Abstract
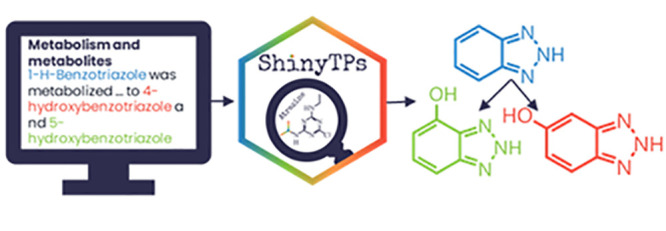
Transformation product (TP) information is essential to accurately evaluate the hazards compounds pose to human health and the environment. However, information about TPs is often limited, and existing data is often not fully Findable, Accessible, Interoperable, and Reusable (FAIR). FAIRifying existing TP knowledge is a relatively easy path toward improving access to data for identification workflows and for machine-learning-based algorithms. ShinyTPs was developed to curate existing transformation information derived from text-mined data within the PubChem database. The application (available as an R package) visualizes the text-mined chemical names to facilitate the user validation of the automatically extracted reactions. ShinyTPs was applied to a case study using 436 tentatively identified compounds to prioritize TP retrieval. This resulted in the extraction of 645 reactions (associated with 496 compounds), of which 319 were not previously available in PubChem. The curated reactions were added to the PubChem Transformations library, which was used as a TP suspect list for identification of TPs using the open-source workflow patRoon. In total, 72 compounds from the library were tentatively identified, 18% of which were curated using ShinyTPs, showing that the app can help support TP identification in non-target analysis workflows.
Keywords: Transformation products, Text mining, Curation, Non-target analysis, FAIR
Introduction
Understanding transformation product (TP) formation is crucial for assessing the compound fate and environmental risks. TPs are generally more polar and thus more mobile in the aquatic environment than their parent compounds.1,2 In addition, the toxicity and persistence of TPs may differ from those of their parents.3−6 However, there is often limited information available about TPs, both due to scarce research and inadequate FAIR7,8 (Findable, Accessible, Interoperable and Reusable) data on TPs, hindering utilization of and advancement on existing knowledge.
Information about TPs is provided by and available in several sources. One source of information is articles that contribute a large part of the known transformation reactions. However, the reactions are often depicted in non-machine-readable formats, such as detailed reaction schemes depicted in figures, making them difficult and time-consuming to curate and make machine-readable. Another source of transformation reaction information is databases. Some examples include MetXBioDB9 and SWISSPEST1910 as well as other NORMAN suspect lists11 (S66, S68 S74, S78, S79, and S81). All of these are combined within the PubChem Transformations library12 together with transformation reactions from ChEMBL,13 totalling 5923 unique reactions by CID. The Rhea14 database also contains balanced transformation reactions of biological interest, with over 15,000 registered reactions. These databases can be used further, including as training sets for machine learning and as suspect lists for screening for TPs.15 However, the number of available reactions in all databases is low in comparison to the >350,000 compounds and mixtures registered for production and use on the global market,16 thus showing the need to expand these resources to include more of the known transformation reactions. Looking beyond reactions, several databases also contain text-based information about transformation reactions. Examples include PharmaGKB,17,18 DrugBank,19 the Toxin and Toxin Target Database,20 and the Hazardous Substance Data Bank (HSDB).22 Several of these data sources are included within the PubChem database. These data sets often facilitate finding metabolic information for specific compounds. However, finding information for large data set purposes remains a time-consuming task.
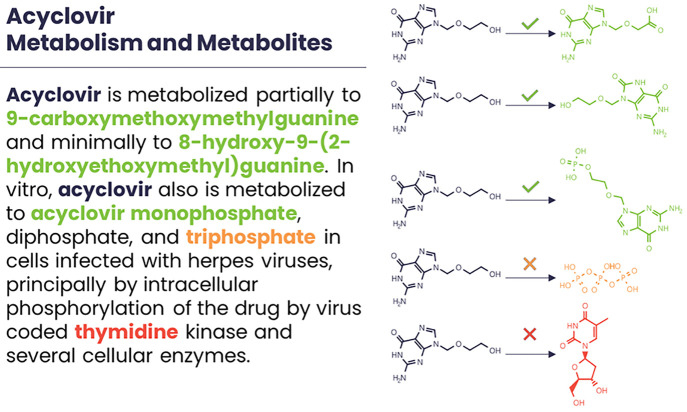
Several text mining tools for chemical name recognition have been developed to support the extraction of chemical information from text. Still, manual validation is often needed to ensure that the chemical names were recognized correctly and that no information has been omitted. Even the most accurate software packages used for text mining of chemical names report having precision and recall around 90%, indicating that without manual validation approximately 10% of the curated information would contain errors.21 This becomes even more complicated when using recognition of chemical names to extract potential TPs as observed by Krier et al.22 for the PubChem Metabolism and Metabolites section. This is due to the fact that even correctly recognized chemical names may be irrelevant to the transformations occurring and thus lead to incorrect reactions if not manually validated. An example of this can be seen in Figure 1.
