complementation of function was observed both in vitro and in vivo for the monoxygenases squalene epoxidase, CYP51, and CYP61 in the ergosterol biosynthesis pathway with which CPR is coupled. Overexpression of CYP51 and FUS produced different levels of ketoconazole resistance in wild-type cells, indicating that the availability of CPR may limit the potential of overproduction of CYP51 as a mechanism of resistance to azole antifungal agents.
NADPH cytochrome P-450 oxidoreductase (CPR; EC 1.6.2.4) is required for microsomal eukaryotic cytochrome P-450 (CYP) monooxygenase activity, transferring either both or sometimes the first electron for these reactions (25). The CYP enzymes are involved in the metabolism of foreign compounds such as lipophilic pollutants, pesticides, and drugs as well as in many biosynthetic reactions, for instance, in steroid, alkaloid, and terpenoid biosynthesis. Although in plants there is CPR diversity, in animal and fungal systems only one has been identified, and this one functions with the many members of the microsomal P-450 superfamily in a particular organism (15).
Within the CYP superfamily, the cytochrome P-45051 (CYP51) family is the only family found in animal, plant, and fungal kingdoms and represents an ancient metabolic role for CYP in sterol biosynthesis, undertaking C-14 demethylation via three sequential hydroxylations (1). This enzyme in fungi is the target of azole antifungal agents which are selective in their inhibition, are central to antifungal chemotherapy, and represent about one-third of the agricultural fungicides used. Two other enzymes of fungal ergosterol biosynthesis require CPR, CYP61 (a sterol 22-desaturase) (3, 10, 17), and a non-CYP monooxygenase, squalene epoxidase (29). The former is likely also to be present in plants and algae, in which 22-desaturation is observed, unlike in animals, but squalene epoxidase is present in all organisms producing sterols (27).
As expected for an antifungal target, disruption of the CYP51 gene was observed to be lethal, but the strain could be rescued by providing an ergosterol supplement which could be taken up only anaerobically (7). In contrast, disruption of the yeast CPR gene produced viable mutants and ergosterol was still produced at an unquantified level (unpublished observation; 18), although no further CPR genes could be detected or appear now to be present in the yeast genome. In reconstituted assays with purified enzyme, cytochrome b5 can act as alternative donor system for the second electron required (4) and may have been supporting catalytic activity in the disruptant, although it has been observed to provide the first electron with poor efficiency. Supporting this concept was the observation that the gene encoding cytochrome b5 can act as a suppressor of the cpr mutant gene disruption phenotype noted by Sutter and Loper (18), namely, hypersensitivity to the CYP51 inhibitor ketoconazole (20). This observation suggested that a large reduction in ergosterol biosynthesis might have occurred.
Here we present the biochemical characterization of a yeast strain containing a disrupted CPR gene and address the role of the enzyme in determining azole sensitivity and resistance. The latter is a significant practical problem, e.g., for resistant strains causing candidiasis in >10% of patients with late-stage AIDS treated with fluconazole (2).
MATERIALS AND METHODS
Strains.
Escherichia coli DH5α (purchased from Gibco-BRL) was used for plasmid maintenance and manipulation, and Saccharomyces cerevisiae JL20 (MATa leu2-3 leu2-112 his4-519 ade1-100 ura3-52; a gift from J. L. Loper, University of Cincinnati) was used for expression and gene disruption. The nomenclature for yeasts is used to describe the genes involved (italics; e.g., cpr for mutant recessive gene), but the proteins are described in capital letters without italics (e.g., CPR).
Chemicals.
All chemicals were purchased from Sigma Chemicals (Poole, United Kingdom) unless otherwise specified. [32-3H]-3β-hydroxylanost-7-en-32-ol (13.3 mCi/mmol) was provided by M. Akhtar, Department of Biochemistry, University of Southampton (Southampton, United Kingdom). Restriction endonucleases were purchased from NBL (Northumbria, United Kingdom). Ketoconazole (molecular weight, 531.438) and organic solvents were obtained from Janssen Pharmaceuticals (Beerse, Belgium) and Fisons Chemicals (Loughborough, United Kingdom), respectively.
Construction of expression vectors.
The expression vectors containing CPR, CYP51, and CYP51 fused with CPR(Δ33) (FUS) were constructed in YEp51, a galactose-inducible yeast expression plasmid. The CPR and CYP51 genes were isolated by PCR with pFBY4 (28) and pVK1 (8) containing as templates CPR and CYP51 with their promoters, respectively. The 5′ sense oligonucleotide primer (5′-CCCGTCGACATCATGCCGTTTGGAATAGACAAC-3′ for CPR and 5′-CCCGTCGACAATATGTCTGCTACCAAGTCAATC-3′ for CYP51 were used and contained a SalI site (underlined sequence) at their 5′ ends. The 3′ antisense oligonucleotide primers 5′-CCCAAGCTTTTACCAGACATCTTCTTGGTA-3′ for CPR and 5′-CCCAAGCTTTTAGATCTTTTGTTCTGGATT-3′ for CYP51 encoded a HindIII site (underlined sequence) at their 3′ ends. The reaction conditions were as follows: 94°C (denaturation) for 1 min, 45°C (annealing) for 1 min, and 72°C (extension) for 5 min with a 2-min ramp time for the first 5 cycles and 94°C for 1 min and 72°C (extension) for 5 min with a 2-min ramp time for the first 5 cycles and 94°C for 1 min and 72°C for 5 min for the remaining cycles in a 30-cycle reaction. PCR was carried out with Pfu polymerase (Stratagene) and a Perkin-Elmer DNA thermal cycler. The target fragments were gel purified, digested with SalI and HindIII, and cloned into YEp51. The transformants were screened by restriction digestion and were confirmed by sequencing. All DNA manipulations and transformations were done by standard protocols (14).
