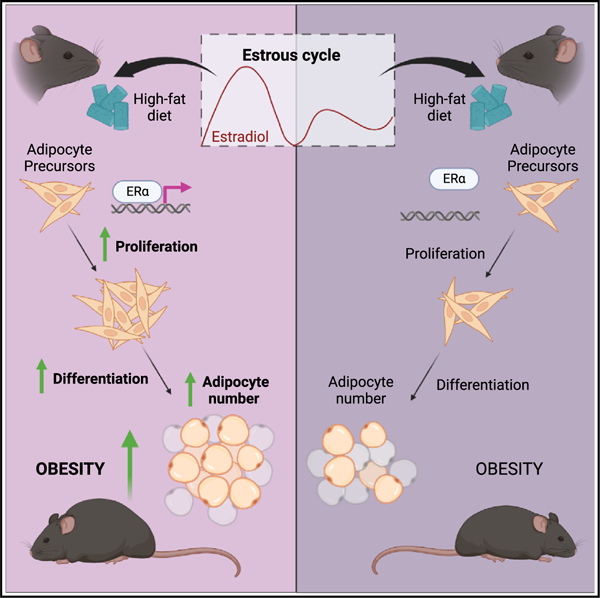
SUMMARY
White adipose tissue (WAT) distribution is sex dependent. Adipocyte hyperplasia contributes to WAT distribution in mice driven by cues in the tissue microenvironment, with females displaying hyperplasia in subcutaneous and visceral WAT, while males and ovariectomized females have visceral WAT (VWAT)-specific hyperplasia. However, the mechanism underlying sex-specific hyperplasia remains elusive. Here, transcriptome analysis in female mice shows that high-fat diet (HFD) induces estrogen signaling in adipocyte precursor cells (APCs). Analysis of APCs throughout the estrous cycle demonstrates increased proliferation only when proestrus (high estrogen) coincides with the onset of HFD feeding. We further show that estrogen receptor α (ERα) is required for this proliferation and that estradiol treatment at the onset of HFD feeding is sufficient to drive it. This estrous influence on APC proliferation leads to increased obesity driven by adipocyte hyperplasia. These data indicate that estrogen drives ERα-dependent obesogenic adipocyte hyperplasia in females, exacerbating obesity and contributing to the differential fat distribution between the sexes.
Graphical abstract
In brief
Saavedra-Peña et al. show that estradiol cycling regulates adipocyte formation in female mice by stimulating ERα-dependent proliferation of adipocyte precursor cells (APCs) at the onset of high-fat diet feeding. This estradiol-stimulated mechanism results in exacerbated obesity due to an increase in adipocyte formation.
INTRODUCTION
In both mice and humans, females of reproductive age are less prone to obesity and metabolic dysfunction compared with males and postmenopausal females.1–3 This protection has been attributed to the cycling of hormones, particularly estradiol, the most potent and abundant circulating estrogen.4–6 Estrogens are considered anti-obesogenic hormones, as they regulate multiple aspects of energy balance including promoting physical activity, reducing food intake, and protecting from hepatic steatosis via signaling through estrogen receptor α (ERα; Esr1).7–11 At the same time, increased fat accumulation occurs simultaneously with elevations in estrogen during puberty and pregnancy,12–14 which suggests a more nuanced relationship between estrogens and adiposity. These nuances are challenging to study due to the tissue-specific roles of estrogens, the altered metabolism of sex steroids in obesity, and compensatory hormonal responses in many experimental models.15–19
In obesity, white adipose tissue (WAT) growth occurs via hypertrophy (increase in adipocyte size) and hyperplasia (increase in adipocyte number). Hyperplastic growth requires adipocyte precursor cell (APC) proliferation and differentiation, as mature adipocytes are postmitotic.20–23 APCs proliferate rapidly and transiently at the onset of a high-fat diet (HFD) feeding, leading to increased adipocyte formation in a process that is specific to obesity.21,24,25 There is a striking difference in the depot patterning of obesogenic hyperplasia between the sexes, with females displaying both visceral (VWAT) and subcutaneous (SWAT) hyperplasia, while males exhibit visceral-specific hyperplasia,19,21,22,25 which mirrors the sex-specific WAT distribution in humans.22,25 While APCs arise from distinct developmental lineages in a depot- and sex-specific manner,26–28 transplant experiments demonstrate it is the microenvironment of the fat depot, and not the developmental lineage, that controls obesogenic hyperplasia.25 Sex hormones are known to play a role in the adipose microenvironment. Androgens influence depot patterning during adipose development,29 and estrogens influence the depot patterning of hyperplasia in obesity. Females lose obesogenic hyperplasia in SWAT after ovariectomy (OVX), when gonadal production of estrogens is lost,25 while males gain SWAT hyperplasia upon estradiol treatment.25
Elucidating the mechanisms underlying sex-specific obesogenic hyperplasia is important, as the distribution of fat impacts metabolic health.30–33 Furthermore, it is believed that hyperplasia leads to a sustained increase in WAT mass, as increased adipocyte number is maintained even after weight loss,34 whereas adipocyte size can rapidly change to meet current energy demands.35–37 While previous studies of hyperplasia in mice and humans have shed light on the hierarchy and complexity of APCs,38–45 APC response during the onset of HFD feeding, when APCs are the most proliferative,21,25 has not been assessed. Here, we perform RNA sequencing in APCs from the VWAT and SWAT of female mice at the onset of HFD feeding to identify molecular pathways involved in the hyperplastic response. We identify estradiol as a major regulator of APC proliferation in both depots of female mice in a process that is dependent of ERα signaling. Furthermore, we demonstrate that the cycling of systemic estrogens determines obesogenic hyperplasia in females, which contributes to increased obesity without benefiting glucose tolerance, thus indicating that this mechanism of adipocyte hyperplasia is not metabolically protective. Overall, these data uncover that the timing of HFD feeding in females relative to the estrous cycle impacts the long-term development of obesity.
RESULTS
Estrogen pathways are upregulated during obesogenic proliferation in females
Although several mechanisms of hyperplasia have been uncovered,46 most of the in vivo studies have been done in male mice.20–22,47 Given the sexually dimorphic patterning of adipose hyperplasia and the potential role of estrogens in female APC proliferation,25 we used RNA sequencing (RNA-seq) to help determine the mechanisms underlying VWAT and SWAT hyperplasia in APCs of female mice (GEO: GSE209663). Bulk RNA-seq was performed to ensure sufficient read depth for mechanistic analysis on APCs from VWAT and SWAT on standard chow (SD) or after 3 days of HFD feeding, when APC proliferation is highest21 (Figure 1A). DESeq2 analysis reveals that an HFD has a has a greater impact on gene expression in VWAT (5,828 genes) than SWAT (2,368) (Figures 1B, 1C, and 1E). Of note, 78.8% and 23.9% of the differentially expressed genes (DEGs) are exclusive to VWAT and SWAT, respectively (Figure 1B). Gene Ontology (GO) analysis reveals processes involving cell cycle progression, mitosis, and protein transport and modification are commonly upregulated in both depots (Figures S1A and S1C). In addition, KEGG pathway analysis reveals upregulation of PI3K-Akt and FoxO signaling, both of which are involved in metabolism and nutrient sensing (Figures S1B and S1D). Next, we used Ingenuity Pathway Analysis (IPA) to predict the pathways and upstream regulators likely involved in the gene expression changes upon HFD feeding. Most of the pathways identified have previously been implicated in adipogenesis, including VEGF,48–50 ERBB2,51,52 PTGER2,53 CSF2,54 FOXM1,55 and CD24,56 which supports that this experiment assesses adipogenic mechanisms. Interestingly, both ERα (Esr1) and 17β-estradiol are among the top positive regulators induced by HFD (Figures 1D and 1F), suggesting that estrogen signaling plays a role in adipogenesis. Further supporting this finding, several of the upstream regulators identified here are modulated by ERα and estrogen signaling, including FOXM1,57,58 VEGF,59 and ERBB2.60,61 Analysis of canonical molecular pathways via IPA shows estrogen-mediated S-phase entry is upregulated in both fat depots on an HFD (Figure 1G). Remarkably, all other shared upregulated pathways, including mitotic roles of Polo-like kinase, GP6, TREM1, and BRCA1 signaling, have been directly linked to estrogen signaling62–66 (Figure 1G). The results of this genomic study in combination with previous results25 highlight a central role for estrogen signaling in obesogenic adipocyte hyperplasia in female mice.
Figure 1. Estrogen pathways are upregulated in female APCs at the onset of HFD.
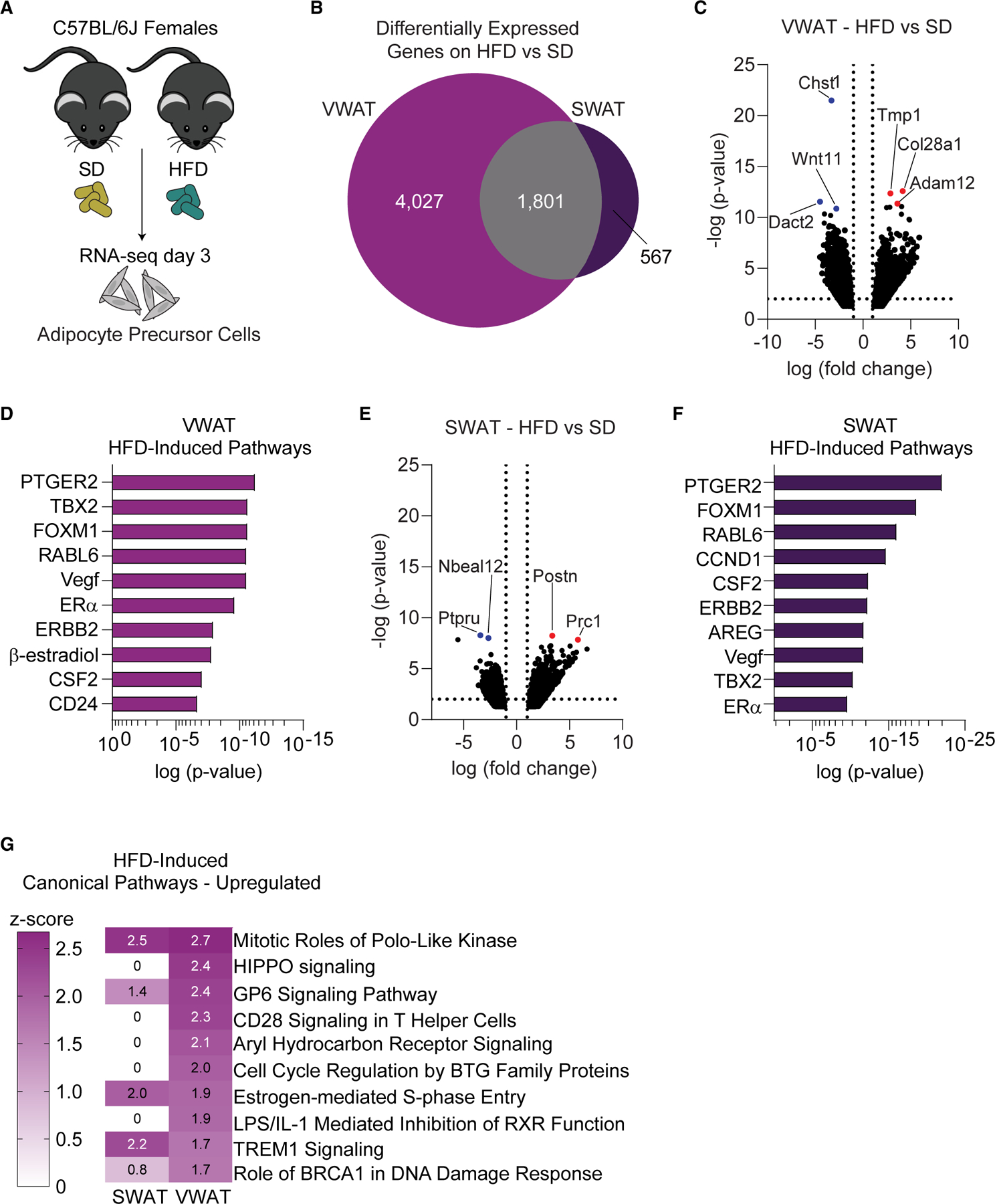
(A) Schematic representation of RNA sequencing experiment in female mice.