Figure 1.
An example of the text-mined chemical names in the PubChem Metabolism and Metabolites section illustrating the true and false reactions. Transformation products (TPs) are highlighted in green; incompletely recognized chemical names of TPs are highlighted in orange, and correctly recognized chemical names that are not TPs are highlighted in red.
This study introduces “ShinyTPs”, an application designed to visualize and curate text-mined chemical reactions from PubChem. Its purpose is to aid in curating TPs and their associated reactions sourced from the Metabolism and Metabolites section of PubChem, and thus to enable their inclusion in downstream workflows or to serve as highly curated data sets for machine learning models.
Materials and Methods
Data Extraction and Processing
The transformation reaction information used in this study was from the Metabolism and Metabolites section of PubChem with HSDB as the data source. This source was selected since it was the largest data source in the Metabolism and Metabolites sections and also has high reliability, as the information was manually curated over 40 years by a scientific review panel.23 The text excerpts are text-mined by PubChem using LeadMine and an internal synonym dictionary to extract all chemical names and link them to their PubChem compound identifiers (CIDs). The HSDB excerpts with accompanying markup and CIDs are available as a JavaScript Object Notation (JSON) file. This JSON file was processed in R to extract the chemical names, together with the CID and other information. Other chemical identifiers, such as the Simplified Molecular-Input Line-Entry System (SMILES), were added. The resulting file was exported as a .csv file together with the original text and source descriptions. The code to create this file is available on GitLab.24
In addition, the PubChem Transformations table is available in full on Zenodo12 and was used to retrieve known reactions. The Transformations library (version 0.1.4) and the HSDB Metabolism and Metabolites data sets include information for 6157 and 3088 compounds, respectively. The Transformations data set is being updated periodically, and the code to do this is likewise available on GitLab.25 HSDB was discontinued in 2019 and the underlying data set is no longer being updated, although the chemical mappings used may change over time with updates to PubChem.
Software and Packages
ShinyTPs is an R package centered around a Shiny application for the curation of text-mined transformation reactions from the Metabolism and Metabolites section in PubChem.26 The app was developed in R27 (version 4.2.1). The user interface and server were constructed using shiny28 (a package for developing web applications) as well as the packages shinydashboard,29 shinyjs,30 shinythemes,31 bslib,32 and miniUI.33 The app also utilizes functions from the DT34 and dplyr35 packages to process data, as well as chemdoodle36 to display and draw chemical structures.
Using ShinyTPs
ShinyTPs is used through R by downloading the package available on GitLab.24 The first step is to read in a file containing the names, CIDs, and SMILES of the compounds of interest, which will create a list of tables that are used inside the app. The app is then launched with a separate function in the ShinyTPs package. Full instructions with code examples and detailed screenshots are available in the app documentation found on the GitLab repository.24
The ShinyTPs user interface consists of an “About” page and three main tabs: “PubChem Transformations”, “Transformation reaction curation”, and “Adding missing entries”. The “PubChem Transformations” tab (screenshots available in the documentation) allows viewing and filtering of the reactions that are already available in the PubChem Transformations library, while the remaining two tabs handle the curation. “Transformation reaction curation” is used to select and save the reactions obtained from the LeadMine text mining. It displays the structures of the input compound and chemical name found in the Metabolism and Metabolites text as well as the original text from HSDB. This facilitates determining whether the text-mined chemical name is a TP or parent of the input compound. Each reaction is then saved in a table in the app, which can be downloaded at the end. It is also possible to include information about the biosystem, enzyme, and transformation type (e.g., hydroxylation) for the reactions if this is found in the text.
