This cross-sectional study measures the association of lipoprotein(a) concentrations in pairs of first- and second-degree relatives of participants in the UK Biobank with high lipoprotein(a) levels.
Key Points
Question
What is the prevalence of high lipoprotein(a) concentrations (≥125 nmol/L) in first- and second-degree relatives of individuals with high lipoprotein(a) levels?
Findings
In this cross-sectional study, 1607 of 3420 (47.0%) first-degree relatives of UK Biobank participants with a lipoprotein(a) concentration at least 125 nmol/L were similarly affected, compared with 4974 of 30 258 (16.4%) unrelated individuals.
Meaning
Cascade screening of first-degree relatives of individuals with high lipoprotein(a) levels, if implemented at scale, is likely to identify a substantial number of additional high-risk individuals.
Abstract
Importance
Lipoprotein(a) (Lp[a]) concentrations are a highly heritable and potential causal risk factor for atherosclerotic cardiovascular disease (ASCVD). Recent consensus statements by the European Atherosclerosis Society and American Heart Association recommend screening of relatives of individuals with high Lp(a) concentrations, but the expected yield of this approach has not been quantified in large populations.
Objective
To measure the prevalence of high Lp(a) concentrations among first- and second-degree relatives of individuals with high Lp(a) concentrations compared with unrelated participants.
Design, Setting, and Participants
In this cross-sectional analysis, pairs of first-degree (n = 19 899) and second-degree (n = 9715) relatives with measured Lp(a) levels from the UK Biobank study and random pairs of unrelated individuals (n = 184 764) were compared. Data for this study were collected from March 2006 to August 2010 and analyzed from December 2021 to August 2023.
Exposure
Serum Lp(a) levels, with a high Lp(a) level defined as at least 125 nmol/L.
Main Outcome and Measure
Concordance of clinically relevant high Lp(a) levels in first- and second-degree relatives of index participants with high Lp(a) levels.
Results
A total of 52 418 participants were included in the analysis (mean [SD] age, 57.3 [8.0] years; 29 825 [56.9%] women). Levels of Lp(a) were correlated among pairs of first-degree (Spearman ρ = 0.45; P < .001) and second-degree (Spearman ρ = 0.22; P < .001) relatives. A total of 1607 of 3420 (47.0% [95% CI, 45.3%-48.7%]) first-degree and 514 of 1614 (31.8% [95% CI, 29.6%-34.2%]) second-degree relatives of index participants with high Lp(a) levels also had elevated concentrations compared with 4974 of 30 258 (16.4% [95% CI, 16.0%-16.9%]) pairs of unrelated individuals. The concordance in high Lp(a) levels was generally consistent among subgroups (eg, those with prior ASCVD, postmenopausal women, and statin users). The odds ratios for relatives to have high Lp(a) levels if their index relative had a high Lp(a) level compared with those whose index relatives did not have high Lp(a) levels were 7.4 (95% CI, 6.8-8.1) for first-degree relatives and 3.0 (95% CI, 2.7-3.4) for second-degree relatives.
Conclusions and Relevance
The findings of this cross-sectional study suggest that the yield of cascade screening of first-degree relatives of individuals with high Lp(a) levels is over 40%. These findings support recent recommendations to use this approach to identify additional individuals at ASCVD risk based on Lp(a) concentrations.
Introduction
Individuals with elevated levels of lipoprotein(a) (Lp[a]) are at increased risk of atherosclerotic cardiovascular disease (ASCVD) and calcific aortic stenosis.1 Lipoprotein(a) is a low-density lipoprotein (LDL)–like particle with an apolipoprotein(a) moiety covalently attached to apolipoprotein B, and trials investigating efficacy of selective and potent therapeutics for lowering Lp(a) levels and reducing cardiovascular disease risk are ongoing.2,3
Lipoprotein(a) concentrations are highly heritable, with up to 90% of observed variance determined by genetic factors, and thus remain relatively constant over a person’s lifetime.4,5,6 The correlation of Lp(a) concentrations across relatives was described in 1992 in a study of 284 siblings,6 where a correlation as high as Spearman ρ = 0.95 was reported in those who shared both LPA (OMIM 152200) alleles as confirmed by the number of kringle IV repeats vs no association if no shared alleles were detected. Consistent with these findings, professional societies advocate measuring Lp(a) levels in relatives of individuals with high Lp(a) concentrations, with the most recent European Atherosclerosis Society (EAS) consensus statement7 and American Heart Association (AHA) scientific statement8 recommending Lp(a) cascade screening in relatives of affected individuals.
Data, however, on the estimated diagnostic yield of such an approach are largely limited to Lp(a) assessment within cascade screening programs for familial hypercholesterolemia or other dyslipidemias. These studies have found that the yield of systematic screening for high Lp(a) levels in first-degree relatives of probands with high Lp(a) levels ranged from 42%, if high Lp(a) level was defined as greater than 50 mg/dL, to 68% if high Lp(a) level was defined as greater than 100 mg/dL (to convert to nmol/L, multiply by 2.15).9,10 Data on Lp(a) cascade screening in the general population are lacking as well as the diagnostic yield for the definition of high Lp(a) level (eg, ≥125 nmol/L) as proposed in the EAS consensus statement7 and AHA and American College of Cardiology (ACC) guideline on the management of blood cholesterol levels.11 Herein, we measure the association of Lp(a) concentrations in pairs of first- and second-degree family members of participants of the UK Biobank with high Lp(a) concentrations to quantify concordance and thereby estimate the expected diagnostic yield of cascade screening.
Methods
Study Population
This cross-sectional study used individual participant data from the UK Biobank, which is a prospective observational study that recruited more than 500 000 participants aged 40 to 69 years from 22 sites across the United Kingdom between March 2006 and August 2010.12 Pairs of first- and second-degree relatives were identified using previously reported genetic kinship matrices.13,14 Relatedness more distant than second degree was considered unrelated and first- and second-degree relatedness were not mutually exclusive for the analyses performed in these 2 groups. For example, if a participant was paired with more than 1 sibling or both a first- and a second-degree relative was present in the study population, this participant was used in multiple analyses (eFigure 1 in Supplement 1). Monozygotic twins were excluded, as well as participants with missing Lp(a) concentrations. Participants who had no relatives in the study (twin, first-degree, or second-degree) were randomly paired with another such participant of the same race and ethnicity to generate a control group of unrelated individual pairs. Race and ethnicity were based on self-identification and are a social construct. Participants provided written informed consent for participation in the UK Biobank and the use of their data for subsequent studies. This study was approved by the Massachusetts General Brigham Institutional Review Board and followed the Strengthening the Reporting of Observational studies in Epidemiology (STROBE) reporting guideline.
