Abstract
The alteration of metabolism is essential for the initiation and progression of numerous types of cancer, including colorectal cancer (CRC). Metabolomics has been used to study CRC. At present, the reprogramming of the metabolism in CRC remains to be fully elucidated. In the present study, comprehensive untargeted metabolomics analysis was performed on the paired CRC tissues and adjacent normal tissues from patients with CRC (n=35) using ultra-high-performance liquid chromatography-mass spectrometry. Subsequently, bioinformatic analysis was performed on the differentially expressed metabolites. The changes in these differential metabolites were compared among groups of patients based on sex, anatomical tumor location, grade of tumor differentiation and stage of disease. A total of 927 metabolites were detected in the tissue samples, and 24 metabolites in the CRC tissue were significantly different compared with the adjacent normal tissue. The present study revealed that the levels of three amino acid metabolites were increased in the CRC tissue, specifically, N-α-acetyl-ε-(2-propenal)-Lys, cyclo(Glu-Glu) and cyclo(Phe-Glu). The metabolites with decreased levels in the CRC tissue included quinaldic acid (also referred to as quinoline-2-carboxilic acid), 17α- and 17β-estradiol, which are associated with tumor suppression activities, as well as other metabolites such as, anhydro-β-glucose, Asp-Arg, lysophosphatidylcholine, lysophosphatidylethanolamine (lysoPE), lysophosphatidylinositol, carnitine, 5′-deoxy-5′-(methylthio) adenosine, 2′-deoxyinosine-5′-monophosphate and thiamine monophosphate. There was no difference in the levels of the differential metabolites between male and female patients. The differentiation of CRC also showed no impact on the levels of the differential metabolites. The levels of lysoPE were increased in the right side of the colon compared with the left side of the colon and rectum. Analysis of the different tumor stages indicated that 2-aminobenzenesulfonic acid, P-sulfanilic acid and quinoline-4-carboxylic acid were decreased in stage I CRC tissue compared with stage II, III and IV CRC tissue. The levels of N-α-acetyl-ε-(2-propenal)-Lys, methylcysteine and 5′-deoxy-5′-(methylthio) adenosine varied at different stages of tumorigenesis. These differential metabolites were implicated in multiple metabolism pathways, including carbohydrate, amino acid, lipid, nucleotide and hormone. In conclusion, the present study demonstrated that CRC tumors had altered metabolites compared with normal tissue. The data from the metabolic profile of CRC tissues in the present study provided supportive evidence to understand tumorigenesis.
Keywords: colorectal cancer, mass spectrometry, metabolites, tissue metabolomics, ultra-high-performance liquid chromatography
Introduction
Colorectal cancer (CRC) is a highly prevalent malignant cancer. It is the third most commonly diagnosed type of cancer and causes the second highest rate of mortality from cancer worldwide (1,2). It was recently indicated that 10% of new cancer cases globally were due to CRC and that CRC led to 9.4% of the mortalities due to cancer (1,3). In China, there has been an increasing trend in the incidence of CRC for from 2000 to 2020 (4). Early diagnosis of CRC is important for the survival of patients. It was revealed that the 5-year survival rate for patients with CRC was up to 90% when diagnosed before metastasis took place, while the 5-year survival rate for patients with CRC with distant metastases was only ~12% (5). Although invasive colonoscopy is a popular method used for the detection of CRC, it can result in false negative colonoscopy findings in a number of patients (6). Therefore, more accurate and less invasive screening methods are needed for CRC detection. The risk of CRC carcinogenesis is multifactorial and may be associated with genetic and epigenetic genomic alterations, age, chronic inflammation, as well as environmental and lifestyle factors (7). During cancer initiation and progression, the cancer cells undergo a metabolic reprogramming, such as an altered energy metabolism and an increased biosynthesis of macromolecules (8). The cellular metabolism regulates the epigenetic characteristics of the tumor cells and these cells interact with their surrounding microenvironment. These metabolic interactions are necessary in the regulation of tumor progression (9).
Metabolomics is a branch of systematic biology that examines the changes of metabolites in numerous types of cancer and other diseases (10). Metabolomics offers understanding of carcinogenesis and potentially less invasive methods of cancer diagnosis (11). Different specimens, including serum, plasma, urine and stool collected from patients with CRC, are commonly used in CRC metabolomic studies (11–13). Alterations of multiple metabolites, including metabolites of carbohydrates, amino acids, lipids, nucleotides and hormones, were revealed in these CRC specimens. However, these results mainly reflected indirect metabolic alterations in CRC cells (11,13,14). By contrast, analysis of the CRC tissue could provide direct metabolic profiling dates, and thus reflect higher specificity to the metabolic consequences of the disease. This would eliminate the non-tumor and/or body responses detected in the serum or other specimens (15–17). It has been reported that the alteration of metabolites in CRC tissue was implicated in energy metabolism, amino acid metabolism, glutathione metabolism and fatty acid metabolism and these observed metabolic changes were relevant to the anatomical location and prognosis of CRC (15,16,18).
In the present study, ultra-high-performance liquid chromatography (UHPLC)-mass spectrometry (MS)-based comprehensive metabolomics analysis of metabolic profiles was performed on resected cancer tissues from patients with CRC. Marked changes were revealed in a number of metabolites that are involved in different pathways in CRC. The present study showed that a number of metabolites, which have been associated with tumorigenesis and may provide insight for the clinical management of CRC.
Materials and methods
Patients and tissue samples
The paired CRC tissues and adjacent normal tissues (5 cm away from the tumor margin) were collected from patients with CRC undergoing surgical treatment in Beijing Rehabilitation Hospital of Capital Medical University (Beijing, China) from June 2020 to January 2022. Metabolomic analyses were performed on these 35 paired samples. The characteristics of the patients with CRC have been presented in Table I. The patients had not received any radio- or chemotherapy before surgery. The present study was approved by the ethics committee of Beijing Rehabilitation Hospital of Capital Medical University (Beijing, China) and all participants signed written informed consent.
Table I.
Demographics of patients with colorectal cancer (n=35).
| Characteristics | Value |
|---|---|
| Sex, n (%) | |
| Male | 23 (65.7) |
| Female | 12 (34.2) |
| Age, years (mean ± SD) | 70.0±14.0 |
| Anatomic location in the colon, n (%) | |
| Right side | 10 (28.6) |
| Left side | 13 (37.1) |
| Rectum | 12 (34.2) |
| Disease stage, n (%) | |
| I | 6 (17.1) |
| II | 14 (37.1) |
| III | 9 (22.9) |
| IV | 6 (17.1) |
| Tumor differentiation, n (%) | |
| Poor | 5 (14.2) |
| Moderate | 25 (71.4) |
| Good | 5 (14.2) |
Tissue procurement
The tissue sample preparation and metabolite extraction was conducted according to the methods described by Romisch-Margl et al (19). After surgical resection, tissue samples were immediately stored at −80°C until used for metabolomics analyses. Tissue samples (20 µg) were thawed on ice for 30 min. After homogenization, the metabolite extraction procedure was carried out as follows: A total of 400 µl 70% methanol-water and internal standard extractant (Table II) were added to each sample, the mixture was vortexed for 2 min, frozen in liquid nitrogen for 5 min, placed on dry ice for 5 min, and then thawed on ice for 5 min. After the sample was vortexed for 2 min, it was inverted three times, then centrifuged at 16,113.6 × g at 4°C for 10 min. Subsequently, 300 µl of the supernatant was transferred to a new tube and stored at −20°C for 30 min to precipitate proteins to avoid any interference of the proteins in the analytic procedure. After final centrifugation at 16,113.6 × g at 4°C for 3 min, 200 µl supernatant was collected for analysis by Allwegene Technology Co., Ltd. The molecules of internal standards are presented in Table II.
Table II.