The “Adding missing entries” tab handles curation of TPs or parent compounds that were not identified by the LeadMine text mining (e.g., “acyclovir diphosphate” in Figure 1). In this case, the structure of the compound can be drawn using chemdoodle, which returns the SMILES of the drawn structure. The compound name and SMILES of the compound are then entered as text inputs to save the reaction. Information about the biosystem, enzyme, and transformation type may also be saved as described for the “Transformation reaction curation” tab described above. Further explanation and screenshots are included in the ShinyTPs documentation on the GitLab repository.24
Uploading the Curated Data
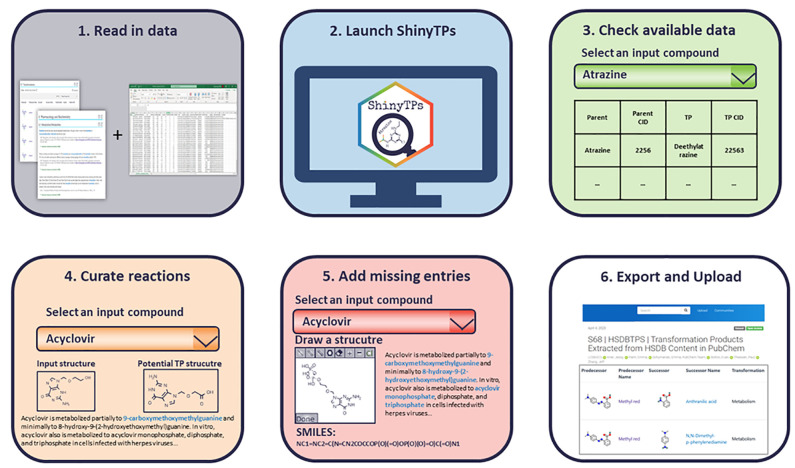
The output of the ShinyTPs app is designed to be included easily in the existing S68 HSDBTPS37 list on the NORMAN Suspect List Exchange (NORMAN-SLE).11 For the material prepared in this article, the information in the downloaded output files was cross-checked by the curator/creator and at least one independent reviewer using the automatically generated “reaction SMILES” via CDK Depict. Any remaining issues (e.g., salts, mismatching stereoisomers, adjustments to the descriptive information displayed) were then corrected where possible or otherwise removed from the data set. The finalized reactions were then added to the S68 HSDBTPS collection, which is integrated into the PubChem deposition and annotation cycle as part of the NORMAN-SLE efforts.11 Once the data was available in PubChem, the PubChem Transformations library was updated to version PubChemTrans-0.1.6.38 A full trace of this process is available on GitLab.39 An example workflow for using ShinyTPs is shown in Figure 2.
Figure 2.
General workflow of using the ShinyTPs app to curate transformation reactions.
Results and Discussion
Application of ShinyTPs to Test Compounds
To test the application, ShinyTPs was used to curate transformation reactions for 77 compounds from five different environmentally relevant data sets with information available from HSDB in the Metabolism and Metabolites section using the workflow shown in Figure 2. The full list of compounds is available in Table S1 in the Supporting Information. In total, 582 chemical names were identified in the HSDB text entries in the Metabolism and Metabolites section. Out of those, 114 (∼20%) corresponded to valid transformation reactions where the identified chemical name was either a TP or parent compound of the compound of interest. This demonstrates why manual validation of curated reactions is very important when handling text-mined data, as there would otherwise be a significant number of incorrect reactions generated due to the presence of other chemical names in the text unrelated to transformation processes. A further 60 reactions containing chemicals that were not recognized by the text-mining tool were curated using the “adding missing entries” function based on expert knowledge. This highlights the need for not only approaches that have a higher accuracy and precision in recognizing chemical names but also the development of approaches that are able to recognize complete chemical reactions. In combination, this further demonstrates the importance of manual review and curation in combination with the text-mined entries to capture all the reactions described in the text.
Of the 174 curated reactions, 143 were not already listed in the PubChem Transformations library and 31 were listed but with different associated metadata (e.g., different biosystem, enzyme, or evidence description). In addition, 18 of the curated TP structures were not yet present in PubChem and were deposited as a part of this work (CIDs 167530376–167530379 and 168354689–168354702). This shows that much of the information available for curation in the Metabolism and Metabolites section of PubChem is not already available in the Transformations library, such that curating this information as done here can help fill significant knowledge gaps in the database. This may then further improve the extraction of reaction rules and the development of machine learning models for the prediction of transformation products.
