Abstract
Background
The identification of genetic mosaicism and the genetic counseling needed following its discovery have been challenging problems in the field of prenatal diagnosis. Herein, we describe the clinical phenotypes and various prenatal diagnostic processes used for two rare cases of 9p duplication mosaicism and review the prior literature in the field to evaluate the merits of different methods for diagnosing mosaic 9p duplication.
Methods
We recorded ultrasound examinations, reported the screening and diagnosis pathways, and analyzed the mosaic levels of the two cases of 9p duplication using karyotype analysis, chromosomal microarray analysis (CMA), and fluorescence in situ hybridization analysis (FISH).
Results
Case 1 had a normal clinical phenotype for tetrasomy 9p mosaicism, and Case 2 showed multiple malformations caused by both trisomy 9 and trisomy 9p mosaicism. Both cases were initially suspected after non‐invasive prenatal screening (NIPT) based on cell‐free DNA. The mosaic ratio of 9p duplication found via karyotyping was lower than what was discovered by CMA and FISH, in both cases. Contrary to previous findings, the mosaic level of trisomy 9 found by karyotype analysis was greater than what was found by CMA, in terms of complex mosaicism involving trisomy 9 and trisomy 9p, in Case 2.
Conclusion
NIPT can indicate 9p duplication mosaicism during prenatal screening. Different strengths and limitations existed in terms of diagnosing mosaic 9p duplication by karyotype analysis, CMA, and FISH. The combined use of various methods may be capable of more accurately determining break‐points and mosaic levels of 9p duplication during prenatal diagnosis.
Keywords: 9p duplication, genetic analysis techniques, mosaicism, prenatal diagnosis
We described the clinical phenotypes and various prenatal diagnostic processes used for two rare cases of 9p duplication mosaicism and reviewed the prior literature in the field to evaluate the merits of different methods for diagnosing mosaic 9p duplication.
1. INTRODUCTION
The biological phenomenon known as “mosaicism” is characterized by the coexistence of two or more distinct cell lines in the same individual (Zhang et al., 2021). Genetic material variation can be caused by changes in DNA sequence, copy number, rearrangements, or chromosome number. Mosaicism can occur during either mitosis or meiosis. When the resultant genetic changes occur somatically, the individual is composed of cells with at least two different genotypes and thus is considered to have mosaicism. This condition is related to a variety of chromosomal abnormalities, including trisomy, monosomy, deletion, duplication, translocation, inversion, ring, isochromosome, and other rare aberrations. Mosaicism has a broad clinical impact, ranging from no phenotype to organ‐specific defects, to multi‐systemic abnormalities, to early spontaneous miscarriage (Spinner & Conlin, 2014). The form of mutation, degree of mosaicism, and tissue distribution of aberrant cell lines all play significant roles in determining how severe the clinical symptoms of mosaicism are in each case (Chen et al., 2007).
Karyotype analysis and fluorescence in situ hybridization analysis (FISH) have long been used for the detection of mosaicism, up until the more recent advent of new molecular genetic techniques, such as next‐generation sequencing of noninvasive prenatal testing (NIPT) and chromosomal microarray analysis (CMA). Since 2011, NIPT has become the standard screening method for detecting common fetal chromosomal aneuploidies (i.e., 13/18/21) based on massively parallel sequencing of whole‐genome cell‐free DNA. This technique has false positive and false negative rates in the range of 0.1%–0.2% (Bianchi et al., 2014; Song et al., 2013; Taylor‐Phillips et al., 2016). In recent years, some have advocated for diagnosing chromosomal abnormalities by combining karyotyping with CMA. Both methods have advantages and disadvantages for detecting mosaicism as follows: (1) CMA has a shorter turnaround time than karyotyping because CMA extracts DNA directly from cells in amniotic fluid rather than requiring extensive cell culture; (2) prolonged cell cultivation and manual cell selection may cause fluctuations in mosaic levels by karyotype analysis; (3) CMA can identify chromosomal microdeletions and microduplications with a resolution of 50–100 kbp, but it cannot identify balanced structural abnormalities and regions uncovered by oligonucleotide probes; and (4) conventional karyotyping is limited to detecting only rearrangements involving more than 5 Mbp (Hao et al., 2020).
The advent of new technologies provides additional options for the detection of mosaicism, while also creating significant new uncertainties. Few publications have described the technical potential of NIPT for rare chromosomal aneuploidies and mosaicism, and its accuracy and effectiveness have yet to be well documented (Lee et al., 2018). Different mosaic levels discovered using different techniques have frequently been reported when diagnosing chromosomal aneuploidy mosaicism (Hao et al., 2020). Mosaicism events with segmental chromosomal duplication are rare, and their detection rate is low in clinical practice. The comparability of different assay techniques in terms of diagnosing mosaic segmental chromosome duplication has yet to be reported on in the literature. Herein, we describe the prenatal testing process for two unusual cases of mosaic 9p duplication. We also review the variations observed by others when detecting 9p duplication mosaicism using different assays, in previously reported studies in the literature.
2. MATERIALS AND METHODS
2.1. Ethical compliance
This study was approved by the Ethics Committee of the Zhuhai Center for Maternal and Child Health Care. Informed consent was obtained from the two study subjects.
2.2. Case presentation
Invasive amniocentesis was recommended for two healthy pregnant women with indications of high risks of chromosome 9 (Chr9) duplication according to NIPT results. Both received pre‐test counseling that included topics, such as the merits and drawbacks of invasive prenatal diagnosis, testing procedures, possible results, and the limitations of various approaches.
In Case 1, NIPT was performed at 13 weeks of gestation on a 32‐year‐old woman who was gravida 5, para 1 and had had three prior abortions. Amniocentesis was carried out at 22 weeks of gestation for G‐banding karyotype, CMA, and FISH.
In Case 2, NIPT was performed at 12 + 4 weeks of gestation on a 32‐year‐old woman who was gravida 2, para 1. At 23 + 4 weeks of gestation, amniocentesis was carried out for karyotype analysis and CMA.