Lp(a) and LDL Cholesterol Assessments
Lipoprotein(a) concentrations were measured in 460 482 individuals, with serum samples obtained at study enrollment using an immunoturbidimetric assay (Randox Laboratories) as described previously.15 High levels of Lp(a) were defined as at least 125 nmol/L according to the most recent EAS consensus paper7 and AHA-ACC guideline on the management of blood cholesterol levels.11 Additional analyses were performed with Lp(a) cutoff levels of at least 150 and at least 200 nmol/L, reflecting the approximate inclusion criteria for the phase 3 trials investigating the effect of lowering Lp(a) levels with pelacarsen2 and olpasiran3 on cardiovascular outcomes. Lipoprotein(a) concentrations in milligrams per deciliter can be approximated by dividing the nanomolar concentrations by 2.15 as previously described.15 We estimated untreated LDL cholesterol levels by adjusting measured values in patients who reported use of medications to lower lipid levels by their mean reduction of LDL cholesterol level (eg, for statins, −30%), as done previously.15
Lp(a) Concordance and Subgroup Analysis
Concordance of high Lp(a) levels between relatives was the percentage of first- or second-degree relatives with a high Lp(a) level related to index participants with high Lp(a) levels. These analyses were repeated for estimated untreated LDL cholesterol levels for comparison. Since Lp(a) levels are known to vary according to race and ethnicity, we repeated the concordance analysis in subgroups according to self-reported race and ethnicity (Black, South Asian, White, other race or ethnicity [including more than 1 race or ethnicity, other race or ethnicity, and unknown race or ethnicity], or race or ethnicity not reported). Subgroup analyses were also performed for relative pairs with index participants based on preexisting coronary artery disease or stroke at time of enrollment, use of statins at enrollment, use of any therapy to lower lipid levels at enrollment, and (in women only) menopausal status and reported use of hormone therapy.
LPA Genetic Analysis
Since Lp(a) concentrations are determined in large part by genetic variation in or near the LPA gene, we investigated whether the concordance in high Lp(a) levels was different among participants carrying variants that increase Lp(a) levels. Participants in the UK Biobank were genotyped with custom arrays (UK BiLEVE Axiom or the UK Biobank Axiom; Thermo Fisher Scientific). Details on sample and variant quality control are described elsewhere.12 After quality control, 487 320 participants with genotyping data were available. Variant imputation was performed using IMPUTE4 and the Haplotype Reference Consortium,16 the UK10K,17 and the 1000 Genomes Phase 3.18 A total of 455 350 participants with both genetic data and Lp(a) level measurements were included, and 2 common variants previously associated with Lp(a) concentrations and coronary artery disease were selected (rs1045872 and rs3798220).19 As in that prior study, index participants were classified as carriers of 0, 1, or 2 or more alleles of Lp(a) level–increasing variants.
Statistical Analysis
Data were analyzed from December 2021 to August 2023. Correlation of Lp(a) and estimated untreated LDL cholesterol levels between relative pairs was determined using Spearman rank correlation coefficients; 95% CIs for concordance were determined by a binomial test. Odds ratios for high Lp(a) levels were determined using logistic regression analyses, without adjusting for any covariates. Numbers needed to screen are the inverse of the concordance. Differences in concordance among subgroups were tested using a χ2 test; the subgroup analysis according to the presence of Lp(a) concentration–increasing genetic variants was performed in White participants only, since these variants were identified in a study with predominantly White participants.19 Since Lp(a) concentrations were significantly higher among those with Lp(a) level–increasing variants, we tested whether the concordance according to genotype would remain significant after addition of Lp(a) concentration to a logistic regression model. All analyses were performed with R, version 4.0.2 (R Project for Statistical Computing). Two-sided P < .05 was considered statistically significant.
Results
A total of 59 768 UK Biobank participants had a first- or second- degree relative who was also enrolled in the study. Excluding 4202 pairs with missing Lp(a) values and 179 pairs of monozygotic twins resulted in a total of 52 418 unique participants with first- and second-degree relatives in the final study population. Among these, 19 899 first-degree relative pairs (36 536 unique participants) and 9715 second-degree relative pairs (17 750 unique participants) were identified and used in subsequent analyses (eFigure 1 in Supplement 1). A total of 369 528 participants with no relatives were included in 184 764 randomly selected pairs as a control group. Among all first- and second-degree relative pairs, 140 and 105, respectively, were Black, 251 and 88, respectively, were South Asian, and 19 126 and 9311, respectively, were White. A total of 127 pairs of first-degree and 30 pairs of second-degree relatives were of another self-reported race and ethnicity (ie, reported more than 1 race, did not know their racial background, or did not identify with any of the other provided categories) or did not report theirs.
All first- and second-degree relatives combined had a mean age of 57.3 (SD, 8.0) years, 22 593 (43.1%) were men, and 29 825 (56.9%) were women. Among relatives, 2580 (4.9%) had cardiovascular disease at time of enrollment and 8455 (16.1%) reported use of statins. Separate characteristics for first- and second-degree relatives are reported in Table 1. Of 52 418 unique participants with first- and second-degree relatives, 8816 (16.8%) had high Lp(a) levels. This proportion was considerably higher in Black participants (117 of 439 [26.7%]) and similar in South Asian participants (94 of 582 [16.2%]). Black and South Asian participants had higher Lp(a) levels (median, 75 [IQR, 44-134] and 31 [IQR, 12-69] nmol/L, respectively) compared with White participants (median, 19 [IQR, 7-73] nmol/L; P < .001). The characteristics of participants per self-reported race and ethnicity are reported in eTable 1 in Supplement 1.
