Abstract
N6-methyladenosine (m6A) serves an essential role in RNA modulation and is implicated in multiple malignancies, including colorectal cancer (CRC). Methyltransferase-like 3 (METTL3) is an important writer in m6A modification, however its role in CRC in modifying small nucleolar RNA host gene 1 (SNHG1), an oncogenic long noncoding RNA, remains unclear. In the present study, METTL3 expression in CRC was assessed using online bioinformatics analysis, immunohistochemistry staining, western blotting, reverse transcription (RT)-quantitative PCR (qPCR) and cell transfections. Cell proliferation, migration and invasion were determined using functional Cell Counting Kit-8 (CCK-8) and Transwell assays. SNHG1 expression in CRC was evaluated using online bioinformatics analysis and RT-qPCR. Methylated RNA immunoprecipitation qPCR was performed to assess m6A modification changes of SNHG1 mRNA. The present study demonstrated that METTL3 is upregulated in CRC tissues and cell lines. Moreover, METTL3 expression was associated with several unfavourable clinical features in patients with CRC, including the stage of lymph node metastases and overall survival. Functional Transwell and CCK-8 assays demonstrated that knockdown of METTL3 suppressed CRC cell proliferation and migration. Furthermore, METTL3 was positively correlated with SNHG1 in CRC tissue, as indicated by analysis of data from The Cancer Genome Atlas. Mechanistically, SNHG1 contains 18 m6A modification sites. Through cell transfections and actinomycin D assays, the present study found that METTL3-mediated m6A modification at these sites enhances the stability of SNHG1 in CRC cells. Finally, it was demonstrated that SNHG1 knockdown partially diminished the facilitative effect of METTL3 on CRC cell migration and proliferation. The present study concluded that METTL3, a potential biomarker for assessing overall survival and metastasis in CRC, may serve as an oncogene, promote SNHG1 m6A modification, improve the stability of SNHG1 and enhance SNHG1-mediated oncogenic function in CRC.
Keywords: methyltransferase-like 3, N6-methyladenosine, SNHG1, proliferation, migration, colorectal cancer
Introduction
Colorectal cancer (CRC) is a common type of intestinal tumour with ~2 million new cases and 1 million estimated associated deaths reported in 2020, representing 10.7 and 9.5% of all new cancer cases and deaths globally, respectively (1). Moreover, cancer statistics from 2018 show that CRC resulted in the deaths of ~27,390 male and 23,240 female patients in the United States, which is the highest death toll among digestive tract tumours (2). However, despite the advances in the treatment of CRC, such as surgical removal and systemic chemotherapy, the mortality rate remains high due to recurrence and distant organ metastases (3). Distant metastasis to the liver is responsible for CRC-related death, with a 5-year survival rate of <10% (4). Therefore, exploring new biomarkers that indicate the promotion of tumour initiation and progression, and assessing the underlying molecular mechanisms, are of foremost importance to develop targeted treatments of CRC malignancies.
N6-methyladenosine (m6A), a common RNA modification in epigenetic regulation, affects multiple aspects of RNA metabolism, such as pre-mRNA processing, translation efficiency, transcript stability and microRNA biogenesis (5–8). Methyltransferase-like 3 (METTL3), also called MT-A70, belongs to the class I methyltransferase (MTase) family and is an important catalytic enzyme of m6A MTase systems (9). METTL3 is widely involved in numerous cancer types, such as gastric and bladder cancer, by acting as an oncogene or as a tumour suppressor (10). Yue et al (11) reported that METTL3 was upregulated in gastric cancer and enhanced the stability of zinc finger MYM-type containing 1 mRNA, thereby facilitating the epithelial-mesenchymal transition (EMT) program and metastasis. In another study, Han et al (12) reported that METTL3 was upregulated in bladder cancer and reduced the expression of phosphatase and tensin homolog by accelerating the maturation of pri-miR221/222, which ultimately promoted the proliferation of bladder cancer. Li et al (13) reported that, as an oncogene, METTL3 promoted CRC progression by maintaining sex determining region Y-Box 2 (SOX2) expression in a m6A-insulin-like growth factor 2 mRNA binding protein 2-dependent manner. However, it remains unclear as to whether METTL3-mediated m6A modification affects any long noncoding (lnc) RNAs in CRC.
Small nucleolar RNA host gene 1 (SNHG1) is an oncogenic lncRNA that serves a key role in CRC progression, such as in proliferation, metastasis, EMT and oncogenesis (14–18). In the present study, the association between METTL3-mediated m6A and SNHG1 in CRC was evaluated and the function of the METTL3/SNHG1 axis in CRC was assessed.
Materials and methods
Patients and tissue samples
Pairs of CRC and adjacent tissue specimens (n=74) were obtained from Liaoning Cancer Hospital and Institute (Shenyang, China) during surgical resection from January 2015 to September 2016. All tissue specimens were preserved in liquid nitrogen. The Medical Ethics Committee of Liaoning Cancer Hospital Research Institute approved the study (approval no. LNCANHOS-2018-012) and all patients signed written informed consent forms.