2.3. Noninvasive prenatal testing
NIPT was used for fetal chromosome screening in early pregnancy based on cell‐free fetal DNA in maternal blood, at Zhuhai Center for Maternal and Child Health Care. A two‐step centrifugation procedure was used to isolate the maternal plasma at a temperature of 2–8°C. Cell‐free DNA extraction, library construction, and sequencing were then carried out using a Fetal Chromosome Aneuploidy (T21/T18/T13) Test Kit (Huada Biotech Co.), according to the manufacturer's instructions. A BGISEQ500 high‐throughput sequencing system was used to sequence the cell‐free DNA, with a joint probe anchor polymerization sequencing method and a sequencing depth of 0.1x. Bioinformatic analysis was performed using BGI Halos software.
2.4. Karyotype analysis
Amniotic fluid chromosomal karyotype samples were obtained through cell culture, harvesting, and Giemsa staining with a >550 band resolution. A Zeiss Axio Scope and IKAROS® software (Metasystems Corporation) were used for the karyotype analysis. Twenty cells were counted for non‐mosaicism, and 100 cells were counted for mosaicism.
2.5. Chromosomal microarray analysis
Genomic DNA was extracted from the uncultured amniotic fluid using a QIAamp Mini DNA Extraction Kit (QIAGEN). Chr21 short tandem repeat (STR) markers were analyzed by quantitative fluorescence polymerase chain reaction using a 21/18/13/X/Y Chromosome Aneuploidy Detection Kit (Darui Biotech Co.) to identify maternal DNA contamination. A CytoScan 750 K chip (Affymetrix Inc.) was used for CMA analysis, according to the manufacturer's instructions. The data were analyzed using Chromosome Analysis Suite 4.0 (Chas) software (Affymetrix Inc.) and annotated with genome version GRCH37/hg19.
2.6. Fluorescence in situ hybridization
FISH was utilized to confirm mosaic ratios and abnormal chromosome structures whenever mosaicism was discovered. In Case 1, PD‐L1 and CSP9 probes positioned at 9p24.3 and the centromeric region of Chr9, respectively, were used on uncultured amniocytes for the FISH investigation (Anbiping Inc.). Images were obtained using a BX51 epifluorescence microscope (Olympus, Tokyo, Japan) and Cytovision software (Applied Imaging). Mosaicism levels were evaluated in 100 cells.
3. RESULTS
3.1. Clinical features
Table 1 presents the clinical details of both cases, and Figure 1 shows some of their abnormal ultrasonography readings.
TABLE 1.
Clinical information of the two cases.
| Case | Maternal age (years) | NIPT | Karyotype | The mosaic levels by karyotype analysis | CMA | The mosaic levels by CMA | FISH | The mosaic levels by FISH | Ultrasound findings and outcome |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 32 | arr[hg19]9p24.3p11.2(609414‐46367678, 45.76 Mb) (Z‐score = 4.3) | 47,XX,+der(9)del(9)(q21q34)dup(9)(p12p24)[10]/ 46,XX[90] | 10% | arr[hg19]9p24.3p13.2(208454‐68216577)*2.43 | 21.50% | 47,XX,+der(9)del(9)(q21q34)dup(9)(p12p24)[25]/46,XX[75] | 25% | Strong light spot in left ventricle |
| 2 | 32 | Trisomy 9 (Z‐score = 7.8) | 47,XX,+9[82]/47,XX, +del(9)(q13)[14]/46,XX[4] | 82%/14% | arr[hg19]9p24.3q34.3(208454‐141018648)*2.75 | 50%/50% | NA | NA | IUGR, VSD, MCDK, Oligohydramnios, Single umbilical artery, Bipedal deformity, Rocker bottom clubfoot/TOP |
Abbreviations: CMA, chromosome microarray analysis; FISH, fluorescence in situ hybridization; IUGR, intrauterine growth retardation; MCDK, multicystic dysplastic kidney; NA, not available; NIPT, noninvasive prenatal testing; TOP, termination of pregnancy; VSD, ventricular septal defect.
FIGURE 1.
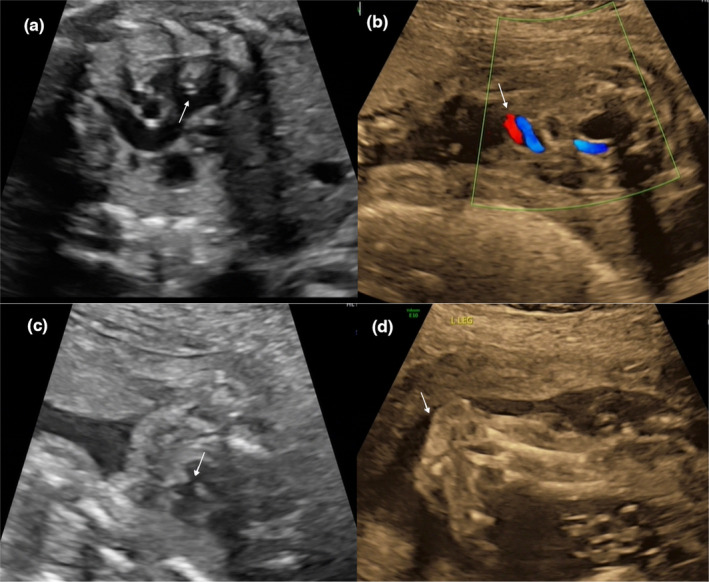
The abnormal ultrasonography readings of the two cases. Arrows indicate the deformity area. (a) Strong light spot in the left ventricle for Case 1. Partial abnormal ultrasound presentations for Case 2: (b) single umbilical artery, (c) ventricular septal defect, and (d) Rocker bottom clubfoot.
In Case 1, except for an abnormal soft marker with a strong bright spot in the left ventricle at 30 + 2 weeks of gestation, no other ultrasound abnormalities were detected throughout the pregnancy. A 2700 g female fetus was delivered at full term with normal physical findings at birth. The baby was phenotypically normal and had normal psychomotor and language development at 2.5 years old, at our last postnatal follow‐up.