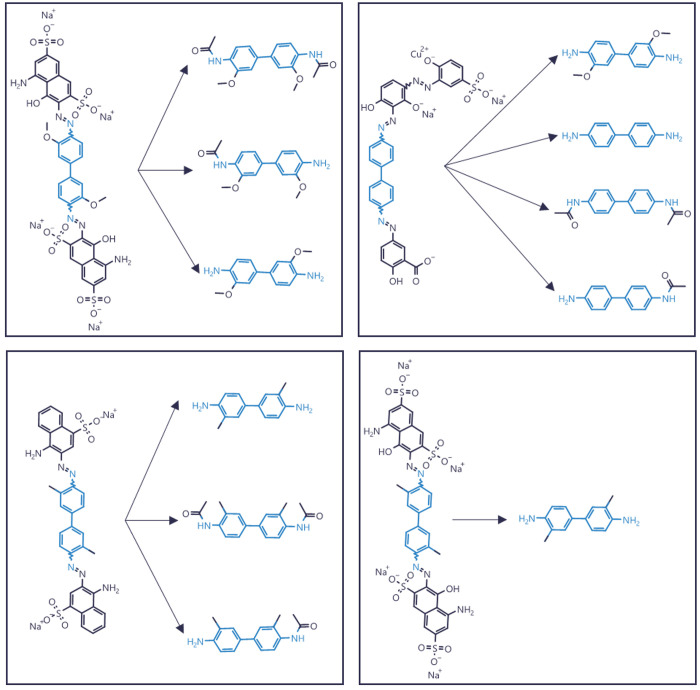
Some examples of TPs curated with ShinyTPs include benzidine and congeners, formed by benzidine-derived azo-dyes, as depicted in Figure 3. Based on these reactions it was possible to derive some reaction rules such as the cleavage of the nitrogen–nitrogen double bond followed by acetylation of the amino-groups. Several of the benzidine congeners have also been found to be carcinogenic and/or mutagenic, indicating that other dyes containing the substructure highlighted in blue in Figure 3 may also form hazardous TPs.
Figure 3.
Transformation reactions of benzidine-derived azo dyes curated using ShinyTPs. The benzidine substructure is highlighted in blue. All reactions can be found on PubChem (e.g., CIDs 8413, 19074, and 626583) and Zenodo.12,37
Application of ShinyTPs in Non-Target Workflows
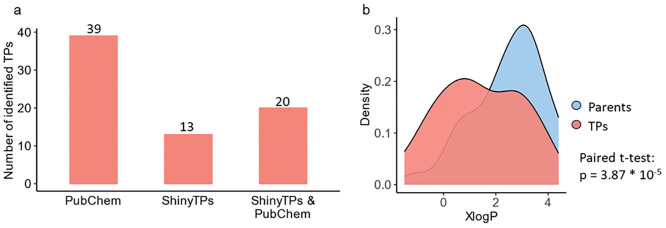
ShinyTPs was also applied to curate TP information for a list of 436 tentatively identified compounds (see Table S2) from a Luxembourgish wastewater treatment plant sample. These tentatively identified compounds were used as inputs to ShinyTPs to curate associated TP information using the “Transformation reaction curation” tab, totalling 645 new reactions associated with 496 compounds. Of these reactions, 319 were not listed for any biosystem in PubChem. The PubChem Transformations library with and without the addition of the new reactions was then used to perform a TP suspect screening in patRoon with the same sample. In total, 72 compounds out of the 436 were labeled as TPs using the TP suspect lists. Out of these, 18% were detected only after curation with ShinyTPs (see Figure 4a). Thus, the additional curation of TPs using ShinyTPs contributed to the number of tentatively identified TPs in this study and demonstrates that expanding the libraries will help support non-target analysis workflows in the community.
Figure 4.
(a) Number of identified TPs only present in the PubChem Transformations library and identified TPs curated with ShinyTPs as well as the identified TPs present in both libraries. (b) Distribution of XlogP values for the identified parent compounds and their corresponding TPs. Density refers to the kernel density.41
Several of the compounds labeled as TPs did not have any identified parent compounds. This may have several causes such as degradation of the parent compound below detection limits, incorrect identification, or direct release of the compound labeled as a TP to the environment. One example of the latter is diclofenac (a common pharmaceutical observed at high intensity in wastewater), which is listed as a potential TP of aceclofenac. Excluding compounds that did not have identified parent compounds resulted in 36 parent and TP pairs. Among these, the TPs were found to have a significantly lower octanol–water partitioning coefficient (logP), via the XlogP40 values calculated in PubChem, compared to their parent compounds (see Figure 4b). This in turn suggests a higher mobility of TPs compared with their parents. Some TPs were also associated with several parent compounds, e.g., terbuthylazine-2-hydroxy (see Table S3), which is a TP of terbuthylazine and terbutryn. This may be an indication of a higher potential for accumulation of these TPs, especially for those with longer half-lives, such as triazines. All tentatively identified TPs and their corresponding parents can be found in Table S3.