Cell culture
The normal human colonic epithelial NCM460 cell line and four human CRC cell lines (LOVO, RKO, SW480 and HT29) were purchased from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. All cells were cultured using RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc.) and supplemented with 10% foetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin (Shanghai Baoman Biotechnology Co., Ltd.) and 100 mg/ml streptomycin (Shanghai Baoman Biotechnology Co., Ltd.). All cells were maintained in a cell incubator with 5% CO2 at 37°C.
Bioinformatics analysis of data from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases
Data from two CR dsxdC-related GEO datasets, GSE41258 (19) and GSE41328 (20) were analysed using GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r) to analyse the expression of METTL3 in CRC (21). Probe nos. 209265_s_at and 213653_at represent METTL3 in datasets GSE41258 and GSE41328, respectively. The expression and relationship of METTL3 and SNHG1 using data from TCGA (https://portal.gdc.cancer.gov/) were analysed using the University of ALabama at Birmingham CANcer data analysis portal (UALCAN; http://ualcan.path.uab.edu/index.html), according to the website's instructions (22). Potential m6A modification sites of SNHG1 were predicted using RMBase (V2.025; http://rna.sysu.edu.cn/rmbase/index.php), according to the website's instructions (23).
Immunohistochemistry (IHC) staining
IHC staining was performed according to a previously described method (24). CRC tissue were fixed with 10% formalin at room temperature for 48 h, dehydrated via gradient alcohol, paraffin-embedded, sliced (4-µm thick), dewaxed in xylene (for 10 min, repeated three times) and then rehydrated using a descending alcohol series, followed by antigen retrieval using Target Retrieval Solution (Dako; Agilent Technologies, Inc.), according to the manufacturer's instructions. Hydrogen peroxide (3%) was applied for 15 min to block endogenous peroxidase activity at room temperature. Sections were sealed using 10% Rangoat serum (Wuhan Servicebio Technology Co., Ltd.) for 5 min at room temperature. Incubation with anti-METTL3 rabbit monoclonal primary antibodies (1:500; cat. no. ab195352; Abcam) was performed overnight at 4°C in the refrigerator and then the next day with biotinylated HRP-conjugated goat anti-rabbit immunoglobulin G secondary antibodies at 37°C for 30 min (1:2,000; cat. no. ab205718; Abcam). This was followed by incubation with 2 µg/ml streptavidin maleate peroxidase for 30 min at 37°C (LSAB kit; Dako; Agilent Technologies, Inc.), staining with 3,3-diaminobenzidine color development kit for 20 min in the dark and then counterstaining with haematoxylin at room temperature for 2 min. The sections were then dehydrated and finally mounted. Control sections of tissue were processed under the same conditions but did not contain primary antibodies. Images were observed using a fluorescence microscope.
RNA extraction and reverse transcription (RT)-quantitative PCR (qPCR)
Total RNA was extracted from CRC tissues and 5×105 cells of each cell line using TRIzol® reagent according to the manufacturer's instructions (Invitrogen; Thermo Fisher Scientific, Inc.). Reverse transcription was performed using PrimeScript RT Master Mix at 37°C for 15 min and 85°C for 5 sec (Takara Biotechnology Co., Ltd.). Takara Biotechnology TB green premix Ex Taq™ II (cat. no. RR820A) was used for the qPCR, and the thermocycling conditions were as follows: 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec, and then 95°C for 15 sec and 60°C for 60 sec. The relative expression of METTL3 or SNHG1 was quantified using the 2−ΔΔCq method (25) and β-actin was used as an internal control (Takara Biotechnology Co., Ltd.). Primer sequences are listed in Table IA.
Table I.
Primer and oligonucleotide sequences used in the present study.
| A, Primer sequences used in the present study | |
|---|---|
|
| |
| Name | Sequence (5′-3′) |
| METTL3 | F: TTGTCTCCAACCTTCCGTAGT |
| R: CCAGATCAGAGAGGTGGTGTAG | |
| SNHG1 | F: GCACGTTGGAACCGAAGAGA |
| R: GCAGCTGAATTCCCCAGGATA | |
| β-actin | F: CTTCTACAATGAGCTGCGTG |
| R: TCATGAGGTAGTCAGTCAGG | |
|
| |
| B, Oligonucleotide sequences used in the present study | |
|
| |
| Name | Sequence (5′-3′) |
|
| |
| METTL3 | F: CTTGGTACCGAGCTCGGATCCATGTCGGACACGTGGAGCTC |
| R: TGCTGGATATCTGCAGAATTCGCTCTGTAAGGAAGTGCTTC | |
| shMETTL3#1 | F: CCGGGCAAGAATTCTGTGACTATGGCTCGAGCCATAGTCACAGAATTCTTGCTTTTTG |
| R: AATTCAAAAAGCAAGAATTCTGTGACTATGGCTCGAGCCATAGTCACAGAATTCTTGC | |
| shMETTL3#2 | F: CCGGGCTGCACTTCAGACGAATTATCTCGAGATAATTCGTCTGAAGTGCAGCTTTTTG |
| R: AATTCAAAAAGCTGCACTTCAGACGAATTATCTCGAGATAATTCGTCTGAAGTGCAGC | |
| shNC | TTCTCCGAACGTGTCACGT |
| siSNHG1-1 | GAAACAGCAGTTGAGGGTTTG |
| siSNHG1-2 | GGTTTGCTGTGTATCACATTT |
| siSNHG1-3 | GCCAATTGTTGATTGAACTTC |
| siNC | UUCUCCGAACGUGUCACGUTT |
METTL3, methyltransferase-like 3; SNHG1, small nucleolar RNA host gene 1; sh, short hairpin RNA; NC, negative control; si, small interfering RNA; F, forward; R, reverse.