In Case 2, a Level III ultrasound at 23 + 4 weeks of gestation revealed intrauterine growth retardation (IUGR) with a fetal biometry equivalent to 20 + 3 weeks of gestation, accompanied by a ventricular septal defect, one multicystic dysplastic kidney, oligohydramnios, a single umbilical artery, a bipedal deformity, and rocker‐bottom clubfoot. The pregnancy was terminated before full genetic results were available.
3.2. Noninvasive prenatal testing
The signal plot of the NIPT is shown in Figure 2. Both cases had Z‐scores for the common chromosomes (13/18/21) within the normal range (−3 < Z < 3).
FIGURE 2.
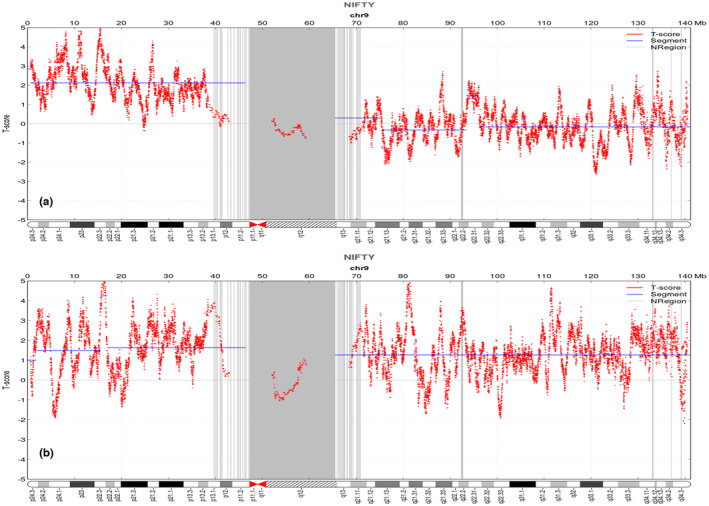
The NIPT signals of the two cases. The baseline value is zero, and the blue line represents the average signal value of corresponding chromosome region. (a) Case 1 demonstrates an increased short arm of Chr9 compared to the baseline. (b) Case 2 shows the whole Chr9 elevated relative to the baseline.
In Case 1, the short arm of Chr9 was duplicated, with a Z‐score of 4.3 based on the NIPT result. The finding on Chr9 suggested a gain of approximately 45.7 Mb in size, encompassing chromosome bands 9p24.3‐9p11.2 (GRCh37:Chr9:609414‐46367678).
Case 2 showed an increased level of DNA for the whole Chr9, with a Z‐score of 7.8, suggesting trisomy 9.
3.3. Karyotyping
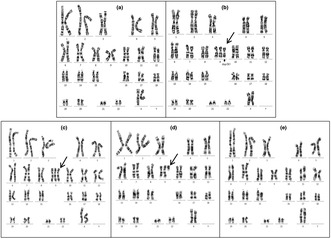
After counting 100 metaphase spreads by karyotyping in Case 1, 10 aberrant cells with extra marker chromosomes and 90 normal cells were noted. They exhibited a karyotype of 47,XX,+der(9)del(9)(q21q34)dup(9)(p12p24)[10]/46,XX[90], implying a 10% (10/100) mosaicism of tetrasomy 9p when the results were cross‐referenced with CMA to determine the origin of the supernumerary marker chromosome (Figure 3).
FIGURE 3.
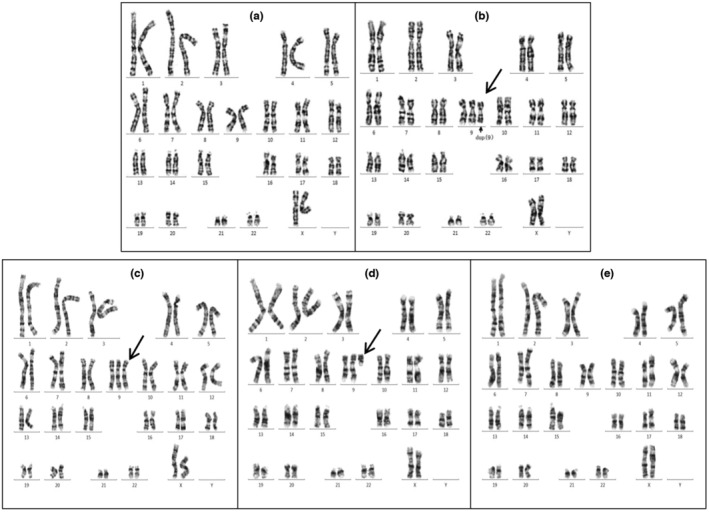
The G banding karyotype results from the two cases with cultured amniotic fluid cells. Arrows indicate derivative Chr9. Case 1 contains two cell lines: (a) 46,XX, (b) 47,XX,+der(9)del(9)(q21q34)dup(9)(p12p24). Case 2 contains three cell lines: (c) 47,XX,+9, (d) 47,XX,+del(9)(q13), (e) 46,XX.
Case 2 showed a mosaic karyotype with three distinct cell lines. Cell line 1 (82/100; 82% cells) had three Chr9, suggesting trisomy 9. Cell line 2 (14/100; 14% cells) had two normal Chr9 and a single centric chromosome with a short arm of Chr9, indicating trisomy 9p. Cell line 3 (4/100; 4% cells) had a normal female karyotype. The karyotype of this fetus was 47,XX,+9[82]/47,XX,+del(9)(q13)[14]/46,XX[4], based on 100 cells from cultured amniocytes, indicating an 82% mosaicism of trisomy 9, a 14% mosaicism of trisomy 9p, and a 4% of normal karyotype (Figure 3).
3.4. Chromosomal microarray analysis
The CMA result of Case 1 revealed that the marker chromosome corresponded to a 64.9 Mb gain at arr[GRCH37]9p24.3p13.2(208454‐68216577)*2.43, indicating a 21.5% mosaicism for 9p24.3p13.2 (Figure 4).
FIGURE 4.