Molecules used as internal standards.
| Name | CAS no. | Concentration |
|---|---|---|
| L-2-chlorophenylalanine | 103616-89-3 | 1 µg/ml |
| (2H3)-L-Carnitine HCl | 350818-62-1 | 1 µg/ml |
| 4-Fluoro-L-α-phenylglycine | 19883-57-9 | 1 µg/ml |
| L-Phenylalanine (2–13C, 99%) | 63-91-2 | 1 µg/ml |
| (2H5)-Hippuric acid | 53518-98-2 | 1 µg/ml |
| (2H5)-Kynurenic acid | 350820-13-2 | 1 µg/ml |
| (2H5)-Phenoxy acetic acid | 154492-74-7 | 1 µg/ml |
A 10-µl 100-ppm mixture of each internal standard was added to 100 µl 70% methanol-water extract.
UHPLC-MS/MS analysis
UHPLC chromatographic separation was performed using an EXIONLC System (SCIEX) installed with a Waters Acquity UHPLC HSS T3 1.8 µm 2.1×100 mm column (Waters Corporation). The details of the internal standard are provided in Table II. The mobile phase A consisted of water (and 0.04% acetic acid), and the mobile phase B consisted of acetonitrile (and 0.04% acetic acid). The following gradient program was used: 98% A, 0 min; 5% A, 11 min; 5% A, 12 min; 5% A, 12.1 min; 95% A, 14 min; and 95% A, 15 min. The flow rate was set to 0.5 ml/min. The injection volume was 2 µl, and the samples were maintained at 4°C in the autosampler.
Linear ion trap (LIT) and triple quadrupole (QQQ) scans were acquired on a triple quadrupole-linear ion trap mass spectrometer (QTRAP), QTRAP® LC-MS/MS System (SCIEX), equipped with an electrospray ionization (ESI) Turbo Ion-Spray interface, operating in positive and negative ion mode and controlled by Analyst software (version 1.6.3; SCIEX). The ESI source operation parameters were as follows: Source temperature 500°C; ion spray voltage 5,500 V (positive), −4,500 V (negative); ion source gas I, ion source gas II and curtain gas were set to 55, 60 and 25 psi, respectively; the collision gas parameter was set at high. Instrument tuning and mass calibration were performed with 10 and 100 µmol/l polypropylene glycol solutions in QQQ and LIT modes, respectively. A specific set of multiple reaction monitoring (MRM) transitions were monitored for each period according to the metabolites eluted within this period. Briefly, in MRM mode, the instrument first selected a parent ion of the target molecule based on specific mass/change ratio (m/z) values, and any initial interference were removed. The parent ion then undergoes collision-induced dissociation. The resultant fragment ions are then filtered through the triple four-pole filter to select the characteristic fragment ions, eliminating non-target ion interference, making the quantification more accurate and improving repeatability.
Data analysis
Based on the metabolome standard database (AllwegeneDB; Allwegene Technology Co., Ltd., qualitative analysis was carried out according to the retention time, parent ion pair information and secondary spectrum data. After obtaining the data of all the tissue samples in the present study, the mass spectrum files of the samples were opened with multiquant software (version 3.0.3; Shanghai AB SCIEX Analytical Instrument Trading Co.) to carry out the integration and correction of the chromatographic peaks. The peak area of each chromatographic peak represents the relative content of the corresponding substance. All of the chromatographic peak area integration data were exported and saved.
Principal component analysis (PCA) was performed using the statistics function prcomp within R (version 3.5.1; www.r-project.org). The data was unit variance scaled before unsupervised PCA. The hierarchical cluster analysis (HCA) results of samples and metabolites were presented as heatmaps with dendrograms, while Pearson correlation coefficients (PCC) between samples were calculated by the cor function in R and presented only as heatmaps. Both HCA and PCC were carried out using the R package ComplexHeatmap (version 1.20.0) (20). For HCA, the normalized signal intensities of the metabolites (unit variance scaling) were visualized as a color spectrum. Significantly regulated metabolites between groups were determined by variable important in projection (VIP) ≥1 and absolute Log2 fold change (FC) ≥1. VIP values were extracted from the orthogonal partial least-squares discriminant analysis (OPLS-DA) results, which also contain score plots and permutation plots that were generated using the R package MetaboAnalystR (version 3.5.1; http://www.metaboanalyst.ca/) (21). The data were log transformed (log2) and mean centered before OPLS-DA. In order to avoid overfitting, a permutation test (200 permutations) was performed. MetaboAnalystR was also used to analyze the significantly different levels of the metabolites between the cancer and adjacent tissues, in which a paired Student's t-test was applied.
Identified metabolites were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Compound database (http://www.kegg.jp/kegg/compound/), and annotated metabolites were then mapped to the KEGG Pathway database (http://www.kegg.jp/kegg/pathway.html). Significantly enriched pathways were identified according to hypergeometric test P-value. All differential metabolites in the present study were examined in PubMed (https://pubmed.ncbi.nlm.nih.gov) and The Human Metabolome Database (https://hmdb.ca/) in order to identify them in previously published studies.
Statistical analysis
With SPSS (version 20; IBM Corp.), statistical analysis was performed for the comparisons of the differential metabolites between cancer tissue and sex, grade of differentiation, tumor location and tumor stage in each group. Non-parametric tests were used to analyze the data of each group after the normality of data was assessed using the Shapiro-Wilk test. The Mann-Whitney U test was used to assessed the different metabolite levels between male and female patients. A Kruskal-Wallis test was used to compare the data among three or four subgroups of patients depending on the grade of differentiation, tumor location and tumor stage, and the Dunn's post hoc test was used for comparison among the subgroups when statistical difference was reached after the Kruskal-Wallis test. P<0.05 was considered to indicate a statistically significant difference.
Results
Metabolic differences between CRC and normal colon tissues
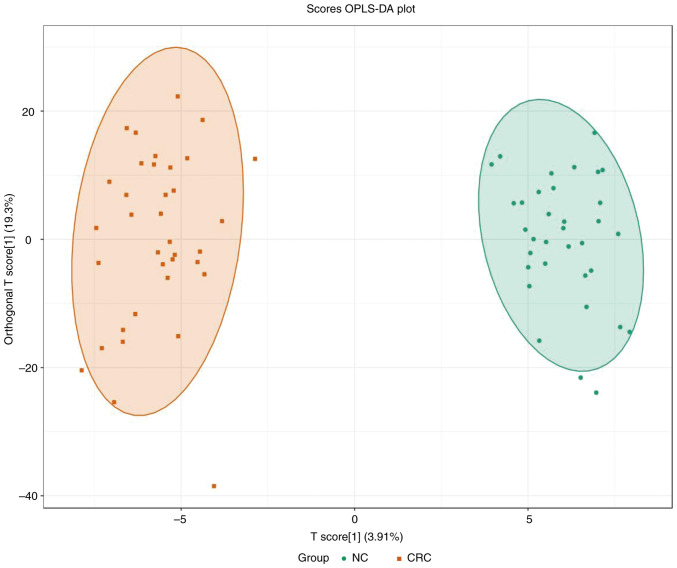
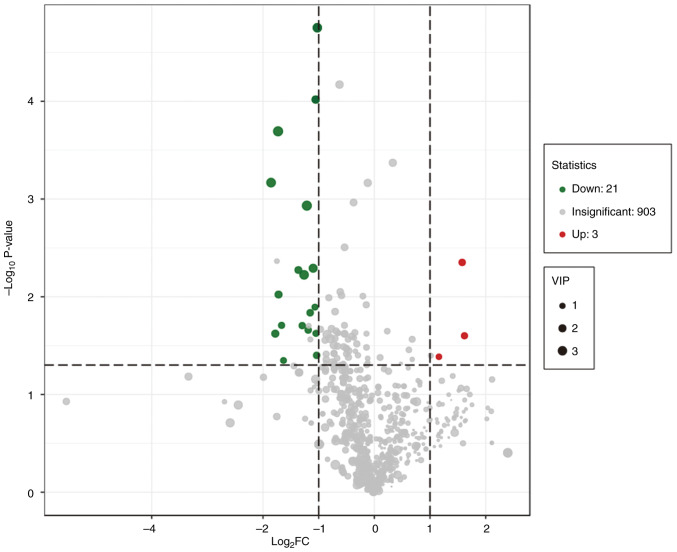
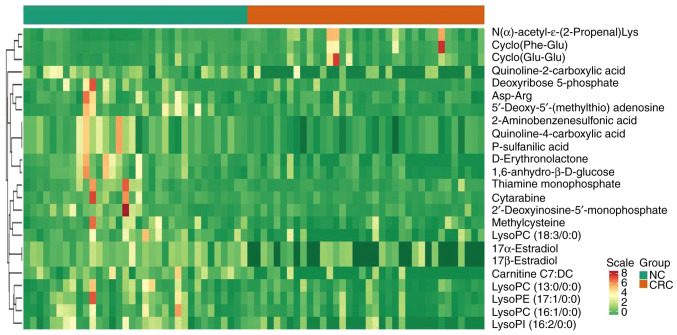
PCA was performed to identify the differences between the paired CRC tissue and normal adjacent colon tissue, which was collected from patients with CRC (Fig. 1). A separation tendency between the normal and the CRC tissues indicated differences in the levels of the metabolites between the two groups. A total of 927 metabolites were detected. Using a univariate analysis, 24 differential metabolites were identified, which had a FC of either ≥2 or <0.5 between normal and CRC tissue (P<0.05). Among them, the levels of three metabolites were upregulated and 21 were downregulated in CRC tissue (Figs. 2 and 3 and Table III). In Fig. 3, the heatmap indicated the changes of all 24 differential metabolites in the 35 patients included in the present study.