Future Perspectives
User contributions will help expand open transformation product libraries, which in turn can help improve screening of environmental contaminants, as illustrated by the application in this article. As the use of ShinyTPs expands (already external users are using the application), it is important to avoid curating duplicate information. This increases the importance of utilizing the “PubChem Transformations” tab to see which reactions are already available. Since HSDB was discontinued in 2019, new compounds will not be added to the HSDB data set and thus the TPs of emerging contaminants are not likely to be present. However, it will also be possible to enhance ShinyTPs to other sections of PubChem (e.g., the “Pharmacodynamics” section) in the future to expand the scope of this effort further. Finally, for current studies describing transformation processes, the inclusion of the results in a standard FAIR reporting template could ease the addition of these TP reactions into public resources.42,43
Acknowledgments
We would like to acknowledge discussions and contributions from various members of the ECI and PubChem teams that were not directly involved in these efforts, as well as the reviewers for their constructive suggestions.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.estlett.3c00537.
Tables S1–S3 containing lists of compounds used in the initial evaluation of ShinyTPs (S1), plus compounds (S2) and TPs (S3) tentatively identified in the wastewater treatment sample (XLSX)
Author Contributions
Emma H. Palm: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software (lead), Validation, Visualization, Writing original draft preparation (lead), Writing review and editing. Parviel Chirsir: Data curation, Investigation, Methodology, Writing review and editing. Jessy Krier: Conceptualization, software. Paul A. Thiessen: Data curation, Methodology, Software, Writing review and editing. Jian Zhang: Data curation, Methodology, Software, Writing review and editing. Evan E. Bolton: Resources, Supervision, Writing review and editing. Emma L. Schymanski: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Resources, Software, Supervision, Writing original draft preparation, Writing review and editing.
The work of P.A.T., J.Z., and E.E.B. was supported by the National Center for Biotechnology Information of the National Library of Medicine (NLM), National Institutes of Health. E.L.S. acknowledges funding support from the Luxembourg National Research Fund (FNR) for project A18/BM/12341006. E.H.P., E.L.S., and P.C. acknowledge funding from the European Union’s Horizon 2020 research and innovation program under Grant Agreement No. 101036756 for project “ZeroPM: Zero pollution of persistent, mobile substances”.
The authors declare no competing financial interest.
Notes
Associated Content: see ref. (44).
Supplementary Material
References
- Kotthoff L.; Keller J.; Lörchner D.; Mekonnen T. F.; Koch M. Transformation Products of Organic Contaminants and Residues—Overview of Current Simulation Methods. Molecules 2019, 24 (4), 753. 10.3390/molecules24040753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsholtz A.; Juarez R.; Svahn O.; Davidsson A.; Cimbritz M.; Falas P. Ozonation of 14C-Labeled Micropollutants - Mineralization of Labeled Moieties and Adsorption of Transformation Products to Activated Carbon. Water Res. 2022, 221, 118738. 10.1016/j.watres.2022.118738. [DOI] [PubMed] [Google Scholar]
- Magdeburg A.; Stalter D.; Schlüsener M.; Ternes T.; Oehlmann J. Evaluating the Efficiency of Advanced Wastewater Treatment: Target Analysis of Organic Contaminants and (Geno-)Toxicity Assessment Tell a Different Story. Water Res. 2014, 50, 35–47. 10.1016/j.watres.2013.11.041. [DOI] [PubMed] [Google Scholar]
- Lee Y.; von Gunten U. Advances in Predicting Organic Contaminant Abatement during Ozonation of Municipal Wastewater Effluent: Reaction Kinetics, Transformation Products, and Changes of Biological Effects. Environmental Science: Water Research & Technology 2016, 2 (3), 421–442. 10.1039/C6EW00025H. [DOI] [Google Scholar]
- Belfroid A. C.; van Drunen M.; Beek M. A.; Schrap S. M.; van Gestel C. A. M.; van Hattum B. Relative Risks of Transformation Products of Pesticides for Aquatic Ecosystems. Science of The Total Environment 1998, 222 (3), 167–183. 10.1016/S0048-9697(98)00298-8. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Ye C.; Li J.; Yu X.; Feng M. Treatment-Driven Removal Efficiency, Product Formation, and Toxicity Evolution of Antineoplastic Agents: Current Status and Implications for Water Safety Assessment. Water Res. 2021, 206, 117729. 10.1016/j.watres.2021.117729. [DOI] [PubMed] [Google Scholar]
- FAIR Principles. GO FAIR. https://www.go-fair.org/fair-principles/ (accessed June 9, 2023).