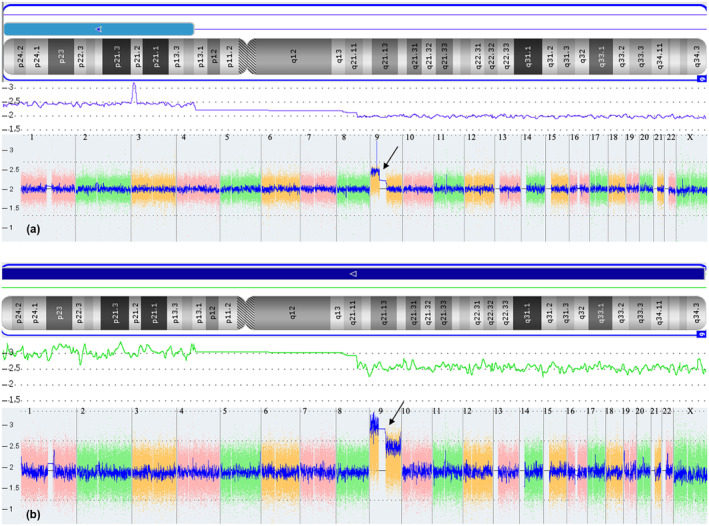
Various signal diagrams by Chas software indicating the results of Chromosomal Microarray Analysis. The arrows indicate the duplicated region of Chr9. (a) Short arm duplication of Chr9 for Case 1. (b) Chr9 duplication with different copy number between short and long arm for Case 2.
According to the whole genome variation (WGV) signal, Case 2 demonstrated a disparity in copy number between the short and long arm of Chr9, with three copies on the short arm and 2.5 copies on the long arm. Based on the karyotype result, the calculated mosaic levels of trisomy 9, trisomy 9p, and 46,XX were determined to be 50%, 50%, and 0%, respectively (Figure 4).
3.5. Fluorescence in situ hybridization
Tetrasomy 9p was detected in 25 of the 100 interphase cells observed on uncultured amniocytes from Case 1 by interphase FISH using PD‐L1 (9p24.3) and CSP9 (centromere) probes, which was indicative of a 25% mosaicism for tetrasomy 9p (Figure 5).
FIGURE 5.
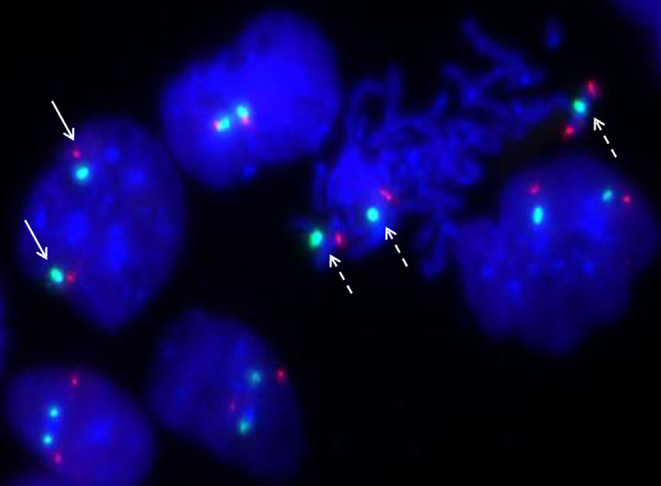
Interphase fluorescence in situ hybridization analysis on uncultured amniocytes using the 9p24.3‐specific probe PD‐L1 (dye: Texas red) and centromere probe CSP9 (dye: FITC, green) shows two red signals and two green signals in a normal cell (solid arrow) and four red signals and three green signals in an abnormal cell with tetrasomy 9p (dotted arrow) for Case 1.
However, because the volume of uncultured amniotic fluid was insufficient for interphase FISH and the patient declined a second amniocentesis, a similar FISH analysis was not performed for Case 2.
4. DISCUSSION
Tetrasomy 9p is often associated with multiple malformations. Nevertheless, few cases of mosaic tetrasomy 9p without clinical symptoms have been reported (Sait & Wetzler, 2003, Bellil et al., 2020; McAuliffe et al., 2005; Shu et al., 2021). Here, we describe an extraordinary case of mosaic tetrasomy 9p without clinical symptoms from conception until 2.5 years of age. To the best of our knowledge, this is the ninth case of mosaic tetrasomy 9p with no clinical symptoms to be reported in the literature, and the first case in which the phenotype was documented from early pregnancy through the postnatal period. The mosaic levels for the eight cases that have been previously described (Table 2) ranged from 6% to non‐mosaic tetrasomy 9p (Baronchelli et al., 2011; Bellil et al., 2020; McAuliffe et al., 2005; Ogino et al., 2007; Papoulidis et al., 2012; Sait & Wetzler, 2003; Shu et al., 2021). The mosaic ratio of our case was within this range by 21.5%. According to Vinksel et al. (2019), the prognosis for mosaic tetrasomy 9p is strongly correlated with the size of the affected segment of the long arm of the chromosome. If the supernumerary chromosome involves 9q, even the heterochromatic region, a worse prognosis is expected, including cardiac malformations, intellectual disability, and death. By contrast, others have argued that this view was biased and based on a small sample size (El Khattabi et al., 2015). In Case 1, there was no clinical phenotype, even though the supernumerary chromosome involved 9p and extended to 9q13, which contradicts the findings of Vinksel et al. The reasons behind the phenotypical heterogeneities observed in mosaic tetrasomy 9p cases are generally unknown. Possible explanations include the size, position, and degree of mosaicism in the duplicated region (Pinto et al., 2018). Arnold et al. considers the timing of non‐disjunction occurrence and tissue distribution to be the most significant prognostic indicators (Arnold et al., 1995). Regardless of the mosaic ratio, neither our case nor any of those shown in Table 2 displayed any anatomical or intellectual issues. We hypothesize that under these situations, aberrant cell lines may not exist in the brain and other vital organs, which may corroborate Arnold's view. Based on the documented tissue‐limited mosaicism propensity of tetrasomy 9p, we were not able to assess the distribution of abnormal cell lines prenatally, so prenatal counseling still should be done with caution, even in cases with normal appearances on ultrasound (Vinksel et al., 2019).