(B) Venn diagram of HFD-induced DEGs in female VWAT and SWAT.
(C) Volcano plot of HFD-induced DEGs in female VWAT. Top 3 genes with lowest p values are colored in red (upregulated) or blue (downregulated).
(D) Top positive upstream regulators on an HFD in VWAT as predicted by Ingenuity Pathway Analysis. Gene predictions are in uppercase.
(E) Volcano plot of HFD-induced DEGs in female SWAT. Top 2 genes with lowest p values are colored in red (upregulated) or blue (downregulated).
(F) Top positive upstream regulators on an HFD in SWAT as predicted by Ingenuity Pathway Analysis. Gene predictions are in uppercase.
(G) Top canonical pathways with a positive Z score induced by HFD as predicted by IPA.
n = 5 samples per group, 3 mice pooled for each sample. VWAT, perigonadal fat; SWAT, inguinal subcutaneous fat; SD, standard diet; HFD, high-fat diet; DEGs, differentially expressed genes.
See also Figure S1.
Timing of obesogenic stimulus in females impacts APC proliferation
Mature female mice undergo hormonal cycling (estrous cycle),67,68 with plasma estradiol peaking during the proestrus stage (Figure 2A).69 As previous studies show that sex hormone signaling influences obesogenic hyperplasia,19,21,25 we assessed if the timing of HFD feeding initiation in the estrous cycle affects APC proliferation. Estrous stages were assessed daily using vaginal smears as previously described70,71 before and during a week of SD or HFD feeding (Figure 2B). APCs were identified via flow cytometry as lineage negative (CD45−, CD31−) and positive for CD29, CD34, and Sca-1 (Figure 2C). Proliferation was determined by measuring BrdU incorporation (Figure 2C). After 1 week of diet, we find that females that experience proestrus at the onset (days 0 to 2) of HFD feeding (termed “early”) have increased APC proliferation in both depots compared with females in other estrous stages at the onset of HFD feeding (termed “late”) (Figures 2D and 2E). APC proliferation was not affected by the estrous cycle on an SD, indicating that the combination of HFD initiation with proestrus is required to drive APC proliferation. Interestingly, CD45+ cells are more proliferative upon HFD feeding but are not influenced by estrous cycling in females (Figure 2F), suggesting that estrous cycle-associated proliferative changes in response to HFD are an APC-specific response. Of note, all females experienced proestrus during the week of the experiment, thus it is the timing of HFD initiation in relation to the estrous cycle that is critical for APC proliferation.
Figure 2. Estrous cycling affects obesogenic APC proliferation.
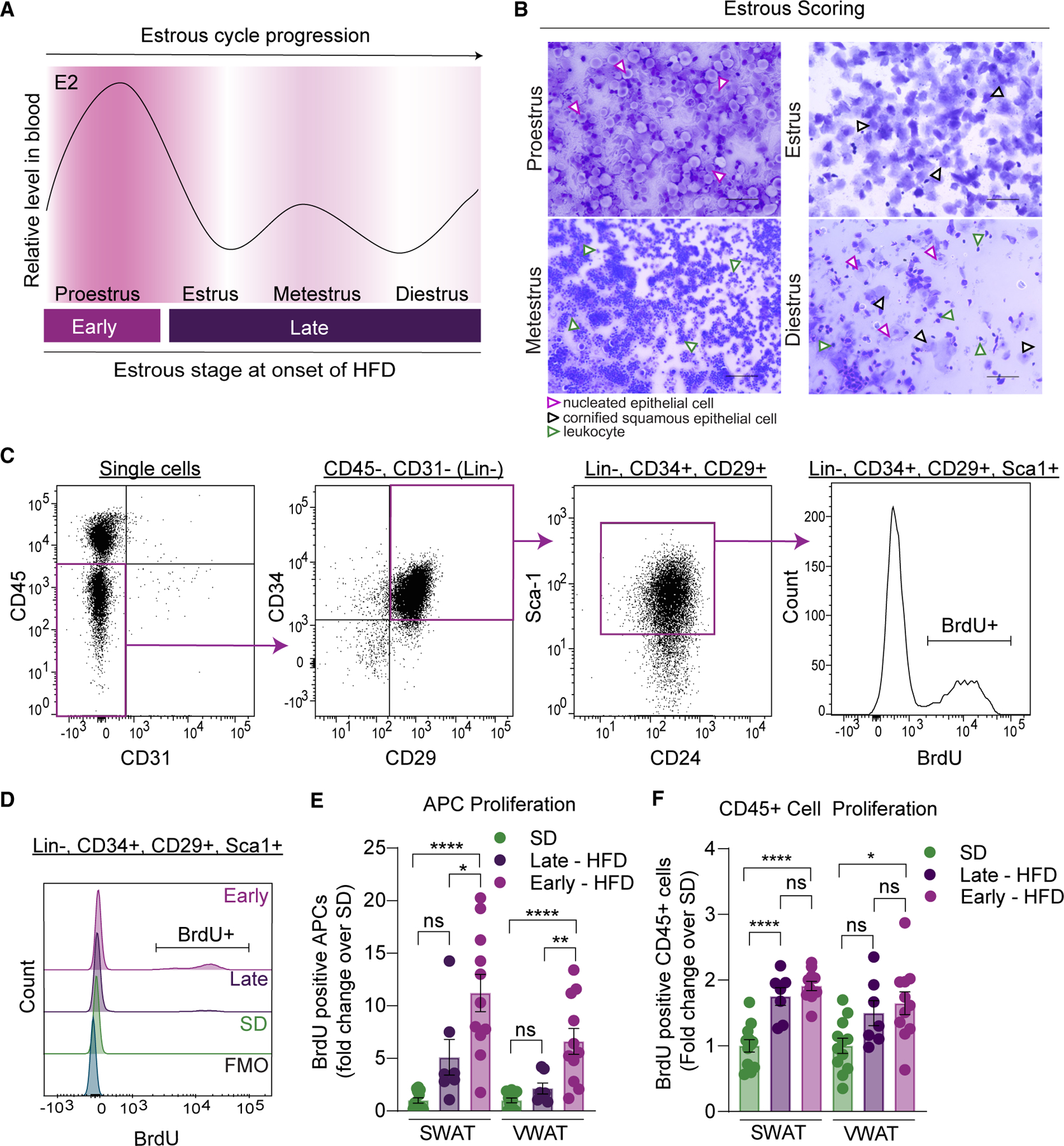
(A) Schematic of circulating estradiol (E2) levels in female mice during the 4 stages of the estrous cycle (adapted from McLean et al.71). Grouping of females based on transition into proestrus happening early (days 0 to 2) or late (days 3 to 7) of a 1-week-long HFD feeding.
(B) Representative images of vaginal smears stained with crystal violet from female mice during the 4 stages of the estrous cycle. Nucleated epithelial cells are highlighted by purple arrows, cornified squamous epithelial cells by black arrows, and leukocytes by green arrows. Scale bar is 100 μm, images taken at 20×.
(C) Representative flow cytometry dot plots to measure BrdU incorporation into APCs. Briefly, APCs are lineage negative (CD45−, CD31−) and positive for CD34, CD29, and Sca-1. BrdU incorporation is measured to assess proliferation.
(D) Representative BrdU histograms from APCs in the different groups including BrdU FMO.
(E) APC proliferation from females after 1 week of SD or HFD feeding.
(F) CD45+ cell proliferation from females after 1 week of SD or HFD feeding.
n = 7–11 mice per group. Statistical significance was determined by ordinary one-way ANOVA with Tukey’s test for (E) and (F). Error bars represent mean ± SEM. ns, not significant, *p < 0.05, **p < 0.01, ****p < 0.0001. APCs, adipocyte precursor cells; VWAT, perigonadal fat; SWAT, inguinal subcutaneous fat; E2, 17β-estradiol; SD, standard diet; HFD, high-fat diet; BrdU, bromodeoxyuridine; FMO, fluorescence minus one.
Estradiol drives female APC proliferation via ERα
As circulating estradiol peaks during proestrus,69 and as APC proliferation is highest at day 3 of HFD feeding,21 the data support a requirement for estrogen signaling in female adipocyte hyperplasia. However, estradiol is only one of the many hormones that fluctuates during the estrous cycle.69 Thus, we assessed if estradiol treatment during the onset of HFD feeding affects APC proliferation. Females in estrus were treated with vehicle or 17β-estradiol (1 mg/kg) daily during days 0–2 of HFD feeding (Figure S2A). As described previously,11,8,72–75 estradiol treatment acutely decreases food intake in females (Figure 3A). But despite decreased consumption of the HFD, estradiol-treated females had increased APC proliferation compared with vehicle controls (Figures 3B and 3C). To determine the mechanisms by which estradiol affects APC proliferation, we assessed the expression of known ERs in APCs. The RNA-seq dataset reveals that only ERα (Esr1), and not ERβ (Esr2) or GPR30 (Gper1), is expressed in female APCs (Figure 3D). ERα is a known regulator of energy homeostasis, influencing both food intake and physical activity.74,76 To determine if ERα is required for APC proliferation in vivo, we used the previously described Pdgfra-cre; Esr1f/f; mTmG strain (AP-ERα-knockout [KO]),19 where APCs lack ERα expression. To mitigate the off-target effects in these KO mice,19 we employed the APC transplant strategy (Figure 3E). We have previously described this method, which does not include Matrigel or impact the proliferative response of the transplanted APCs.19,25 Here, we successfully identify transplanted wild-type APCs in the recipient depot via flow cytometry as RFP+ (Figure S2B) and demonstrate the transplanted cells retain proliferation capacity in the recipient depot similar to endogenous cells (male VWAT) (Figure S2C). To assess the requirement of ERα in APC proliferation, we transplanted GFP+ ERα-KO APCs directly into wild-type female SWAT and identify these in the recipient depot via flow cytometry (Figure S3D). During the experiment, estrous stages were assessed, and only females in proestrus at the onset of HFD feeding were included to ensure a proliferative microenvironment in the tissue. When assessing proliferation of endogenous APCs (GFP−) and ERα-KO APCs (GFP+), we find that APCs lacking ERα do not proliferate in response to HFD feeding compared with endogenous cells in SWAT or VWAT (Figures 3F and 3G). These data demonstrate that APC-intrinsic ERα signaling is required for the obesogenic proliferation that occurs when the onset of HFD feeding coincides with proestrus. Taken together, these data demonstrate that estradiol mediates APC proliferation in female WAT at the onset of HFD feeding, likely contributing to the sexual dimorphism in adipocyte hyperplasia.25
Figure 3. Estradiol drives female APC proliferation in an ERα-dependent manner.