Figure 1.
Metabolic differences between normal and cancerous colon tissues revealed by untargeted metabolome profiling. OPLS-DA scores plot that assessed the variance of the features between normal colon tissue and CRC tissue (n=35/group). The circle in the plot indicates the 95% confidence interval. For the OPLS-DA model, R2Y=0.968 and Q2=0.705. OPLS-DA, orthogonal partial least-squares discriminant analysis; NC, adjacent normal tissue; CRC, colorectal cancer tissue.
Figure 2.
Volcano plot analysis of the 927 metabolites detected in the normal and colorectal cancer tissues. Green dots represent the downregulated metabolites, red dots represent the upregulated metabolites and grey dots represent the metabolites without significant differences in their levels. VIP, variable important in projection; FC, fold change.
Figure 3.
Heatmap indicating differential metabolites from individual pairwise comparisons between CRC and normal tissues (n=35/group). All represented differential metabolites are statistically significant (variable important in projection ≥1 and absolute Log2 FC ≥1, P<0.05), scale indicates z-score. NC, adjacent normal tissue; CRC, colorectal cancer tissue; lysoPC, lysophosphatidylcholine; lysoPI, lysophosphatidylinositol; lysoPE, lysophosphatidylethanolamine.
Table III.
Differential metabolites in colorectal cancer tissue vs. adjacent normal tissues.
| Compounds | Class | VIP | P-value | Fold change |
|---|---|---|---|---|
| Cyclo(Glu-Glu) | Cyclodipeptide | 1.14 | 0.041 | 2.24 |
| Cyclo(Phe-Glu) | Cyclodipeptide | 1.50 | 0.025 | 3.07 |
| N-α-acetyl-ε-(2-propenal)-Lys | Amino acid | 1.63 | 0.004 | 2.98 |
| Quinoline-2-carboxylic acid | Amino acid | 3.51 | 0.001 | −2.33 |
| Quinoline-4-carboxylic acid | Amino acid | 1.99 | 0.001 | −2.08 |
| D-Erythronolactone | Carbohydrate | 3.20 | 0.001 | −3.57 |
| 1,6-anhydro-β-D-glucose | Carbohydrate | 1.83 | 0.005 | −2.56 |
| Asp-Arg | Dipeptide | 1.31 | 0.013 | −2.08 |
| Methylcysteine | Amino acid | 1.60 | 0.022 | −2.27 |
| P-sulfanilic acid | Amino acid | 1.99 | <0.001 | −2.08 |
| Cytarabine | Nucleotide | 2.00 | 0.024 | −3.45 |
| 5′-Deoxy-5′-(methylthio) adenosine | Nucleotide | 1.92 | 0.009 | −3.33 |
| Deoxyribose 5-phosphate | Nucleotide | 1.45 | 0.020 | −3.23 |
| 2′-Deoxyinosine-5′-monophosphate | Nucleotide | 1.28 | 0.045 | −3.13 |
| Carnitine C7:DC | Lipid | 3.52 | <0.001 | −3.33 |
| LysoPC (18:3/0:0) | Lipid | 3.06 | 0.006 | −2.38 |
| LysoPC (16:1/0:0) | Lipid | 1.66 | 0.015 | −2.22 |
| LysoPC (13:0/0:0) | Lipid | 1.40 | 0.024 | −2.08 |
| LysoPI (16:2/0:0) | Lipid | 2.60 | 0.005 | −2.13 |
| LysoPE (17:1/0:0) | Lipid | 1.60 | 0.040 | −2.04 |
| Thiamine monophosphate | Heterocyclic compound | 1.52 | 0.020 | −2.44 |
| 17α-Estradiol | Hormone | 2.90 | <0.001 | −2.04 |
| 17β-Estradiol | Hormone | 2.90 | <0.001 | −2.04 |
| 2-Aminobenzenesulfonic acid | Benzene derivatives | 1.99 | <0.001 | −2.08 |
LysoPC, lysophosphatidylcholine; lysoPI, lysophosphatidylinositol; lysoPE, lysophosphatidylethanolamine; VIP, variable important in projection.
Without stratification, the levels of two carbohydrate metabolites, D-erythronolactone and 1,6-anhydro-β-D-glucose, were decreased in CRC tissue compared with the levels in normal tissue. The levels of the amino acid metabolite, N-α-acetyl-ε-(2-propenal)-Lys, and two cyclodipeptides (CDPs), cyclo(Glu-Glu) and cyclo(Phe-Leu), were increased in CRC tissue, while other amino acid metabolites, quinaldic acid (also referred to quinoline-2-carboxylic acid) and quinoline-4-carboxylic acid were decreased in CRC tissue compared with the levels in normal tissue. The results demonstrated that the levels of nucleotide metabolites were consistently deceased in CRC tissue compared with normal tissue. These included cytarabine, 5′-deoxy-5′-(methylthio) adenosine, deoxyribose 5-phosphate and 2′-deoxyinosine-5′-monophosphate. Similarly, lipid metabolites, such as carnitine, phosphatidylcholine (PC) derivative lysoPC, including two sub-types lysophosphatidylethanolamine (lysoPE) and lysophosphatidylinositol, were decreased in the CRC tissue compared with normal tissue. Furthermore, the levels of 17α- and 17β-estradiol were lower in CRC tissue compared with normal tissue. Lastly, the level of thiamine monophosphate, a phosphorylated form of thiamine (vitamin B1) that is required to maintain sodium and potassium gradients for conducting nerve impulses in the central nervous system (22), was decreased in CRC tissue compared with normal tissue (Table III).
The 24 differential metabolites identified in the present study were examined in public databases, which revealed that 11 of the metabolites had been reported previously (Table IV). Among the 11 metabolites, six were revealed in human CRC tissues, and seven in other human samples, such as plasma, urine or stool samples. A total of four metabolites were reported in mouse samples, including tumor tissue, plasma or stool samples.