Table 1. Characteristics of Study Population.
| Characteristic | First-degree relatives (n = 36 536) | Second-degree relatives (n = 17 750) | Unrelated control group (n = 369 528) |
|---|---|---|---|
| Sex, No. (%) | |||
| Male | 15 380 (42.1) | 7965 (44.9) | 170 724 (46.2) |
| Female | 21 156 (57.9) | 9785 (55.1) | 198 804 (53.8) |
| Age, mean (SD), y | 57.3 (7.4) | 56.3 (9.2) | 57.1 (8.1) |
| BMI, mean (SD) | 27.3 (4.7) | 27.7 (4.9) | 27.4 (4.8) |
| Currently smoking, No. (%) | 3720 (10.2) | 2206 (12.4) | 38 466 (10.4) |
| Hypertension, No. (%) | 10 156 (27.8) | 5039 (28.4) | 105 039 (28.4) |
| Type 2 diabetes, No. (%) | 1866 (5.1) | 961 (5.4) | 20 425 (5.5) |
| Cholesterol level, mean (SD), mg/dL | |||
| Total | 229 (42) | 226 (43) | 228 (42) |
| Estimated untreated LDL | 146 (33) | 145 (34) | 146 (33) |
| LDL | 138 (33) | 137 (34) | 138 (34) |
| HDL | 57 (15) | 55 (14) | 56 (15) |
| Triglyceride level, median (IQR), mg/dL | 134 (94-195) | 137 (95-201) | 135 (95-198) |
| Lp(a) level, median (IQR), nmol/L | 20 (8-75) | 19 (8-73) | 20 (8-75) |
| Statin use, No. (%) | 5774 (15.8) | 3009 (17.0) | 60 440 (16.4) |
| Ezetimibe use, No. (%) | 220 (0.6) | 108 (0.6) | 2111 (0.6) |
| ASCVD at enrollment, No. (%)a | 1674 (4.6) | 1002 (5.6) | 17 935 (4.9) |
Abbreviations: ASCVD, atherosclerotic cardiovascular disease; BMI, body mass index (calculated as weight in kilograms divided by height in meters squared); HDL, high-density lipoprotein; LDL, low-density lipoprotein; Lp(a), lipoprotein(a).
SI conversion factors: To convert cholesterol to mmol/L, multiply by 0.0259; triglyceride to mmol/L, multiply by 0.0113. To approximate levels of Lp(a) in mg/dL, divide by 2.15.
Defined as a composite of coronary artery disease (myocardial infarction and its acute complications, coronary artery bypass graft surgery, or percutaneous angioplasty or stent placement) and ischemic stroke (cerebral infarction due to thrombosis or cerebral atherosclerosis or cerebrovascular syndromes).8
The Lp(a) concentrations were correlated between first-degree relatives (Spearman ρ = 0.45; P < .001) and less so between second-degree relatives (Spearman ρ = 0.22; P < .001) (eFigure 2A and B in Supplement 1). In unrelated pairs, no correlation was noted (Spearman ρ = 0; P = .80) (eFigure 2C in Supplement 1). By comparison, the correlation in estimated untreated LDL cholesterol levels was less pronounced in first-degree (Spearman ρ = 0.25; P < .001) and second-degree (Spearman ρ = 0.09; P < .001) relatives (eFigure 2D and E in Supplement 1) and not present in unrelated pairs (Spearman ρ = 0; P = .30) (eFigure 2F in Supplement 1).
Among first- and second-degree relatives of index individuals with high Lp(a) levels, median Lp(a) levels were 115 (IQR, 19-184) nmol/L and 50 (IQR, 10-152) nmol/L, respectively (Figure 1A). A total of 1607 of 3420 first-degree relatives of individuals with high Lp(a) levels were similarly affected and had high Lp(a) levels (47.0% [95% CI, 45.3%-48.7]). In second-degree relatives, 514 of 1614 relatives of individuals with high Lp(a) levels had high Lp(a) levels (31.8% [95% CI, 29.6%-34.2%]), while 4974 of 30 258 of unrelated individuals paired with an individual with high Lp(a) levels had high Lp(a) levels themselves (16.4% [95% CI, 16.0%-16.9%]) (Figure 1B). The unadjusted odds ratio for high Lp(a) levels in relatives of participants with high Lp(a) levels compared with those with index relatives without high Lp(a) levels were 7.4 (95% CI, 6.8-8.1) in first-degree relatives and 3.0 (95% CI, 2.7-3.4) in second-degree relatives. The estimated number needed to screen for first-degree relatives was 2.1; for second-degree relatives, 3.1; and for unrelated individuals, 6.2 (Figure 1C). By comparison, the concordance for high levels of LDL cholesterol (defined as estimated untreated LDL cholesterol level ≥190 mg/dL) was 17% for first-degree relatives, 12% for second-degree relatives, and 9% for unrelated individuals (Figure 1B). These numbers changed minimally after excluding index participants using therapies to lower lipid levels (eFigure 3 in Supplement 1).
Figure 1. Lipoprotein(a) (Lp[a]) Levels, Concordance in High Lp(a) Levels, and Numbers Needed to Screen Among Relatives of Index Participants With High Lp(a) Levels.