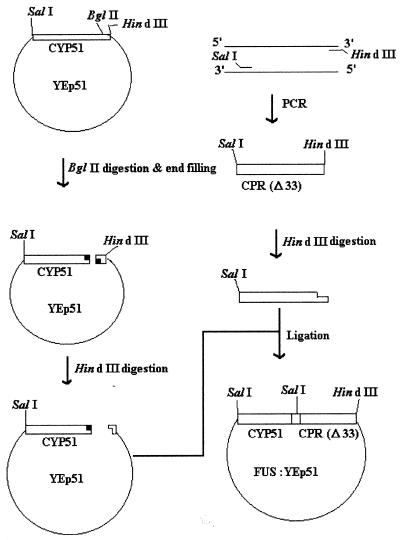
The scheme used for constructing FUS:YEp51 is depicted in Fig. 1. CYP51:YEp51 linearized by digestion with BglII was incubated at 72°C for 30 min with Pfu to fill in the 3′ overhanging ends. The blunt-ended linearized vector was digested with HindIII and was ligated to the HindIII-digested CPR(Δ33) gene, the CPR gene lacking a coding sequence for the N-terminal 33-amino-acid (membrane binding domain) coding sequence, isolated by PCR. BglII digestion, filling in, HindIII digestion, and ligation led to the removal of the termination codon from the CYPcyp51 gene and insertion of the CPR(Δ33 gene in frame at the 3′ end of CYP51 and a coding sequence for four amino acids (Pro, Val, Asp and Ile) as a linker between CYP51 and CPR(Δ33).
FIG. 1.
Schematic representation of the strategy used to make the yeast CYP51 fused with CPR(Δ33) construct.
CPR gene disruption in JL20.
In the JL20 strain the chromosomal CPR gene was disrupted by inserting URA3+ to generate strain JL20D (18). The 0.7-kb BamHI internal fragment of the CPR gene in the construct CPR:YEp51 was replaced by the 1.1-kb URA3+-containing HindIII fragment isolated from plasmid pJJ244 (6) by filling in and blunt-ended ligation to obtain the CPR::URA3:YEp51 construct. Chromosomal CPR in the haploid JL20 yeast strain was then inactivated by transplacement with the SalI-HindIII fragment containing CPR::URA3, and the disruption was confirmed by PCR (by using the cells of Ura3+ colonies of the transformed JL20 strain and the primers and conditions mentioned above).
Sterol isolation and analysis.
The cells harvested from 100 ml of culture were resuspended in 3 ml of methanol, 2 ml of 60% (wt/vol) KOH, and 2 ml of 0.5% (wt/vol) pyrogallol dissolved in methanol and were saponified by heating at 90°C for 1 h. Nonsaponifiable lipids (sterols) were extracted from the saponified mixture three times with 5 ml of hexane, pooled, and dried under nitrogen. The sterols were suspended in 100 μl of toluene and heated at 60°C for an hour for silylation after adding 20 μl of bis(trimethylsilyl)trifluoride. The silyl sterols were analyzed by gas chromatography-mass spectrometry (VG 12-250; VG BIOTECH) by using split injections with a split ratio of 20:1. Sterol identification was by reference to the relative retention times and mass spectra reported previously (13, 23).
Ketoconazole susceptibility tests.
The ketoconazole resistance in strain JL20D transformed with various expression vectors was compared to that of strain JL20. The MICs were determined by inoculating cells obtained from the mid-log-phase cultures at 104 cells/ml in 2 ml of YM-gal, his medium (1.34% [wt/vol] Difco yeast nitrogen base without amino acids but with galactose ([2%; wt/vol] and l-histidine [20 μg/ml]) in 60-ml sterilin sterile pots containing various concentrations of ketoconazole, and the pots were incubated for 2 days at 30°C (growth was assessed microscopically by cell counting) (9). The tests were carried out in triplicate.
Immunoblot analysis.
Sodium dodecyl sulfate-polyacrylamide (10%) gel electrophoresis, nitrocellulose filter transfer, and immunodetection of protein were performed as described previously (11, 19). Anti-CYP51 immunoglobulin G was kindly provided by W. H. Schunk of the Max Delbruck Centre for Molecular Medicine (Berlin, Germany).
Heterologous expression in strain JL20.
The JL20 transformants carrying the CPR:YEp51, CYP51:YEp51, and FUS:YEp51 expression vectors were grown in yeast minimal medium containing Difco yeast nitrogen base without amino acids (1.34%; wt/vol), 20 μg of l-histidine per ml, and glucose (2%; wt/vol) at 30°C until the glucose was completely consumed, and then heterologous expression was induced with galactose (3%; wt/vol) for 20 h (16) as described previously by us for CYP51 of Candida albicans (16).
Preparation of cell extracts, cytosol, and microsomes.