Table IV.
Comparison of the differential metabolites identified in the present study with those reported in other published reports.
| Patient studies (Refs.) | Animal studies (Refs.) | ||
|---|---|---|---|
|
|
|
||
| Compounds | CRC tissues | Other samples | Samplesa |
| Asp-Arg | (56) | - | - |
| Carnitine C7:DC | (57–60) | (61)b,(62,63)c | - |
| LysoPC (18:3/0:0) | (56,64,65) | (66)b | - |
| LysoPC (16:1/0:0) | (58,65,67,68) | (69,70)b | - |
| LysoPE (17:1/0:0) | (65) | - | - |
| Methylcysteine | - | (71)c | - |
| 5′-Deoxy-5′-(methylthio) adenosine | - | (26)d | - |
| P-sulfanilic acid | (72) | - | (73) |
| 1,6-anhydro-β-D-glucose | - | (26)d | (74,75) |
| N-α-acetyl-ε-(2-propenal)-Lys | - | (76)b,(61)d | (57,77)b,(75) |
| Deoxyribose 5-phosphate | - | - | (78)d |
Tumor tissues unless stated otherwise;
plasma;
urine;
stool. CRC, colorectal cancer; lysoPC, lysophosphatidylcholine; lysoPE, lysophosphatidylethanolamine.
Comparison of the differential metabolites based on sex, anatomic location, stage and grade of differentiation
No statistical differences were revealed when comparing the changes in the levels of the metabolites based on sex or on the grade of tumor differentiation (data not shown). The levels of the metabolites were subsequently compared based on the anatomic location of the tumors and it was revealed that the right side of the colon had higher levels of lysoPE (17:1/0:0) (10.45 fold) compared with the left side of the colon (0.56 fold) and rectum (1.07 fold) (P<0.05), there was no difference in the level of lysoPE (17:1/0:0) between the left side of the colon and the rectum (data not shown). The present study subsequently analyzed the levels of the metabolites based on disease stage, and six metabolites were revealed to have different levels at different stages (Table V). In stage I, the levels of three metabolites, 2-aminobenzenesulfonic acid, P-sulfanilic acid and quinoline-4-carboxylic acid, were lower compared with the other stages, while N-α-acetyl-ε-(2-propenal)-Lys was higher in stage III compared with the other stages. The levels of methylcysteine and 5′-deoxy-5′-(methylthio) adenosine varied at the different stages of tumorigenesis.
Table V.
Differential metabolite changes in colorectal cancer at different stages.
| Fold change in different stages | ||||
|---|---|---|---|---|
|
|
||||
| Metabolite name | I | II | III | IV |
| 2-Aminobenzenesulfonic acid | 0.26a | 0.85 | 0.83 | 0.67 |
| P-sulfanilic acid | 0.26a | 0.85 | 0.83 | 0.67 |
| Quinoline-4-carboxylic acid | 0.28a | 0.47 | 0.74 | 0.64 |
| N-α-acetyl-ε-(2-propenal)-Lys | 11.02a | 1.82 | 23.28b | 3.00 |
| Methylcysteine | 0.10b | 143.45c | 0.46 | 1.17 |
| 5′-Deoxy-5′-(methylthio) adenosine | 0.15d | 0.99 | 2.38 | 1.32 |
P<0.05 when compared with stages II, III and IV;
P<0.05 when compared with stages II and IV;
P<0.05 when compared with stages III and IV;
P<0.05 when compared with stage III.
Identification of distinct metabolites in enriched pathways
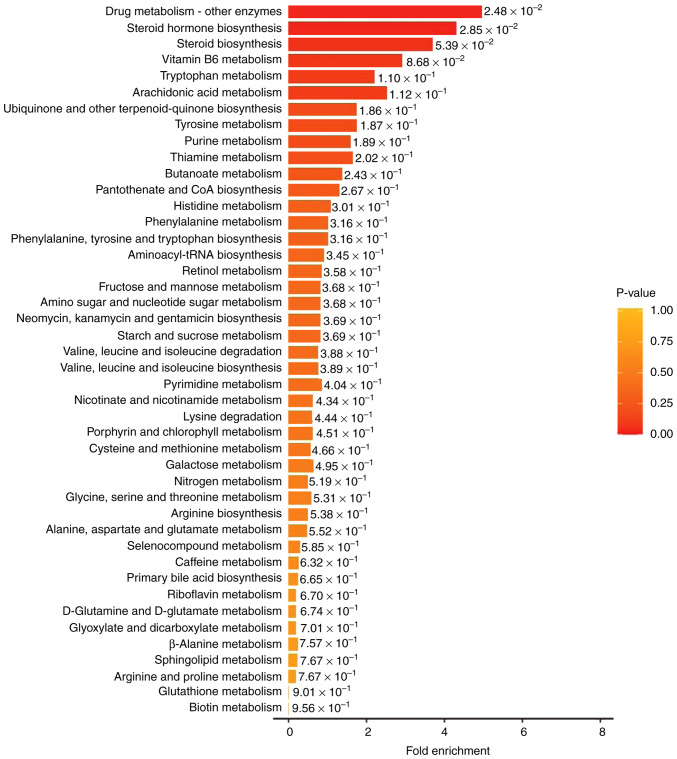
To identify distinct metabolite-associated pathways, the 24 differential metabolites were imported into KEGG for pathway enrichment analysis. KEGG is a public database providing integrated metabolic pathways (23). Using KEGG, the enriched pathways of the metabolites in the present study were identified as the metabolism of amino acids, carbohydrates, lipids, nucleotides, hormones and vitamins, and thermogenesis. The classification annotation and differential abundance score of the differential metabolites analyzed using KEGG are presented in Fig. 4. There was a synergistic or mutually exclusive relationship between the different metabolites.
Figure 4.
Bar graphs of the classification annotation and differential abundance score of the pathways identified between normal and tumor colon tissues using the Kyoto Encyclopedia of Genes and Genomes database (Showing the top 44 pathways, P<1.00). CoA, coenzyme A; tRNA, transfer RNA.
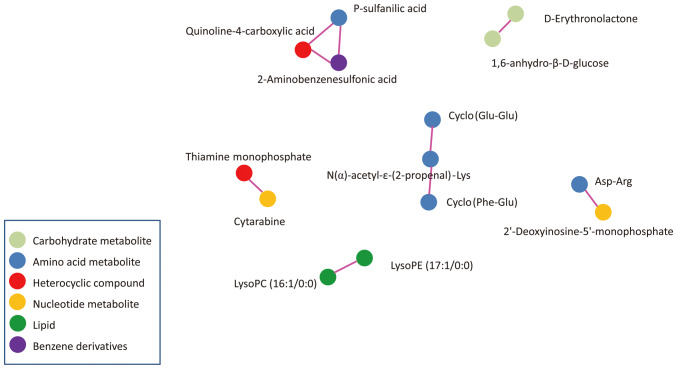
Pearson correlation analysis was used to measure the metabolic proxies between significantly different metabolites (Fig. 5). This analysis is helpful to further understand the relationship between metabolites in the process of biological metabolism. These differential metabolites were carbohydrate metabolites, amino acid metabolites, heterocyclic compounds, nucleotide metabolites, lipids and benzene derivatives.
Figure 5.
Correlation of differential metabolites between CRC and normal tissues. Pearson correlation analysis measuring the metabolic proxies between significantly different metabolites. Each dot represents one metabolite. Lines between the dots represent the metabolites are correlated. Absolute value R>0.8 and P<0.05. lysoPC, lysophosphatidylcholine; lysoPE, lysophosphatidylethanolamine.
