Abstract
Complement offers a first line of defence against infection through the opsonization of microbial pathogens, recruitment of professional phagocytes to the infection site and the coordination of inflammatory responses required for the resolution of infection. Staphylococcus aureus is a successful pathogen that has developed multiple mechanisms to thwart host immune responses. Understanding the precise strategies employed by S. aureus to bypass host immunity will be paramount for the development of vaccines and or immunotherapies designed to prevent or limit infection. To gain a better insight into the specific immune evasion mechanisms used by S. aureus we examined the pathogen’s interaction with the soluble complement inhibitor, C4b-binding protein (C4BP). Previous studies indicated that S. aureus recruits C4BP using a specific cell-wall-anchored surface protein and that bound C4BP limits complement deposition on the staphylococcal surface. Using flow-cytometric-based bacterial-protein binding assays we observed no interaction between S. aureus and C4BP. Moreover, we offer a precautionary warning that C4BP isolated from plasma can be co-purified with minute quantities of human IgG, which can distort binding analysis between S. aureus and human-derived proteins. Combined our data indicates that recruitment of C4BP is not a complement evasion strategy employed by S. aureus .
Keywords: C4b-binding protein, complement, immune evasion, Staphylococcus aureus
Introduction
Staphylococcus aureus exists as a bacterial commensal of the human nares and skin with significant pathogenic potential, frequently causing a suite of diseases ranging from skin and soft tissue infections to life-threatening bacteraemia [1]. Treatment of S. aureus has been made complicated by the worldwide emergence of multiple lineages that have acquired resistance to a multitude of antibiotics [2]. Consequently, the WHO has listed S. aureus as a ‘priority pathogen’, where the development of novel antimicrobials and immunotherapeutics are urgently required to treat infections [3]. To develop effective therapeutic interventions, a greater understanding of the pathophysiological process of S. aureus disease is central.
An immediate host response to S. aureus infection is governed by complement, a sophisticated evolutionary conserved system that promotes the detection of microbes and mediates the coordination of inflammatory processes that serve to eliminate foreign species [4]. The complement system consists of multiple soluble and membrane-bound proteins with specific activator and inhibitor functions. Complement is activated by microbes via three distinct pathways (the classical, lectin and alternative pathways), which join at the level of C3 convertase formation and subsequent cleavage of the central C3 molecule. C3 is cleaved into C3a, an inflammatory modulator component and C3b, the major opsonin that can deposit onto cellular surfaces and promote the uptake of microbes by professional phagocytes. Extended C3b deposition results in the formation of the C5 convertases, which cleave C5 into C5a, a potent anaphylatoxin and C5b, a central initiator of the membrane attack complex (MAC).
To limit damage generated by complement activation, the host has evolved several fluid-phase and immobilized proteins that act as complement inhibitors [5]. C4b-binding protein (C4BP), a large 500 kDa multimeric glycoprotein and Factor H (FH), a 155 kDa serum glycoprotein are two soluble complement inhibitors. C4BP limits classical and lectin complement pathways through its ability to bind and restrict the function of activated complement C4b. C4BP can act as a cofactor to the serine protease Factor I (FI), directing the inactivation of soluble and membrane bound C4b, inhibiting the formation of the classical C3 convertase. In addition, C4BP accelerates the natural decay of the classical C3 convertase and can serve as a cofactor to FI in the cleavage of fluid-phase C3b, thus interfering with the alternative complement pathway [6]. FH represents the master regulator of the alternative pathway; FH acts as a cofactor for FI-mediated C3b cleavage, competes with Factor B for interaction with C3b, preventing the formation of fluid-phase C3 convertase and functions to promote the dissociation of the alternative C3 convertase [7].