Cells harvested from the cultures by centrifugation were resuspended in buffer A (100 mM potassium phosphate containing 20% glycerol, 1 mM reduced glutathione, 0.5 mM EDTA) and were homogenized with glass beads (0.45 to 0.5 mm in diameter) in a Braun disintegrator (Braun GmbH, Mesungen, Germany) operating at 4,000 rpm with 30-s bursts and carbon dioxide cooling. Cell extract was obtained as a supernatant by centrifuging the cell homogenate at 1,500 × g for 10 min. The extract was centrifuged at 10,000 × g for 15 min to remove the mitochondria as a pellet, and the resulting supernatant was centrifuged at 100,000 × g for 90 min to obtain the microsomes as a pellet and the cytosol as the supernatant. The microsomal pellet was resuspended in buffer A by using a Potter-Elvehjem homogenizer. The protein content in the cell extract and microsomes was measured with the bicinchoninic acid protein estimation kit (Sigma) with bovine serum albumin as a standard (16, 23).
In vitro ergosterol biosynthesis and its inhibition by ketoconazole.
The reaction mixture (1 ml) containing 924 μl of cell-free extract, 50 μl of cofactor solution (1 μmol of NADP, 1 μmol of NAD, 1 μmol of NADPH, 3 μmol of glucose-6-phosphate, 5 μmol of ATP, and 3 μmol of reduced glutathione dissolved in distilled water and adjusted to pH 7.0), 15 μl of divalent cation solution (10 μl of 0.5 M MgCl2, 5 μl of 0.4 M MnCl2), 1 μl of ketoconazole at various concentrations, and 10 μl of [2-14C]mevalonate (0.25 μCi of 53 mCi/mmol) was incubated at 37°C and 150 rpm. After 2 h of incubation the reaction was stopped by adding 1 ml of freshly prepared saponification reagent (15% [wt/vol] KOH in 90% [vol/vol] ethanol), and the mixture was saponified by heating at 80°C for 1 h. The nonsaponified sterols were extracted from the mixture twice with 3 ml of petroleum ether (boiling point, 40 to 60°C). The extracts were pooled, dried under nitrogen gas, and redissolved in 100 μl of petroleum ether. The nonsaponifiable sterols were applied to silica gel thin-layer chromatography plates (ART 573; Merck), and the plates were developed with toluene-diethyl ether at a 9:1 ratio (vol/vol). Radioactive sterols were localized by autoradiography and were excised for scintillation counting (24).
Sterol 14α-demethylation assay.
A total of 0.5 ml of buffer A containing NADP+ (2 mg), glucose phosphate (5 mg), and glucose-6-phosphate dehydrogenase (3 units) was incubated at 30°C and 150 rpm to generate NADPH. After 20 min, 1 mg of membrane protein and 12.2 nmol of [32-3H]-3β-hydroxylanost-7-en-32-ol (13.3 mCi/mmol) were added to the NADPH, the total volume was adjusted to 1 ml with buffer A, and incubation was continued at 30°C and 150 rpm. Aliquots of 0.2 ml were removed at intervals of 0, 5, 10, 30, and 60 min after the substrate and enzyme were added to 1 ml of a dichloromethane and water (1:1) mixture and the mixture was vortexed and centrifuged for phase separation. The resulting aqueous phases were washed twice with 0.5 ml of dichloromethane and treated with charcoal for 1 h at 4°C, and then the radioactivity in the aqueous phases was measured by liquid scintillation counting with a Beckman scintillation counter (16). For assays containing cumene hydroperoxide (25 mM), the components of the NADPH-regenerating system and preincubation were omitted.
Spectrophotometric measurements.
A Philips PU8800 UV/VIS scanning spectrophotometer was used for all spectral studies. The cytochrome P-450 content in the microsomes was determined by a reduced CO difference spectrum (12). Type II binding spectra were measured by adding ketoconazole in increments to microsomal cytochrome P-450 (0.4 nmol) in the sample cuvette after adjusting the baseline (23). Cytochrome c reductase activity was measured as described previously (26). The ergosterol content in the nonsaponifiable lipids was calculated on basis of its molar extinction coefficient at 282 nm (Em = 11,900).
RESULTS
Complementation studies of a cpr gene disruptant.
The gene-disrupted strain JL20D generated as described above had the phenotype described previously for a strain with a LEU2 disruption generated in the same way (18). In contrast to strain JL20, this mutant was highly sensitive to ketoconazole, an inhibitor of the sterol 14α-demethylase (CYP51). The increase in ketoconazole sensitivity in the cpr strain and the diminution of that sensitivity after functional complementation with the heterologously expressed proteins was studied by MIC testing (Table 1). The disruption of the CPR gene resulted in a 200-fold increase in ketoconazole sensitivity. The GAL10-mediated expression of CPR in the cpr host strain JL20D completely restored resistance to ketoconazole. In addition, an eightfold increase in resistance compared to the level of resistance of CPR+ strain JL20 was observed for transformants of JL20D expressing the FUS protein. The presence of CYP51:YEp51 or YEp51 in the cpr host strain had no effect upon the drug sensitivity. In contrast, GAL10-mediated expression of CPR in the CPR strain JL20 did not alter its susceptibility to ketoconazole, while hyperexpression of either the CYP51 or the FUS enzyme increased the level of ketoconazole resistance, but to different levels. JL20 transformants expressing the FUS enzyme were more drug resistant than the transformants expressing CYP51, suggesting that limitation of endogenous CPR may be the cause for the relatively lower level of ketoconazole resistance in the transformant expressing CYP51.