TABLE 2.
Previously reported cases with normal phenotypes of mosaic tetrasomy 9p.
| Case | Literature | Age | Sex | Mosaicism level (blood) | Indication | Phenotype |
|---|---|---|---|---|---|---|
| 1 | Sait and Wetzler (2003) | 41 | Male | 43% | Skin lesions | Healthy |
| 2 | Shu et al. (2021) | 33 | Female | 73% | Skin lesions | Healthy |
| 3 | McAuliffe et al. (2005) | 37 | Male | 20% | Oligozoospermia | Healthy |
| 4 | Bellil et al. (2020) | 41 | Male | 50%–60% | Oligozoospermia | Healthy |
| 5 | Ogino et al. (2007) | 10 | Male | 6% | Inconspicuous penis | Healthy |
| 6 | Baronchelli et al. (2011) | Adult | Female | 72% | Premature ovarian failure | Healthy |
| 7 | Papoulidis et al. (2012) | 20 | Female | Full | Familial inv. (7) | Healthy |
| 8 | Papoulidis et al. (2012) | 28 | Female | 80% | IVF for male azoospermia | Healthy |
The fetus in Case 2 was shown to have a mosaic karyotype composed of three cell lines: 47,XX,+9/47,XX,+del(9)(q13)/46,XX. As far as we know, this is the first case to be reported with the mosaic karyotype of both trisomy 9 and trisomy 9p. Trisomy 9 is uncommon in live‐birth babies, manifesting more typically as mosaicism (Pejcic et al., 2018). IUGR, congenital heart abnormality, multi‐organ malformations, and the absence of parietal bone are the most common prenatal ultrasound manifestations of mosaic trisomy 9 (Wang et al., 2020). Trisomy 9p cases display distinctive craniofacial deformities and a broad spectrum of phenotypes involving multiple organs (Cammarata‐Scalisi, 2019). Case 2 had two abnormal cell lines with trisomy 9 and trisomy 9p, and most clinical manifestations were comparable to mosaic trisomy 9 or trisomy 9p. However, the oligohydramnios and single umbilical artery symptoms that presented in Case 2 have never before been reported in mosaic trisomy 9 or trisomy 9p. Our case added new prenatal ultrasound characteristics to the list of mosaic trisomy 9/trisomy 9p fetal anomalies. Whether the interaction of the two aberrant cell lines produced a new phenotype needs to be confirmed in additional cases.
Numerous studies have confirmed the feasibility of NIPT for detecting common chromosomal aneuploidy (13/18/21), and a few publications have described the detection of rare chromosomal aneuploidy, mosaicism, and copy number variation using NIPT (Lee et al., 2018). Cases concerning Chr9 detected by NIPT are sporadic in the literature, and evidence regarding the efficacy of NIPT for the evaluation of Chr9 remains inconclusive. Three cases of fetal trisomy 9 mosaicism that were initially suspected after NIPT and verified by an invasive counterpart were reported by Lee et al (Lee et al., 2018). In a similar report, Ma et al. predicted the approximate mosaic ratio of trisomy 9 using NIPT (Ma et al., 2015). NIPT once unintentionally discovered a maternal mosaic tetrasomy 9p case during pregnancy, in a report by Shu et al (Shu et al., 2021). In our study, the gain of genetic material on Chr9 was first discovered by NIPT, then verified by amniocentesis. Both prior reports and our findings indicate the potential usefulness of NIPT for screening Chr9 mosaicism during early gestation. However, the zygote undergoes a series of cleavage divisions that lead to two functionally distinct cell masses, the inner cell mass and the trophectoderm, which differentiate into the embryo and placenta, respectively (Molè et al., 2020). Cell‐free fetal DNA in the maternal plasma is mainly released from placental trophoblast cells and thus only serves as an indirect indicator of fetal genetic information. Vanishing twins, maternal aneuploidy, maternal mosaicism, maternal copy number variations, and maternal malignancy may all interfere with the accuracy of NIPT as well (Renga, 2018). Therefore, invasive prenatal testing is necessary to confirm any positive NIPT results, due to the latter's limitations as a prenatal diagnosis technique, as well as the risk of confined placental mosaicism (Ma et al., 2015).
The difference in mosaic levels between uncultured and cultivated amniocytes has been confirmed previously (Chen et al., 2014). Trisomic mosaic levels have been reported to be lower on karyotype analyses of cultured amniocytes than on CMA or FISH analyses of uncultured amniocytes. Monomeric mosaic levels have been reported to be higher on karyotype analyses than on CMA (Chen et al., 2013; Chen et al., 2015; Chen, Chang, et al., 2012; Chen, Su, et al., 2012; Hao et al., 2020; Prakash et al., 2014; Tian et al., 2019; Wu et al., 2020). This difference may be attributable to poor genetic stability and the loss of trisomic cells during long‐term culture of amniotic fluid cells, which contrasts with the case of monomeric cells. We reviewed the mosaic levels of 9p duplication detected by various testing methods that have been previously published (Table 3) (Bellil et al., 2020; Chen et al., 2014; Pinto et al., 2018). According to our findings, mosaic levels of 9p duplication decreased during prolonged culture, which may explain one previous observation of possible false‐negative diagnoses of mosaic 9p duplication when testing cultured amniocytes (Chen et al., 2014). Along with our case, a total of three cases have shown mosaic levels on uncultured amniocytes by FISH, with equivalent mosaic levels between FISH and CMA in two cases, but lower ones seen on FISH compared to CMA in the third case. We speculate that the third case may have been the result of artificial selection. This study provided evidence that CMA and FISH analyses on uncultured amniocytes may accurately detect mosaic levels of 9p duplication. Additionally, we hypothesize that the mosaic ratio discrepancy may be more significant between uncultured and cultivated amniocytes because segmental Chr9 is more unstable and prone to loss during mitosis than whole Chr9.
TABLE 3.