Discussion
In the present study, a total of 24 differential metabolites were identified between CRC tissue and adjacent normal tissues. These metabolites were associated with multi-pathways, including energy, carbohydrate, amino acid, lipid, nucleotide, vitamin metabolism and hormone. In CRC tissue two carbohydrate metabolites were downregulated compared with normal tissue, indicating an alteration of carbohydrate metabolism. Previous evidence suggests that the energy of CRC cells may rely on the reversed Warburg effect (24,25). In other reports, 1,6-anhydro-β-D-glucose levels were revealed to be normal in the feces of patients with CRC (26) and in the urine of patients with lung cancer (27). This discordance might be related to the different specimens used.
The present data demonstrated that there were increases and decreases in different amino acid metabolites in CRC tissue compared with normal tissue. Tumor cells increase both the synthesis and degradation of proteins. These changes may indicate the active metabolism of protein for cell proliferation, as well as the degradation of protein in tumor cells (28). Notably, two CDPs, cyclo(Glu-Glu) and cyclo(Phe-Glu), were increased in CRC tissues. To the best of our knowledge, this is the first time that CDPs were found in CRC tissue, and compared between tumor tissue and adjacent normal tissues in patients with CRC. CDPs are the smallest of the cyclic peptides and occur naturally, forming a large class of secondary metabolites produced by microorganisms, plants and mammals. CDPs possess specific chemical and diverse biological functions such as antitumor, antimicrobial and radical-scavenging activities (29). Although the source and effects of CDPs in tissues are unknown, they are likely either endogenous or microbial metabolites, since bacteria that exist in CRC tissue serve a role in the tumorigenesis of CRC (30,31). Asp-Arg is a structural unit of cyanophycin, a biopolymer of long chains of Asp-Arg. Cyanophycin is synthesized by numerous types of bacteria, of which cyanobacteria is the most well-known (32). It is possible that the changes in the levels of Asp-Arg reflect the microbiota changes in the colon. Future studies will compare the microbiota between the tumor samples and normal samples to verify the source of CDPs and Asp-Arg.
The alterations of lipid metabolism in cancer cells are well known (33). Cancer progression is associated with de novo lipogenesis and supply from the microenvironment. The high energy requirement of CRC during processes of malignant transformation and cancer cell proliferation lead to the dysfunction of lipid metabolism (33). In agreement with previous studies, lysoPCs, the product of the degradation of PC, was markedly decreased in CRC tissue compared with normal tissue, and it has been proposed that the increased breakdown of the hemolytic metabolic product of phosphatidylcholine increased cancer malignancy (11,34). The reason for the reduction of lipid metabolites could be due to an increased degradation and reduced supply from the microenvironment (9). Cleavage of lysoPC may increase free fatty acid, and subsequently this free fatty acid can be used by cancer cells for energy via fatty acid oxidation (33). However, carnitine, a major transporter for fatty acid acyl moieties entering the mitochondrial matrix for oxidation (35), was decreased in CRC tissue compared with normal tissue in the present study, which was not in agreement with previous reports (11,36). Carnitine is also considered to inhibit cell proliferation and increase apoptosis (37). The decrease of carnitine indicated the reduction of lipid metabolism in the mitochondria. Although lipid metabolites have been proposed as diagnostic and prognostic predictors for patients with CRC (38,39), the alteration of lipids in CRC tissue is inconsistent, and as such, the influence of lipids on tumorigenesis is inconclusive (40).
Nucleotide metabolism serves a role in carcinogenesis and cancer progression. It not only meets the requirements of cell proliferation but also affects a variety of activities beyond proliferation, such as, tumor immunity, antitumor drug resistance and interactions of the tumor cell with the microenvironment (41). Cancer cells tend to use the de novo nucleotide synthesis pathway (42). In the present study, the levels of nucleotide metabolites, including cytarabine, 5′-deoxy-5′-(methylthio) adenosine, deoxyribose 5-phosphate and 2′-deoxyinosine-5′-monophosphate, were decreased in CRC tissues compared with normal tissue. It was hypothesized that the decrease of these metabolites may indicate the insufficient replenishment of nucleotide metabolites for active nucleotide synthesis in CRC cells. Unlike lipids and proteins, cancer cells may not be obtaining sufficient amounts of nucleotides and their components from the tumor microenvironment (9). The decreased levels of 5′-deoxy-5′-(methylthio) adenosine in CRC tissues, indicated alterations in the cysteine and methionine metabolism pathways.
Decreases in the 17α- and 17β-estradiol levels in the tumor tissues regardless of sex is consistent with the notion that estrogen has an anti-tumor effect in CRC, presumably by activating the estrogen receptor (ER)-β (43,44). ER-β-modulated transcription events resulted in the stimulation of proapoptotic signaling, regulation of mismatch repair proteins as well as the modulation of the antitumor immune response (45). Quinaldic acid, a tryptophan metabolite and a derivative of kynurenic acid, induced apoptosis in colon cancer cells via the activation of the p53 tumor suppressor but had no toxicity to non-cancerous colon cell lines (46,47). Therefore, decreased levels of quinaldic acid contributed to colon cancer cell growth. Notably, the levels of 2-aminobenenesulfonic acid, quinoline-4-carboxylic acid and p-sulfanilic acid were revealed to be correlated to one another, even though they are in three different KEGG pathways. In fact, sulfanilic acid (also referred to as 4-aminobenzenesulfonic acid) is an intermediate product of azo dyes, plant protectives and detergents (48). In the present study, it was revealed that the levels of these metabolites were correlated to the stage of CRC (Table V). It is unclear the resource of sulfanilic acid. Since sulfanilic acid is degraded by Pseudomonas paucimobilis, an aerobic bacterium, perhaps bacterial colonization may be increased in the tumor.
In the present study, the level of lysoPE (17:1/0:0) was increased on the right-side of the colon compared with the left-side of the colon and rectum. Additionally, there were metabolite profile differences between the right- and left-side colon tissues in healthy individuals (49). Cai et al (16) revealed that there were differences in metabolites in early-stage CRC tissues between the right- and left-side of the colon. The anatomic location of the CRC was relevant to the development and prognosis of the patients with CRC (50). In the present study, the changes of six differential metabolites in different stages of CRC were significant, including 2-aminobenzenesulfonic acid, p-sulfanilic acid, quinoline-4-carboxylic acid, N-α-acetyl-ε-(2-propenal)-Lys, methylcysteine and 5′-deoxy-5′-(methylthio) adenosine. These metabolites are involved in amino acid and nucleotide metabolism. Although previous studies compared the changes of differential metabolites in different stages of CRC tissues, there were various results in different studies (11,16,51,52). The data of the present study demonstrated that the level of differential metabolites were regardless of sex and differentiations of CRC. Since the sample size in the present study was small, further studies are required to confirm the results of the present study.
The detection of metabolites within cancer tissue directly reflects the true metabolic state of the cancer cell unlike the use of other specimens, such as serum, urine and stool. However, unlike proteomics, genomics or transcriptomics, metabolomics only represents a transient phenotypic state that may vary rapidly (11). The results of the present study agreed with the findings of a number of previous studies, while they differed from others (11,36,53). The differential metabolic profile identified in the present study was unique and different from other studies (Table IV). There were multiple factors that could have caused the variations in the metabolites between the different studies, such as different preparation methods, samples collected from patients of different ages, weight, diet and even circadian rhythm (54), or different analytic platforms and processing software (11,36,55). Further conclusive studies are required before any clear explanations can be offered for the different changes in the metabolites.
The present study had a number of limitations. Firstly, the present study had a small sample size. The verification of the results in another set of patients compared with the one used for metabolomics analysis could not be performed because of the lack of available samples. Further investigation using a larger sample size would increase the accuracy and reproducibility of the study. Secondly, the present study is cross-sectional. Further longitudinal studies or experimental studies could provide further evidence to confirm the relationship between metabolite levels and CRC, and may elucidate the mechanisms of the alterations in the tumorigenesis of CRC. Thirdly, the difference between cancer tissues and adenomatous polyps, a precursor lesion for the majority of CRC, could have been compared in order to investigate the metabolic changes in the transition to malignancy. Fourthly, if the metabolite changes could be confirmed using blood samples from patients, it could aid the establishment of biomarkers for CRC. The patients of the present study are being followed-up for further investigation of the relationship between the alteration of metabolism and prognosis. Additionally, future study will compare the alteration of metabolites in tissue and blood.