Western blotting
Western blotting was conducted as described previously (26). Total proteins were extracted from NCM460, HT29, LOVO and PKO cell lines using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA) and protein concentrations were then semi-quantified using a BCA protein assay kit (Santa Cruz Biotechnology, Inc.) Proteins (10 µl/lane) were separated by 10% SDS-PAGE and then transferred to PVDF membranes (Amresco, LLC). The membranes were blocked with 5% BSA (Sigma-Aldrich; Merck KGaA) for 1 h at room temperature, after which the blocking solution was removed and anti-METTL3 rabbit monoclonal (1:1,000; cat. no. ab195352; Abcam) and anti-GAPDH mouse monoclonal (1:500; cat. no. ab8245; Abcam) primary antibodies were refrigerated at 4°C and incubated with the membranes overnight. Primary antibodies were then washed away with TBST (0.1% Tween) and goat anti-mouse immunoglobulin G HRP-conjugated secondary antibodies (1:2,000; cat. no. ab205719; Abcam) were added at room temperature for 1 h. The ECL Western Blotting substrate kit (cat. no. ab65623; Abcam) was utilized to visualise the target proteins and ImageJ software (v2; National Institutes of Health) was used to analyse the protein bands.
Cell Counting Kit-8 (CCK-8) assay
HT29 and LOVO cells were seeded in 96-well plates at a density of 2×103 cells/well and incubated at 37°C with 5% CO2. On days 1–5, 10 µl CCK-8 solution (Dojindo Molecular Technologies, Inc.) was added to each well and incubated at 37°C. After 2 h, the 96-well plates were removed and a microplate reader (Bio-Rad Laboratories, Inc.) was used to measure the absorbance at 450 nm. Experiments were performed in triplicate.
Transwell assay
The assay was performed as previously published (27). LOVO and HT29 cells (5×104 for migration assays and 1×105 for invasion assays) were seeded into uncoated or Matrigel-precoated (BD Biosciences; 4 h at 37°C) upper chambers (Corning, Inc.). Serum-free medium was added to both upper chambers and medium containing 10% FBS was added to the lower cell chambers. The cells were incubated at 37°C with 5% CO2 for 12 h, after which the cells remaining in the upper chamber were removed and those in the lower chamber were fixed using anhydrous ethanol at room temperature for 30 min, stained with 1% crystal violet for 1 h and counted using an inverted microscope (Leica Microsystems GmbH).
Plasmid and oligonucleotide transfection
Specific lentivirus short hairpin (sh) RNA targeting METTL3, negative control shRNA (shNC) and METTL3 overexpression plasmids were synthesized by Shanghai GenePharma Co., Ltd. Specific small interfering (si) RNAs targeting SNHG1 and negative control (NC) siRNA were chemically synthesized by Guangzhou RiboBio Co., Ltd. To obtain cells with stable knockdown or overexpression of METTL3, 3×105 HT29 and LOVO cells were transfected with the aforementioned constructed plasmids (140 µg/ml; 2 µl; siSHNG1) and selected with Geneticin (G418; 800 µg/ml; Procell Life Science & Technology Co., Ltd.) for 4 weeks. METTL3 expression levels in selected cell clones were confirmed using RT-qPCR (as per the aforementioned method) and the constructed cell clones with stable overexpressed or knocked down METTL3 were used for further functional assays. To determine the effect of SNHG1 on METTL3, HT29 and LOVO cells with stable overexpression of METTL3 were transfected with SNHG1 siRNAs at 37°C for 24 husing the RiboFECT™ Transfection Kit (166T; Guangzhou RiboBio Co., Ltd.), performed according to the manufacturer's protocols. RT-qRCR assay was performed 24 h after transfection to determine knockdown efficiency, and then follow-up experiments were performed. The sequences of siRNAs are listed in Table IB.
Methylated RNA immunoprecipitation (MeRIP) qPCR
MeRIP-qPCR assays were performed to determine the m6A modification level of SNHG1 as previously reported (13). All procedures were carried out using the Magna MeRIP™ m6A Kit (cat. no. 1710499; Merck KGaA), according to the manufacturer's instructions. In brief, total RNA was isolated as described above and fragmented using 2 µl RNA fragmentation buffer. The input control was one-tenth of the isolated RNA saved. The magnetic bead A/G blend was washed three times, resuspended and incubated with 500 µl MeRIP reaction fluid (containing fragmental RNAs, RNase inhibitor and IP buffer) at 4°C for 2 h. The beads were then harvested by The beads were then harvested by Magnetic rack adsorption to remove superessence and eluted with elution buffer (containing IP buffer, 20 mM m6A and RNase inhibitor; from the MeRIP kit) in a vertical mixer (4°C, 5 min, 10 rpm/min). Eluted RNA fragments were harvested and purified using the A&D Pure TRIzol Total RNA Purification Kit (A&D Co, Ltd). Enrichment levels were determined using qPCR (as per the aforementioned method) and the corresponding m6A enrichment level of each sample was calculated by normalizing the input data (RNA from target sample without m6A antibodies).