Previously reported mosaic levels of 9p duplication using different methods.
| Authors | Sample type | Karyotype | The mosaic ratio by karyotype analysis | CMA/CGH | The mosaic ratio by CMA/CGH | FISH | The mosaic ratio by FISH |
|---|---|---|---|---|---|---|---|
| Pinto et al. (2018) | peripheral blood | 47,XY,i(9p)[45]/46,XY[5] | 90% | arr[hg19] 9p24.3q21.11(203,861‐70,974,662)*4 | 100% | NA | NA |
| Bellil et al. (2020) | peripheral blood | 47,XY,+i(9)(p11)[6]/46,XY[18] | 25% | arr[hg19] 9p24.2p13.2 | 60% | 47,XY,+i(9)(p11)/46,XY | 50%–60% |
| Chen et al. (2014) | amniocytes | 47,XX,i(9p)[6]/46,XX[22] | 21.40% | arr [hg19] 9p24.3p13.1(0‐40,450,202)*3.4 | 70% | 47,XX,i(9p)[49]/46,XX[55] | 47.10% |
Abbreviations: CMA, chromosome microarray; CGH, comparative genomic hybridization; FISH, fluorescence in situ hybridization; NA, not available.
It is noteworthy that, contrary to an earlier finding that the level of trisomic mosaicism observed on karyotype analysis was lower than what was seen on CMA, the mosaic level of trisomy 9 seen on karyotype analysis was greater than what was apparent from CMA in our Case 2. What made Case 2 unique was the complicated mosaicism that included two distinct abnormal cell lines with both trisomy 9 and trisomy 9p. We investigated the potential causes of this disagreement with earlier findings as follows: (1) Case 2 was a mixture of three cell lines, with a predominance of abnormal cell lines, wherein the two aberrant cells may have fought for proliferative space and trisomy 9 had a growth advantage and/or higher genetic stability over trisomy 9p; and (2) the amniotic fluid was a combination of cells from the embryo's three germ layers. The three cell lines may have come from different tissues, accounting for the differences on the mosaic level. As no similar examples have been recorded in the literature, additional evidence is required in the future to support one or both hypotheses.
This study could serve as an essential reference for clinicians who need to evaluate the merits of different techniques used to diagnose 9p duplication mosaicism in the future.
However, some key limitations of this study should be acknowledged as well. First, more cases are required to fully assess the use of various approaches for identifying mosaic 9p duplication in the future, because the number of relevant cases, both in this study and in the literature, is limited. Second, the patient in Case 2 declined a second amniocentesis, which meant that there was insufficient amniotic fluid for FISH, so we were unable to evaluate the differences between FISH and the other procedures for that embryo. Third, the neonatal outcome for Case 2 is unknown since the pregnancy was terminated at 24 weeks.
We recommend NIPT as a routine chromosomal screening technique during early pregnancy. Invasive amniocentesis is required for CMA and karyotype testing when NIPT indicates abnormalities. If either CMA or karyotype analysis suggests the possibility of mosaicism, further verification of the mosaic ratio and abnormal structure by FISH should be considered. Once abnormal structure and mosaic levels have been determined, continuous ultrasound monitoring of their genetic effects is essential.
5. CONCLUSION
Tetrasomy 9p mosaicism can have no clinical appearance during pregnancy and postpartum. Unlike trisomy 9 or trisomy 9p, complicated mosaicism of trisomy 9p and trisomy 9 has a novel ultrasound appearance. This study demonstrated the potential of using NIPT to suggest 9p duplication in early pregnancy. Discrepancies were present in the mosaic levels of 9p duplication between approaches. The comprehensive use of NIPT, karyotype analysis, CMA, and FISH was helpful for early screening, identifying the origin of the marker chromosome, evaluating the mosaic level, and preparing effective genetic counseling.
AUTHOR CONTRIBUTIONS
Sufen Zhang performed the non‐invasive prenatal screening, the chromosomal microarray analysis and drafted the manuscript. Yuqiu Zhou assisted with comment on the manuscript. Gefei Xiao contributed to study design and revised the manuscript. Xianrong Qiu performed the karyotype analysis, the fluorescence in situ hybridization analysis and follow‐up.
FUNDING INFORMATION
Open access funding is provided by Zhuhai Center for Maternal and Child Health Care.
CONFLICT OF INTEREST STATEMENT
All the authors declare no conflicts of interest.
ACKNOWLEDGMENTS
We thank our patients and their families for supporting this study, and we are grateful to ultrasonographer Xiaojun Meng of the Zhuhai Center for Maternal and Child Health Care for providing the ultrasound images. We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Zhang, S. , Zhou, Y. , Xiao, G. , & Qiu, X. (2023). Application of various genetic analysis techniques for detecting two rare cases of 9p duplication mosaicism during prenatal diagnosis. Molecular Genetics & Genomic Medicine, 11, e2229. 10.1002/mgg3.2229
Contributor Information
Gefei Xiao, Email: xgf8111_cn@hotmail.com.