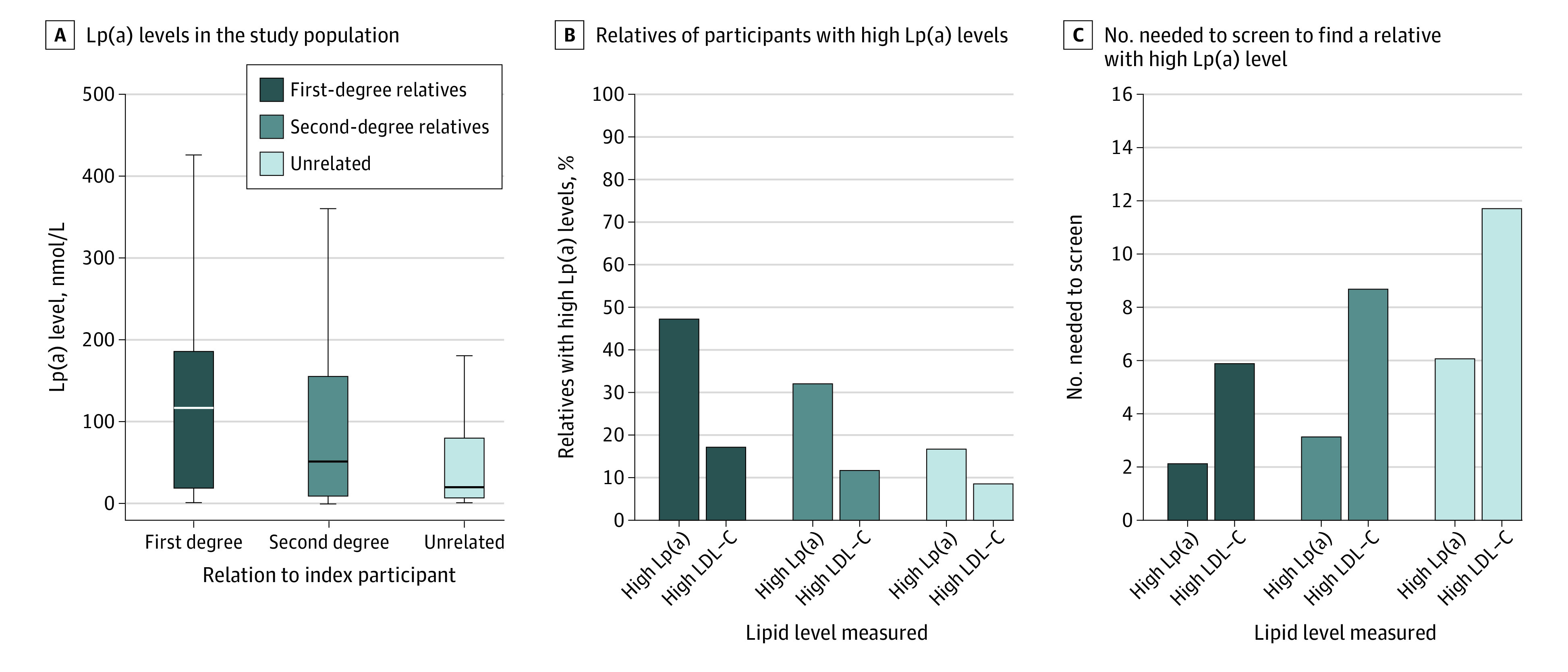
Concentrations of Lp(a) in relatives of index participants with high Lp(a) levels (≥125 nmol/L [to approximate levels in mg/dL, divide by 2.15]) for first- and second-degree relatives (A) resulted in a high percentage of relatives who also have high Lp(a) levels (B) and a low number needed to screen to identify 1 relative with high Lp(a) levels (C). As a comparator, Lp(a) concentrations in unrelated randomly matched pairs of individuals with high Lp(a) are included in all panels, and the concordance and number needed to screen among relatives is also shown for high levels of low-density lipoprotein cholesterol (LDL-C) (≥190 mg/dL [to convert to mmol/L, multiply by 0.0529]). In panel A, vertical lines indicate medians, boxes indicate IQRs, and error bars indicate the largest and smallest data point, but not further than 1.5 times the IQR.
We hypothesized that the concordance in high Lp(a) levels would be higher if a participant had more than 1 relative with a high Lp(a) level. Among 955 trios of first-degree relatives, the concordance for high Lp(a) levels was numerically higher at 57.5% (95% CI, 46.5%-68.0%) compared with 47.0% (95% CI, 45.2%-48.8%) for trios with or without 2 relatives with high Lp(a) levels, respectively, but this difference was not statistically significant (P = .07).
Since Lp(a) concentrations are known to vary according to race and ethnicity, we also investigated the concordance in Black, South Asian, and White individuals separately (Table 2). The concordance was similar among different subgroups: 50.0% (95% CI, 32.4%-67.6%) in Black individuals and 63.2% (95% CI, 46.0%-78.2%) in South Asian individuals compared with 46.8% (95% CI, 45.1%-48.6%) in White individuals (P = .13).
Table 2. Concordance of High Lipoprotein(a) Levels Among Relatives of Participants With High Lp(a) Levels by Different Definitions of High Level and Self-Reported Race and Ethnicitya.
| Concordance group | Definition of high Lp(a) level | Total, % (95% CI) | By race and ethnicity, % (95% CI) | |||
|---|---|---|---|---|---|---|
| Black | South Asian | White | Otherb | |||
| First-degree relatives | ≥125 nmol/L | 47.0 (45.3-48.7) | 50.0 (32.4-67.6) | 63.2 (46.0-78.2) | 46.8 (45.1-48.6) | 28.6 (8.4-58.1) |
| ≥150 nmol/L | 41.6 (39.7-43.5) | 44.4 (25.5-64.7) | 62.5 (43.7-78.9) | 41.5 (39.6-43.5) | 15.4 (1.9-45.4) | |
| ≥200 nmol/L | 34.3 (31.7-36.9) | 35.7 (12.8-64.9) | 43.8 (19.8-70.1) | 34.2 (31.6-36.9) | NA | |
| Second-degree relatives | ≥125 nmol/L | 31.8 (29.6-34.2) | 36.7 (19.9-56.1) | 20.0 (2.5-55.6) | 31.7 (29.4-34.1) | 33.3 (4.3-77.7) |
| ≥150 nmol/L | 27.7 (25.2-30.3) | 25.0 (9.8-46.7) | 22.2 (2.8-60.0) | 27.7 (25.2-30.4) | 66.7 (9.4-99.2) | |
| ≥200 nmol/L | 19.4 (16.5-22.7) | 23.5 (6.8-49.9) | 100 (2.5-100) | 19.1 (16.1-22.4) | NA | |
| Unrelated individuals | ≥125 nmol/L | 16.4 (16.0-16.9) | 26.9 (23.8-30.1) | 9.3 (6.7-12.4) | 16.8 (16.4-17.2) | 13.5 (10.8-16.6) |
| ≥150 nmol/L | 12.7 (12.2-13.1) | 23.8 (20.4-27.4) | 7.5 (4.9-11.0) | 12.6 (12.1-13.0) | 11.5 (8.7-14.9) | |
| ≥200 nmol/L | 6.7 (6.2-7.1) | 11.9 (8.5-16.0) | 3.4 (1.1-7.7) | 6.6 (6.1-7.0) | NA | |
Abbreviations: Lp(a), lipoprotein(a); NA, not applicable.
The number (%) of index participants with high Lp(a) underlying the data in this table are reported in eTable 5 in Supplement 1.
Includes more than 1 race or ethnicity, other race or ethnicity, unknown race or ethnicity, and race or ethnicity not reported.