TABLE 1.
MICs of ketoconazole for the yeast transformants harboring different expression vectors
| Plasmid | Ketoconazole MIC (μM)
|
|
|---|---|---|
| JL20 | JL20D (CPR::URA3) | |
| None | 4.0 | 0.02 |
| YEp51 | 4.0 | 0.02 |
| CPR:YEp51 | 8.0 | 8.00 |
| CYP51:YEp51 | 16.0 | 0.02 |
| FUS:YEp51 | 64.0 | 32.00 |
Deletion of the CPR gene led to a decrease in the levels of ergosterol. The ergosterol content in the disrupted strain JL20D was fourfold lower than that in the undisrupted strain (JL20). However, ergosterol was the major sterol in both strains and accounted for about 90% of the total sterols. In the cpr host strain the ergosterol levels were restored to the levels in the CPR strain JL20 upon CPR and FUS expression (Table 2). Expression of these proteins in JL20 (CPR) did not alter the ergosterol content (data not shown).
TABLE 2.
Levels of ergosterol in various yeast transformantsa
| Transformant | Concn of ergosterol (mg/g [dry wt] of cells) | Ergosterol as % of total sterols |
|---|---|---|
| JL20 | 19.7 ± 2.2 | 89.0 ± 4.8 |
| JL20D | 5.9 ± 0.3 | 89.9 ± 3.7 |
| YEp51/JL20D | 5.5 ± 0.6 | 89.3 ± 5.6 |
| CPR:YEp51/JL20D | 21.2 ± 3.3 | 86.4 ± 5.5 |
| CYP51:YEp51/JL20D | 5.4 ± 0.9 | 90.6 ± 3.9 |
| FUS:YEp51/JL20D | 25.6 ± 1.3 | 87.8 ± 5.8 |
Values are means ± standard deviations.
Expression of CPR, CYP51, and FUS proteins in the transformed strains.
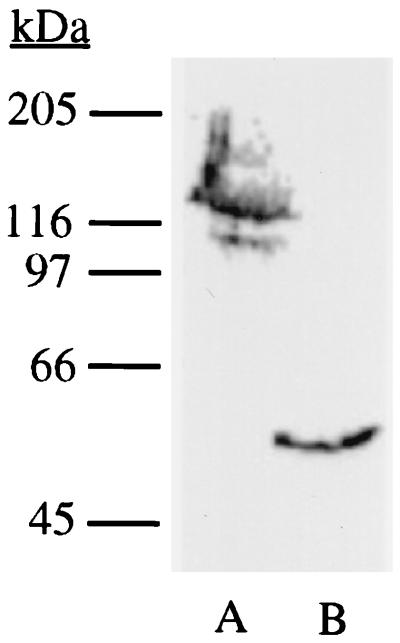
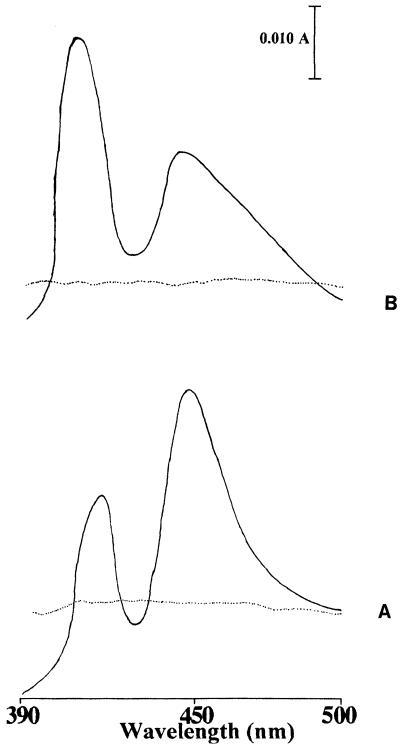
The results of Western blot analysis of the microsomal fractions containing the CYP51 and FUS proteins are presented in Fig. 2. Anti-CYP51 immunoreactive bands with molecular masses of 130 and 55 kDa were detected in the microsomal proteins of transformants expressing FUS and CYP51, respectively. The migration of the bands was consistent with the predicted molecular masses of the FUS and CYP51 enzymes. In the Western blot the immunoreactive band corresponding to the endogenous CYP51 (at about 55 kDa) was not seen in FUS-expressing transformant microsomes. It is probably due to either a very low level of expression of endogenous CYP51 or a low specificity of the antibodies, or a combination of both. Similarly, probing of microsomal fractions of the cpr transformants expressing CPR with anti-CPR by Western blotting suggested that the mobility of the expressed microsomal protein was consistent with the reported molecular mass of 78 kDa for CPR (data not shown). The cytochrome P-450 and CPR contents were determined by using the reduced carbon monoxide spectrum and cytochrome c reduction assay, respectively. FUS and CYP51 containing microsomal fractions produced a typical reduced CO spectrum with a Soret peak at 448 nm (Fig. 3). The cytochrome P-450 and CPR contents in the cytosolic and microsomal fractions of the transformants containing different expression vectors are presented in Table 3. The cytochrome P-450-specific content and yields from transformants expressing the CYP51 were slightly higher than those from transformants expressing the FUS protein. In addition, no CPR activity was detected in the microsomal fractions of the cpr strain or the strain transformed with YEp51 (data not shown).