Microbes have evolved multiple strategies to evade the complement system, promoting host colonization and access to resources [8]. One strategy used by several bacteria is the recruitment of host complement inhibitors, such as C4BP, to the microbial surface, limiting complement activation and subsequent eradication [6, 8]. Previous work has indicated that S. aureus recruits C4BP via the cell-wall-anchored serine-aspartate repeat (Sdr) protein E (SdrE) and bone sialoprotein binding protein (Bbp), an allelic variant of SdrE with 75 % sequence identity [9], resulting in C4BP being localized to the bacterial surface, disrupting complement activation [10, 11]. The distribution of the sdrE/bbp gene has been examined previously; Sabat et al. reported that the sdrE gene was present in 89.5 % (445/497) of strains from a genetically diverse, clinical collection (MSSA n=382; MRSA n=115) where sdrE distribution did not differ between MSSA and MRSA isolates [12]. Another study investigated the carriage of the sdr locus among diverse S. aureus lineages revealing that the sdrE gene was present in 68.1 % (196/288) isolates [13]. The SdrE sequence exhibits significant variation between different S. aureus lineages compared to within lineages, with higher levels of variation observed in the protein domains at the host interface [14].
One aim of this study was to determine if additional methods of C4BP recruitment existed and to investigate to what degree C4BP recruitment is conserved across genetically distinct S. aureus isolates. Understanding the conservation and existence of alternative staphylococcal C4BP binding proteins is important to enhance our understanding of S. aureus immune evasion and to direct future therapeutic intervention strategies.
Employing flow-cytometry-based, bacteria-protein binding assays and defined S. aureus isogenic mutants, we conclude that S. aureus does not recruit C4BP under standard laboratory conditions. Our data also provides a precautionary warning that minute quantities of human IgG can be associated with C4BP following purification from human plasma, which can distort results, particularly if bacteria under question strongly bind IgG.
Methods
Bacterial isolates and growth conditions
S. aureus strains were grown in tryptic soy broth (TSB) at 37 °C with shaking (180 r.p.m.). S. aureus transposon mutants were obtained from the Nebraska Transposon Mutant Library [15] and were grown in TSB containing erythromycin (5 µg ml−1). Newman sdrE::Tn and Newman spa::Tn were generated through transduction using ϕ11. Transductants containing the inserted transposon were examined on tryptic soy agar containing erythromycin (10 µg ml−1). SdrE and Spa transposon mutants were verified for correct Tn insertion by colony PCR using gene specific primers; sdrE_FW: 5′-CCAACTACACCTCAAGAATCTAC-3′; sdrE_RV: 5′-GGCTTGTTTCTTTACCTGCTG-3′ and spa_FW: 5′-CCTCAGCACATTCAAAGCC-3′; spa_RV: 5′-AGCCGTTACGTTGTTCTTC-3′. Lactococcus lactis MG1363 was used to express S. aureus proteins, protein A and SdrE, the genes of which had been previously cloned into the expression vector pKS80 [16]. L. lactis strains were grown in M17 broth containing 1 % glucose and erythromycin (5 µg ml−1) at 30 °C without shaking. Streptococcus pyogenes strain AP1 was grown in Todd–Hewitt broth at 37 °C with 5 % CO2 without shaking. Moraxella catarrhalis strain RH4 was cultured on chocolate agar plates for 24 h at 37 °C with 5 % CO2.
Human serum preparation, C4BP protein purification and labelling
Normal human serum (NHS) was harvested from freshly obtained blood from eight healthy volunteers using vacutainer serum CAT collection tubes (BD). Blood samples were allowed to clot at room temperature for 30 min and then incubated on ice for 1 h. After two rounds of centrifugations at 800 g at 4 °C for 8 min, serum samples were collected and pooled at 4 °C and then immediately aliquoted and stored at −80 °C. Healthy volunteers provided written informed consent according to the recommendations of the University of Bath, Research Ethics Approval Committee for Health (reference: EP 18/19 108). Heat-inactivated serum (HI-S) was prepared by heating serum at 56 °C for 30 min using a water bath.