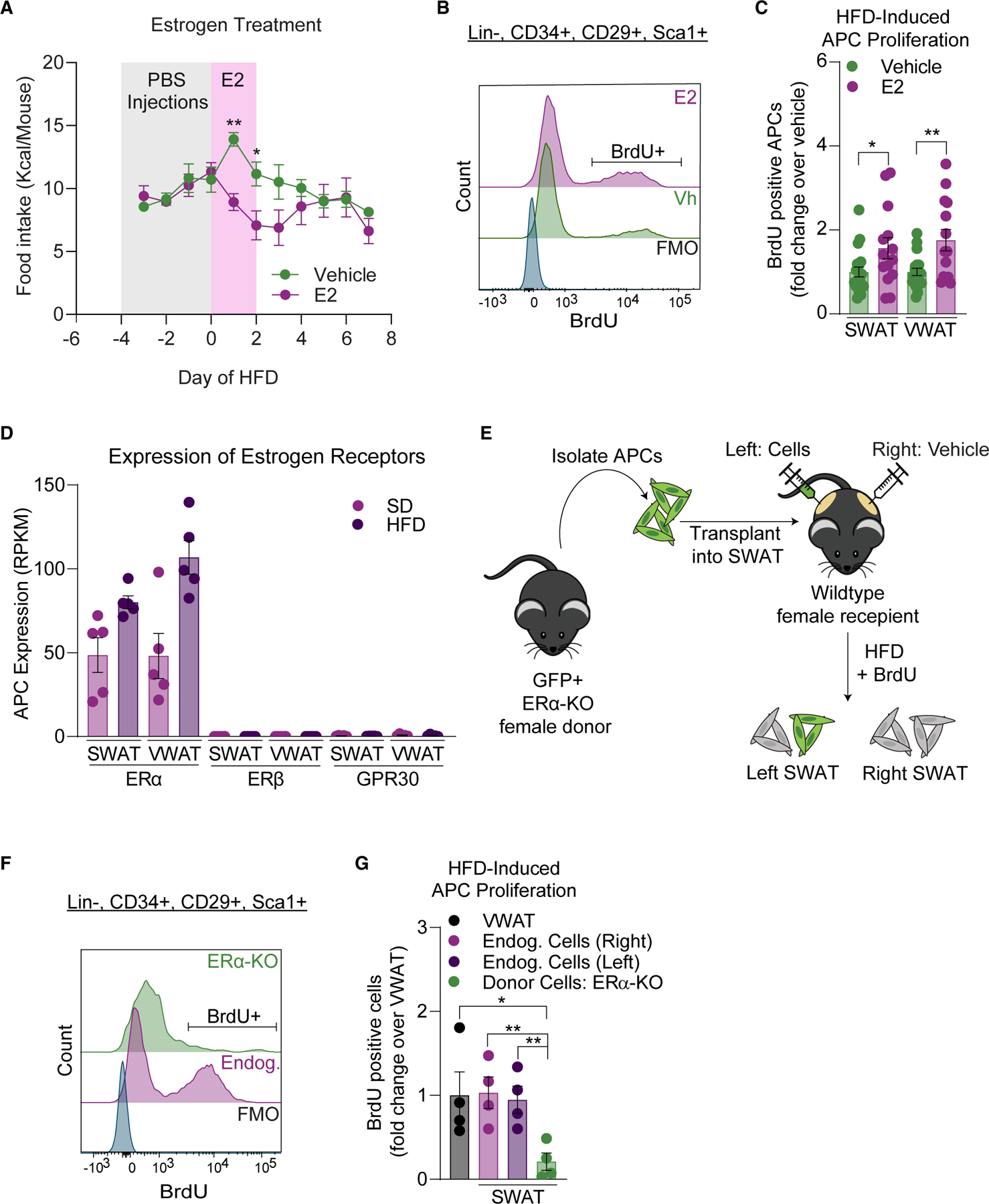
(A) Food intake (cage average) during vehicle or E2 treatment (days 0–2 of an HFD) in female mice (n = 14–19 mice per group).
(B) Representative BrdU histograms in APCs from estradiol treatment experiment including BrdU FMO.
(C) APC proliferation after 1 week of HFD feeding in vehicle or E2 treated female mice.
(D) Expression of estrogen receptors in female APCs from RNA-seq data (n = 5 samples per group, 3 mice pooled per sample).
(E) Schematic of APC transplantation assay into female SWAT.
(F) Representative BrdU histograms in APCs from APC-ERαKO transplant experiment including BrdU FMO.
(G) APC proliferation of transplanted ERα-KO and endogenous APCs after 1 week of HFD feeding in females (n = 4 mice per group).
Statistical significance was determined by one-way ANOVA with Šidák’s tests for (A) and unpaired t tests for (C) and (F). Error bars represent mean ± SEM. ns, not significant, *p < 0.05, **p < 0.01. APCs, adipocyte precursor cells; VWAT, perigonadal fat; SWAT, inguinal subcutaneous fat; BrdU, bromodeoxyuridine; E2, 17β-estradiol; Vh, vehicle; SD, standard diet; HFD, high-fat diet; BrdU, bromodeoxyuridine; Endog, endogenous; FMO, fluorescence minus one.
See also Figure S2.
Timing of obesogenic stimulus influences female obesity
As APC proliferation is impacted by the timing of an HFD feeding in female mice, we next determined if the timing of an HFD feeding also affects the development of obesity. We assessed estrous cycle stages during the first week of an SD or HFD feeding, at which point the females on an HFD were classified into early (APC proliferative) and late (non-APC proliferative) groups (Figure 2A). To measure adipocyte hyperplasia, BrdU was given during the first week of the experiment and chased for 12 weeks (Figure 4A). Adipocyte hyperplasia was measured using immunohistochemistry to assess BrdU incorporation specifically into the nuclei of mature adipocytes. In this assay, we identify mature adipocyte nuclei by staining the adipocyte plasma membranes with caveolin-1 (Figures 4B and 4C), as previously described.21,46 Consistent with increased APC proliferation, adipocyte hyperplasia is increased in females undergoing proestrus at the onset of HFD feeding (Figure 4D). Importantly, we find that females undergoing proestrus at the onset of HFD feeding have increased body weight long term (Figure S3A), driven by a 22% increase in fat mass accumulation (Figure 4E), with no impact on lean mass (Figure S3B). In line with increased total fat mass, these females have 21% increased SWAT and 23% increased VWAT accumulation (Figure 4F) with no differences in liver or brown fat weight (Figure S3C), indicating that the increase in weight is indeed due to the increased white adipose mass. Remarkably, adipocyte sizing analysis reveals that upon HFD feeding, hypertrophy is not affected by the timing of the estrous cycle in VWAT with minimal impact in SWAT (Figures 4G and 4H), thus the 22% increased fat mass is driven almost exclusively by increased number of mature adipocytes. Not surprisingly, as estrous-cycle-driven hyperplasia impacts both depots, the distribution of WAT accumulation is not altered between the HFD groups (Figure S3D). However, hyperplastic females have a slight but significant worsening of glucose tolerance after 12 weeks of HFD feeding (Figure S3E), suggesting that in contrast to previous findings on the metabolic role of adipocyte hyperplasia,77,78 this estradiol-driven adipocyte hyperplasia is not metabolically protective, likely as this hyperplasia is not accompanied by reduced adipocyte hypertrophy. Lastly, as alternative activation of APCs can also contribute to adipose fibrosis,44,79–81 we measured the expression of fibrotic genes in WAT after 12 weeks of SD or HFD feeding. Although there are diet-driven changes in fibrotic gene expression, no differences are seen between the HFD groups (Figures S4A and S4B). Of note, the expression of several adipogenic genes after 12 weeks of HFD were also not different between the diets (Figures S4C and S4D), which supports the transient nature of these genes in obesogenic adipose hyperplasia.21 These data indicate that in female mice, the estrous cycle at the onset of HFD feeding influences hyperplastic WAT growth, thereby contributing to obesity. Taken together with previous findings, these data highlight the complexity of the role of estrogens in energy balance and female physiology, effectively driving acute anti-obesogenic effects in the brain via ERα (decreased food intake and increased physical activity) and long-term pro-obesogenic effects in WAT via hyperplasia.23,82,83
Figure 4. Timing of HFD determines hyperplasia in female mice.
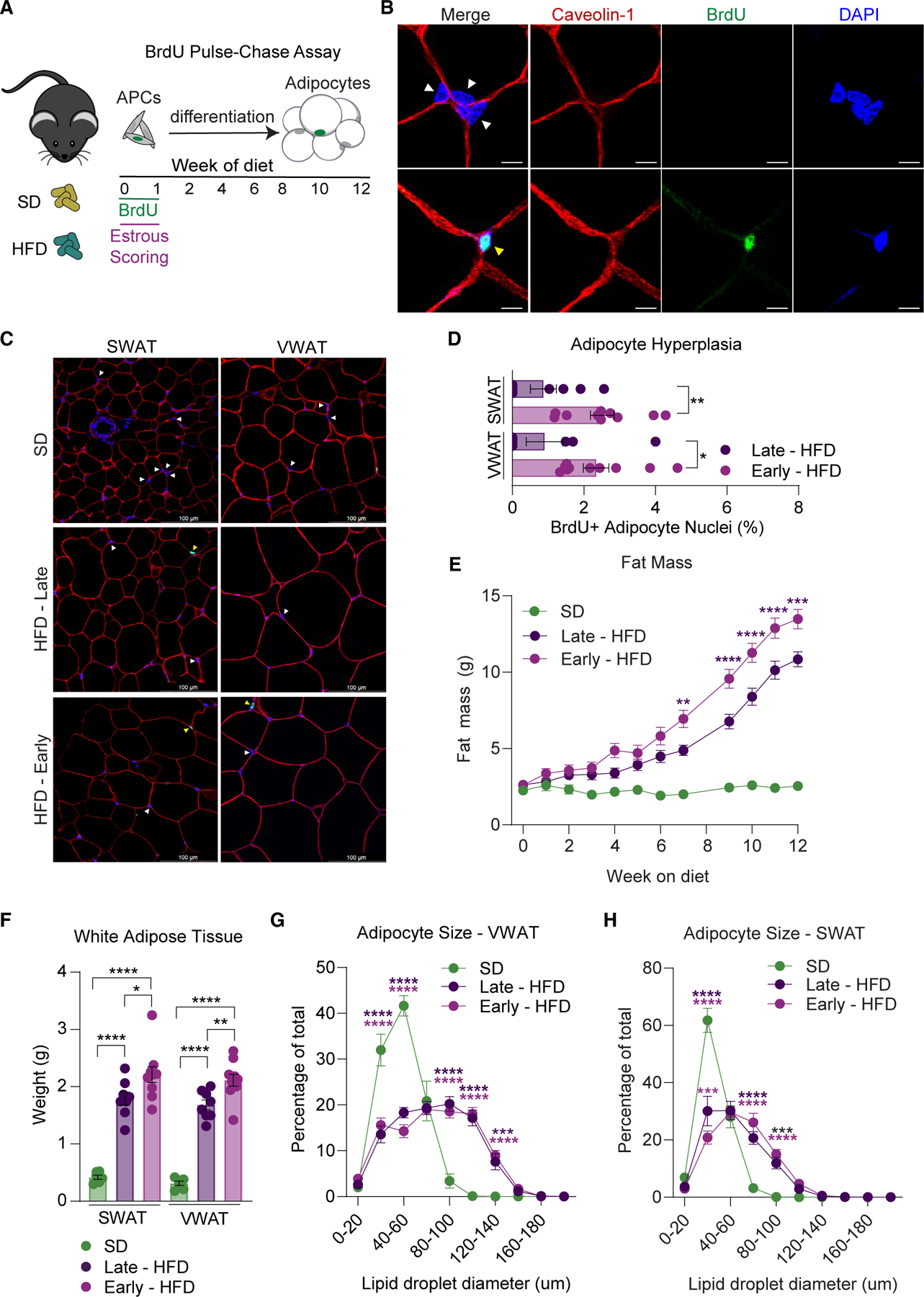
(A) Schematic of BrdU pulse-chase assay in combination with estrous scoring. Briefly, BrdU was given during the first week of diet along with estrous scoring and then chased for 12 weeks to allow APCs to differentiate into adipocytes.