Next, we investigated the yield of high Lp(a) levels in all relatives for any given Lp(a) level, reflecting Lp(a) levels on a continuous scale found in clinical care. Higher Lp(a) concentrations in the index participant were associated with a gradual increase in the proportion of first- and second-degree relatives with high Lp(a) levels (Figure 2 and eTables 2 and 3 in Supplement 1). For example, if an index individual had an Lp(a) level of at least 327 nmol/L (≥99th percentile), 62.1% of first-degree relatives, 44.2% of second-degree relatives, and 1.3% of unrelated individuals had high Lp(a) levels.
Figure 2. Percentage of First- and Second-Degree Relatives With High Lipoprotein(a) (Lp[a]) Levels for Any Index Cutoff Lp(a) Concentration.
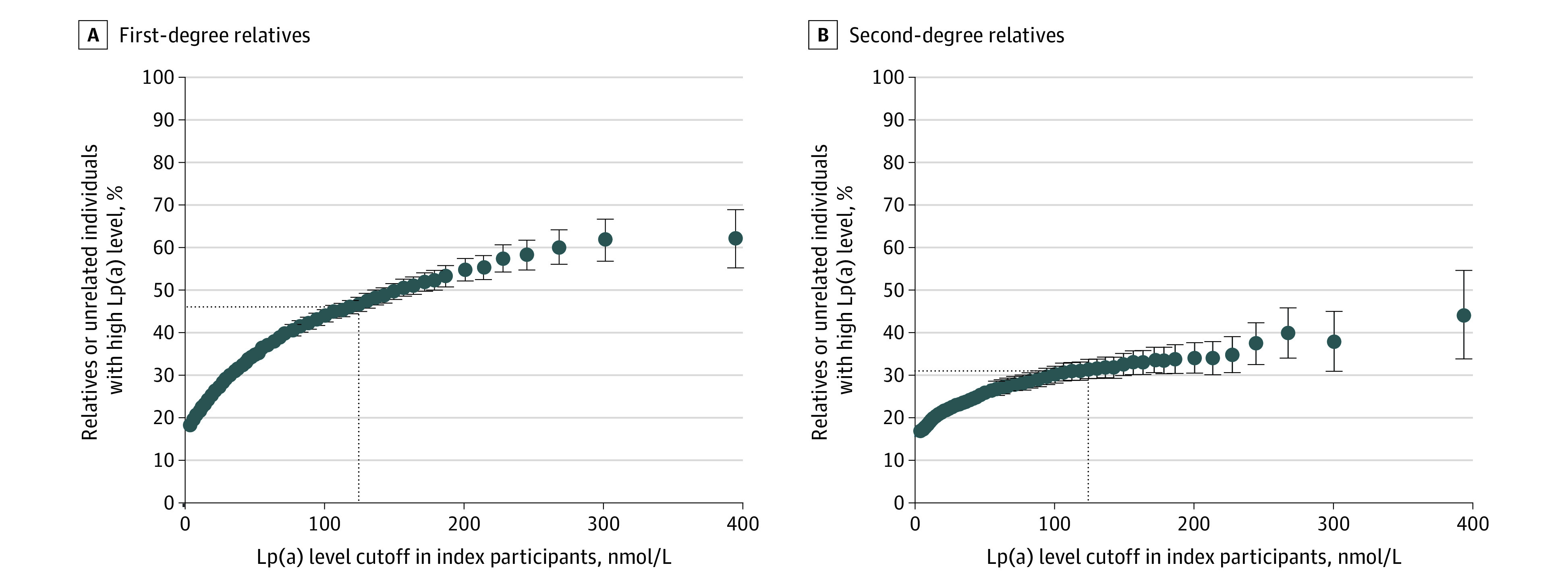
High Lp(a) levels were at least 125 nmol/L (to approximate levels in mg/dL, divide by 2.15). The dotted lines indicate the concordance between high Lp(a) (≥125 nmol/L) in index participants and their relatives. Error bars indicate 95% CIs.
Additional analyses were performed for 2 other cutoffs of high Lp(a) levels that are currently used as inclusion criteria for 2 cardiovascular outcome trials with Lp(a) medications: at least 150 nmol/L and at least 200 nmol/L. This yielded a concordance for the cutoff of at least 150 nmol/L of 41.6% (95% CI, 39.7%-43.5%) for first-degree relatives, 27.7% (95% CI, 25.2%-30.3%) for second-degree relatives, and 12.7% (95% CI, 12.2%-13.1%) for unrelated individuals (Table 2). The concordance for a cutoff of at least 200 nmol/L was 34.3% (95% CI, 31.7%-36.9%) for first-degree relatives, 19.4% (95% CI, 16.5%-22.7%) for second-degree relatives, and 6.7% (95% CI, 6.2%-7.1%) for unrelated individuals (Table 2). The concordance for these cutoffs was numerically higher if a participant had 2 first-degree relatives with high Lp(a) levels compared with participants with only 1 first-degree relative with a high Lp(a) level: 55.4% (95% CI, 41.5%-68.7%) vs 41.8% (95% CI, 39.8%-43.8; P = .06), respectively, for at least 150 nmol/L, and 52.0% (95% CI, 31.3%-72.2%) vs 34.4% (95% CI, 31.6%-37.2%; P = .11), respectively, for at least 200 nmol/L. Similar to high Lp(a) levels defined as at least 125 nmol/L, higher Lp(a) levels in the index participant resulted in a higher proportion of first- and second-degree relatives with an Lp(a) level of at least 150 nmol/L (eFigure 4 and eTables 2 and 3 in Supplement 1) and an Lp(a) level of at least 200 nmol/L (eFigure 5 and eTables 2 and 3 in Supplement 1).
Next, we investigated whether the concordance in high Lp(a) levels was higher among index participants who also had a quantifiable genetic predisposition for high Lp(a) levels. To this end, we compared the concordance in high Lp(a) levels among first-degree relatives of index participants with either 0, 1, or at least 2 LPA variants that were previously associated with Lp(a) concentrations.19 Indeed, the concordance increased from 43.8% (95% CI, 40.6%-47.0%) if 0 variants were present to 46.7% (95% CI, 44.6%-48.8%) if 1 variant was present and 68.8% (95% CI, 60.5%-76.3%) if at least 2 variants were present in the index participant (P < .001 for trend) (Figure 3). This stepwise increase was also seen if high Lp(a) level was defined as at least 150 nmol/L or at least 200 nmol/L but remained significant for at least 125 nmol/L only after adjustment for Lp(a) concentration (eTable 4 and eFigures 6 and 7 in Supplement 1).