FIG. 2.
Western blot analysis of heterologous expression of FUS (lane A) and CYP51 (lane B) proteins in JL20 transformants. The microsomal proteins isolated from the galactose-induced cells were analyzed by immunoblotting with polyclonal anti-immunoglobulin G recognizing CYP51.
FIG. 3.
Reduced CO difference spectra of strain JL20 transformant microsomes containing CYP51:YEp51 (A) and FUS:YEp51 (B) expression plasmids. The dotted line is the baseline.
TABLE 3.
Levels of cytochrome P-450 and CPR in microsomal fractions of the transformants containing various expression plasmidsa
| Transformant | Cytochrome P-450
|
CPR
|
||
|---|---|---|---|---|
| Concn as nmol/liter | Concn as nmol/mg of protein | Concn as nmol/liter | Concn as nmol/mg of protein | |
| YEp51/JL20 | NDb | ND | 2.0 ± 0.3 | 0.015 ± 0.004 |
| CYP51:YEp51/JL20 | 15.9 ± 1.6 | 0.12 ± 0.03 | 1.9 ± 0.4 | 0.014 ± 0.003 |
| CPR:YEp51/JL20 | ND | ND | 13.3 ± 1.2 | 0.104 ± 0.013 |
| FUS:YEp51/JL20 | 13.0 ± 1.1 | 0.11 ± 0.02 | 11.6 ± 1.7 | 0.1 ± 0.01 |
Values are means ± standard deviations.
ND, not determined.
Catalytic activities of the expressed proteins.
In vitro studies on ergosterol biosynthesis and its inhibition by ketoconazole were carried out by using the cell-free extracts of the various transformants of the cpr host (Table 4), an assay routinely applied for assessment of ergosterol biosynthesis inhibitors. The amount of ergosterol synthesized in vitro was reduced by about 3-fold, and 50% inhibition of its synthesis by ketoconazole was achieved at an approximately 200-fold lower concentration upon CPR disruption compared to that for the parental strain. However, these changes were reverted when the CPR or the FUS protein was expressed in the cpr strain. The presence of either the YEp51 or the CYP51:YEp51 expression plasmid in the cpr strain did not alter the in vitro ergosterol biosynthesis or inhibition by ketoconazole. The 50% inhibitory concentration (IC50) of ketoconazole for in vitro ergosterol synthesis for transformants expressing the FUS protein was about ninefold higher than that for strain JL20 (CPR), although there was not much change in the amount of ergosterol synthesized. The results obtained for in vitro ergosterol biosynthesis and its inhibition by ketoconazole correlated with the functional complementation and MIC results.
TABLE 4.
Amount of ergosterol synthesized in vitro and the IC50 of ketoconazole for in vitro ergosterol biosynthesis for the various yeast transformantsa
| Transformant | Amt of ergosterol synthesized (dpm) | IC50 (nM) for in vitro ergosterol biosynthesis |
|---|---|---|
| JL20 | 8,950 ± 429 | 22.12 ± 3.73 |
| JL20D | 3,345 ± 324 | 0.11 ± 0.06 |
| YEp51/JL20D | 3,178 ± 648 | 0.12 ± 0.04 |
| CPR:YEp51/JL20D | 9,256 ± 482 | 25.49 ± 4.67 |
| CYP51/JL20D | 3,542 ± 486 | 0.14 ± 0.05 |
| FUS/JL20D | 9,453 ± 756 | 200.43 ± 9.87 |
Values are means ± standard deviations.
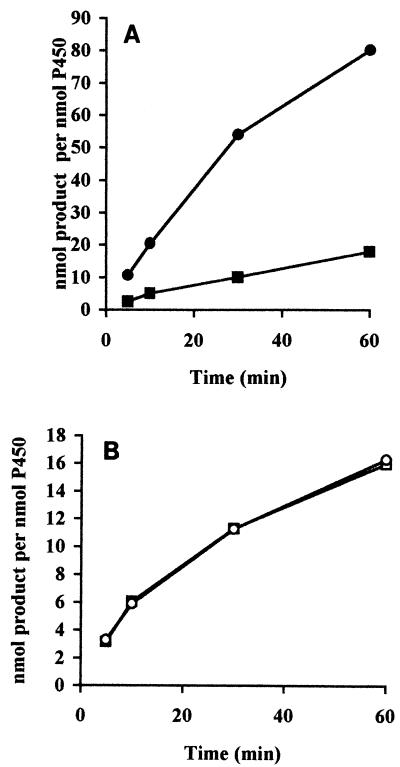
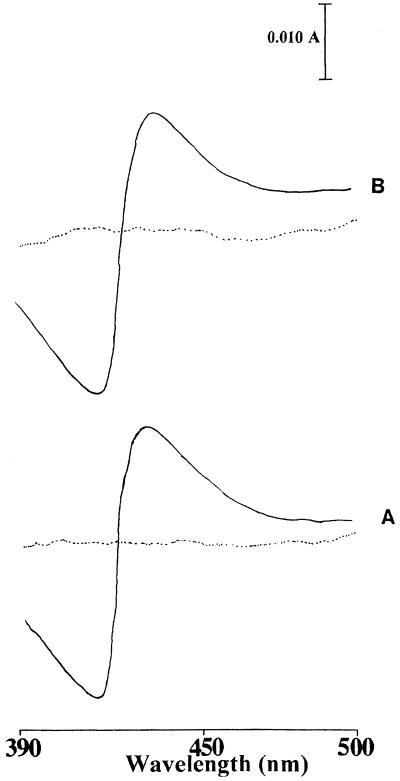
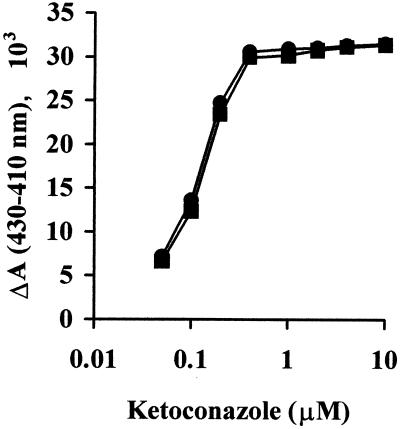