(B) Representative images of immunofluorescence staining in SWAT to quantify BrdU incorporation in adipocyte nuclei. Tissue is stained for caveolin-1 to visualize adipocyte plasma membranes, DAPI to visualize nuclei, and BrdU. Adipocyte nuclei are indicated with arrowheads (yellow indicates BrdU+ and white indicates BrdU−) and are identified by their location inside the adipocyte plasma membrane. Scale bar is 25 μm.
(C) Representative images of WAT from females fed an SD or HFD for 12 weeks stained with caveolin-1, DAPI, and BrdU. Examples of adipocyte nuclei (white arrows) and BrdU+ adipocyte nuclei (yellow arrows) are shown. 30–35 images were taken per tissue to ensure at least 200 adipocyte nuclei were counted. Scale bar is 100 μm, images taken at 40×.
(D) Adipocyte hyperplasia analysis from female SWAT and VWAT after 12 weeks of HFD feeding.
(E) Total fat mass during 12 weeks of SD or HFD feeding in female mice.
(F) SWAT and VWAT weight after 12 weeks of SD or HFD feeding.
(G and H) Histogram of VWAT (G) and SWAT (H) adipocyte size from females after 12 weeks of feeding.
n = 6–10 mice per group. Statistical significance was determined by unpaired t test for (D), ordinary one-way ANOVA with Tukey’s test for (F), and ordinary two-way ANOVA with Tukey’s test for (E), (G), and (H). Error bars represent mean ± SEM. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. APCs, adipocyte precursor cells; VWAT, perigonadal fat; SWAT, inguinal subcutaneous fat; SD, standard diet; HFD, high-fat diet; BrdU, bromodeoxyuridine.
See also Figures S3 and S4 and Table S1.
DISCUSSION
Previous studies demonstrate that female WAT undergoes significant gene expression changes during the estrous cycle.84 Our findings clearly show that peak physiological levels of estradiol during the estrous cycle contribute to diet-induced obesity by affecting hyperplastic expansion of both visceral and subcutaneous adipose tissue in female mice. Thus, the effect of estradiol in APC response to an HFD is a time-sensitive event dependent on the estrous cycle. Importantly, APC proliferation occurred despite an estradiol-induced decrease in food intake (Figure 3A), indicating that this mechanism is dependent on dietary composition instead of increased caloric intake. As adipocyte number is maintained after weight loss,34,36 our data suggest that the timing of obesogenic stimuli in relation to estradiol exposure may influence long-term weight gain, thereby contributing to obesity.
There are numerous associations between estrogen and adipose mass in women; therefore, these findings have several potential clinical implications. Women with obesity are more likely to have irregular cycles85,86 and higher estradiol than non-obese women.87 In addition, estradiol is elevated during puberty in women,88 which is when there is also a physiological increase in fat mass12,89 and exacerbation of pre-existing obesity.90,91 Similarly, estrogen levels are increased during pregnancy, which is also a major risk factor for developing obesity, with many women retaining significant adipose mass long after giving birth.92 Consistent with our findings here, weight gain during pregnancy does not correlate with changes in caloric intake.13 To date, adipocyte hyperplasia has not been studied in women during key developmental or reproductive events. However, in mice, adipocyte hyperplasia contributes to the establishment of fat mass during puberty.93 Mice also have increased obesity postpartum,94 and HFD feeding during pregnancy exacerbates long-term obesity and metabolic defects.95 As adipocyte hyperplasia is induced by pregnancy in mice,96 it is possible that sustained postpartum weight gain in women is also mediated in part through estrogen-induced adipocyte hyperplasia. Thus, our findings here highlight the need for mechanistic studies in humans to explore the contribution of adipocyte hyperplasia in fat mass expansion during periods of increased estrogen.
Limitations of the study
Due to the nature of this study, all female mice were group housed, and the HFD experimental groups were mixed; therefore, we could not measure precise food intake throughout the study for each group. Although the increased fat mass is due to increase in adipocyte number and not size, other contributors like energy expenditure and nutrient uptake were not measured. Therefore, if differences in food intake or these other parameters are contributing to the difference in fat mass remains unknown and should be further explored. As C57BL/6J female mice lack a menstrual period, the connection to human menstrual cycles and this work is not direct. Lastly, HFD feeding and obesity can impact estrous cycle length. However, to avoid unnecessary stress in this study, vaginal smears were only done for 2 weeks at the beginning of the HFD feeding. Thus, any impact of the increased obesity from estrous-timed HFD initiation on estrogen signal and estrous length is not known.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Matthew S. Rodeheffer (matthew.rodeheffer@yale.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Bulk RNA-seq data have been deposited at GEO with accession number GSE209663 and are publicly available as of the date of publication. Accession number is also listed in the key resources table. All other data reported in this paper will be shared by the lead contact upon request.
This article does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| CD45 APC-eFluor 780 | eBioscience | Cat# 47–0451–80; RRID: AB_1548790 |
| CD31 PE-Cy7 | eBioscience | Cat# 25–0311–82; RRID: AB_2716949 |
| CD29 Alexa Fluor 700 | BioLegend | Cat# 102218; RRID: AB_493711 |
| Sca-1 V500 | BD Horizon | Cat# 561228; RRID: AB_10584334 |
| BrdU- Alexa Fluor 647 | Phoenix Flow Systems | Cat# AX647 |
| CD34 Brilliant Violet 421 | BioLegend | Cat# 119321; RRID: AB_10900980 |
| CD24 PerCP-Cyanine 5.5 | eBioscience | Cat# 45–0242-80; RRID: AB_1210702 |
| rat anti-BrdU | Abcam | Cat# ab6326; RRID: AB_305426 |
| rabbit anti-Caveolin-1 | Cell Signaling | Cat# 3238; RRID: AB_2072166 |
| anti-rabbit rhodamine-X-red | Jackson ImmunoResearch Labs | Cat# 111–295-144; RRID: AB_2338028 |
| goat anti-rat-A488 | Jackson ImmunoResearch Labs | Cat# 111–295-144; RRID: AB_2338028 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| 60% Lard High-Fat Diet (HFD) | Research Diets | Cat# D12492 |
| Chow diet (SD) | Harlan Laboratories | Cat# 2018S |
| DPBS | Life Technologies | Cat# 14190144 |
| TRIzol™ Reagent | Invitrogen | Cat# 15596018 |
| Collagenase Type II | Worthington Biochemical | Cat# LS004174 |
| Bovine Serum Albumin | AmericanBio | Cat# AB01088 |
| HBSS | Gibco | Cat# 14185–052 |
| Phosflow™ Lyse/Fix Buffer | BD Biosciences | Cat# 558049 |
| Perm Buffer III | BD Biosciences | Cat# 558050 |
| Deoxyribonuclease I | Worthington | Cat# LS002007 |
| DPBS with MgCl2 and CaCl2 | Sigma | Cat# D8662 |
| NucRed™ Dead 647 | Invitrogen | Cat# R37113 |
| 0.1% Crystal Violet Aqueous Solution | Ward’s Science | Cat# 470300–938 |
| BrdU | US Biological | Cat# B2850 |
| 17β-estradiol | Sigma | Cat# E2758 |
| Hydroxypropyl-β-cyclodextrin | Cayman | Cat# 16169 |
| Isoflurane | Covetrus | Cat# 11695067772 |
| DAPI Fluoromount-G | Southern Biotech | Cat# 0100–20 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Direct-zol RNA Miniprep Kit | Zymo Research Corporation | Cat# R2052 |
| KAPA mRNA HyperPrep Kit | Roche | Cat# KR1352 |
| KAPA Library Quantification Kit | Roche | Cat# KK4854 |
| High-Capacity cDNA reverse Transcription Kit | Applied Biosystems | Cat# 4368814 |
| SYBR FAST quantitative PCR kit | Kapa Biosystems | Cat# KK4611 |
|
| ||
| Deposited data | ||
|
| ||
| Raw bulk RNA-sequencing data | GEO | GSE209663 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6J | Jackson Laboratories | Cat# 000664; RRID: IMSR_JAX:000664 |
| C57BL/6-Tg(Pdgfra-cre)1Clc/J | Jackson Laboratories | Cat# 013148; RRID: IMSR_JAX:013148 |
| B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | Jackson Laboratories | Cat# 007676; RRID: IMSR_JAX:007676 |
| Esr1flox/flox | Feng et al.97 | https://doi.org/10.1073/pnas.0706933104 |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S1 for qPCR primer sequences. | ||
|
| ||
| Software and algorithms | ||
|
| ||
| Prism version 9.3.1 | GraphPad | N/A |
| Echo MRI | Echo Medical System | N/A |
| TopHat | Trapnell et al.98 | https://doi.org/10.1093/bioinformatics/btp120 |
| HTSeq package | Anders et al.99 | https://doi.org/10.1093/bioinformatics/btu638 |
| DESeq2 | Love et al.100 | https://doi.org/10.1186/s13059-014-0550-8 |
| Ingenuity Pathway Analysis | QIAGEN Inc. | https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis |
| Gene Ontology | Ashburner et al.101 | https://doi.org/10.1038/75556 |
| KEGG | Kanehisa Laboratories102 | https://doi.org/10.1093/nar/28.1.27 |
| FlowJo™ Software v10.8.1 | BD Life Sciences | N/A |
| CellProfiler™ | Carpenter et al.103 | https://doi.org/10.1186/gb-2006-7-10-r100 |
| BD FACSDiva Software | BD Life Sciences | N/A |
|
| ||
| Other | ||
|
| ||
| 40μm filters | BD Falcon | Cat# 352340 |
| EasyTouch Glucose Monitoring System | EasyTouch | Cat# 807001 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The Institutional Animal Care and Use Committee (IACUC) at Yale University approved all animal studies. All animals in this study were group housed in temperature and humidity-controlled rooms on a 12-h:12-h light:dark cycle, with lights on from 7:00 a.m. to 7:00 p.m. Unless otherwise noted, mice were kept on chow diet from Harlan Laboratories (2018S). All mice used for these studies were on the C57BL/6J (no. 000664; RRID: IMSR_JAX:000664) genetic background. Pdgfrα-cre mice (no. 013148; RRID: IMSR_JAX:013148) and mTmG mice (no. 007676; RRID: IMSR_JAX:007676) were purchased from Jackson Laboratories. Esr1fl mice97 was a generous gift from Dr. Sean Morrison (UT Southwestern, Dallas, TX, USA). Breeding was done in the Yale Animal Resource Center and mice were weaned at p21. Unless otherwise noted, mice were females 6–8 weeks of age at the start of experiments. VWAT refers to the perigonadal WAT and SWAT refers to the inguinal WAT in mice. Body composition measures were done with NMR using the Echo MRI whole body composition analyzer (Echo Medical System, Houston, TX). High-fat diet (HFD) is from Research Diets (no. D12492). Standard diet (SD) is from Harlan Laboratories (no. 2018S).