In conclusion, the present study revealed that the alteration of metabolism in CRC cells could provide novel insights to decode an unknown aspect of colon carcinogenesis. The differential levels of the metabolites may serve as biomarkers in CRC. With collective efforts, further studies on CRC metabolites may lead to an improved diagnosis or treatment of CRC.
Acknowledgements
The authors would like to thank Dr. Chuixiong Chen from Key Tech Statistics and Science Company (Beijing, China) for his work on the statistical analysis.
Funding Statement
This study was supported by National Natural Science Foundation of China (grant no. 81370487), Scientific Research Fund of Beijing Rehabilitation Hospital, Capital Medical University (grant nos. 2017-004, 2021-018, 2019-043, 2019R-001 and 2020-056 and 2021-015) and Scientific Research Fund of Beijing Anorectal Society (grant no. 2020ABCP002). These funds were used for the design of the study, sample collection, data analysis, interpretation of the data and in writing the manuscript.
Availability of data and materials
The datasets generated during the current study are available in the MetaboLights repository, https://www.ebi.ac.uk/metabolights/ (study no. MTBLS8090).
Authors' contributions
QG, CK, JZ, SJ and FC conceived and designed the study. CK, JZ, MZ, XL and MX recruited patients and collected samples. JZ, MZ, MX, YW, XL and DD collected data. QG, FC, MZ CK, SJ and XL were involved in data analysis and the literature search. JZ, MZ, YW, DD and MX were involved in project administration. QG, SJ, CK and MZ wrote the manuscript. QG and CK confirm the authenticity of all the raw data. QG, CK, MX and MZ acquired funding. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
All procedures used in this research were approved by the Ethics Committee of Beijing Rehabilitation Hospital (approval no. 2020bkkyLW003). Written informed consent was obtained from all study participants. All methods performed in this study were in accordance with the Declaration of Helsinki.
Patient consent for publication
All study participants gave consent to publish the data of the study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M, Saad A. Colorectal cancer epidemiology: Recent trends and impact on outcomes. Curr Drug Targets. 2021;22:998–1009. doi: 10.2174/13894501MTExCNTkBy. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 4.Xia C, Dong X, Li H, Cao M, Sun D, He S, Yang F, Yan X, Zhang S, Li N, Chen W. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin Med J (Engl) 2022;135:584–590. doi: 10.1097/CM9.0000000000002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, Lin H, Li S. Prognoses of different pathological subtypes of colorectal cancer at different stages: A population-based retrospective cohort study. BMC Gastroenterol. 2019;19:164. doi: 10.1186/s12876-019-1083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanth P, Inadomi JM. Screening and prevention of colorectal cancer. BMJ. 2021;374:n1855. doi: 10.1136/bmj.n1855. [DOI] [PubMed] [Google Scholar]
- 7.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Reyes I, Chandel NS. Cancer metabolism: Looking forward. Nat Rev Cancer. 2021;21:669–680. doi: 10.1038/s41568-021-00378-6. [DOI] [PubMed] [Google Scholar]
- 9.Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klassen A, Faccio AT, Canuto GA, da Cruz PL, Ribeiro HC, Tavares MF, Sussulini A. Metabolomics: Definitions and significance in systems biology. Adv Exp Med Biol. 2017;965:3–17. doi: 10.1007/978-3-319-47656-8_1. [DOI] [PubMed] [Google Scholar]
- 11.Gold A, Choueiry F, Jin N, Mo X, Zhu J. The Application of metabolomics in recent colorectal cancer studies: A state-of-the-art review. Cancers (Basel) 2022;14:725. doi: 10.3390/cancers14030725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erben V, Bhardwaj M, Schrotz-King P, Brenner H. Metabolomics biomarkers for detection of colorectal neoplasms: A systematic review. Cancers (Basel) 2018;10:246. doi: 10.3390/cancers10080246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashim NAA, Ab-Rahim S, Suddin LS, Saman MSA, Mazlan M. Global serum metabolomics profiling of colorectal cancer. Mol Clin Oncol. 2019;11:3–14. doi: 10.3892/mco.2019.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Giovanni N, Meuwis MA, Louis E, Focant JF. Specificity of metabolic colorectal cancer biomarkers in serum through effect size. Metabolomics. 2020;16:88. doi: 10.1007/s11306-020-01707-w. [DOI] [PubMed] [Google Scholar]
- 15.Shen Y, Sun M, Zhu J, Wei M, Li H, Zhao P, Wang J, Li R, Tian L, Tao Y, et al. Tissue metabolic profiling reveals major metabolic alteration in colorectal cancer. Mol Omics. 2021;17:464–471. doi: 10.1039/D1MO00022E. [DOI] [PubMed] [Google Scholar]
- 16.Cai Y, Rattray NJW, Zhang Q, Mironova V, Santos-Neto A, Muca E, Vollmar AKR, Hsu KS, Rattray Z, Cross JR, et al. Tumor tissue-specific biomarkers of colorectal cancer by anatomic location and stage. Metabolites. 2020;10:257. doi: 10.3390/metabo10060257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao P, Zhou C, Zhao L, Zhang G, Zhang Y. Tissue amino acid profile could be used to differentiate advanced adenoma from colorectal cancer. J Pharm Biomed Anal. 2016;118:349–355. doi: 10.1016/j.jpba.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 19.Romisch-Margl W, Prehn C, Bogumil R, Röhring C, Suhre K, Adamski J. Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics. 2012;8:133–142. doi: 10.1007/s11306-011-0293-4. [DOI] [Google Scholar]
- 20.Xu Y, Wang X, Han D, Wang J, Luo Z, Jin T, Shi C, Zhou X, Lin L, Shan J. Revealing the mechanism of Jiegeng decoction attenuates bleomycin-induced pulmonary fibrosis via PI3K/Akt signaling pathway based on lipidomics and transcriptomics. Phytomedicine. 2022;102:154207. doi: 10.1016/j.phymed.2022.154207. [DOI] [PubMed] [Google Scholar]
- 21.Xiao Y, Liu H, Li H, Liu Q, Lu Q, Varshney RK, Chen X, Honget Y. Widely targeted metabolomics characterizes the dynamic changes of chemical profile in postharvest peanut sprouts grown under the dark and light conditions. Food Sci Technol. 2021;152:112283. [Google Scholar]
- 22.McCann A, Midttun Ø, Whitfield KC, Kroeun H, Borath M, Sophonneary P, Ueland PM, Green TJ. Comparable performance characteristics of plasma thiamine and erythrocyte thiamine diphosphate in response to thiamine fortification in rural cambodian women. Nutrients. 2017;9:676. doi: 10.3390/nu9070676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chekulayev V, Mado K, Shevchuk I, Koit A, Kaldma A, Klepinin A, Timohhina N, Tepp K, Kandashvili M, Ounpuu L, et al. Metabolic remodeling in human colorectal cancer and surrounding tissues: Alterations in regulation of mitochondrial respiration and metabolic fluxes. Biochem Biophys Rep. 2015;4:111–125. doi: 10.1016/j.bbrep.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satoh K, Yachida S, Sugimoto M, Oshima M, Nakagawa T, Akamoto S, Tabata S, Saitoh K, Kato K, Sato S, et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc Natl Acad Sci USA. 2017;114:E7697–E7706. doi: 10.1073/pnas.1710366114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R. Fecal metabolomics: Assay performance and association with colorectal cancer. Carcinogenesis. 2014;35:2089–2096. doi: 10.1093/carcin/bgu131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stretch C, Eastman T, Mandal R, Eisner R, Wishart DS, Mourtzakis M, Prado CM, Damaraju S, Ball RO, Greiner R, Baracos VE. Prediction of skeletal muscle and fat mass in patients with advanced cancer using a metabolomic approach. J Nutr. 2012;142:14–21. doi: 10.3945/jn.111.147751. [DOI] [PubMed] [Google Scholar]