To characterize the FUS and CYP51 proteins biochemically in further detail, sterol 14α-demethylation assays were carried out with the microsomal fractions of the CPR strain (strain JL20) expressing the FUS and CYP51 proteins and with [32-3H]3β-hydroxylanost-7-en-32-ol as a substrate. The demethylation of the substrate was quantified by measuring the amount of radioactive formic acid formed, since the 14α-methyl group of the substrate is released by this reaction as formic acid. The time-dependent demethylation of substrate by the CYP51 and FUS enzymes in the presence of either NADPH or cumene hydroperoxide, an artificial electron donor, is shown in Fig. 4. In the presence of NADPH, the FUS protein showed about a fourfold higher level of activity compared to that of the CYP51 protein, with values of 2.07 ± 0.12 and 0.51 ± 0.07 nmol/nmol of P-450/min, respectively. This difference was not observed for reactions supported by cumene hydroperoxide, for which the values were 0.6 ± 0.04 and 0.59 ± 0.02 nmol/nmol of P-450/min for the CYP51 and FUS proteins, respectively. This result suggests that limitation of endogenous CPR may be the cause for the relatively low level of activity of CYP51 in the presence of NADPH. Microsomes containing expressed CYP51 and FUS proteins produced typical type II binding spectra with ketoconazole that was indicative of a low spin state of the heme iron of cytochrome P-450 that results from the binding of inhibitor to CYP (5). The type II binding spectra obtained with both the proteins showed an absorbance maximum at 430 nm and an absorbance minimum at 410 nm (Fig. 5). A ketoconazole concentration-dependent change in the magnitude of the type II binding spectra of the CYP51 and FUS protein-expressing microsomes was saturated when an equimolar concentration of ketoconazole was added to the microsomal CYP proteins (Fig. 6). These data suggest that both CYP51 and FUS proteins showed similar affinities for ketoconazole.
FIG. 4.
Time-dependent conversion of substrate by CYP51 and FUS proteins by using NADPH-generating system (A) and cumene hydroperoxide (B). □ and ▪, CYP51::YEp51; ○ and •, FUS::YEp51.
FIG. 5.
Type II binding spectra obtained by adding equimolar concentration of ketoconazole to the microsomal cytochrome P-450 isolated from the JL20 transformants containing CYP51:YEp51 (A) or FUS:YEp51 (B) expression plasmids. The dotted line is the baseline.
FIG. 6.
Ketoconazole concentration-dependent change in the magnitude of type II binding spectra of CYP51 and FUS proteins. ΔA is the difference in absorbance between the maximum at 430 nm and the minimum at 410 nm. ▪, CYP51; •, FUS.
DISCUSSION
One of the earliest roles of CPR, as for the CYP superfamily, may have been in sterol biosynthesis, although it presumably participated in electron transport and reductive metabolism prior to this. Besides supporting cytochrome P-450-mediated reactions, CPR also functions in the squalene epoxidation reaction (29), which precedes the sterol 14α-demethylation step undertaken by CYP51 (1), as well as the later sterol 22-desaturation step undertaken by CYP61 during ergosterol biosynthesis (17).
The previous finding of hypersensitivity to ketoconazole on disruption of the gene encoding CPR was in many ways consistent with the concept of inefficient sterol biosynthesis, so that only a low dose of CYP51 inhibitor would arrest growth. We confirmed the observation of Sutter and Loper (18) of a 200-fold increased sensitivity to ketoconazole in such a strain but observed only a relatively small reduction in the amount of ergosterol synthesized to about 25% of that synthesized by the parent strain when the amounts were compared by using the dry weights of the cells. Analysis of sterols by gas chromatography and thin-layer chromatography did not reveal the accumulation of intermediates such as lanosterol or ergosta-5,7-dienol indicative of a block to enzyme activity at the steps in which CPR participates. This suggests that the electron donor system remaining in the cells is efficient, delivering both electrons for CYP activity.