METHOD DETAILS
RNA-sequencing and differential gene expression analysis
Female mice were fed an SD or HFD for 3 days after which VWAT and SWAT were excised for APC isolation via FACS. Each of the 5 samples was pooled from 3 mice. Sorted APCs were washed 2X with DPBS (Life Technologies no. 14190144) and the pellet resuspended in TRIzol Reagent (Invitrogen no. 15596018), vortexed, and flash-frozen in liquid nitrogen. RNA was isolated using Direct-zol RNA Miniprep Kit (Zymo Research no. R2052), according to manufacturers’ instructions. RNA was quantified by a nanodrop spectrophotometer (Thermo Fisher Scientific). RNA samples were sent to the Yale Center for Genome Analysis (YCGA). Total RNA quality was determined by estimating the A260/A280 and A260/A230 ratios by nanodrop. RNA integrity was determined by running an Agilent Bioanalyzer gel, which measures the ratio of the ribosomal peaks. Samples with RIN values of 7 or greater were used for library prep. For library prep, mRNA was purified from approximately 200ng of total RNA with oligo-dT beads and sheared by incubation at 94C in the presence of Mg2+ (Roche Kapa mRNA Hyperprep no. KR1352). Following first-strand synthesis with random primers, second strand synthesis and A-tailing was performed with dUTP for generating strand-specific sequencing libraries. Adapter ligation with 3′ dTMP overhangs were ligated to library insert fragments. Library amplification amplified fragments carrying the appropriate adapter sequences at both ends. Strands marked with dUTP were not amplified. Indexed libraries were quantified by qRT-PCR using a commercially available kit (Roche KAPA Biosystems no. KK4854) and insert size distribution determined by either the Agilent Bioanalyzer. Samples with a yield of ≥0.5 ng/ul and a size distribution of 150–300bp were used for sequencing. For flow cell preparation and sequencing, sample concentrations were normalized to 15 pM and loaded onto an Illumina HiSeq 2500 V4 flow cell at a concentration that yields 25 million passing filter clusters per sample. Samples were sequenced using 75 bp single-end sequencing according to Illumina protocols. The 6 bp index is read during additional sequencing reads that automatically follow the completion of read 1. Data generated during sequencing runs are simultaneously transferred to the YCGA high-performance computing cluster. A positive control (prepared bacteriophage Phi X library) provided by Illumina was spiked into every lane at a concentration of 0.3% to monitor sequencing quality in real time. Signal intensities were converted to individual base calls during a run using the system’s Real Time Analysis (RTA) software. Base calls were transferred from the machine’s dedicated personal computer to the Yale High Performance Computing cluster via a 1 Gigabit network mount for downstream analysis. Primary analysis - sample de-multiplexing and alignment to the mouse genome - was performed using Illumina’s CASAVA 1.8.2 software suite. Only samples with an error rate less than 2% and with a distribution of reads per sample in a lane within reasonable tolerance were further processed. Reads were aligned to reference genome mm9 with TopHat.98 The python module HTSeq99 was used with preset parameters to quantify gene expression. Genes that had less than 20 reads in all samples were filtered out. DESeq2100 was used with preset parameters to identify differentially expressed genes (DEGs) between standard diet (SD) and high-fat diet (HFD) conditions within each fat depot. Diet-induced DEGs were identified by adjusted p value <0.01 and log2FC > 1 or < −1. Canonical pathways and upstream regulator analysis was done with Ingenuity Pathway Analysis (IPA) software (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis) using FDR <0.05 and FC > 2 or < −2. Gene Ontology101 and KEGG102 were also used to analyze biological functions and pathways from gene lists.
Flow cytometry
Flow cytometry was performed as described previously.19–21 All centrifugation was done for 3 min at 300 × g. Briefly, mice were euthanized by cervical dislocation and adipose tissues were excised, minced, and digested for 1 h in a shaking water bath (130 rpm) at 37°C with 0.8 mg/mL collagenase type II (Worthington Biochemical no. LS004174) in buffer (3% BSA in 1X HBSS (Gibco no. 14185–052) pH 7.2) supplemented with 0.8 mM ZnCl2, 1.0 mM MgCl2 and 1.2 mM CaCl2 (ion supplementation is only used during digestion). After digestion, samples were passed through 40μm filters (BD Falcon no. 352340), centrifuged, and lipid layer aspirated to remove adipocytes and further process the stromal-vascular fraction (SVF). SVF was then incubated protected from light for 30 min on ice with buffer solution the following antibodies: CD45 APC-eFluor 780 (eBioscience no. 47–0451–80) at 1:1000, CD31 PE-Cy7 (eBioscience no. 25–0311–82) at 1:500, CD29 Alexa Fluor 700 (BioLegend no. 102218) at 1:400, and Sca-1 V500 (BD Horizon no. 561228) at 1:300. SVF was washed with buffer, centrifuged, then fixed and permeabilized using Phosflow Lyse/Fix (BD Biosciences no. 558049) and Perm Buffer III (BD Biosciences no. 558050) according to the manufacturer’s recommendations. Cells were then treated with DNase (deoxyribonuclease I; Worthington no. LS002007; × 200 units/ml) in DPBS (Sigma no. D8662; with CaCl2 and MgCl2) for 2 h in a shaking water bath 37 °C at 40 rpm. Cells were then washed in buffer, centrifuged, and washed again with DBPS (no ions). Cells were then stained with BrdU- Alexa Fluor 647 antibody (Phoenix Flow Systems no. AX647) at 1:30 in buffer overnight in the dark at 4°C. Cells were then washed in buffer and incubated with the same antibodies as pre-fixation plus CD34 Brilliant Violet 421 (BioLegend no. 119321) at 1:400 and CD24 PerCP-Cyanine 5.5 (eBioscience no. 45–0242–80) at 1:250. Following antibody incubation, samples were washed with buffer and analyzed on a BD LSRII analyzer. Data analysis was performed using BD FACS Diva software and FlowJo Software (BD Life Sciences).
FACS analysis
Samples were collected and prepped as described for flow cytometry with the following changes: after digestion, samples were passed through a 200μm filter, centrifuged, and lipid layer aspirated to remove adipocytes. Cells were resuspended in buffer (no ions), passed through 40 μm filters and centrifuged. Cells were incubated on ice for 30–60 min in buffer containing the following antibodies: CD45 APC-eFluor 780 at 1:1000, CD31 PE-Cy7 at 1:500, CD29 Alexa Fluor 700 at 1:400, CD34 Brilliant Violet 421 at 1:400, CD24 PerCP-Cyanine 5.5 at 1:250, and Sca-1 V500 at 1:300. Cells were washed in buffer, centrifuged, and resuspended in buffer solution with 1:600 NucRed™ Dead 647 (Invitrogen no. R37113) to identify live cells.
Vaginal smears
Smears were taken daily at the start of the light cycle (7 am). Female mice were acclimated to the procedure for one week before the start of experiments. To collect the sample, a latex bulb fitted with a filter tip containing 100 μL of ddH2O was used to repeatedly flush the vaginal canal of female mice with 25–50 μL of volume at a time. The tip was placed right at the opening but not inserted into the vaginal canal to avoid potential pseudopregnancy. The fluid was placed on a glass slide and allowed to dry completely at room temperature. Once dried, the slides were stained with 0.1% crystal violet aqueous solution (Ward’s Science no. 470300–938) for 1 min at room temperature and washed 2X with ddH2O. Once dried, glycerol was used to mount the coverslip for immediate imaging in a Keyence brightfield microscope at 20X. Estrous cycle stage was determined by assessing the ratios of nucleated epithelial cells, cornified squamous epithelial cells, and leukocytes present in the smear: Proestrus (predominant nucleated epithelial cells), Estrus (predominant cornified squamous epithelial cells), Metestrus (predominant leukocytes), and Diestrus (predominant leukocytes but nucleated and cornified squamous epithelial cells are present).
BrdU treatments
For BrdU experiments, BrdU (US Biological no. B2850) was given in the drinking water at 0.8 mg/mL for experiments lasting one week or less and 0.4 mg/mL for experiments lasting more than one week. BrdU water was kept in dark bottles and replenished every 48 h.
Estradiol treatment
17β-estradiol (Sigma no. E2758) was dissolved in 45% Hydroxypropyl-β-cyclodextrin (Cayman no. 16169) in DPBS. 17β-estradiol was given at 1 mg/kg daily with intraperitoneal injection during day 0–2 of the HFD feeding. Females were acclimated to IP injections with PBS before the start of the experiment. Food was weighed daily and calories calculated as an average per mice per cage (SD: 3.1 kcal per gram, HFD: 5.2 kcal per gram).
APC transplant assay
For cell transplant assays, VWAT and SWAT of AP-ERα-KO animals were pooled and APCs were isolated by FACS and transplanted in SWAT as previously described.19,25 Recipient female C57BL/6J mice were anesthetized with isoflurane (Covetrus no. 11695067772) and surgeries performed using sterile technique. 0.5–1 million ERα-KO APCs were re-suspended in 15 μL of DPBS and injected into the left SWAT of 4–5-week-old congenic wildtype female mice. A control DPBS injection was given in the right SWAT. Mice were allowed to recover for 2 weeks. Vaginal smears were done daily one week prior to the addition of an HFD to acclimate them. Estrous cycle was tracked as described previously and only females in proestrus at the onset of the HFD were included. Left and right SWAT tissues were collected and analyzed separately via flow cytometry for incorporation with BrdU. ERα-KO APCs were identified by GFP fluorescence. Results were counted only for transplants in which more than 100 individual donor APCs were recovered in the recipient depot.
Confocal microscopy
Adipose tissue was collected and prepared for paraffin-embedded tissue as previously described.21,104 Briefly, tissues were fixed in zinc formaldehyde for 24–48 hours at 4°C. Tissues were washed in DPBS and dehydrated in increasing concentrations of ethanol over 6 hours and embedded in paraffin wax. Sections (5 μm) from the paraffin blocks were deparaffinized, rehydrated, and incubated under pressure (2100 Retriever from PickCell Laboratories) in 10 mM sodium citrate (pH 6.0) for antigen retrieval. Tissues were blocked and stained with 2 % BSA in DPBS. Blocking was done at RT for 30 minutes. Primary antibodies used were rat anti-BrdU (Abcam no. ab6326, clone no. BU1/75 (ICR1)) at 1:350 and rabbit anti-Caveolin-1 (Cell Signaling no. 3238) at 1:400 overnight at 4°C. Tissues were then washed and stained with secondary antibodies (1:250) including goat anti-rabbit rhodamine-X-red (Jackson Immunoresearch no. 111–295-144) and goat anti-rat-A488 (Jackson Immunoresearch no. 112-545-167) for 3 hours at RT. Slides were mounted with DAPI Fluoromount-G mounting media (Southern Biotech no. 0100–20) and imaged with Leica TCS SP5 or Leica STELLARIS 5 confocal microscopes. For adipocyte nuclei analysis, 20–30 images for every tissue section were acquired at 40X. Quantification of BrdU in adipocyte nuclei was done as previously described21 where adipocyte nuclei were identified as those inside intact membranes (stained with Caveolin-1) as shown in Figure 4B. At least 50 adipocyte nuclei were scored for each animal. For adipocyte diameter measurements, the area of each adipocyte (in square pixels) was measured using CellProfiler™.103 The diameter of each adipocyte was calculated using the measured area, assuming each adipocyte is a perfect circle. At least 200 adipocytes were measured for each animal.
Glucose tolerance test
Mice were fasted overnight and fasting blood glucose measured by tail nick incision using the EasyTouch glucose monitoring system (no. 807001). 2 g/kg of glucose was given as a 20 % glucose solution in 0.9 % saline via intraperitoneal injection. Blood (<5 μl) was collected via the tail nick incision 10, 20, 30, 60, and 120-minutes post glucose injection. This was performed after 12 weeks of SD or HFD.