- Wilkinson M. D.; Dumontier M.; Aalbersberg Ij. J.; Appleton G.; Axton M.; Baak A.; Blomberg N.; Boiten J.-W.; da Silva Santos L. B.; Bourne P. E.; Bouwman J.; Brookes A. J.; Clark T.; Crosas M.; Dillo I.; Dumon O.; Edmunds S.; Evelo C. T.; Finkers R.; Gonzalez-Beltran A.; Gray A. J. G.; Groth P.; Goble C.; Grethe J. S.; Heringa J.; ’t Hoen P. A. C.; Hooft R.; Kuhn T.; Kok R.; Kok J.; Lusher S. J.; Martone M. E.; Mons A.; Packer A. L.; Persson B.; Rocca-Serra P.; Roos M.; van Schaik R.; Sansone S.-A.; Schultes E.; Sengstag T.; Slater T.; Strawn G.; Swertz M. A.; Thompson M.; van der Lei J.; van Mulligen E.; Velterop J.; Waagmeester A.; Wittenburg P.; Wolstencroft K.; Zhao J.; Mons B. The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 2016, 3 (1), 160018. 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumbou-Feunang Y.; Fiamoncini J.; Gil-de-la-Fuente A.; Greiner R.; Manach C.; Wishart D. S. BioTransformer: A Comprehensive Computational Tool for Small Molecule Metabolism Prediction and Metabolite Identification. Journal of Cheminformatics 2019, 11 (1), 2. 10.1186/s13321-018-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer K.; Müller A.; Singer H.; Hollender J. New Relevant Pesticide Transformation Products in Groundwater Detected Using Target and Suspect Screening for Agricultural and Urban Micropollutants with LC-HRMS. Water Res. 2019, 165, 114972. 10.1016/j.watres.2019.114972. [DOI] [PubMed] [Google Scholar]
- Mohammed Taha H.; Aalizadeh R.; Alygizakis N.; Antignac J.-P.; Arp H. P. H.; Bade R.; Baker N.; Belova L.; Bijlsma L.; Bolton E. E.; Brack W.; Celma A.; Chen W.-L.; Cheng T.; Chirsir P.; Čirka L.; D’Agostino L. A.; Djoumbou Feunang Y.; Dulio V.; Fischer S.; Gago-Ferrero P.; Galani A.; Geueke B.; Głowacka N.; Glüge J.; Groh K.; Grosse S.; Haglund P.; Hakkinen P. J.; Hale S. E.; Hernandez F.; Janssen E. M.-L.; Jonkers T.; Kiefer K.; Kirchner M.; Koschorreck J.; Krauss M.; Krier J.; Lamoree M. H.; Letzel M.; Letzel T.; Li Q.; Little J.; Liu Y.; Lunderberg D. M.; Martin J. W.; McEachran A. D.; McLean J. A.; Meier C.; Meijer J.; Menger F.; Merino C.; Muncke J.; Muschket M.; Neumann M.; Neveu V.; Ng K.; Oberacher H.; O’Brien J.; Oswald P.; Oswaldova M.; Picache J. A.; Postigo C.; Ramirez N.; Reemtsma T.; Renaud J.; Rostkowski P.; Rüdel H.; Salek R. M.; Samanipour S.; Scheringer M.; Schliebner I.; Schulz W.; Schulze T.; Sengl M.; Shoemaker B. A.; Sims K.; Singer H.; Singh R. R.; Sumarah M.; Thiessen P. A.; Thomas K. V.; Torres S.; Trier X.; Van Wezel A. P.; Vermeulen R. C. H.; Vlaanderen J. J.; Von Der Ohe P. C.; Wang Z.; Williams A. J.; Willighagen E. L.; Wishart D. S.; Zhang J.; Thomaidis N. S.; Hollender J.; Slobodnik J.; Schymanski E. L. The NORMAN Suspect List Exchange (NORMAN-SLE): Facilitating European and Worldwide Collaboration on Suspect Screening in High Resolution Mass Spectrometry. Environ. Sci. Eur. 2022, 34 (1), 104. 10.1186/s12302-022-00680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymanski E.; Bolton E.; Cheng T.; Thiessen P.; Zhang J.; Helmus R.; Blanke G.. Transformations in PubChem. - Full Dataset; 2023. 10.5281/zenodo.7838005. [DOI]
- ChEMBL Database. https://www.ebi.ac.uk/chembl/ (accessed May 12, 2023).