The in vitro egosterol biosynthesis studies showed that inhibition with ketoconazole corresponded to the changes in the MIC, with the extract from the cpr host JL20D being about 200 times more sensitive than the parent strain to ketoconazole. The altered sensitivity was despite the observation that ergosterol biosynthesis in untreated samples showed only a small reduction in activity to about 25% of that of the parent strain (as observed for whole cells). Previous studies with purified CYP51 have not observed a change, compared to the interaction of microsomal CYP51, when purified CYP51 interacts with azole antifungal agents (30), which excludes an altered azole affinity for the enzyme in the absence of CPR. These results indicate that the alternative mechanism of electron transport is more sensitive to azole antifungal agents than the CPR-based system, implicating a mechanism of action of ketoconazole other than binding to CYP51, at least in a cpr strain. Such cpr strains show similar hypersensitivities to fluconazole (unpublished observation), and these findings may have general relevance for azole antifungal agents because the cytochrome b5 system supports other cellular reactions such as sterol C-5 desaturation and C-4 demethylation.
Studies on the sensitivity of transformants of the cpr host to ketoconazole when the CPR and FUS proteins are expressed had all demonstrated restoration of the ergosterol content and the ketoconazole sensitivity to levels similar to those for the parental strain (complementation). As observed for other CYP-CPR fusions, the coupling of the CPR in FUS to CYP51 does not block catalytic ability, but more surprising was the ability of the coupling to allow full ergosterol biosynthesis. The results indicate that transfer of electrons to the other enzymes from the CPR of FUS can occur, indicating a great degree of flexibility in this protein-protein interaction.
Data from in vitro ergosterol biosynthesis assays with transformants did not suggest that CPR had a large effect on ketoconazole sensitivity under normal conditions of growth. Expression of CPR caused a marginal increase in the MICs for CPR transformants, which might indicate that a limitation exists in control cells or that the increase in this microsomal enzyme had other effects. However, expression of CYP51 increased the level of resistance to ketoconazole in the CPR host through a gene dosage effect, as expected, but a further increase was observed with expression of FUS although the amount of hemoprotein expressed was lower. It was clearly suggested from these data and others (22) that gene dosage mechanisms of azole antifungal resistance which have been proposed as playing a role in some clinical isolates (21) would be limited in their effect by the availability of CPR. This was supported in activity studies in which enhanced activity for FUS-containing microsomes was observed and no change in affinity for azoles in comparison to that of CYP51 could be detected. The absence of a gene dosage effect for cpr transformants expressing CYP51 was surprising, and it is possible that the increased level of CYP51 was offset by other changes to ergosterol biosynthesis. The ranked order of ketoconazole sensitivities seen in the MICs for the transformants was again mirrored by evaluation of the IC50 of ketoconazole for inhibition of in vitro sterol biosynthesis.
In conclusion, the necessity of CPR for ergosterol biosynthesis has been shown to be less important than was previously envisaged, and the alternative electron donor system appears to deliver both electrons efficiently to all three enzymes known to require such a transport system. This system is hypersensitive to the effects of ketoconazole, implying an additional effect of ketoconazole besides that on CYP-CPR-based activity. Finally, the CPR component of FUS is able to provide an electron donor for squalene epoxidase and sterol 22-desaturase, despite being fused to CYP51, and results in restored levels of ergosterol in a cpr host.
ACKNOWLEDGMENT
One of us (K.V.) was supported by a Commonwealth Scholarship.
REFERENCES
- 1.Aoyama Y, Yoshida Y, Sonoda Y, Sato Y. Metabolism of 32-hydroxy-24,25-dihydrolanosterol by purified cytochrome P45014DM from yeast. J Biol Chem. 1989;264:18502–18505. [PubMed] [Google Scholar]
- 2.Baily G G, Perry F M, Denning D W, Mandal B K. Fluconazole-resistant candidosis in an HIV cohort. AIDS. 1994;8:787–792. doi: 10.1097/00002030-199406000-00010. [DOI] [PubMed] [Google Scholar]
- 3.Hata S, Nishino T, Katsuki H, Aoyama Y, Yoshida Y. Characterization of Δ22-desaturation in ergosterol biosynthesis of yeast. Agric Biol Chem. 1987;51:1349–1354. [Google Scholar]
- 4.Imai Y. The roles of cytochrome-b5 in reconstituted mono-oxygenase systems containing various forms of hepatic microsomal cytochrome P-450. J Biochem. 1981;89:351–362. doi: 10.1093/oxfordjournals.jbchem.a133209. [DOI] [PubMed] [Google Scholar]
- 5.Jefcoate J R. Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol. 1979;52:258–279. doi: 10.1016/s0076-6879(78)52029-6. [DOI] [PubMed] [Google Scholar]
- 6.Jones J S, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- 7.Kalb V F, Woods C W, Turi T G, Dey C R, Sutter T R, Loper J C. Primary structure of the P450 lanosterol demethylase gene from Saccharomyces cerevisiae. DNA. 1987;6:529–537. doi: 10.1089/dna.1987.6.529. [DOI] [PubMed] [Google Scholar]
