Dinet and Michelot discuss work from Wang et al. demonstrating that pollen tube pH gradients regulate the localization and activity of two ADF/cofilin isoforms, providing spatial control over actin networks.
Abstract
In a recent study, Wang et al. (https://doi.org/10.1083/jcb.202206074) demonstrate that subtle differences between two ADF/cofilin isoforms allow fine spatial regulation of the actin cytoskeleton in pollen tubes. This article illustrates how two similar proteins have progressively evolved to adapt their localization and activity according to the cellular environment.
All cells need control over their spatial organization. This regulation enables them to perform specific functions at precise locations rather than randomly throughout the cytoplasmic volume. This is particularly true of the eukaryotic actin cytoskeleton, a three-dimensional network of biological polymers that allows cells to exert forces in a controlled manner. Cells use actin monomers to build locally specialized filament networks of precise architectures (1).
How cells generate such network diversity from identical filaments is somehow established; they express multiple families of regulatory proteins that contribute to network assembly in different ways. To add to this molecular complexity, each protein family is often represented by multiple isoforms, which are often expressed simultaneously within a single cell type. What is less clear is how these different regulatory proteins act locally in cells. While some families of proteins (for example, the Arp2/3 complex or formins) are activated at precise locations by specific signaling pathways (1), others (e.g., ADF/cofilins or tropomyosins) are highly sensitive to the geometric organization of actin networks, leading to their segregation into different networks (2). But the question of how similar, yet distinct, isoforms of the same protein family segregate and provide localized activity remains unclear.
The pollen tube is an excellent cellular model to address this question. These large and elongated cells grow from their tips to deliver non-motile sperm cells. Tip growth depends on ion dynamics, and cytosolic gradients along the principal axis of the cell are maintained by the action of specific transporters (3). Notably, ions such as calcium and protons regulate various cellular processes and structures critical for pollen tube growth, including the actin cytoskeleton (4, 5). In Arabidopsis, a pH gradient is maintained along the pollen tube, with pH gradually increasing from the apex to the base (Fig. 1). Additionally, actin is abundant in pollen tubes and is organized into structurally distinct domains (4, 5). This model system is therefore well suited to determine how the chemical environment of a cell can influence the assembly and dynamics of the actin cytoskeleton.
Figure 1.
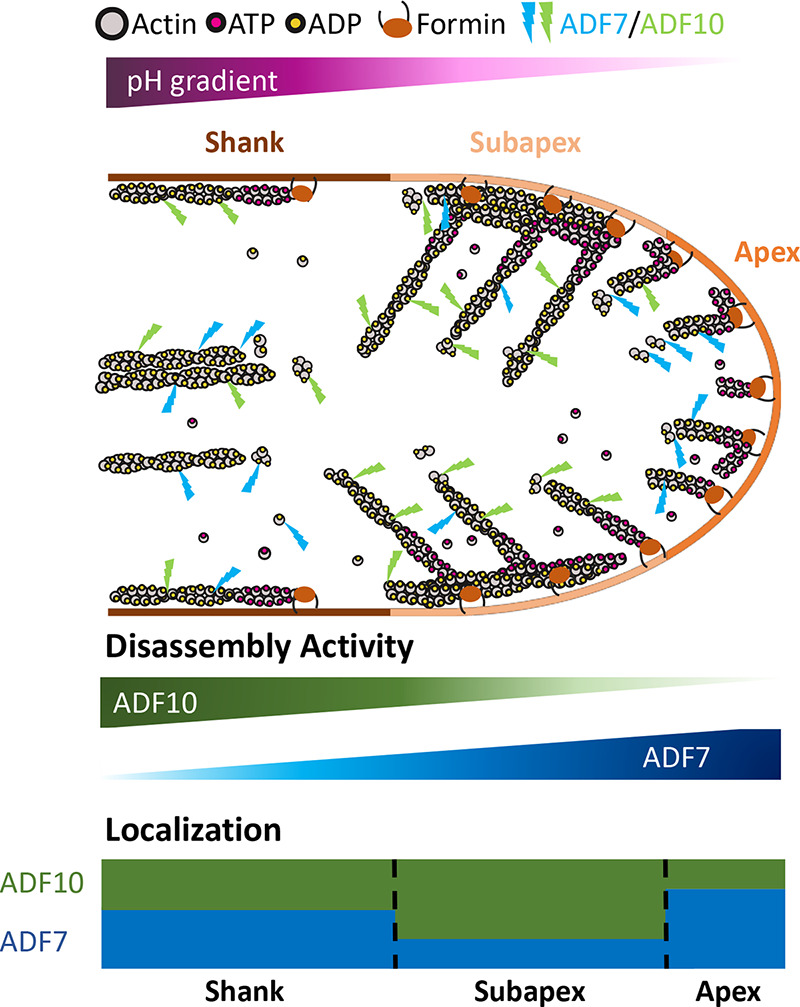
Schematic representation of the regulation of ADF7 and ADF10 in Arabidopsis thaliana pollen tubes. Pollen tube growth in A. thaliana depends on the formation of pH gradients along its main axis. The gradual increase in pH from tip to base is important for the regulation of several cellular processes, including the assembly and dynamics of the actin cytoskeleton. ADF7 and ADF10, two predominantly expressed isoforms of the ADF/cofilin protein family, are sensitive to these pH differences. While ADF7 binds to filamentous actin and functions at low pH, ADF10 prefers high pH. As a consequence, ADF7 and ADF10 partially segregate to different regions of the pollen tube and act on different actin networks. In particular, ADF7 plays an important role in enhancing actin filament turnover in the apical region of low pH, while ADF10 acts primarily on the denser actin networks of a subapical region.