- Bansal P.; Morgat A.; Axelsen K. B.; Muthukrishnan V.; Coudert E.; Aimo L.; Hyka-Nouspikel N.; Gasteiger E.; Kerhornou A.; Neto T. B.; Pozzato M.; Blatter M.-C.; Ignatchenko A.; Redaschi N.; Bridge A. Rhea, the Reaction Knowledgebase in 2022. Nucleic Acids Res. 2022, 50 (D1), D693–D700. 10.1093/nar/gkab1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmus R.; ter Laak T. L.; van Wezel A. P.; de Voogt P.; Schymanski E. L. PatRoon: Open Source Software Platform for Environmental Mass Spectrometry Based Non-Target Screening. Journal of Cheminformatics 2021, 13 (1), 1. 10.1186/s13321-020-00477-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Walker G. W.; Muir D. C. G.; Nagatani-Yoshida K. Toward a Global Understanding of Chemical Pollution: A First Comprehensive Analysis of National and Regional Chemical Inventories. Environ. Sci. Technol. 2020, 54 (5), 2575–2584. 10.1021/acs.est.9b06379. [DOI] [PubMed] [Google Scholar]
- Whirl-Carrillo M.; Huddart R.; Gong L.; Sangkuhl K.; Thorn C. F.; Whaley R.; Klein T. E. An Evidence-Based Framework for Evaluating Pharmacogenomics Knowledge for Personalized Medicine. Clinical Pharmacology & Therapeutics 2021, 110 (3), 563–572. 10.1002/cpt.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whirl-Carrillo M.; McDonagh E. M.; Hebert J. M.; Gong L.; Sangkuhl K.; Thorn C. F.; Altman R. B.; Klein T. E. Pharmacogenomics Knowledge for Personalized Medicine. Clinical Pharmacology & Therapeutics 2012, 92 (4), 414–417. 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Feunang Y. D.; Guo A. C.; Lo E. J.; Marcu A.; Grant J. R.; Sajed T.; Johnson D.; Li C.; Sayeeda Z.; Assempour N.; Iynkkaran I.; Liu Y.; Maciejewski A.; Gale N.; Wilson A.; Chin L.; Cummings R.; Le D.; Pon A.; Knox C.; Wilson M. DrugBank 5.0: A Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.; Arndt D.; Pon A.; Sajed T.; Guo A. C.; Djoumbou Y.; Knox C.; Wilson M.; Liang Y.; Grant J.; Liu Y.; Goldansaz S. A.; Rappaport S. M. T3DB: The Toxic Exposome Database. Nucleic Acids Res. 2015, 43 (D1), D928–D934. 10.1093/nar/gku1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krallinger M.; Leitner F.; Rabal O.; Vazquez M.; Oyarzabal J.; Valencia A. CHEMDNER: The Drugs and Chemical Names Extraction Challenge. Journal of Cheminformatics 2015, 7 (1), S1. 10.1186/1758-2946-7-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krier J.; Singh R. R.; Kondić T.; Lai A.; Diderich P.; Zhang J.; Thiessen P. A.; Bolton E. E.; Schymanski E. L. Discovering Pesticides and Their TPs in Luxembourg Waters Using Open Cheminformatics Approaches. Environ. Int. 2022, 158, 106885. 10.1016/j.envint.2021.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonger G. C.; Hakkinen P.; Jordan S.; Publicker S. The National Library of Medicine’s (NLM) Hazardous Substances Data Bank (HSDB): Background, Recent Enhancements and Future Plans. Toxicology 2014, 325, 209–216. 10.1016/j.tox.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm E.; Krier J.; Schymanski E.. ShinyTPs; 2023. https://gitlab.lcsb.uni.lu/eci/shinytps (accessed June 20, 2023).
- Schymanski E. annotations/tps/Transformations.R · master · Environmental Cheminformatics/pubchem · GitLab. GitLab. https://gitlab.lcsb.uni.lu/eci/pubchem/-/blob/master/annotations/tps/Transformations.R (accessed July 7, 2023).
- Kim S.; Chen J.; Cheng T.; Gindulyte A.; He J.; He S.; Li Q.; Shoemaker B. A.; Thiessen P. A.; Yu B.; Zaslavsky L.; Zhang J.; Bolton E. E. PubChem 2023 Update. Nucleic Acids Res. 2023, 51, D1373. 10.1093/nar/gkac956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing; 2022. https://www.R-project.org/ (accessed May 17, 2023).
- Chang W.; Cheng J.; Allaire J. J.; Sievert C.; Schloerke B.; Xie Y.; Allen J.; McPherson J.; Dipert A.; Borges B.; Posit Software, PBC; jQuery Foundation; jQuery contributors; jQuery UI contributors; Otto M.; Thornton J.; Bootstrap contributors; Twitter, Inc; Nawaz Khan P.; Tsaran V.; Lembree D.; Chakravarthula S.; O’Connor C.; PayPal, Inc; Petre S.; Rowls A.; Reavis B.; Bejaoui S.; Ineshin D.; Samhuri S.; SpryMedia Limited; Fraser J.; Gruber J.; Sagalaev I.; R Core Team. . Shiny: Web Application Framework for R; 2022. https://cran.r-project.org/web/packages/shiny/index.html (accessed May 17, 2023).