Statistical analysis
Data were analysed and evaluated using GraphPad Prism V7.0 (GraphPad Software; Dotmatics) software and SPSS 19.0 statistical software (IBM Corp.). Differences between groups were analysed using unpaired Student's t test or one-way ANOVA followed by a post hoc LSD or Tukey's test. Kaplan-Meier analysis was used to estimate overall survival using the log-rank test. P<0.05 was considered to indicate a statistically significant difference. All data were from three independent repeated experiments and are expressed as the mean ± standard deviation.
Results
METTL3 is upregulated in patients with CRC
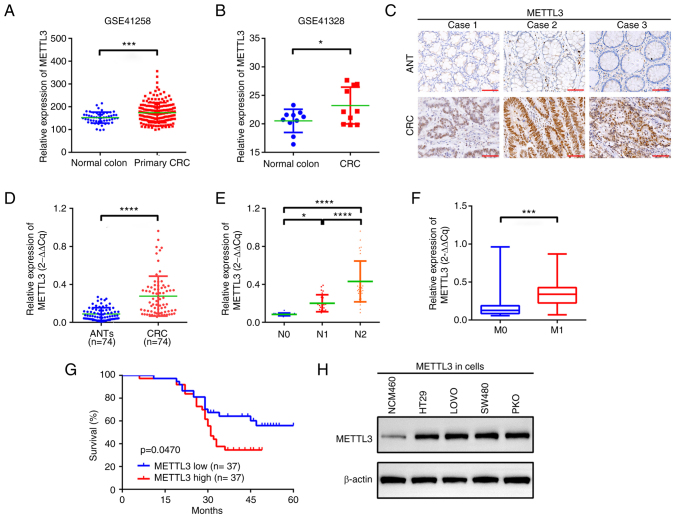
METTL3 mRNA expression was initially analysed using the GEO datasets GSE41258 and GSE41328. CRC tissues demonstrated significantly higher METTL3 protein expression compared with that in normal colonic tissues (Fig. 1A and B). Additionally, METTL3 expression was significantly increased in CRC tissue samples compared with that in paired adjacent non-tumour tissue samples (Fig. 1C and D). METTL3 protein expression was also demonstrated to be upregulated in CRC cells (Fig. 1H). High METTL3 expression was significantly associated with lymph node and metastasis stages and a significantly shorter overall survival in patients with CRC, compared with those with low METTL3 expression (Fig. 1E-G). However, other factors were not considered. These findings indicated that METTL3 may be a promising biomarker of CRC.
Figure 1.
METTL3 is upregulated in patients with CRC. METTL3 expression in CRC, demonstrated by analysis of datasets from two genome-wide studies, (A) GSE41258 and (B) GSE41328. (C) METTL3 expression in CRC and in paired ANTs, assessed using immunohistochemistry. Scale bar, 20 µm. (D) Differential expression of METTL3 in CRC tissues and ANTs, evaluated using reverse transcription-quantitative PCR. (E) The expression of METTL3 in patients with advanced N stage compared with those with low N stage. (F) METTL3 expression in patients with M1 stage compared with those with M0 stage. (G) Kaplan-Meier analysis indicating overall survival of patients with high METTL3 expression compared with that of patients with low METTL3 expression. n=37 for each group. (H) Expression of METTL3 in NCM460, LOVO, RKO, SW480 and HT29 cells, measured using western blotting. *P<0.05; ***P<0.001; ****P<0.0001. METTL3, methyltransferase-like 3; CRC, colorectal cancer; ANT, adjacent nontumor tissue; N stage, lymph node stage; M stage; metastasis stage.
The data above showed that METTL3 knockdown inhibits CRC migration and proliferation
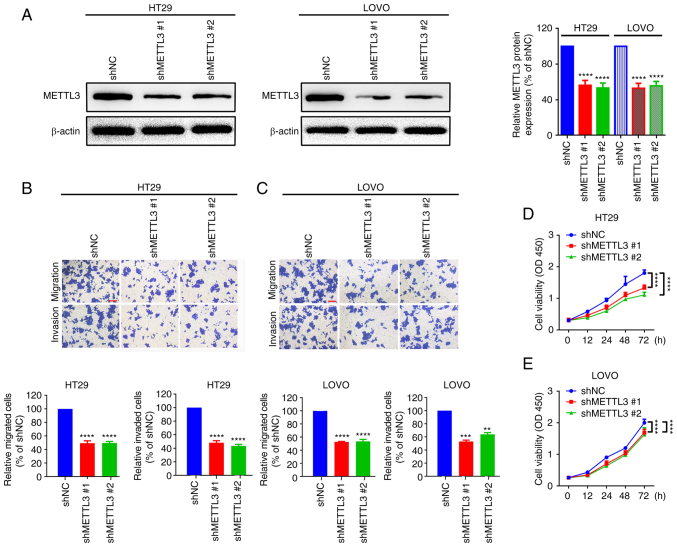
METTL3 shRNAs were transfected into two CRC cell lines, HT29 and LOVO, to determine the functional role of METTL3 in CRC cells. Western blotting demonstrated that METTL3 was successfully knocked down in HT29 and LOVO cells (Fig. 2A). The migration of HT29 and LOVO cells was then assessed and was demonstrated to have significantly decreased after METTL3 knockdown, compared with that in the shNC groups (Fig. 2B and C). Finally, a CCK-8 assay was performed, and compared with that of the shNC group, the proliferation of HT29 and LOVO cells was significantly decreased when METTL3 was knocked down (Fig. 2D and E). The data showed that METTL3 knockdown inhibits CRC cell migration and proliferation.