Xianrong Qiu, Email: 13229755901@163.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
REFERENCES
- Arnold, G. L. , Kirby, R. S. , Stern, T. P. , & Sawyer, J. R. (1995). Trisomy 9: Review and report of two new cases. American Journal of Medical Genetics, 56(3), 252–257. 10.1002/ajmg.1320560303 [DOI] [PubMed] [Google Scholar]
- Baronchelli, S. , Conconi, D. , Panzeri, E. , Bentivegna, A. , Redaelli, S. , Lissoni, S. , Saccheri, F. , Villa, N. , Crosti, F. , Sala, E. , Martinoli, E. , Volontè, M. , Marozzi, A. , & Dalprà, L. (2011). Cytogenetics of premature ovarian failure: An investigation on 269 affected women. Journal of Biomedicine & Biotechnology, 2011, 370195. 10.1155/2011/370195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellil, H. , Herve, B. , Herzog, E. , Ayoubi, J. M. , Vialard, F. , & Poulain, M. (2020). A high level of tetrasomy 9p mosaicism but no clinical manifestations other than moderate oligozoospermia with chromosomally balanced sperm: A case report. Journal of Assisted Reproduction and Genetics, 37(3), 573–577. 10.1007/s10815-020-01690-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, D. W. , Rava, R. P. , & Sehnert, A. J. (2014). DNA sequencing versus standard prenatal aneuploidy screening. The New England Journal of Medicine, 371(6), 578. 10.1056/NEJMc1405486 [DOI] [PubMed] [Google Scholar]
- Cammarata‐Scalisi, F. (2019). Trisomy 9p. A brief clinical, diagnostic and therapeutic description. Arch Argent Pediatr, 117(5), e473–e476. 10.5546/aap.2019.eng.e473 (Trisomia 9p. Una breve descripcion clinica, diagnostica y terapeutica.). [DOI] [PubMed] [Google Scholar]
- Chen, C. P. , Chang, S. D. , Chueh, H. Y. , Su, Y. N. , Su, J. W. , Chern, S. R. , Chen, Y. T. , Lee, C. C. , Town, D. D. , Chen, W. L. , Chen, L. F. , Lee, M. S. , Pan, C. W. , & Wang, W. (2012). Rapid positive confirmation of trisomy 21 mosaicism at amniocentesis by interphase FISH, QF‐PCR and aCGH on uncultured amniocytes. Taiwanese Journal of Obstetrics & Gynecology, 51(3), 475–480. 10.1016/j.tjog.2012.07.035 [DOI] [PubMed] [Google Scholar]
- Chen, C. P. , Chang, T. Y. , Chern, S. R. , Lee, C. C. , Town, D. D. , Lee, M. S. , & Wang, W. (2007). Prenatal diagnosis of low‐level mosaic tetrasomy 9p by amniocentesis. Prenatal Diagnosis, 27(4), 383–385. 10.1002/pd.1678 [DOI] [PubMed] [Google Scholar]
- Chen, C. P. , Chern, S. R. , Chen, Y. N. , Wu, P. S. , Yang, C. W. , Chen, L. F. , & Wang, W. (2015). Mosaic trisomy 15 at amniocentesis: Prenatal diagnosis, molecular genetic analysis and literature review. Taiwanese Journal of Obstetrics & Gynecology, 54(4), 426–431. 10.1016/j.tjog.2015.06.002 [DOI] [PubMed] [Google Scholar]
- Chen, C. P. , Su, Y. N. , Chern, S. R. , Chen, Y. T. , Wu, P. S. , Su, J. W. , Pan, C. W. , & Wang, W. (2012). Mosaic trisomy 2 at amniocentesis: Prenatal diagnosis and molecular genetic analysis. Taiwanese Journal of Obstetrics & Gynecology, 51(4), 603–611. 10.1016/j.tjog.2012.09.016 [DOI] [PubMed] [Google Scholar]
- Chen, C. P. , Su, Y. N. , Su, J. W. , Chern, S. R. , Chen, Y. T. , Chen, L. F. , & Wang, W. (2013). Mosaic trisomy 12 at amniocentesis: Prenatal diagnosis and molecular genetic analysis. Taiwanese Journal of Obstetrics & Gynecology, 52(1), 97–105. 10.1016/j.tjog.2013.01.012 [DOI] [PubMed] [Google Scholar]
- Chen, C. P. , Wang, L. K. , Chern, S. R. , Wu, P. S. , Chen, Y. T. , Kuo, Y. L. , Chen, W. L. , Lee, M. S. , & Wang, W. (2014). Mosaic tetrasomy 9p at amniocentesis: Prenatal diagnosis, molecular cytogenetic characterization, and literature review. Taiwanese Journal of Obstetrics & Gynecology, 53(1), 79–85. 10.1016/j.tjog.2013.12.002 [DOI] [PubMed] [Google Scholar]
- El Khattabi, L. , Jaillard, S. , Andrieux, J. , Pasquier, L. , Perrin, L. , Capri, Y. , Benmansour, A. , Toutain, A. , Marcorelles, P. , Vincent‐Delorme, C. , Journel, H. , Henry, C. , De Barace, C. , Devisme, L. , Dubourg, C. , Demurger, F. , Lucas, J. , Belaud‐Rotureau, M. A. , Amiel, J. , … Verloes, A. (2015). Clinical and molecular delineation of Tetrasomy 9p syndrome: Report of 12 new cases and literature review. American Journal of Medical Genetics. Part A, 167(6), 1252–1261. 10.1002/ajmg.a.36932 [DOI] [PubMed] [Google Scholar]
- Hao, M. , Li, L. , Zhang, H. , Li, L. , Liu, R. , & Yu, Y. (2020). The difference between karyotype analysis and chromosome microarray for mosaicism of aneuploid chromosomes in prenatal diagnosis. Journal of Clinical Laboratory Analysis, 34(12), e23514. 10.1002/jcla.23514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. Y. , Su, H. J. , Cheng, Y. T. , Ku, Y. L. , Ngo, Y. G. , Chen, C. M. , Ou, Y. C. , Lee, M. C. , & Shaw, S. W. S. (2018). Detection of fetal trisomy 9 mosaicism by noninvasive prenatal testing through maternal plasma DNA sequencing. Taiwanese Journal of Obstetrics & Gynecology, 57(4), 594–597. 10.1016/j.tjog.2018.06.021 [DOI] [PubMed] [Google Scholar]
- Ma, J. , Cram, D. S. , Zhang, J. , Shang, L. , Yang, H. , & Pan, H. (2015). Birth of a child with trisomy 9 mosaicism syndrome associated with paternal isodisomy 9: Case of a positive noninvasive prenatal test result unconfirmed by invasive prenatal diagnosis. Molecular Cytogenetics, 8, 44. 10.1186/s13039-015-0145-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe, F. , Winsor, E. J. , & Chitayat, D. (2005). Tetrasomy 9p mosaicism associated with a normal phenotype. Fetal Diagnosis and Therapy, 20(3), 219–222. 10.1159/000083909 [DOI] [PubMed] [Google Scholar]