Figure 3. Subgroup Analysis for Concordance in High Lipoprotein(a) (Lp[a]) Levels Among First-Degree Relatives.
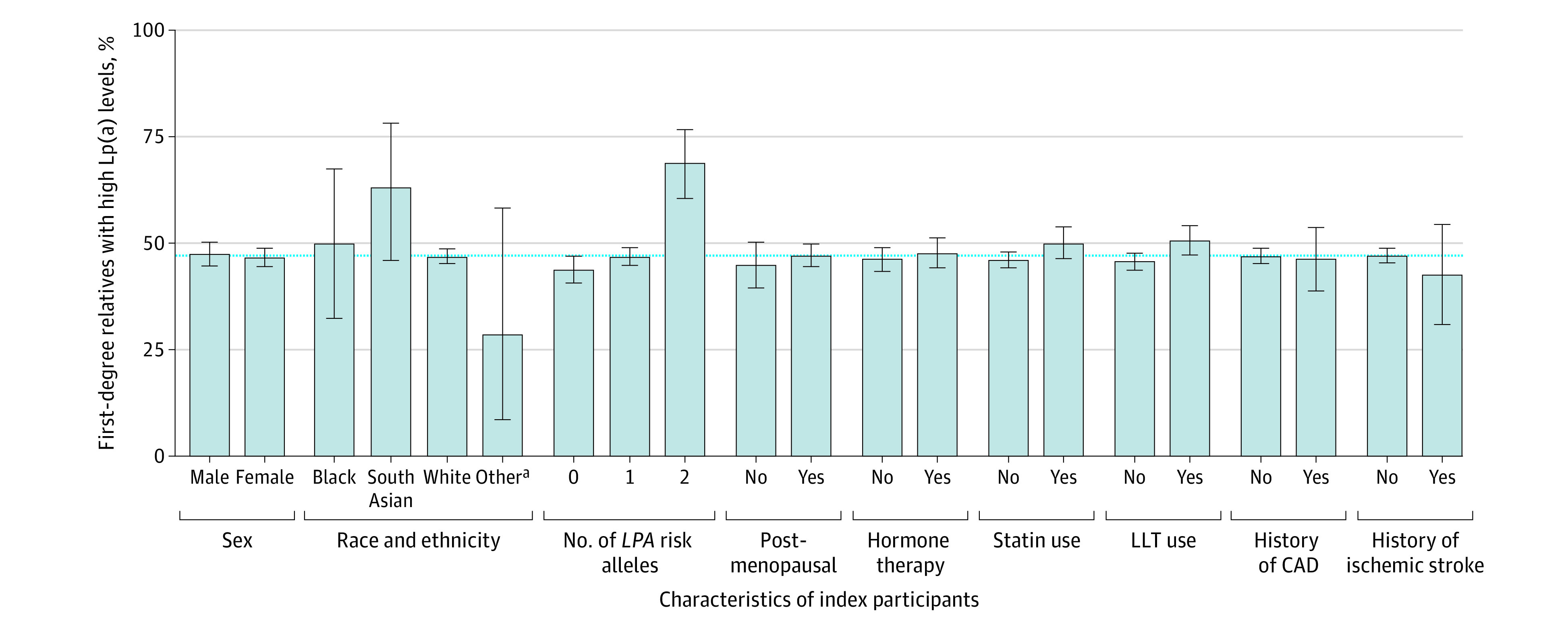
High Lp(a) levels were at least 125 nmol/L (to approximate levels in mg/dL, divide by 2.15). Subgroup analysis for high Lp(a) level concordance in first-degree relatives was stratified by the presence or absence of certain traits in index participants. The concordance of high Lp(a) among relatives was largely the same among subgroups and ranged from 28.6% (95% CI, 8.4%-58.1%) in those with no self-reported or other race to 68.8% (95% CI, 60.5%-76.3%) in those with a strong genetic predisposition to high Lp(a). Number of LPA risk alleles is defined as the number of variants (rs1045872 and rs3798220) in the LPA gene, which were previously associated with Lp(a) concentrations and coronary artery disease (CAD).19 Genetic subgroup analyses were only performed in White participants. For the subgroup analyses including menopause and hormone therapy, only first-degree relative pairs with female index participants were included. The blue dotted line represents the concordance for high Lp(a) levels among all first-degree relatives (47.0%). Error bars indicate 95% CIs. LLT indicates use of therapy to lower lipid levels.
aIncludes more than 1 race or ethnicity, other race or ethnicity, unknown race or ethnicity, and race or ethnicity not reported.
Last, since Lp(a) concentrations are known to be influenced by menopause, hormone therapy use, and statin use, subgroup analyses were performed for patients with or without presence of these traits. The concordance was largely consistent among first- and second-degree relatives in these subgroups except for statin and use of any therapy to lower lipid levels in first-degree relatives (Figure 3 and eFigures 6 and 7 in Supplement 1). A modestly increased concordance was observed among statin users vs nonusers if high Lp(a) levels were defined as at least 150 nmol/L (45.8% [95% CI, 41.8%-49.8%] for statin users vs 40.3% [95% CI, 38.5%-42.5%] for nonusers; P = .02) and among users of any therapy to lower lipid levels vs nonusers in first-degree relatives at both high Lp(a) concentrations of at least 125 nmol/L (50.7% [95% CI, 47.3%-54.1%] vs 45.8% [95% CI, 43.8%-47.7%]; P = .01) and at least 150 nmol/L (46.0% [95% CI, 42.3%-49.8%] vs 40.0% [95% CI, 37.8%-42.3%]; P = .007).
Discussion
In this cross-sectional study, we investigated the yield of screening for high Lp(a) levels in first- and second-degree relative pairs in the UK Biobank study and found that Lp(a) levels are correlated between relatives. This resulted in a high concordance in high Lp(a) level: 47.0% and 31.8% among first- and second-degree relatives of index individuals with high Lp(a) levels, respectively. These observations have, to our interpretation, at least 5 implications.
First, cascade screening of family members for high Lp(a) levels has a high diagnostic yield when the index patient has a high Lp(a) level. Cascade screening programs for familial hypercholesterolemia have been successful in the past, with reported yields ranging from 34% to 55%, depending on the exact study methodology used, which is similar to the yield for high Lp(a) level screening as demonstrated herein.21 Importantly, familial hypercholesterolemia is a rare monogenic disorder with a prevalence of approximately 0.4%, while high Lp(a) levels are present in 12% to 25%, depending on the exact threshold and studied population.7,15 Thus, Lp(a) screening has the potential to identify a greater number of individuals at high risk for ASCVD than current familial hypercholesterolemia screening programs.