Figure 2.
Knockdown of METTL3 suppresses the migration and proliferation of HT29 and LOVO cells. (A) METTL3 protein expression after transfection of specific METTL3 short hairpin RNAs, measured using western blotting. ****P<0.0001. Cell motility changes in (B) HT29 and (C) LOVO cells after knockdown of METTL3, assessed using Transwell assays. Scale bar, 200 µm. Changes in the proliferation of (D) HT29 and (E) LOVO cells after knockdown of METTL3, evaluated using Cell Counting Kit-8 assays. **P<0.01 vs. shNC; ***P<0.001 vs. shNC; ****P<0.0001 vs. shNC. METTL3, methyltransferase-like 3; NC, negative control; sh, short hairpin RNA; OD, optical density.
METTL3-mediated m6A modification is associated with SNHG1 stability in HT29 and LOVO cells
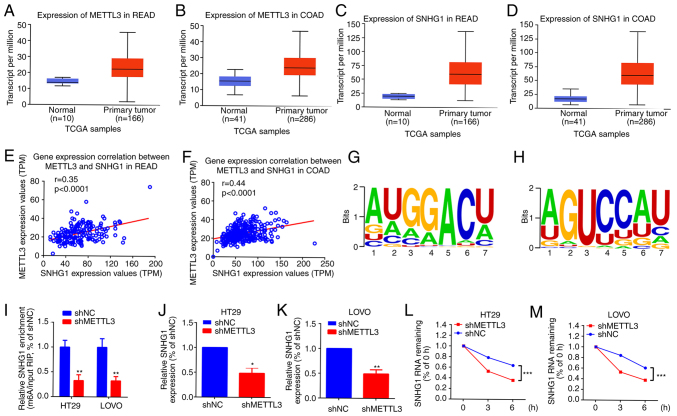
Previous studies reported that METTL3 is closely implicated in certain lncRNA m6A modifications (28–30). SNHG1 is an oncogenic lncRNA in certain cancers, including CRC (16,31,32). Analysis of METTL3 and SNHG1 expression data from TCGA using the online software UALCAN (22), both METTL3 and SNHG1 were demonstrated to be notably upregulated in CRC primary tumour samples, compared with that in normal samples (Fig. 3A-D). Additionally, a significant positive correlation was demonstrated between METTL3 and SNHG1 expression in CRC (Fig. 3E and F). Moreover, SNHG1 contained 18 m6A modification sites, demonstrated using RMBase 2.0 (23) (Table SI) and METTL3 provided six m6A binding sites for SNHG1 (Table SII; Fig. 3G and H). The m6A level of SNHG1 in cells with different METTL3 expression levels was then assessed and compared with that in the shNC groups, the m6A level of SNHG1 was significantly decreased in the METTL3 knockdown groups in both HT29 and LOVO cells (Fig. 3I). SNHG1 expression was also significantly reduced in METTL3 knockdown groups in both HT29 and LOVO cells, compared with that in shNC groups (Fig. 3J and K). Furthermore, to evaluate the relationship between METTL3-mediated m6A modification and SNHG1 upregulation, 2 µmol/l actinomycin D was added to HT29 and LOVO cells with knocked down METTL3 and RT-qPCR was used to assess the half-life of SNHG1. It was demonstrated that the half-life of SNHG1 was significantly reduced in groups with downregulated METTL3 in HT29 and LOVO cells, compared with that in the shNC groups (Fig. 3L and M). These results indicated that METTL3 affected the stability of SNHG1 in an m6A-dependent manner.
Figure 3.
METTL3-mediated m6A modification is associated with SNHG1 upregulation in HT29 and LOVO cells. The expression of METTL3 in (A) READ and (B) COAD and the expression of SNHG1 in (C) READ and (D) COAD, using data from TCGA, analysed using the online software UALCAN (http://ualcan.path.uab.edu/index.html). The correlation between the expression of METTL3 and SNHG1 in (E) READ r=0.35 and (F) COAD r=0.44, assessed using Spearman correlation analysis. (G) and (H) METTL3 binding motifs in the exon region of SNHG1, predicted using RMBase (version 2.0; http://rna.sysu.edu.cn/rmbase/). (I) m6A level of SNHG1 in different METTL3-expressing cells, assessed using methylated RNA immunoprecipitation quantitative PCR. The expression of SNHG1 after METTL3 knockdown in (J) HT29 and (K) LOVO cells, measured using RT-qPCR. The level of SNHG1 in METTL3-expressing cells (L) HT29 and (M) LOVO after actinomycin D intervention, quantified using RT-qPCR. *P<0.05; **P<0.01; ***P<0.001. METTL3, methyltransferase-like 3; m6A, N6-methyladenosine; SNHG1, small nucleolar RNA host gene 1; READ, rectal adenocarcinoma; COAD, colon adenocarcinoma; TCGA, The Cancer Genome Atlas; UALCAN, University of ALabama at Birmingham CANcer data analysis portal; RT-qPCR, reverse transcription-quantitative PCR; TPM, transcripts per million; RIP, RNA immunoprecipitation; NC, negative control; sh, short hairpin RNA.