Since actin polymerization itself is not very sensitive to small pH changes, an attractive hypothesis is that certain actin regulators may be more sensitive to pH differences. A particularly interesting candidate is the ADF/cofilin family. Indeed, it has been established that some ADF/cofilins are strongly pH dependent (for example, human ADF/cofilins; 6), while others are not (for example, Saccharomyces cerevisiae’s unique ADF/cofilin Cof1p; 7). Under acidic conditions, pH-sensitive ADF/cofilins do not efficiently disassemble actin filaments, whereas this activity is enhanced at both neutral and high pH levels (8). In pollen tubes, the severing activity of ADF/cofilin is thought to increase the number of filament ends, thereby promoting actin polymerization at the subapex (5).
The reported differences in pH sensitivity of ADF/cofilins motivated Wang et al. (9) to compare two isoforms predominantly expressed in pollen tubes, namely ADF7 and ADF10 (10). Consistent with previous studies, they show that both proteins are capable of inducing actin filament fragmentation. However, these proteins disassemble actin filaments optimally at different pH levels. While ADF7 binds to actin and functions optimally at low pH, ADF10 binds to actin and is more efficient at higher pH (Fig. 1).
To understand the physiological significance of these differences, Wang et al. examined the spatial distribution of both proteins within the apical and subapical regions of pollen tubes. They found that ADF7 is comparatively more abundant than ADF10 in the apical region, which is precisely a zone of lower pH. Conversely, ADF10 is more abundant in the subapical region, where the pH is higher (Fig. 1). While these variations are subtle, they are significant and consistent with the in vitro data.
The authors then examined the loss-of-function phenotypes for each isoform. They found that ADF7 promotes actin remodeling at the apex, whereas ADF10 is important for the turnover and spatial organization of subapical actin filaments. Differences were also detected in the transport of intracellular vesicles at the pollen tube tips, which is dependent on proper actin organization. A loss of function of ADF7 led to a decrease in vesicle density at the tip of pollen tubes. This effect is probably due to excessive polymerization of less dynamic actin filaments in the apical region, which prevent vesicle accumulation by a crowding effect. In contrast, ADF10's deficiency led to an accumulation of vesicles in the apical region. Such an accumulation is characteristic of excessive actin polymerization in the subapical region, which acts as a physical barrier to prevent vesicle backward diffusion.
While this study clearly shows that ADF7 and ADF10 have quite distinct localizations and functions in pollen tubes, it does not yet reveal how these proteins contribute to the establishment of distinct actin networks in apical and subapical regions. Nonetheless, this study is a striking reminder of the fact that the cellular cytoplasm is far from homogeneous, and that many biophysical and biochemical parameters can locally influence protein activity. This article is also a perfect illustration of the importance of the vast molecular diversity inherent to living organisms and shows that a protein family should not be systematically reduced to the study of a limited number of isoforms. Even isoforms like ADF7 and ADF10, which share a high degree of sequence homology, similar folding, comparable activities, and partially redundant functions, still exhibit significant differences. It is only by carefully and thoroughly exploring all this diversity that we will be able to unravel the full range of molecular mechanisms that living organisms have evolved.
Acknowledgments
This work was supported by the Fondation pour la Recherche Médicale to A. Michelot, grant Équipe FRM EQU202103012764.
References
- 1.Skau, C.T., and Waterman C.M.. 2015. Annu. Rev. Biophys. 10.1146/annurev-biophys-060414-034308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gressin, L., et al. 2015. Curr. Biol. 10.1016/j.cub.2015.04.011 [DOI] [PubMed] [Google Scholar]
- 3.Michard, E., et al. 2017. Plant Physiol. 10.1104/pp.16.01561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bascom, C.S., Jr, et al. 2018. Plant Physiol. 10.1104/pp.17.01466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang, R., et al. 2023. Plant Physiol. 10.1093/plphys/kiad203 [DOI] [PubMed] [Google Scholar]
- 6.Hawkins, M., et al. 1993. Biochemistry. 10.1021/bi00089a014 [DOI] [Google Scholar]
- 7.Pavlov, D., et al. 2006. Cell Motil. Cytoskeleton. 10.1002/cm.20142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wioland, H., et al. 2019. Biochemistry. 10.1021/acs.biochem.8b01001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, J., et al. 2023. J. Cell Biol. 10.1083/jcb.202206074 [DOI] [Google Scholar]
- 10.Jiang, Y., et al. 2022. Plant J. 10.1111/tpj.15723 [DOI] [Google Scholar]