- 28.Vettore L, Westbrook RL, Tennant DA. New aspects of amino acid metabolism in cancer. Br J Cancer. 2020;122:150–156. doi: 10.1038/s41416-019-0620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra AK, Choi J, Choi SJ, Baek KH. Cyclodipeptides: An overview of their biosynthesis and biological activity. Molecules. 2017;22:1796. doi: 10.3390/molecules22101796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng Y, Ling Z, Li L. The intestinal microbiota and colorectal cancer. Front Immunol. 2020;11:615056. doi: 10.3389/fimmu.2020.615056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharon I, Grogg M, Hilvert D, Schmeing TM. Structure and Function of the β-Asp-Arg Polymerase Cyanophycin Synthetase 2. ACS Chem Biol. 2022;17:670–679. doi: 10.1021/acschembio.1c01007. [DOI] [PubMed] [Google Scholar]
- 33.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Z, Xiao Y, Elson P, Tan H, Plummer SJ, Berk M, Aung PP, Lavery IC, Achkar JP, Li L, et al. Plasma lysophosphatidylcholine levels: Potential biomarkers for colorectal cancer. J Clin Oncol. 2007;25:2696–2701. doi: 10.1200/JCO.2006.08.5571. [DOI] [PubMed] [Google Scholar]
- 35.Console L, Scalise M, Mazza T, Pochini L, Galluccio M, Giangregorio N, Tonazzi A, Indiveri C. Carnitine traffic in cells. Link with cancer. Front Cell Dev Biol. 2020;8:583850. doi: 10.3389/fcell.2020.583850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F, Zhang Y, Zhao W, Deng K, Wang Z, Yang C, Ma L, Openkova MS, Hou Y, Li K. Metabolomics for biomarker discovery in the diagnosis, prognosis, survival and recurrence of colorectal cancer: A systematic review. Oncotarget. 2017;8:35460–35472. doi: 10.18632/oncotarget.16727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dionne S, Elimrani I, Roy MJ, Qureshi IA, Sarma DR, Levy E, Seidman EG. Studies on the chemopreventive effect of carnitine on tumorigenesis in vivo, using two experimental murine models of colon cancer. Nutr Cancer. 2012;64:1279–1287. doi: 10.1080/01635581.2012.722247. [DOI] [PubMed] [Google Scholar]
- 38.Sun Y, Liu B, Chen Y, Xing Y, Zhang Y. Multi-Omics prognostic signatures based on lipid metabolism for colorectal cancer. Front Cell Dev Biol. 2021;9:811957. doi: 10.3389/fcell.2021.811957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurabe N, Hayasaka T, Ogawa M, Masaki N, Ide Y, Waki M, Nakamura T, Kurachi K, Kahyo T, Shinmura K, et al. Accumulated phosphatidylcholine (16:0/16:1) in human colorectal cancer; possible involvement of LPCAT4. Cancer Sci. 2013;104:1295–1302. doi: 10.1111/cas.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pakiet A, Kobiela J, Stepnowski P, Sledzinski T, Mika A. Changes in lipids composition and metabolism in colorectal cancer: A review. Lipids Health Dis. 2019;18:29. doi: 10.1186/s12944-019-0977-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma J, Zhong M, Xiong Y, Gao Z, Wu Z, Liu Y, Hong X. Emerging roles of nucleotide metabolism in cancer development: Progress and prospect. Aging. 2021;13:13349–13358. doi: 10.18632/aging.202962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lane AN, Fan TW. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015;43:2466–2485. doi: 10.1093/nar/gkv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haziman AA, Ravinderan S, Thangavelu T, Thomas W. A novel role for estrogen-induced signaling in the colorectal cancer gender bias. Ir J Med Sci. 2019;188:389–395. doi: 10.1007/s11845-018-1867-1. [DOI] [PubMed] [Google Scholar]
- 44.Ditonno I, Losurdo G, Rendina M, Pricci M, Girardi B, Ierardi E, Di Leo A. Estrogen receptors in colorectal cancer: Facts, novelties and perspectives. Curr Oncol. 2021;28:4256–4263. doi: 10.3390/curroncol28060361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caiazza F, Ryan EJ, Doherty G, Winter DC, Sheahan K. Estrogen receptors and their implications in colorectal carcinogenesis. Front Oncol. 2015;5:19. doi: 10.3389/fonc.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langner E, Jeleniewicz W, Turski WA, Plech T. Quinaldic acid induces changes in the expression of p53 tumor suppressor both on protein and gene level in colon cancer LS180 cells. Pharmacol Rep. 2019;71:189–193. doi: 10.1016/j.pharep.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 47.Langner E, Walczak K, Jeleniewicz W, Turski WA, Rajtar G. Quinaldic acid inhibits proliferation of colon cancer ht-29 cells in vitro: Effects on signaling pathways. Eur J Pharmacol. 2015;757:21–27. doi: 10.1016/j.ejphar.2015.02.053. [DOI] [PubMed] [Google Scholar]
- 48.Perei K, Rakhely G, Kiss I, Polyák B, Kovács KL. Biodegradation of sulfanilic acid by Pseudomonas paucimobilis. Appl Microbiol biotechnol. 2001;55:101–107. doi: 10.1007/s002530000474. [DOI] [PubMed] [Google Scholar]
- 49.Baxter BA, Parker KD, Nosler MJ, Rao S, Craig R, Seiler C, Ryan EP. Metabolite profile comparisons between ascending and descending colon tissue in healthy adults. World J Gastroenterol. 2020;26:335–352. doi: 10.3748/wjg.v26.i3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong MCS, Huang J, Lok V, Wang J, Fung F, Ding H, Zheng ZJ. Differences in incidence and mortality trends of colorectal cancer worldwide based on sex, age, and anatomic location. Clin Gastroenterol Hepatol. 2021;19:955–966.e61. doi: 10.1016/j.cgh.2020.02.026. [DOI] [PubMed] [Google Scholar]
- 51.Mirnezami R, Jimenez B, Li JV, Kinross JM, Veselkov K, Goldin RD, Holmes E, Nicholson JK, Darzi A. Rapid diagnosis and staging of colorectal cancer via high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy of intact tissue biopsies. Ann Surg. 2014;259:1138–1149. doi: 10.1097/SLA.0b013e31829d5c45. [DOI] [PubMed] [Google Scholar]
- 52.Jimenez B, Mirnezami R, Kinross J, Cloarec O, Keun HC, Holmes E, Goldin RD, Ziprin P, Darzi A, Nicholson JK. 1H HR-MAS NMR spectroscopy of tumor-induced local metabolic ‘field-effects’ enables colorectal cancer staging and prognostication. J Proteome Res. 2013;12:959–968. doi: 10.1021/pr3010106. [DOI] [PubMed] [Google Scholar]