SNHG1 knockdown partially attenuates the facilitative effect of METTL3 on migration and proliferation in HT29 and LOVO cells
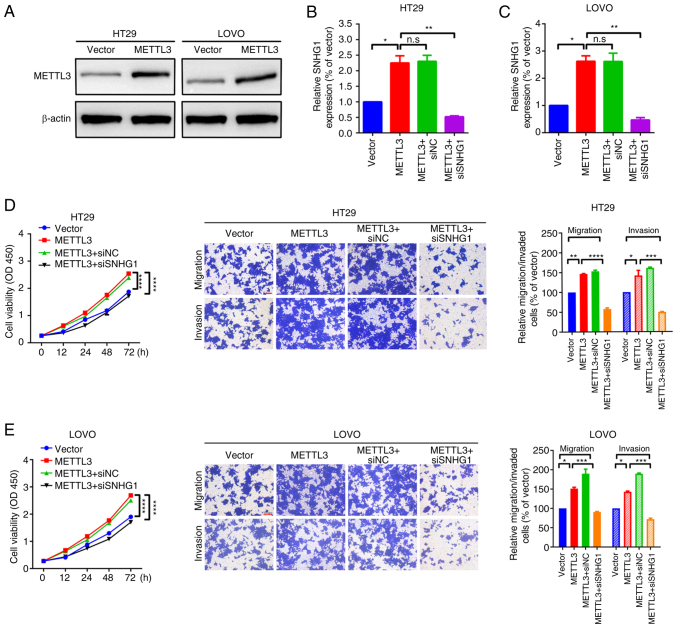
SNHG1-related loss-of-function assays were performed to further assess the relationship between SNHG1- and METTL3-mediated promotion of cell migration and proliferation. Initially, HT29 and LOVO cell models with stable METTL3 overexpression were constructed (Fig. 4A). Specific SNHG1 siRNAs were transfected into METTL3-overexpressing HT29 and LOVO cells and SNHG1 expression was quantified by RT-qPCR (Fig. S1A and B). Overexpression of METTL3 significantly increased the level of SNHG1 expression, compared with vector group, whilst specific SNHG1 siRNAs significantly reduced SNHG1 expression, compared with METTL3-siNC group (Fig. 4B and C). Functional CCK-8 and Transwell assays were then performed to evaluate the role of SNHG1 in METTL3-mediated cell proliferation and cell motility. Overexpression of METTL3 significantly promoted proliferation and migration compared with the empty vector control group; however, this facilitative effect was significantly reversed by knockdown of SNHG1 in HT29 and LOVO cells when compared with the METTL3-siNC group (Fig. 4D and E). These findings indicated that knockdown of SNHG1 partially attenuated the facilitative effect of METTL3 on HT29 and LOVO cell migration and proliferation.
Figure 4.
Knockdown of SNHG1 partially attenuated the facilitative effect of METTL3 on migration and proliferation in HT29 and LOVO cells. (A) The expression of METTL3 protein in HT29 and LOVO cells, measured using western blotting. The expression level of SNHG1 in (B) HT29 and (C) LOVO cells, assessed using RT-qPCR. Cell proliferation, invasion and migration of (D) HT29 and (E) LOVO cells, determined using CCK-8 assays (left panel) and Transwell assays (right panel). Scale bar, 200 µm. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. SNHG1, small nucleolar RNA host gene 1; METTL3, methyltransferase-like 3; CCK-8; Cell Counting Kit-8; NC, negative control; si, small interfering RNA; n.s, non-significance; OD, optical density.
Discussion
RNA modifications are common features in epigenetic regulation in numerous human diseases, including cancer (33). One of the most common RNA modifications is m6A, accounting for ~50% of total methylated ribonucleotides and 0.1-0.4% of all adenosine in total cellular RNA (34). m6A regulates cellular processes, including cell self-renewal, differentiation, invasion and apoptosis, and it is also extensively implicated in neoplastic diseases, including CRC, osteosarcoma, lung cancer and ovarian cancer (29,35–37). m6A methyltransferases, demethylases and reader proteins write, remove and recognise m6A, respectively (38).