- Molè, M. A. , Weberling, A. , & Zernicka‐Goetz, M. (2020). Comparative analysis of human and mouse development: From zygote to pre‐gastrulation. Current Topics in Developmental Biology, 136, 113–138. 10.1016/bs.ctdb.2019.10.002 [DOI] [PubMed] [Google Scholar]
- Ogino, W. , Takeshima, Y. , Nishiyama, A. , Yagi, M. , Oka, N. , & Matsuo, M. (2007). Mosaic tetrasomy 9p case with the phenotype mimicking Klinefelter syndrome and hyporesponse of gonadotropin‐stimulated testosterone production. The Kobe Journal of Medical Sciences, 53(4), 143–150 https://www.ncbi.nlm.nih.gov/pubmed/17932453 [PubMed] [Google Scholar]
- Papoulidis, I. , Kontodiou, M. , Tzimina, M. , Saitis, I. , Hamid, A. B. , Klein, E. , Kosyakova, N. , Kordass, U. , Kunz, J. , Siomou, E. , Nicolaides, P. , Orru, S. , Thomaidis, L. , Liehr, T. , Petersen, M. B. , & Manolakos, E. (2012). Tetrasomy 9p mosaicism associated with a normal phenotype in two cases. Cytogenetic and Genome Research, 136(4), 237–241. 10.1159/000337520 [DOI] [PubMed] [Google Scholar]
- Pejcic, L. , Stankovic, T. , Ratkovic‐Jankovic, M. , Vasic, K. , & Nikolic, I. (2018). Clinical manifestations in trisomy 9 mosaicism. The Turkish Journal of Pediatrics, 60(6), 729–734. 10.24953/turkjped.2018.06.015 [DOI] [PubMed] [Google Scholar]
- Pinto, I. P. , Minasi, L. B. , Steckelberg, R. , da Silva, C. C. , & da Cruz, A. D. (2018). Mosaic Tetrasomy of 9p24.3q21.11 postnatally identified in an infant born with multiple congenital malformations: A case report. BMC Pediatrics, 18(1), 298. 10.1186/s12887-018-1275-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash, S. , Guo, D. , Maslen, C. L. , Silberbach, M. , Milewicz, D. , & Bondy, C. A. (2014). Single‐nucleotide polymorphism array genotyping is equivalent to metaphase cytogenetics for diagnosis of Turner syndrome. Genetics in Medicine, 16(1), 53–59. 10.1038/gim.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renga, B. (2018). Non invasive prenatal diagnosis of fetal aneuploidy using cell free fetal DNA. European Journal of Obstetrics, Gynecology, and Reproductive Biology, 225, 5–8. 10.1016/j.ejogrb.2018.03.033 [DOI] [PubMed] [Google Scholar]
- Sait, S. N. J. , & Wetzler, M. (2003). Tetrasomy 9p with no apparent phenotype characteristics (abstract), 53th ASHG Annual Meeting: Nr 678.
- Shu, W. , Cheng, S. S. W. , Xue, S. , Chan, L. W. , Soong, S. I. , Kan, A. S. Y. , Cheung, S. W. H. , & Choy, K. W. (2021). First case report of maternal mosaic Tetrasomy 9p incidentally detected on non‐invasive prenatal testing. Genes (Basel), 12(3), 370. 10.3390/genes12030370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. , Liu, C. , Qi, H. , Zhang, Y. , Bian, X. , & Liu, J. (2013). Noninvasive prenatal testing of fetal aneuploidies by massively parallel sequencing in a prospective Chinese population. Prenatal Diagnosis, 33(7), 700–706. 10.1002/pd.4160 [DOI] [PubMed] [Google Scholar]
- Spinner, N. B. , & Conlin, L. K. (2014). Mosaicism and clinical genetics. American Journal of Medical Genetics. Part C, Seminars in Medical Genetics, 166C(4), 397–405. 10.1002/ajmg.c.31421 [DOI] [PubMed] [Google Scholar]
- Taylor‐Phillips, S. , Freeman, K. , Geppert, J. , Agbebiyi, A. , Uthman, O. A. , Madan, J. , Clarke, A. , Quenby, S. , & Clarke, A. (2016). Accuracy of non‐invasive prenatal testing using cell‐free DNA for detection of Down, Edwards and Patau syndromes: A systematic review and meta‐analysis. BMJ Open, 6(1), e010002. 10.1136/bmjopen-2015-010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Y. , Zhang, L. , Tian, W. , Wu, Y. , Zheng, Q. , Zhang, Z. , & Jia, L. (2019). A case of maternal 45,X/46,XX mosaicism detected by non‐invasive prenatal testing. Zhonghua Yi Xue Yi Chuan Xue Za Zhi, 36(11), 1120–1122. 10.3760/cma.j.issn.1003-9406.2019.11.016 [DOI] [PubMed] [Google Scholar]
- Vinksel, M. , Volk, M. , Peterlin, B. , & Lovrecic, L. (2019). A systematic clinical review of prenatally diagnosed Tetrasomy 9p. Balkan Journal of Medical Genetics: BJMG, 22(1), 11–20. 10.2478/bjmg-2019-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Chen, Y. , Zhao, J. , & Xia, Y. (2020). Prenatal diagnosis and genetic counseling of low‐level trisomy 9 mosaicism with a favorable outcome. Taiwanese Journal of Obstetrics & Gynecology, 59(5), 786–787. 10.1016/j.tjog.2020.07.032 [DOI] [PubMed] [Google Scholar]
- Wu, X. , An, G. , Xie, X. , Su, L. , Cai, M. , Chen, X. , Li, Y. , Lin, N. , He, D. , Wang, M. , Huang, H. , & Xu, L. (2020). Chromosomal microarray analysis for pregnancies with or without ultrasound abnormalities in women of advanced maternal age. Journal of Clinical Laboratory Analysis, 34(4), e23117. 10.1002/jcla.23117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhong, M. , & Zheng, D. (2021). Chromosomal mosaicism detected by karyotyping and chromosomal microarray analysis in prenatal diagnosis. Journal of Cellular and Molecular Medicine, 25(1), 358–366. 10.1111/jcmm.16080 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.