- 53.Tian JS, Xue WN, Yin HH, Zhang N, Zhou J, Long Z, Wu C, Liang Z, Xie K, Li S, et al. Differential metabolic alterations and biomarkers between gastric cancer and colorectal cancer: A systematic review and meta-analysis. Onco Targets Ther. 2020;13:6093–6108. doi: 10.2147/OTT.S247393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kosmides AK, Kamisoglu K, Calvano SE, Corbett SA, Androulakis IP. Metabolomic fingerprinting: Challenges and opportunities. Crit Rev Biomed Eng. 2013;41:205–221. doi: 10.1615/CritRevBiomedEng.2013007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yusof HM, Ab-Rahim S, Suddin LS, Saman MSA, Mazlan M. Metabolomics profiling on different stages of colorectal cancer: A systematic review. Malays J Med Sci. 2018;25:16–34. doi: 10.21315/mjms2018.25.5.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long Z, Zhou J, Xie K, Wu Z, Yin H, Daria V, Tian J, Zhang N, Li L, Zhao Y, et al. Metabolomic markers of colorectal tumor with different clinicopathological features. Front Oncol. 2020;10:981. doi: 10.3389/fonc.2020.00981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manna SK, Tanaka N, Krausz KW, Haznadar M, Xue X, Matsubara T, Bowman ED, Fearon ER, Harris CC, Shah YM, Gonzalez FJ. Biomarkers of coordinate metabolic reprogramming in colorectal tumors in mice and humans. Gastroenterology. 2014;146:1313–1324. doi: 10.1053/j.gastro.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen X, Cai Y, Lu L, Huang H, Yan H, Paty PB, Muca E, Ahuja N, Zhang Y, Johnson CH, Khan SA. Asparagine metabolism in tumors is linked to poor survival in females with colorectal cancer: A cohort study. Metabolites. 2022;12:164. doi: 10.3390/metabo12020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ong ES, Zou L, Li S, Cheah PY, Eu KW, Ong CN. Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. Mol Cell Proteomics. 2010 Feb 10; doi: 10.1074/mcp.M900551-MCP200. doi: 10.1074/mcp.M900551-MCP200 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 60.Li Z, Deng X, Luo J, Lei Y, Jin X, Zhu J, Lv G. Metabolomic comparison of patients with colorectal cancer at different anticancer treatment stages. Front Oncol. 2021;11:574318. doi: 10.3389/fonc.2021.574318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016;4:11. doi: 10.1186/s40170-016-0151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liesenfeld DB, Habermann N, Toth R, Owen RW, Frei E, Staffa J, Schrotz-King P, Klika KD, Ulrich CM. Changes in urinary metabolic profiles of colorectal cancer patients enrolled in a prospective cohort study (ColoCare) Metabolomics. 2015;11:998–1012. doi: 10.1007/s11306-014-0758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Udo R, Katsumata K, Kuwabara H, Enomoto M, Ishizaki T, Sunamura M, Nagakawa Y, Soya R, Sugimoto M, Tsuchida A. Urinary charged metabolite profiling of colorectal cancer using capillary electrophoresis-mass spectrometry. Sci Rep. 2020;10:21057. doi: 10.1038/s41598-020-78038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams MD, Zhang X, Park JJ, Siems WF, Gang DR, Resar LM, Reeves R, Hill HH., Jr Characterizing metabolic changes in human colorectal cancer. Anal Bioanal Chem. 2015;407:4581–4595. doi: 10.1007/s00216-015-8662-x. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Hinz S, Uckermann O, Hönscheid P, von Schönfels W, Burmeister G, Hendricks A, Ackerman JM, Baretton GB, Hampe J, et al. Shotgun lipidomics-based characterization of the landscape of lipid metabolism in colorectal cancer. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865:158579. doi: 10.1016/j.bbalip.2019.158579. [DOI] [PubMed] [Google Scholar]
- 66.Shen S, Yang L, Li L, Bai Y, Cai C, Liu H. A plasma lipidomics strategy reveals perturbed lipid metabolic pathways and potential lipid biomarkers of human colorectal cancer. J Chromatogr B Analyt Technol Biomed Life Sci. 2017:1068–1069. 41–48. doi: 10.1016/j.jchromb.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 67.Cai Y, Rattray NJW, Zhang Q, Mironova V, Santos-Neto A, Hsu KS, Rattray Z, Cross JR, Zhang Y, Paty PB, et al. Sex differences in colon cancer metabolism reveal a novel subphenotype. Sci Rep. 2020;10:4905. doi: 10.1038/s41598-020-61851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ecker J, Benedetti E, Kindt ASD, Höring M, Perl M, Machmüller AC, Sichler A, Plagge J, Wang Y, Zeissig S, et al. The colorectal cancer lipidome: Identification of a robust Tumor-Specific lipid species signature. Gastroenterology. 2021;161:910–923.e19. doi: 10.1053/j.gastro.2021.05.009. [DOI] [PubMed] [Google Scholar]
- 69.Kuhn T, Floegel A, Sookthai D, Johnson T, Rolle-Kampczyk U, Otto W, von Bergen M, Boeing H, Kaaks R. Higher plasma levels of lysophosphatidylcholine 18:0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med. 2016;14:13. doi: 10.1186/s12916-016-0552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holowatyj AN, Gigic B, Herpel E, Scalbert A, Schneider M, Ulrich CM, MetaboCCC Consortium; ColoCare Study Distinct molecular phenotype of sporadic colorectal cancers among young patients based on multiomics analysis. Gastroenterology. 2020;158:1155–1158.e2. doi: 10.1053/j.gastro.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zarei I, Baxter BA, Oppel RC, Borresen EC, Brown RJ, Ryan EP. Plasma and urine metabolite profiles impacted by increased dietary navy bean intake in colorectal cancer survivors: A randomized-controlled trial. Cancer Prev Res (Phila) 2021;14:497–508. doi: 10.1158/1940-6207.CAPR-20-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin L, Zeng X, Liang S, Wang Y, Dai X, Sun Y, Wu Z. Biomarkers of coordinate metabolic reprogramming and the construction of a co-expression network in colorectal cancer. Ann Transl Med. 2022;10:1115. doi: 10.21037/atm-22-4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li M, Wu S, Zhuang C, Shi C, Gu L, Wang P, Guo F, Wang Y, Liu Z. Metabolomic analysis of circulating tumor cells derived liver metastasis of colorectal cancer. Heliyon. 2023;9:e12515. doi: 10.1016/j.heliyon.2022.e12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Montrose DC, Zhou XK, Kopelovich L, Yantiss RK, Karoly ED, Subbaramaiah K, Dannenberg AJ. Metabolic profiling, a noninvasive approach for the detection of experimental colorectal neoplasia. Cancer Prev Res (Phila) 2012;5:1358–1367. doi: 10.1158/1940-6207.CAPR-12-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoshie T, Nishiumi S, Izumi Y, Sakai A, Inoue J, Azuma T, Yoshida M. Regulation of the metabolite profile by an APC gene mutation in colorectal cancer. Cancer Sci. 2012;103:1010–1021. doi: 10.1111/j.1349-7006.2012.02262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Long Y, Sanchez-Espiridion B, Lin M, White L, Mishra L, Raju GS, Kopetz S, Eng C, Hildebrandt MAT, Chang DW, et al. Global and targeted serum metabolic profiling of colorectal cancer progression. Cancer. 2017;123:4066–4074. doi: 10.1002/cncr.30829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo Z, Wang H, Lin S, Liao L, Cai L, Zhang X, Tan Y, Shen M. Study on the levels of N-nitrosamine compounds and untargeted metabolomics in patients with colorectal cancer. Anal Bioanal Chem. 2022;414:3483–3496. doi: 10.1007/s00216-022-03969-w. [DOI] [PubMed] [Google Scholar]
- 78.Yang F, DeLuca JAA, Menon R, Garcia-Vilarato E, Callaway E, Landrock KK, Lee K, Safe SH, Chapkin RS, Allred CD, Jayaraman A. Effect of diet and intestinal AhR expression on fecal microbiome and metabolomic profiles. Microb Cell Fact. 2020;19:219. doi: 10.1186/s12934-020-01463-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are available in the MetaboLights repository, https://www.ebi.ac.uk/metabolights/ (study no. MTBLS8090).