METTL3, first purified in 1994 from HeLa cell nuclei (39), is the core component of the m6A methyltransferase complex (MTC) and contains 580 amino acids; it functions as the catalytic subunit in the MTC, using adenosylmethionine as the methyl donor (9,40). METTL3 acts as an m6A methyltransferase in numerous cancers, including liver, gastric, lung, pancreatic, bladder, prostate and breast cancers, CRC and acute myeloid leukaemia (10). In line with previously reported findings, analysis of GEO datasets GSE41258 and GSE41328 and experimental research in the present study demonstrated that METTL3 was upregulated and correlated with poor features such as lymph node and metastasis stages, and a significantly shorter overall survival time in patients with CRC. Functional CCK-8 and Transwell assays demonstrated that METTL3 knockdown suppressed the migration and proliferation of HT29 and LOVO cells, suggesting that METTL3 serves as an oncogene in CRC. The downstream targets of METTL3 in CRC include SOX2, chromobox 8, hexokinase 2 and solute carrier family 2 member 1 (13,41,42). Additionally, METTL3 participates in the m6A modification of several noncoding RNAs, including lncRNA RP11-138 J23.1, primary-microRNA-1246 and circular RNA NOP2/Sun RNA methyltransferase 2 (43–45). In the present study, only the relationship between METTL3 and SNHG1 was evaluated.
SNHG1, located in the 11q12.3 region of the chromosome, contains 11 exons and is a host to eight small nucleolar RNAs from its spliced intron (46). As an oncogenic lncRNA, SNHG1 is aberrantly expressed in numerous malignancies, including colorectal, liver, lung and prostate cancer, and promotes tumourigenesis via diverse signalling pathways (16,31). SNHG1 upregulation promotes CRC progression through multiple mechanisms, such as microRNA sponging, Wnt/β-catenin signal activation and p53 pathway modulation (14,18,32,47). Previous studies have reported that m6A modification affects the stability of several lncRNAs, such as X inactive-specific transcript, growth arrest-specific 5 and metastasis associated lung adenocarcinoma transcript 1 (48–50). In the present study, it was demonstrated that SNHG1 was positively correlated with METTL3. Additionally, it was demonstrated that METTL3 affected the m6A level of SNHG1 and the half-life of SNHG1. Furthermore, it was demonstrated via online bioinformatics analysis that SNHG1 supplied 18 m6A modification sites. Furthermore, knockdown of SNHG1 was demonstrated to have partially attenuated the facilitative effect of METTL3 on migration and proliferation in CRC cells. The findings from the present study together suggest that SNHG1 is a downstream target of METTL3 in CRC.
CRC carcinogenesis is an intricate biological process involving numerous factors and molecules. In the present study, it was demonstrated that METTL3 promotes the proliferation and migration of CRC cells via SNHG1 m6A modification. The findings of the present study therefore suggest that the inhibition of METTL3 may provide a new therapeutic target in the molecular treatment of CRC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Science and Technology Innovation Fund for Master's Students of Shenyang Medical College (grant no. Y20210515).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XW conceived and designed the study, and revised the manuscript. YX, YB and GQ performed the experiments, prepared the figures, and designed Table I, SI and SII. YB and GQ confirm the authenticity of all the raw data. YX, HY and MH developed the methods and performed data analysis. YX and MH acquired and interpreted the data, and wrote and revised the paper. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
All experiments were approved by the Animal Ethics and Laboratory Committee of Affiliated Central Hospital of Shenyang Medical Science (approval no. 20220528). All individuals provided written informed consent for the use of their tissues in the present study
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Morgan E, Arnold M, Gini A, Lorenzoni V, Cabasag CJ, Laversanne M, Vignat J, Ferlay J, Murphy N, Bray F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut. 2023;72:338–344. doi: 10.1136/gutjnl-2022-327736. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.De Rosa M, Pace U, Rega D, Costabile V, Duraturo F, Izzo P, Delrio P. Genetics, diagnosis and management of colorectal cancer (Review) Oncol Rep. 2015;34:1087–1096. doi: 10.3892/or.2015.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244:254–259. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 10.Zeng C, Huang W, Li Y, Huang W. Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. J Hematol Oncol. 2020;13:117. doi: 10.1186/s13045-020-00951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, Zhao G. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18:142. doi: 10.1186/s12943-019-1065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, Chen ZH, Zeng ZL, Wang F, Zheng J, et al. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, Li B, Liu Z, Jiang L, Wang G, Lv M, Li D. Up-regulation of lncRNA SNHG1 indicates poor prognosis and promotes cell proliferation and metastasis of colorectal cancer by activation of the Wnt/β-catenin signaling pathway. Oncotarget. 2017;8:111715–111727. doi: 10.18632/oncotarget.22903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avazpour N, Hajjari M, Kazemi Nezhad SR, Tahmasebi Birgani M. SNHG1 long noncoding RNA is potentially up-regulated in colorectal adenocarcinoma. Asian Pac J Cancer Prev. 2020;21:897–901. doi: 10.31557/APJCP.2020.21.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thin KZ, Tu JC, Raveendran S. Long non-coding SNHG1 in cancer. Clin Chim Acta. 2019;494:38–47. doi: 10.1016/j.cca.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Sun X, Wang Z, Yuan W. Down-regulated long non-coding RNA SNHG1 inhibits tumor genesis of colorectal carcinoma. Cancer Biomark. 2017;20:67–73. doi: 10.3233/CBM-170112. [DOI] [PubMed] [Google Scholar]