- 8.Kalb V F, Loper J C, Dey C R, Woods C W, Sutter T R. Isolation of a cytochrome P-450 structural gene from Saccharomyces cerevisiae. Gene. 1986;45:237–245. doi: 10.1016/0378-1119(86)90021-1. [DOI] [PubMed] [Google Scholar]
- 9.Kelly S L, Lamb D C, Corran A J, Baldwin B C, Kelly D E. Mode of action and resistance of azole antifungals associated with the formation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol. Biochem Biophys Res Commun. 1995;207:910–905. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 10.Kelly S L, Lamb D C, Corran A J, Baldwin B C, Parks L W, Kelly D E. Purification and reconstitution of activity of Saccharomyces cerevisiae P450 61, a sterol Δ22-desaturase. FEBS Lett. 1995;377:217–220. doi: 10.1016/0014-5793(95)01342-3. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:333–346. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 13.Quail M A, Arnoldi A, Moore D J, Goosey M W, Kelly S L. Ketoconazole mediated growth inhibition in Botrytis cinera and Saccharomyces cerevisiae. Phytochemistry. 1993;32:273–280. [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Sevrioukova I F, Peterson J A. NADPH-P-450 reductase: structural and functional comparisons of the eukaryotic and prokaryotic isoforms. Biochimie. 1995;77:562–572. doi: 10.1016/0300-9084(96)88172-7. [DOI] [PubMed] [Google Scholar]
- 16.Shyadehi A Z, Lamb D C, Kelly S L, Kelly D E, Schunck W-H, Wright N, Corina D, Akhtar M. The mechanism of the acyl-carbon bond-cleavage reaction catalyzed by recombinant sterol 14α-demethylase of Candida albicans (other names are lanosterol 14α-demethylase, P-450[14DM], and CYP51) J Biol Chem. 1996;271:12445–12450. doi: 10.1074/jbc.271.21.12445. [DOI] [PubMed] [Google Scholar]
- 17.Skaggs B A, Alexander J F, Pierson C A, Schweitzer K S, Chun K T, Koegel C, Barbuch R, Bard M. Cloning and characterization of the Saccharomyces cerevisiae C-22 sterol desaturase gene, encoding a 2nd cytochrome P-450 involved in ergosterol biosynthesis. Gene. 1996;169:105–109. doi: 10.1016/0378-1119(95)00770-9. [DOI] [PubMed] [Google Scholar]
- 18.Sutter T R, Loper J C. Disruption of the Saccharomyces cerevisiae gene for NADPH-cytochrome P-450 reductase causes increased sensitivity to ketoconazole. Biochem Biophys Res Commun. 1989;160:1257–1266. doi: 10.1016/s0006-291x(89)80139-1. [DOI] [PubMed] [Google Scholar]
- 19.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Traun G, Epinat J-C, Rougeulle C, Cullin C, Pompon D. Cloning and characterization of a yeast cytochrome b5-encoding gene which suppresses ketoconazole hypersensitivity in a NADPH–P-450 reductase-deficient strain. Gene. 1994;149:123–127. doi: 10.1016/0378-1119(94)90366-2. [DOI] [PubMed] [Google Scholar]
- 21.Vanden Bossche H, Marichal P, Odds F C, Le Jeune L, Coene M C. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1992;36:2602–2610. doi: 10.1128/aac.36.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van den Brink J M H, van Nistelrooy J G M H, de Waard M A, van den Hondel A M J J C, van Gorcom R F M. Increased resistance to 14α-demethylase inhibitors (DMIs) in Aspergillus niger by coexpression of the Penicillium italicum eburicol 14α-demethylase (cyp51) and the A. niger cytochrome P450 reductase (cprA) genes. J Biotechnol. 1996;49:13–18. doi: 10.1016/0168-1656(96)01403-4. [DOI] [PubMed] [Google Scholar]
- 23.Venkateswarlu K, Denning D W, Manning N J, Kelly S L. Comparison of D0870, a new triazole antifungal agent, to fluconazole for inhibition of Candida albicans cytochrome P-450 by using in vitro assays. Antimicrob Agents Chemother. 1996;40:1382–1386. doi: 10.1128/aac.40.6.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venkateswarlu K, Denning D W, Manning N J, Kelly S L. Resistance to fluconazole in Candida albicans from AIDS patients correlated with reduced intracellular accumulation of drug. FEMS Microbiol Lett. 1995;131:337–341. doi: 10.1111/j.1574-6968.1995.tb07797.x. [DOI] [PubMed] [Google Scholar]
- 25.Vermillion J L, Ballou D P, Massey V, Coon M J. Separate roles for FMN and FAD in catalysis by liver microsomal NADPH-cytochrome P-450 reductase. J Biol Chem. 1981;256:266–277. [PubMed] [Google Scholar]
- 26.Vermillion J L, Coon M J. Purified liver microsomal NADPH-cytochrome P-450 reductase. J Biol Chem. 1978;253:2694–2704. [PubMed] [Google Scholar]
- 27.Weete J D. Structure and functions of sterols in fungi. Adv Lipid Res. 1989;23:115–167. [Google Scholar]
- 28.Yabusaki Y, Murakami H, Ohkawa H. Primary structure of Saccharomyces cerevisiae NADPH-cytochrome P450 reductase deduced from nucleotide sequence of its cloned gene. J Biochem. 1988;103:1004–1010. doi: 10.1093/oxfordjournals.jbchem.a122370. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida Y. Cytochrome P450 of fungi: primary target for azole antifungal agents. Curr Top Med Mycol. 1988;2:388–418. doi: 10.1007/978-1-4612-3730-3_11. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida Y, Aoyama Y. Interaction of azole antifungal agents with cytochrome P45014DM purified from Saccharomyces cerevisiae microsomes. Biochem Pharmacol. 1987;36:229–235. doi: 10.1016/0006-2952(87)90694-0. [DOI] [PubMed] [Google Scholar]