RNA extraction and cDNA synthesis
For gene expression analysis, whole tissues were collected and stored at −80C until RNA extraction was performed. RNA was isolated using Direct-zol RNA Miniprep Kit (Zymo Research Corporation no. R2052), according to manufacturers’ instructions. RNA was quantified by a nanodrop spectrophotometer (Thermo Fisher Scientific) and single stranded cDNA was synthesized from total RNA using the High-Capacity cDNA reverse Transcription Kit (Applied Biosystems, Life Technologies no. 4368814) according to the manufacturer’s instructions.
Quantitative real-time PCR
Quantitative RT-PCR (qPCR) was performed on the cDNA using the Sybr green method of quantification on a Roche Lightcycler 480 using a SYBR FAST quantitative PCR kit (Kapa Biosystems no. KK4611). Gene expression was analyzed for each sample in triplicate using the primers described in Table S1. Tbk1 was used as housekeeping gene. For each experiment, cDNA samples were pooled and a standard curve was generated to quantify relative mRNA transcript levels.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses are described in each figure legend. All tests were performed using GraphPad Prism version 9.3.1. Data are presented as mean ± S.E.M. and p<0.05 was considered statistically significant. A minimum of 5 animals were used for each experiment, unless statistical significance was reached with fewer animals. Sample size is indicated in each figure legend (“n”). Experiments were not blinded, as groupings and genotypes of mice were known prior to analysis.
Supplementary Material
Highlights.
HFD upregulates estrogen pathways in female APCs
Timing of HFD initiation in relation to the estrous cycle affects APC proliferation
ERα signaling is necessary for HFD-induced APC proliferation
Estrous cycle stage at the onset of HFD impacts adipocyte formation and obesity
ACKNOWLEDGMENTS
We thank Jasper Martin Anton de Jong for critically reviewing the manuscript. We thank Ewa Menet and Zhao Zhao at the Yale Flow Cytometry Facility for their technical assistance. The graphical abstract was created with BioRender.com. This work was supported by NIDDK grants DK090489 and DK132563 and the Naratil Pioneer Award from the Women’s Health Research at Yale to M.S.R.; NICHD grant HD097368 to C.F.; the Science, Technology and Research Scholars (STARS) Program at Yale to N.T.; and the National Science Foundation Graduate Research Fellowship (NSF-GRFP) and Ford Foundation Predoctoral Fellowship to R.d.M.S.-P.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112390.
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research. One or more of the authors of this paper received support from a program designed to increase minority representation in their field of research.
REFERENCES
- 1.Hong J, Stubbins RE, Smith RR, Harvey AE, and Núñez NP (2009). Differential susceptibility to obesity between male, female and ovariectomized female mice. Nutr. J 8, 11. 10.1186/1475-2891-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stubbins RE, Holcomb VB, Hong J, and Núñez NP (2012). Estrogen modulates abdominal adiposity and protects female mice from obesity and impaired glucose tolerance. Eur. J. Nutr 51, 861–870. 10.1007/s00394-011-0266-4. [DOI] [PubMed] [Google Scholar]
- 3.Toth MJ, Tchernof A, Sites CK, and Poehlman ET (2000). Effect of menopausal status on body composition and abdominal fat distribution. Int. J. Obes. Relat. Metab. Disord 24, 226–231. 10.1038/sj.ijo.0801118. [DOI] [PubMed] [Google Scholar]
- 4.Mauvais-Jarvis F, Clegg DJ, and Hevener AL (2013). The role of estrogens in control of energy balance and glucose homeostasis. Endocr. Rev 34, 309–338. 10.1210/er.2012-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baird DT, and Fraser IS (1974). Blood production and ovarian secretion rates of estradiol-17 beta and estrone in women throughout the menstrual cycle. J. Clin. Endocrinol. Metab 38, 1009–1017. 10.1210/jcem-38-6-1009. [DOI] [PubMed] [Google Scholar]
- 6.Gruber CJ, Tschugguel W, Schneeberger C, and Huber JC (2002). Production and actions of estrogens. N. Engl. J. Med 346, 340–352. 10.1056/NEJMra000471. [DOI] [PubMed] [Google Scholar]
- 7.Clegg DJ, Brown LM, Woods SC, and Benoit SC (2006). Gonadal hormones determine sensitivity to central leptin and insulin. Diabetes 55, 978–987. 10.2337/diabetes.55.04.06.db05-1339. [DOI] [PubMed] [Google Scholar]
- 8.Barr SI, Janelle KC, and Prior JC (1995). Energy intakes are higher during the luteal phase of ovulatory menstrual cycles. Am. J. Clin. Nutr 61, 39–43. 10.1093/ajcn/61.1.39. [DOI] [PubMed] [Google Scholar]
- 9.Heine PA, Taylor JA, Iwamoto GA, Lubahn DB, and Cooke PS (2000). Increased adipose tissue in male and female estrogen receptor-alpha knockout mice. Proc. Natl. Acad. Sci. USA 97, 12729–12734. 10.1073/pnas.97.23.12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villa A, Della Torre S, Stell A, Cook J, Brown M, and Maggi A (2012). Tetradian oscillation of estrogen receptor alpha is necessary to prevent liver lipid deposition. Proc. Natl. Acad. Sci. USA 109, 11806–11811. 10.1073/pnas.1205797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Correa SM, Newstrom DW, Warne JP, Flandin P, Cheung CC, Lin-Moore AT, Pierce AA, Xu AW, Rubenstein JL, and Ingraham HA (2015). An estrogen-responsive module in the ventromedial hypothalamus selectively drives sex-specific activity in females. Cell Rep 10, 62–74. 10.1016/j.celrep.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogol AD, Roemmich JN, and Clark PA (2002). Growth at puberty. J. Adolesc. Health 31, 192–200. 10.1016/S1054-139X(02)00485-8. [DOI] [PubMed] [Google Scholar]
- 13.Jebeile H, Mijatovic J, Louie JCY, Prvan T, and Brand-Miller JC (2016). A systematic review and metaanalysis of energy intake and weight gain in pregnancy. Am. J. Obstet. Gynecol 214, 465–483. 10.1016/j.ajog.2015.12.049. [DOI] [PubMed] [Google Scholar]
- 14.Catov JM, Sun B, Lewis CE, Bertolet M, and Gunderson EP (2022). Prepregnancy weight change associated with high gestational weight gain. Obesity 30, 524–534. 10.1002/oby.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtis Hewitt S, Couse JF, and Korach KS (2000). Estrogen receptor transcription and transactivation: estrogen receptor knockout mice: what their phenotypes reveal about mechanisms of estrogen action. Breast Cancer Res 2, 345–352. 10.1186/bcr79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gustafsson KL, Farman H, Henning P, Lionikaite V, Movérare-Skrtic S, Wu J, Ryberg H, Koskela A, Gustafsson JÅ, Tuukkanen J, et al. (2016). The role of membrane ERalpha signaling in bone and other major estrogen responsive tissues. Sci. Rep 6, 29473. 10.1038/srep29473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kauffman AS, Thackray VG, Ryan GE, Tolson KP, Glidewell-Kenney CA, Semaan SJ, Poling MC, Iwata N, Breen KM, Duleba AJ, et al. (2015). A novel letrozole model recapitulates both the reproductive and metabolic phenotypes of polycystic ovary syndrome in female mice. Biol. Reprod 93, 69. 10.1095/biolreprod.115.131631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Nedungadi TP, Zhu L, Sobhani N, Irani BG, Davis KE, Zhang X, Zou F, Gent LM, Hahner LD, et al. (2011). Distinct hypothalamic neurons mediate estrogenic effects on energy homeostasis and reproduction. Cell Metab 14, 453–465. 10.1016/j.cmet.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saavedra Pena RDM, Taylor N, and Rodeheffer MS (2022). Insights of the role of estrogen in obesity from two models of ERalpha deletion. J. Mol. Endocrinol 68, 179–194. 10.1530/JME-21-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berry R, and Rodeheffer MS (2013). Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol 15, 302–308. 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffery E, Church CD, Holtrup B, Colman L, and Rodeheffer MS (2015). Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat. Cell Biol 17, 376–385. 10.1038/ncb3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang QA, Tao C, Gupta RK, and Scherer PE (2013). Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med 19, 1338–1344. 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arner P, Bernard S, Salehpour M, Possnert G, Liebl J, Steier P, Buchholz BA, Eriksson M, Arner E, Hauner H, et al. (2011). Dynamics of human adipose lipid turnover in health and metabolic disease. Nature 478, 110–113. 10.1038/nature10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang QA, Tao C, Jiang L, Shao M, Ye R, Zhu Y, Gordillo R, Ali A, Lian Y, Holland WL, et al. (2015). Distinct regulatory mechanisms governing embryonic versus adult adipocyte maturation. Nat. Cell Biol 17, 1099–1111. 10.1038/ncb3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeffery E, Wing A, Holtrup B, Sebo Z, Kaplan JL, Saavedra-Peña R, Church CD, Colman L, Berry R, and Rodeheffer MS (2016). The adipose tissue microenvironment regulates depot-specific adipogenesis in obesity. Cell Metab 24, 142–150. 10.1016/j.cmet.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sebo ZL, Jeffery E, Holtrup B, and Rodeheffer MS (2018). A mesodermal fate map for adipose tissue. Development 145, dev166801. 10.1242/dev.166801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez-Gurmaches J, and Guertin DA (2014). Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nat. Commun 5, 4099. 10.1038/ncomms5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Gurmaches J, and Guertin DA (2014). Adipocyte lineages: tracing back the origins of fat. Biochim. Biophys. Acta 1842, 340–351. 10.1016/j.bbadis.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sebo ZL, and Rodeheffer MS (2022). Prepubertal androgen signaling is required to establish male fat distribution. Stem Cell Rep 17, 1081–1088. 10.1016/j.stemcr.2022.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmer BF, and Clegg DJ (2015). The sexual dimorphism of obesity. Mol. Cell. Endocrinol 402, 113–119. 10.1016/j.mce.2014.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zamboni M, Armellini F, Milani MP, De Marchi M, Todesco T, Robbi R, Bergamo-Andreis IA, and Bosello O (1992). Body fat distribution in pre- and post-menopausal women: metabolic and anthropometric variables and their inter-relationships. Int. J. Obes. Relat. Metab. Disord 16, 495–504. [PubMed] [Google Scholar]
- 32.Björkelund C, Lissner L, Andersson S, Lapidus L, and Bengtsson C (1996). Reproductive history in relation to relative weight and fat distribution. Int. J. Obes. Relat. Metab. Disord 20, 213–219. [PubMed] [Google Scholar]
- 33.Yonezawa R, Wada T, Matsumoto N, Morita M, Sawakawa K, Ishii Y, Sasahara M, Tsuneki H, Saito S, and Sasaoka T (2012). Central versus peripheral impact of estradiol on the impaired glucose metabolism in ovariectomized mice on a high-fat diet. Am. J. Physiol. Endocrinol. Metab 303, E445–E456. 10.1152/ajpendo.00638.2011. [DOI] [PubMed] [Google Scholar]