- 18.Bai J, Xu J, Zhao J, Zhang R. lncRNA SNHG1 cooperated with miR-497/miR-195-5p to modify epithelial-mesenchymal transition underlying colorectal cancer exacerbation. J Cell Physiol. 2020;235:1453–1468. doi: 10.1002/jcp.29065. [DOI] [PubMed] [Google Scholar]
- 19.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, Paty PB, Gerald WL, Notterman DA, Domany E. Association of survival and disease progression with chromosomal instability: A genomic exploration of colorectal cancer. Proc Natl Acad Sci USA. 2009;106:7131–7136. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin G, He X, Ji H, Shi L, Davis RW, Zhong S. Reproducibility Probability Score-incorporating measurement variability across laboratories for gene selection. Nat Biotechnol. 2006;24:1476–1477. doi: 10.1038/nbt1206-1476. [DOI] [PubMed] [Google Scholar]
- 21.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res 41(Database issue) 2013:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xuan JJ, Sun WJ, Lin PH, Zhou KR, Liu S, Zheng LL, Qu LH, Yang JH. RMBase v2.0: Deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res 46(D1) 2018:D327–D334. doi: 10.1093/nar/gkx934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haderk F, Olivas V, Bivona TG. Immunohistochemistry to study YAP in human tissue samples. Methods Mol Biol. 2019;1893:89–95. doi: 10.1007/978-1-4939-8910-2_7. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Zeng X, Wang N, Zhao W, Zhang X, Teng S, Zhang Y, Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p-TGF-β1 axis. Mol Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J, Wang J, Cao M, Cai J, Wu J, Wang X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13:5. doi: 10.1186/s13045-019-0839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue L, Li J, Lin Y, Liu D, Yang Q, Jian J, Peng J. m6 A transferase METTL3-induced lncRNA ABHD11-AS1 promotes the Warburg effect of non-small-cell lung cancer. J Cell Physiol. 2021;236:2649–2658. doi: 10.1002/jcp.30023. [DOI] [PubMed] [Google Scholar]
- 30.Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, Di W, Hu B, An J, Kong L, et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12:135. doi: 10.1186/s13045-019-0830-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Huang L, Jiang X, Wang Z, Zhong X, Tai S, Cui Y. Small nucleolar RNA host gene 1: A new biomarker and therapeutic target for cancers. Pathol Res Pract. 2018;214:1247–1252. doi: 10.1016/j.prp.2018.07.033. [DOI] [PubMed] [Google Scholar]
- 32.Fu Y, Yin Y, Peng S, Yang G, Yu Y, Guo C, Qin Y, Zhang X, Xu W, Qin Y. Small nucleolar RNA host gene 1 promotes development and progression of colorectal cancer through negative regulation of miR-137. Mol Carcinog. 2019;58:2104–2117. doi: 10.1002/mc.23101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20:303–322. doi: 10.1038/s41568-020-0253-2. [DOI] [PubMed] [Google Scholar]
- 34.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang Z, Hu Y, Hu J, Huang Y, Zheng S, Guo C. The crucial roles of N6-methyladenosine (m6A) modification in the carcinogenesis and progression of colorectal cancer. Cell Biosci. 2021;11:72. doi: 10.1186/s13578-021-00583-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C, Meng Y, Zhao J, Ma J, Zhao Y, Gao R, Liu W, Zhou X. Deubiquitinase USP13 regulates glycolytic reprogramming and progression in osteosarcoma by stabilizing METTL3/m6A/ATG5 axis. Int J Biol Sci. 2023;19:2289–2303. doi: 10.7150/ijbs.82081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bi X, Lv X, Liu D, Guo H, Yao G, Wang L, Liang X, Yang Y. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 2021;28:335–349. doi: 10.1038/s41417-020-00222-3. [DOI] [PubMed] [Google Scholar]
- 38.Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269:17697–17704. doi: 10.1016/S0021-9258(17)32497-3. [DOI] [PubMed] [Google Scholar]
- 40.Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X, Zhang X, Cao Y, Ma D, Zhu X, et al. m6A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19:72. doi: 10.1186/s12943-020-01190-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Kang M, Zhang B, Meng F, Song J, Kaneko H, Shimamoto F, Tang B. m6A modification-mediated CBX8 induction regulates stemness and chemosensitivity of colon cancer via upregulation of LGR5. Mol Cancer. 2019;18:185. doi: 10.1186/s12943-019-1116-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, Ji D, Wang Q, Zhang Z, Tang J, Sun Y. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38:393. doi: 10.1186/s13046-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, Han K, Chen JW, Judde JG, Deas O, et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, Li J, An P, Lu L, Luo N, et al. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18:87. doi: 10.1186/s12943-019-1014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tycowski KT, Shu MD, Steitz JA. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y, Qin ZS, Feng Y, Tang XJ, Zhang T, Yang L. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) promote cell proliferation in colorectal cancer by affecting P53. Eur Rev Med Pharmacol Sci. 2018;22:976–984. doi: 10.26355/eurrev_201802_14379. [DOI] [PubMed] [Google Scholar]
- 48.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, Liu J, Che L, Li J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer. 2019;18:143. doi: 10.1186/s12943-019-1079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, Sloan KE, Bohnsack MT. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–2014. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.