- Chang W.; Ribeiro B. B.; RStudio; Almasaeed Studio; Adobe Systems Incorporated. . shinydashboard: Create Dashboards with “Shiny”; 2021. https://cran.r-project.org/web/packages/shinydashboard/index.html (accessed May 17, 2023).
- Attali D.shinyjs R package | by Dean Attali | Overview. https://deanattali.com/shinyjs/overview (accessed May 17, 2023).
- Chang W.; RStudio; Park T.; Dziedzic L.; Willis N.; Google Corporation; McInerney M.; Adobe Systems Incorporated; Canonical Ltd. . shinythemes: Themes for Shiny; 2021. https://cran.r-project.org/web/packages/shinythemes/index.html (accessed May 17, 2023).
- Sievert C.; Cheng J.; Aden-Buie G.; Posit Software, PBC; Bootstrap contributors; Twitter, Inc; Aguilar J.; Park T.; PayPal. . bslib: Custom “Bootstrap” “Sass” Themes for “Shiny” and “rmarkdown”; 2022. https://cran.r-project.org/web/packages/bslib/index.html (accessed May 17, 2023).
- Cheng J.; RStudio. . miniUI: Shiny UI Widgets for Small Screens; 2018. https://cran.r-project.org/web/packages/miniUI/index.html (accessed May 17, 2023).
- Xie Y.; Cheng J.; Tan X.; Allaire J. J.; Girlich M.; Ellis G. F.; Rauh J.; SpryMedia Limited; Reavis B.; Gersen L.; Szopka B.; Pickering A.; Holmes W.; Marttila M.; Quintero A.; Laurent S.; Posit Software, PBC. . DT: A Wrapper of the JavaScript Library “DataTables”; 2023. https://cran.r-project.org/web/packages/DT/index.html (accessed May 17, 2023).
- Wickham H.; François R.; Henry L.; Müller K.; Vaughan D.; Posit Software, PBC. . dplyr: A Grammar of Data Manipulation; 2023. https://cran.r-project.org/web/packages/dplyr/index.html (accessed May 17, 2023).
- Charlop-Powers Z.Chemdoodle; 2023. https://github.com/zachcp/chemdoodle (accessed May 17, 2023).
- LCSB-ECI; Krier J.; Schymanski E.; PubChem Team; Bolton E.; Thiessen P.; Zhang J.. S68 | HSDBTPS | Transformation Products Extracted from HSDB Content in PubChem; 2020. 10.5281/zenodo.3830987. [DOI]
- Schymanski E.; Bolton E.; Cheng T.; Thiessen P.; Zhang J.; Helmus R.; Blanke G.. Transformations in PubChem - Full Dataset; 2023. 10.5281/zenodo.8117741. [DOI]
- Schymanski E.Environmental Cheminformatics/pubchem · GitLab. GitLab. https://gitlab.lcsb.uni.lu/eci/pubchem (accessed July 4, 2023).
- Cheng T.; Zhao Y.; Li X.; Lin F.; Xu Y.; Zhang X.; Li Y.; Wang R.; Lai L. Computation of Octanol-Water Partition Coefficients by Guiding an Additive Model with Knowledge. J. Chem. Inf. Model. 2007, 47 (6), 2140–2148. 10.1021/ci700257y. [DOI] [PubMed] [Google Scholar]
- Wickham H.; Chang W.; Henry L.; Pedersen T. L.; Takahashi K.; Wilke C.; Woo K.; Yutani H.; Dunnington D.. Smoothed density estimates — geom_density. https://ggplot2.tidyverse.org/reference/geom_density.html (accessed July 20, 2023).
- Schymanski E. L.; Bolton E. E. FAIR Chemical Structures in the Journal of Cheminformatics. Journal of Cheminformatics 2021, 13 (1), 50. 10.1186/s13321-021-00520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymanski E. L.; Bolton E. E. FAIRifying the Exposome Journal: Templates for Chemical Structures and Transformations. Exposome 2022, 2 (1), osab006. 10.1093/exposome/osab006. [DOI] [Google Scholar]
- Palm E. H.; Chirsir P.; Krier J.; Thiessen P. A.; Zhang J.; Bolton E. E.; Schymanski E. L.. ShinyTPs: Curating transformation products from text mining results. ChemRxiv. 2023. 10.26434/chemrxiv-2023-xm41h-v2 (accessed September 12, 2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.