- 34.Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, Bergmann O, Blomqvist L, Hoffstedt J, Näslund E, Britton T, et al. (2008). Dynamics of fat cell turnover in humans. Nature 453, 783–787. 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 35.Hansson B, Morén B, Fryklund C, Vliex L, Wasserstrom S, Albinsson S, Berger K, and Stenkula KG (2019). Adipose cell size changes are associated with a drastic actin remodeling. Sci. Rep 9, 12941. 10.1038/s41598-019-49418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arner P, and Rydén M (2022). Human white adipose tissue: a highly dynamic metabolic organ. J. Intern. Med 291, 611–621. 10.1111/joim.13435. [DOI] [PubMed] [Google Scholar]
- 37.Hansson B, Wasserstrom S, Morén B, Periwal V, Vikman P, Cushman SW, Göransson O, Storm P, and Stenkula KG (2018). Intact glucose uptake despite deteriorating signaling in adipocytes with high-fat feeding. J. Mol. Endocrinol 60, 199–211. 10.1530/JME-17-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao M, Hepler C, Zhang Q, Shan B, Vishvanath L, Henry GH, Zhao S, An YA, Wu Y, Strand DW, and Gupta RK (2021). Pathologic HIF1alpha signaling drives adipose progenitor dysfunction in obesity. Cell Stem Cell 28, 685–701.e7. 10.1016/j.stem.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwalie PC, Dong H, Zachara M, Russeil J, Alpern D, Akchiche N, Caprara C, Sun W, Schlaudraff KU, Soldati G, et al. (2018). A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature 559, 103–108. 10.1038/s41586-018-0226-8. [DOI] [PubMed] [Google Scholar]
- 40.Sun W, Dong H, Balaz M, Slyper M, Drokhlyansky E, Colleluori G, Giordano A, Kovanicova Z, Stefanicka P, Balazova L, et al. (2020). snRNA-seq reveals a subpopulation of adipocytes that regulates thermogenesis. Nature 587, 98–102. 10.1038/s41586-020-2856-x. [DOI] [PubMed] [Google Scholar]
- 41.Nahmgoong H, Jeon YG, Park ES, Choi YH, Han SM, Park J, Ji Y, Sohn JH, Han JS, Kim YY, et al. (2022). Distinct properties of adipose stem cell subpopulations determine fat depot-specific characteristics. Cell Metab 34, 458–472.e6. 10.1016/j.cmet.2021.11.014. [DOI] [PubMed] [Google Scholar]
- 42.Emont MP, Jacobs C, Essene AL, Pant D, Tenen D, Colleluori G, Di Vincenzo A, Jørgensen AM, Dashti H, Stefek A, et al. (2022). A single-cell atlas of human and mouse white adipose tissue. Nature 603, 926–933. 10.1038/s41586-022-04518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merrick D, Sakers A, Irgebay Z, Okada C, Calvert C, Morley MP, Percec I, and Seale P (2019). Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 364, eaav2501. 10.1126/science.aav2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hepler C, Shan B, Zhang Q, Henry GH, Shao M, Vishvanath L, Ghaben AL, Mobley AB, Strand D, Hon GC, and Gupta RK (2018). Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife 7, e39636. 10.7554/eLife.39636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vijay J, Gauthier MF, Biswell RL, Louiselle DA, Johnston JJ, Cheung WA, Belden B, Pramatarova A, Biertho L, Gibson M, et al. (2020). Single-cell analysis of human adipose tissue identifies depot and disease specific cell types. Nat. Metab 2, 97–109. 10.1038/s42255-019-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berry R, Jeffery E, and Rodeheffer MS (2014). Weighing in on adipocyte precursors. Cell Metab 19, 8–20. 10.1016/j.cmet.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jo J, Gavrilova O, Pack S, Jou W, Mullen S, Sumner AE, Cushman SW, and Periwal V (2009). Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth. PLoS Comput. Biol 5, e1000324. 10.1371/journal.pcbi.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fukumura D, Ushiyama A, Duda DG, Xu L, Tam J, Krishna V, Chatterjee K, Garkavtsev I, and Jain RK (2003). Paracrine regulation of angiogenesis and adipocyte differentiation during in vivo adipogenesis. Circ. Res 93, e88–e97. 10.1161/01.RES.0000099243.20096.FA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao Y (2007). Angiogenesis modulates adipogenesis and obesity. J. Clin. Invest 117, 2362–2368. 10.1172/JCI32239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Claffey KP, Wilkison WO, and Spiegelman BM (1992). Vascular endothelial growth factor. Regulation by cell differentiation and activated second messenger pathways. J. Biol. Chem 267, 16317–16322. [PubMed] [Google Scholar]
- 51.Vazquez-Martin A, Ortega-Delgado FJ, Fernandez-Real JM, and Menendez JA (2008). The tyrosine kinase receptor HER2 (erbB-2): from oncogenesis to adipogenesis. J. Cell. Biochem 105, 1147–1152. 10.1002/jcb.21917. [DOI] [PubMed] [Google Scholar]
- 52.Pagano E, Coso O, and Calvo JC (2008). Down-modulation of erbB2 activity is necessary but not enough in the differentiation of 3T3-L1 preadipocytes. J. Cell. Biochem 104, 274–285. 10.1002/jcb.21621. [DOI] [PubMed] [Google Scholar]
- 53.Noack C, Hempel U, Preissler C, and Dieter P (2015). Prostaglandin E2 impairs osteogenic and facilitates adipogenic differentiation of human bone marrow stromal cells. Prostaglandins Leukot. Essent. Fatty Acids 94, 91–98. 10.1016/j.plefa.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 54.Pamir N, Liu NC, Irwin A, Becker L, Peng Y, Ronsein GE, Bornfeldt KE, Duffield JS, and Heinecke JW (2015). Granulocyte/macrophage colony-stimulating factor-dependent dendritic cells restrain lean adipose tissue expansion. J. Biol. Chem 290, 14656–14667. 10.1074/jbc.M115.645820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakae J, Kitamura T, Kitamura Y, Biggs WH 3rd, Arden KC, and Accili D (2003). The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell 4, 119–129. 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- 56.Rodeheffer MS, Birsoy K, and Friedman JM (2008). Identification of white adipocyte progenitor cells in vivo. Cell 135, 240–249. 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 57.Millour J, Constantinidou D, Stavropoulou AV, Wilson MSC, Myatt SS, Kwok JMM, Sivanandan K, Coombes RC, Medema RH, Hartman J, et al. (2010). FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene 29, 2983–2995. 10.1038/onc.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanders DA, Ross-Innes CS, Beraldi D, Carroll JS, and Balasubramanian S (2013). Genome-wide mapping of FOXM1 binding reveals cobinding with estrogen receptor alpha in breast cancer cells. Genome Biol 14, R6. 10.1186/gb-2013-14-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Applanat MP, Buteau-Lozano H, Herve MA, and Corpet A (2008). Vascular endothelial growth factor is a target gene for estrogen receptor and contributes to breast cancer progression. Adv. Exp. Med. Biol 617, 437–444. 10.1007/978-0-387-69080-3_42. [DOI] [PubMed] [Google Scholar]
- 60.Read LD, Keith D Jr., Slamon DJ, and Katzenellenbogen BS (1990). Hormonal modulation of HER-2/neu protooncogene messenger ribonucleic acid and p185 protein expression in human breast cancer cell lines. Cancer Res 50, 3947–3951. [PubMed] [Google Scholar]
- 61.Rajendran RR, Nye AC, Frasor J, Balsara RD, Martini PGV, and Katzenellenbogen BS (2003). Regulation of nuclear receptor transcriptional activity by a novel DEAD box RNA helicase (DP97). J. Biol. Chem 278, 4628–4638. 10.1074/jbc.M210066200. [DOI] [PubMed] [Google Scholar]
- 62.Lim CL, Or YZ, Ong Z, Chung HH, Hayashi H, Shrestha S, Chiba S, Lin F, and Lin VCL (2020). Estrogen exacerbates mammary involution through neutrophil-dependent and -independent mechanism. Elife 9, e57274. 10.7554/eLife.57274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bray PF, Howard TD, Vittinghoff E, Sane DC, and Herrington DM (2007). Effect of genetic variations in platelet glycoproteins Ibalpha and VI on the risk for coronary heart disease events in postmenopausal women taking hormone therapy. Blood 109, 1862–1869. 10.1182/blood-2006-03-013151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolf G, Hildenbrand R, Schwar C, Grobholz R, Kaufmann M, Stutte HJ, Strebhardt K, and Bleyl U (2000). Polo-like kinase: a novel marker of proliferation: correlation with estrogen-receptor expression in human breast cancer. Pathol. Res. Pract 196, 753–759. 10.1016/S0344-0338(00)80107-7. [DOI] [PubMed] [Google Scholar]
- 65.Sasanuma H, Tsuda M, Morimoto S, Saha LK, Rahman MM, Kiyooka Y, Fujiike H, Cherniack AD, Itou J, Callen Moreu E, et al. (2018). BRCA1 ensures genome integrity by eliminating estrogen-induced pathological topoisomerase II-DNA complexes. Proc. Natl. Acad. Sci. USA 115, E10642–E10651. 10.1073/pnas.1803177115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scherbakov AM, Shestakova EA, Galeeva KE, and Bogush TA (2019). BRCA1 and estrogen receptor alpha expression regulation in breast cancer cells. Mol. Biol 53, 442–451. 10.1134/S0026898419030169. [DOI] [PubMed] [Google Scholar]
- 67.Allen E (1922). The oestrous cycle in the mouse. Am. J. Anat 30, 297–371r. [Google Scholar]
- 68.Bronson FHD, Dagg CP, and Snell GD (1966). Reproduction. In Biology of the Laboratory Mouse, Green EL, ed. (Dover Publications, Inc.), pp. 187–204. [Google Scholar]
- 69.Nilsson ME, Vandenput L, Tivesten Å., Norlén AK, Lagerquist MK, Windahl SH, Börjesson AE, Farman HH, Poutanen M, Benrick A, et al. (2015). Measurement of a comprehensive sex steroid profile in rodent serum by high-sensitive gas chromatography-tandem mass spectrometry. Endocrinology 156, 2492–2502. 10.1210/en.2014-1890. [DOI] [PubMed] [Google Scholar]
- 70.Ajayi AF, and Akhigbe RE (2020). Staging of the estrous cycle and induction of estrus in experimental rodents: an update. Fertil. Res. Pract 6, 5. 10.1186/s40738-020-00074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McLean AC, Valenzuela N, Fai S, and Bennett SA (2012). Performing vaginal lavage, crystal violet staining, and vaginal cytological evaluation for mouse estrous cycle staging identification. J. Vis. Exp, e4389. 10.3791/4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lissner L, Stevens J, Levitsky DA, Rasmussen KM, and Strupp BJ (1988). Variation in energy intake during the menstrual cycle: implications for food-intake research. Am. J. Clin. Nutr 48, 956–962. 10.1093/ajcn/48.4.956. [DOI] [PubMed] [Google Scholar]
- 73.Tarttelin MF, and Gorski RA (1971). Variations in food and water intake in the normal and acyclic female rat. Physiol. Behav 7, 847–852. 10.1016/0031-9384(71)90050-3. [DOI] [PubMed] [Google Scholar]