Second, previous studies have investigated the diagnostic yield of Lp(a) cascade screening within existing familial hypercholesterolemia screening programs or among patients with common dyslipidemias and found a number needed to screen of 1.5 to 2.8, depending on the definition of high Lp(a) level.9,10 The current results extend these prior results to the general population, with a similar number needed to screen of approximately 2.
Third, we showed that the concordance in high Lp(a) levels among relatives was higher in those with a genetic predisposition for high Lp(a) levels. The genetic determinants that drive Lp(a) concentrations are complex. Common variants in the LPA gene are associated with Lp(a) concentrations, but the latter is primarily influenced by the number of kringle IV repeats at the LPA locus.5,6,7 Unfortunately, determination of kringle IV repeats was not possible in the current study, and a concordance analysis according to these repeats or other genetic instruments might yield different results.
Fourth, the challenge in investigating Lp(a) concentrations rather than a dichotomous risk factor such as familial hypercholesterolemia-causing variants lies in pinpointing the optimal Lp(a) threshold for cascade screening. Here we observe that the current consensus definition of high Lp(a) levels (≥125 nmol/L) might be feasible for cascade screening with a reasonable number needed to screen in first-degree relatives, but that other commonly used cutoffs (≥150 nmol/L and ≥200 nmol/L) yield similar results of 41.6% and 34.3% concordance in first-degree relatives, respectively. Of note, small differences in concordance exist among different subgroups of index participants (eg, those using statins), but these differences are likely to have only a minor effect on the number needed to screen in Lp(a) cascade screening programs.
Last, despite guidelines advocating the measurement of Lp(a) at least once in an individual’s lifetime, routine measurement of Lp(a) in current clinical practice remains uncommon.22 For example, in Germany, Lp(a) was measured in fewer than 1% of patients with known ASCVD, and in Israel only 0.1% of individuals within a health maintenance organization had their Lp(a) measured.20,23 The poor uptake of routine Lp(a) testing lends further support to advocate cascade screening for high Lp(a) levels. Fortunately, the clustering of high Lp(a) levels within families makes it an easy target for risk stratification in those families. Efficient means of prioritizing screening for Lp(a) are likely to become increasingly important if 2 clinical trials investigating therapies to lower Lp(a) levels—pelacarsen and olpasiran—demonstrate reductions in cardiovascular events.2,3
Limitations
We used cross-sectional UK Biobank data to estimate the yield of diagnoses of high Lp(a) levels within families, and thus our results do not reflect the true feasibility of a family cascade screening program. Our results should not be extrapolated to children, as it has been suggested that Lp(a) levels are lower in children.24 Last, we tested 3 Lp(a) thresholds based on professional society consensus statements and inclusion criteria for ongoing cardiovascular outcome trials of therapies to lower Lp(a) levels, but other values might become clinically relevant in the future as well.7,8
Conclusions
In this cross-sectional study, Lp(a) levels are correlated between first- and second-degree relatives, and the concordance in high Lp(a) levels among first- and second-degree relatives is high. Similar to familial hypercholesterolemia screening, these data support the implementation of cascade screening for high Lp(a) levels in relatives of individuals with high Lp(a) levels to identify and treat patients at increased Lp(a)-attributable ASCVD risk early and effectively.
eFigure 1. Flowchart of Participants and Relative Pair Selection
eFigure 2. Correlation in Lp(a) and Estimated Untreated LDL Cholesterol Plasma Levels Between First- and Second-Degree Relatives
eFigure 3. Sensitivity Analysis for Lp(a) Levels, Concordance in High Lp(a), and Numbers Needed to Screen Among Relatives of Index Participants With High Lp(a) After Excluding Index Participants Using Therapies to Lower Lipid Levels
eFigure 4. Percentage of First- and Second-Degree Relatives With Lp(a) ≥150 nmol/L for Any Index Cutoff Lp(a) Concentration
eFigure 5. Percentage of First- and Second-Degree Relatives With Lp(a) ≥200 nmol/L for Any Index Cutoff Lp(a) Concentration
eFigure 6. Subgroup Analysis for Concordance in High Lp(a) (≥150 nmol/L) Among First-Degree Relatives
eFigure 7. Subgroup Analysis for Concordance in High Lp(a) (≥200 nmol/L) Among First-Degree Relatives
eTable 1. Characteristics of Cohort by Self-Reported Race and Ethnicity
eTable 2. Concordance and Number Needed to Screen (NNS) for High Lp(a) in First-Degree Relatives
eTable 3. Concordance and Number Needed to Screen (NNS) for High Lp(a) in Second-Degree Relatives
eTable 4. Concordance in High Lp(a) in Index Participants With LPA Variants (rs1045872 or rs3798220)
eTable 5. Number of Index Participants With High Lp(a) According to Different Definitions of High Lp(a) and Self-Reported Race and Ethnicity
eReferences
Data Sharing Statement
References
- 1.Tsimikas S, Fazio S, Ferdinand KC, et al. NHLBI Working Group recommendations to reduce lipoprotein(a)-mediated risk of cardiovascular disease and aortic stenosis. J Am Coll Cardiol. 2018;71(2):177-192. doi: 10.1016/j.jacc.2017.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assessing the Impact of Lipoprotein (a) Lowering With Pelacarsen (TQJ230) on Major Cardiovascular Events in Patients With CVD (Lp[a]HORIZON). ClinicalTrials.gov Identifier: NCT04023552. Updated August 8, 2023. Accessed July 4, 2023. https://classic.clinicaltrials.gov/ct2/show/NCT04023552
- 3.Olpasiran Trials of Cardiovascular Events and Lipoprotein(a) Reduction (OCEAN[a])—Outcomes Trial. ClinicalTrials.gov Identifier: NCT05581303. Updated July 27, 2023. Accessed July 4, 2023. https://classic.clinicaltrials.gov/ct2/show/NCT05581303