- 74.Krause WC, Rodriguez R, Gegenhuber B, Matharu N, Rodriguez AN, Padilla-Roger AM, Toma K, Herber CB, Correa SM, Duan X, et al. (2021). Oestrogen engages brain MC4R signalling to drive physical activity in female mice. Nature 599, 131–135. 10.1038/s41586-021-04010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Musatov S, Chen W, Pfaff DW, Mobbs CV, Yang XJ, Clegg DJ, Kaplitt MG, and Ogawa S (2007). Silencing of estrogen receptor alpha in the ventromedial nucleus of hypothalamus leads to metabolic syndrome. Proc. Natl. Acad. Sci. USA 104, 2501–2506. 10.1073/pnas.0610787104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Butera PC (2010). Estradiol and the control of food intake. Physiol. Behav 99, 175–180. 10.1016/j.physbeh.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu Q, Li M, Zou Y, and Cao T (2014). Induction of adipocyte hyperplasia in subcutaneous fat depot alleviated type 2 diabetes symptoms in obese mice. Obesity 22, 1623–1631. 10.1002/oby.20705. [DOI] [PubMed] [Google Scholar]
- 78.Shao M, Vishvanath L, Busbuso NC, Hepler C, Shan B, Sharma AX, Chen S, Yu X, An YA, Zhu Y, et al. (2018). De novo adipocyte differentiation from Pdgfrbeta(+) preadipocytes protects against pathologic visceral adipose expansion in obesity. Nat. Commun 9, 890. 10.1038/s41467-018-03196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iwayama T, Steele C, Yao L, Dozmorov MG, Karamichos D, Wren JD, and Olson LE (2015). PDGFRalpha signaling drives adipose tissue fibrosis by targeting progenitor cell plasticity. Genes Dev 29, 1106–1119. 10.1101/gad.260554.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marcelin G, Ferreira A, Liu Y, Atlan M, Aron-Wisnewsky J, Pelloux V, Botbol Y, Ambrosini M, Fradet M, Rouault C, et al. (2017). A PDGFRalpha-mediated switch toward CD9(high) adipocyte progenitors controls obesity-induced adipose tissue fibrosis. Cell Metab 25, 673–685. 10.1016/j.cmet.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 81.Sarvari AK, Van Hauwaert EL, Markussen LK, Gammelmark E, Marcher AB, Ebbesen MF, Nielsen R, Brewer JR, Madsen JGS, and Mandrup S (2021). Plasticity of epididymal adipose tissue in response to diet-induced obesity at single-nucleus resolution. Cell Metab 33, 437–453.e5. 10.1016/j.cmet.2020.12.004. [DOI] [PubMed] [Google Scholar]
- 82.Björntorp P, Carlgren G, Isaksson B, Krotkiewski M, Larsson B, and Sjöström L (1975). Effect of an energy-reduced dietary regimen in relation to adipose tissue cellularity in obese women. Am. J. Clin. Nutr 28, 445–452. 10.1093/ajcn/28.5.445. [DOI] [PubMed] [Google Scholar]
- 83.Kral JG, Björntorp P, Scherstén T, and Sjöström L (1977). Body composition and adipose tissue cellularity before and after jejuno-ileostomy in severely obese subjects. Eur. J. Clin. Invest 7, 413–419. 10.1111/j.1365-2362.1977.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 84.Zhou Y, Yan H, Liu W, Hu C, Zhou Y, Sun R, Tang Y, Zheng C, Yang J, and Cui Q (2022). A multi-tissue transcriptomic landscape of female mice in estrus and diestrus provides clues for precision medicine. Front. Cell Dev. Biol 10, 983712. 10.3389/fcell.2022.983712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roman Lay AA, Pereira A, and Garmendia Miguel ML (2021). Association between obesity with pattern and length of menstrual cycle: the role of metabolic and hormonal markers. Eur. J. Obstet. Gynecol. Reprod. Biol 260, 225–231. 10.1016/j.ejogrb.2021.02.021. [DOI] [PubMed] [Google Scholar]
- 86.Wei S, Schmidt MD, Dwyer T, Norman RJ, and Venn AJ (2009). Obesity and menstrual irregularity: associations with SHBG, testosterone, and insulin. Obesity 17, 1070–1076. 10.1038/oby.2008.641. [DOI] [PubMed] [Google Scholar]
- 87.Yeung EH, Zhang C, Albert PS, Mumford SL, Ye A, Perkins NJ, Wactawski-Wende J, and Schisterman EF (2013). Adiposity and sex hormones across the menstrual cycle: the BioCycle Study. Int. J. Obes 37, 237–243. 10.1038/ijo.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bates K, and Herzog ED (2020). Maternal-fetal circadian communication during pregnancy. Front. Endocrinol 11, 198. 10.3389/fendo.2020.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karastergiou K, Smith SR, Greenberg AS, and Fried SK (2012). Sex differences in human adipose tissues - the biology of pear shape. Biol. Sex Differ 3, 13. 10.1186/2042-6410-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCartney CR, Blank SK, Prendergast KA, Chhabra S, Eagleson CA, Helm KD, Yoo R, Chang RJ, Foster CM, Caprio S, and Marshall JC (2007). Obesity and sex steroid changes across puberty: evidence for marked hyperandrogenemia in pre- and early pubertal obese girls. J. Clin. Endocrinol. Metab 92, 430–436. 10.1210/jc.2006-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jasik CB, and Lustig RH (2008). Adolescent obesity and puberty: the “perfect storm”. Ann. N. Y. Acad. Sci 1135, 265–279. 10.1196/annals.1429.009. [DOI] [PubMed] [Google Scholar]
- 92.Endres LK, Straub H, McKinney C, Plunkett B, Minkovitz CS, Schetter CD, Ramey S, Wang C, Hobel C, Raju T, et al. (2015). Postpartum weight retention risk factors and relationship to obesity at 1 year. Obstet. Gynecol 125, 144–152. 10.1097/AOG.0000000000000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holtrup B, Church CD, Berry R, Colman L, Jeffery E, Bober J, and Rodeheffer MS (2017). Puberty is an important developmental period for the establishment of adipose tissue mass and metabolic homeostasis. Adipocyte 6, 224–233. 10.1080/21623945.2017.1349042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ladyman SR, Khant Aung Z, and Grattan DR (2018). Impact of pregnancy and lactation on the long-term regulation of energy balance in female mice. Endocrinology 159, 2324–2336. 10.1210/en.2018-00057. [DOI] [PubMed] [Google Scholar]
- 95.Qiao L, Chu K, Wattez JS, Lee S, Gao H, Feng GS, Hay WW Jr., and Shao J (2019). High-fat feeding reprograms maternal energy metabolism and induces long-term postpartum obesity in mice. Int. J. Obes 43, 1747–1758. 10.1038/s41366-018-0304-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zwick RK, Rudolph MC, Shook BA, Holtrup B, Roth E, Lei V, Van Keymeulen A, Seewaldt V, Kwei S, Wysolmerski J, et al. (2018). Adipocyte hypertrophy and lipid dynamics underlie mammary gland remodeling after lactation. Nat. Commun 9, 3592. 10.1038/s41467-018-05911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Feng Y, Manka D, Wagner KU, and Khan SA (2007). Estrogen receptor-alpha expression in the mammary epithelium is required for ductal and alveolar morphogenesis in mice. Proc. Natl. Acad. Sci. USA 104, 14718–14723. 10.1073/pnas.0706933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Anders S, Pyl PT, and Huber W (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 25, 25–29. 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kanehisa M, and Goto S (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100. 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Berry R, Church CD, Gericke MT, Jeffery E, Colman L, and Rodeheffer MS (2014). Imaging of adipose tissue. Methods Enzymol 537, 47–73. 10.1016/B978-0-12-411619-1.00004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk RNA-seq data have been deposited at GEO with accession number GSE209663 and are publicly available as of the date of publication. Accession number is also listed in the key resources table. All other data reported in this paper will be shared by the lead contact upon request.
This article does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| CD45 APC-eFluor 780 | eBioscience | Cat# 47–0451–80; RRID: AB_1548790 |
| CD31 PE-Cy7 | eBioscience | Cat# 25–0311–82; RRID: AB_2716949 |
| CD29 Alexa Fluor 700 | BioLegend | Cat# 102218; RRID: AB_493711 |
| Sca-1 V500 | BD Horizon | Cat# 561228; RRID: AB_10584334 |
| BrdU- Alexa Fluor 647 | Phoenix Flow Systems | Cat# AX647 |
| CD34 Brilliant Violet 421 | BioLegend | Cat# 119321; RRID: AB_10900980 |
| CD24 PerCP-Cyanine 5.5 | eBioscience | Cat# 45–0242-80; RRID: AB_1210702 |
| rat anti-BrdU | Abcam | Cat# ab6326; RRID: AB_305426 |
| rabbit anti-Caveolin-1 | Cell Signaling | Cat# 3238; RRID: AB_2072166 |
| anti-rabbit rhodamine-X-red | Jackson ImmunoResearch Labs | Cat# 111–295-144; RRID: AB_2338028 |
| goat anti-rat-A488 | Jackson ImmunoResearch Labs | Cat# 111–295-144; RRID: AB_2338028 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| 60% Lard High-Fat Diet (HFD) | Research Diets | Cat# D12492 |
| Chow diet (SD) | Harlan Laboratories | Cat# 2018S |
| DPBS | Life Technologies | Cat# 14190144 |
| TRIzol™ Reagent | Invitrogen | Cat# 15596018 |
| Collagenase Type II | Worthington Biochemical | Cat# LS004174 |
| Bovine Serum Albumin | AmericanBio | Cat# AB01088 |
| HBSS | Gibco | Cat# 14185–052 |
| Phosflow™ Lyse/Fix Buffer | BD Biosciences | Cat# 558049 |
| Perm Buffer III | BD Biosciences | Cat# 558050 |
| Deoxyribonuclease I | Worthington | Cat# LS002007 |
| DPBS with MgCl2 and CaCl2 | Sigma | Cat# D8662 |
| NucRed™ Dead 647 | Invitrogen | Cat# R37113 |
| 0.1% Crystal Violet Aqueous Solution | Ward’s Science | Cat# 470300–938 |
| BrdU | US Biological | Cat# B2850 |
| 17β-estradiol | Sigma | Cat# E2758 |
| Hydroxypropyl-β-cyclodextrin | Cayman | Cat# 16169 |
| Isoflurane | Covetrus | Cat# 11695067772 |
| DAPI Fluoromount-G | Southern Biotech | Cat# 0100–20 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Direct-zol RNA Miniprep Kit | Zymo Research Corporation | Cat# R2052 |
| KAPA mRNA HyperPrep Kit | Roche | Cat# KR1352 |
| KAPA Library Quantification Kit | Roche | Cat# KK4854 |
| High-Capacity cDNA reverse Transcription Kit | Applied Biosystems | Cat# 4368814 |
| SYBR FAST quantitative PCR kit | Kapa Biosystems | Cat# KK4611 |
|
| ||
| Deposited data | ||
|
| ||
| Raw bulk RNA-sequencing data | GEO | GSE209663 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6J | Jackson Laboratories | Cat# 000664; RRID: IMSR_JAX:000664 |
| C57BL/6-Tg(Pdgfra-cre)1Clc/J | Jackson Laboratories | Cat# 013148; RRID: IMSR_JAX:013148 |
| B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | Jackson Laboratories | Cat# 007676; RRID: IMSR_JAX:007676 |
| Esr1flox/flox | Feng et al.97 | https://doi.org/10.1073/pnas.0706933104 |
|
| ||
| Oligonucleotides | ||
|
| ||
| See Table S1 for qPCR primer sequences. | ||
|
| ||
| Software and algorithms | ||
|
| ||
| Prism version 9.3.1 | GraphPad | N/A |
| Echo MRI | Echo Medical System | N/A |
| TopHat | Trapnell et al.98 | https://doi.org/10.1093/bioinformatics/btp120 |
| HTSeq package | Anders et al.99 | https://doi.org/10.1093/bioinformatics/btu638 |
| DESeq2 | Love et al.100 | https://doi.org/10.1186/s13059-014-0550-8 |
| Ingenuity Pathway Analysis | QIAGEN Inc. | https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis |
| Gene Ontology | Ashburner et al.101 | https://doi.org/10.1038/75556 |
| KEGG | Kanehisa Laboratories102 | https://doi.org/10.1093/nar/28.1.27 |
| FlowJo™ Software v10.8.1 | BD Life Sciences | N/A |
| CellProfiler™ | Carpenter et al.103 | https://doi.org/10.1186/gb-2006-7-10-r100 |
| BD FACSDiva Software | BD Life Sciences | N/A |
|
| ||
| Other | ||
|
| ||
| 40μm filters | BD Falcon | Cat# 352340 |
| EasyTouch Glucose Monitoring System | EasyTouch | Cat# 807001 |