- 4.Trinder M, Paruchuri K, Haidermota S, et al. Repeat measures of lipoprotein(a) molar concentration and cardiovascular risk. J Am Coll Cardiol. 2022;79(7):617-628. doi: 10.1016/j.jacc.2021.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukamel RE, Handsaker RE, Sherman MA, et al. Protein-coding repeat polymorphisms strongly shape diverse human phenotypes. Science. 2021;373(6562):1499-1505. doi: 10.1126/science.abg8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boerwinkle E, Leffert CC, Lin J, Lackner C, Chiesa G, Hobbs HH. Apolipoprotein(a) gene accounts for greater than 90% of the variation in plasma lipoprotein(a) concentrations. J Clin Invest. 1992;90(1):52-60. doi: 10.1172/JCI115855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kronenberg F, Mora S, Stroes ESG, et al. Lipoprotein(a) in atherosclerotic cardiovascular disease and aortic stenosis: a European Atherosclerosis Society consensus statement. Eur Heart J. 2022;43(39):3925-3946. doi: 10.1093/eurheartj/ehac361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reyes-Soffer G, Ginsberg HN, Berglund L, et al. ; American Heart Association Council on Arteriosclerosis, Thrombosis and Vascular Biology; Council on Cardiovascular Radiology and Intervention; and Council on Peripheral Vascular Disease . Lipoprotein(a): a genetically determined, causal, and prevalent risk factor for atherosclerotic cardiovascular disease: a scientific statement from the American Heart Association. Arterioscler Thromb Vasc Biol. 2022;42(1):e48-e60. doi: 10.1161/ATV.0000000000000147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis KL, Pérez de Isla L, Alonso R, Fuentes F, Watts GF, Mata P. Value of measuring lipoprotein(a) during cascade testing for familial hypercholesterolemia. J Am Coll Cardiol. 2019;73(9):1029-1039. doi: 10.1016/j.jacc.2018.12.037 [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty A, Chan DC, Ellis KL, et al. Cascade testing for elevated lipoprotein(a) in relatives of probands with high lipoprotein(a). Am J Prev Cardiol. 2022;10:100343. doi: 10.1016/j.ajpc.2022.100343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundy SM, Stone NJ, Bailey AL, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;139(25):e1082-e1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203-209. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867-2873. doi: 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid NJ, Brockman DG, Elisabeth Leonard C, Pelletier R, Khera AV. Concordance of a high polygenic score among relatives: implications for genetic counseling and cascade screening. Circ Genom Precis Med. 2021;14(2):e003262. doi: 10.1161/CIRCGEN.120.003262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel AP, Wang M, Pirruccello JP, et al. Lp(a) (lipoprotein[a]) concentrations and incident atherosclerotic cardiovascular disease: new insights from a large national biobank. Arterioscler Thromb Vasc Biol. 2021;41(1):465-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarthy S, Das S, Kretzschmar W, et al. ; Haplotype Reference Consortium . A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279-1283. doi: 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter K, Min JL, Huang J, et al. ; UK10K Consortium . The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82-90. doi: 10.1038/nature14962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium . A global reference for human genetic variation. Nature. 2015;526(7571):68-74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke R, Peden JF, Hopewell JC, et al. ; PROCARDIS Consortium . Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361(26):2518-2528. doi: 10.1056/NEJMoa0902604 [DOI] [PubMed] [Google Scholar]
- 20.Stürzebecher PE, Schorr JJ, Klebs SHG, Laufs U. Trends and consequences of lipoprotein(a) testing: cross-sectional and longitudinal health insurance claims database analyses. Atherosclerosis. 2023;367:24-33. doi: 10.1016/j.atherosclerosis.2023.01.014 [DOI] [PubMed] [Google Scholar]
- 21.Lee C, Rivera-Valerio M, Bangash H, Prokop L, Kullo IJ. New case detection by cascade testing in familial hypercholesterolemia: a systematic review of the literature. Circ Genom Precis Med. 2019;12(11):e002723. doi: 10.1161/CIRCGEN.119.002723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catapano AL, Daccord M, Damato E, et al. How should public health recommendations address Lp(a) measurement, a causative risk factor for cardiovascular disease (CVD)? Atherosclerosis. 2022;349:136-143. doi: 10.1016/j.atherosclerosis.2022.02.013 [DOI] [PubMed] [Google Scholar]
- 23.Zafrir B, Aker A, Saliba W. Lipoprotein(a) testing in clinical practice: real-life data from a large healthcare provider. Eur J Prev Cardiol. 2022;29(14):e331-e333. doi: 10.1093/eurjpc/zwac124 [DOI] [PubMed] [Google Scholar]
- 24.De Boer LM, Hof MH, Wiegman A, Stroobants AK, Kastelein JJP, Hutten BA. Lipoprotein(a) levels from childhood to adulthood: data in nearly 3,000 children who visited a pediatric lipid clinic. Atherosclerosis. 2022;349:227-232. doi: 10.1016/j.atherosclerosis.2022.03.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure 1. Flowchart of Participants and Relative Pair Selection
eFigure 2. Correlation in Lp(a) and Estimated Untreated LDL Cholesterol Plasma Levels Between First- and Second-Degree Relatives
eFigure 3. Sensitivity Analysis for Lp(a) Levels, Concordance in High Lp(a), and Numbers Needed to Screen Among Relatives of Index Participants With High Lp(a) After Excluding Index Participants Using Therapies to Lower Lipid Levels
eFigure 4. Percentage of First- and Second-Degree Relatives With Lp(a) ≥150 nmol/L for Any Index Cutoff Lp(a) Concentration
eFigure 5. Percentage of First- and Second-Degree Relatives With Lp(a) ≥200 nmol/L for Any Index Cutoff Lp(a) Concentration
eFigure 6. Subgroup Analysis for Concordance in High Lp(a) (≥150 nmol/L) Among First-Degree Relatives
eFigure 7. Subgroup Analysis for Concordance in High Lp(a) (≥200 nmol/L) Among First-Degree Relatives
eTable 1. Characteristics of Cohort by Self-Reported Race and Ethnicity
eTable 2. Concordance and Number Needed to Screen (NNS) for High Lp(a) in First-Degree Relatives
eTable 3. Concordance and Number Needed to Screen (NNS) for High Lp(a) in Second-Degree Relatives
eTable 4. Concordance in High Lp(a) in Index Participants With LPA Variants (rs1045872 or rs3798220)
eTable 5. Number of Index Participants With High Lp(a) According to Different Definitions of High Lp(a) and Self-Reported Race and Ethnicity
eReferences
Data Sharing Statement