C4BP was purified from pooled plasma using barium chloride, anion exchange chromatography and gel filtration as described previously [17]. In addition C4BP was expressed from the pcDNA3 vector in a human kidney 293 cell line as outlined by Blom et al. [18]. Lastly C4BP was also purchased (Complement Technologies; A109), aliquoted and stored at −80 °C until use. C4BP was labelled with Dylight 488 using DyLight 488 NHS Ester (ThermoFisher) according to the manufacturer’s instructions. Labelled C4BP were separated from uncoupled dye via buffer exchange using Zeba Spin Desalting Columns (ThermoFisher). The degree of labelling was determined following the manufacturer's instructions. Briefly, for each C4BP preparation, absorbance at 280 nm and at 493 nm was measured by spectrometry, and used in the following calculation:
Plasma purified C4BP (4.33), recombinantly produced C4BP (3.74) and C4BP purchased from CompTech (4.36), exhibited similar labelling degrees defined as moles dye per mole protein using this protocol. Labelled C4BP was eluted in PBS and stored at 4 °C. IgM/IgG depleted serum was purchased from Pel-Freez Biologicals, aliquoted and stored at −80 °C until required. Human IgG contamination of C4BP was measured by SDS-PAGE and western blot. Purified C4BP was loaded at 1 and 2 µg and subjected to electrophoresis using a 5 % SDS gel under non-reducing conditions. The gel was transferred onto a PVDF membrane using a Trans blot turbo transfer system (BioRad). The membrane was blocked overnight in TBST containing 5 % semi skinned milk before being incubated with goat anti-human IgG-HRP conjugate (1 : 1000; Life Technologies) for 45 min at room temperature before being visualised using an Amersham-ECL kit (Cytiva).
Examining bacterial interaction with C4b-binding protein
For S. aureus , overnight cultures were 1 : 200 diluted in fresh TSB and sub-cultured to OD600=0.5–0.6. For S. pyogenes , overnight culture was normalized to OD600=0.1 and sub-cultured to OD600=0.3–0.4 in Todd–Hewitt broth. For M. catarrhalis , bacteria were cultured on chocolate agar plates overnight and streaked onto new chocolate agar plates grown for 7 h. M. catarrhalis were scraped from plates and resuspended into 25 % BHI with glycerol. Glycerol stocks were kept in −80 °C and thawed at 37 °C for 30 min before use. For L. lactis expressing S. aureus surface proteins, bacteria were normalized from stationary phase culture.
Bacteria were harvested via centrifugation at 13 000 g for 5 min following PBS wash. S. aureus, S. pyogenes and M. catarrhalis were normalized to OD600=2 in PBS. Bacteria were stained using 2 µM Cell Trace Far Red (ThermoFisher) for 20 min at 37 °C with shaking. Unbound dye was removed by washing cells in 1 % BSA/PBS. Bacteria were resuspended in either PBS for labelled C4BP binding experiments, or in GVB++ for binding of C4BP from NHS. L. lactis were normalized to OD600=1 in PBS and stained using 1 µM of Cell Trace Far Red and prepared as above.
To detect bacterial binding of labelled C4BP, 50 µl stained bacteria cells were mixed with 50 µl labelled C4BP in 96-well plates at 37 °C for 30 min. Bacteria were centrifuged and washed once in 1 % BSA/PBS and resuspended in 100 µl PBS. Bacteria incubated without labelled C4BP was used as negative control. In IgG-C4BP competition experiments, bacteria were pre-incubated with purified human IgG (Sigma) for 30 min, washed three times and then incubated with labelled C4BP for 30 min. Following incubation, bacteria were washed once in 1 % BSA/PBS and resuspended in 100 µl PBS.
To detect binding of C4BP from NHS, 50 µl stained bacteria cells were mixed with 50 µl NHS in 96-well plates at 37 °C for 30 min. Bacteria were centrifuged and washed once in 1 % BSA/PBS. F(ab’)2 monoclonal mouse anti-human C4BP MK104 [19] antibodies were prepared using the F(ab’)2 preparation kit (Pierce). Bacteria were resuspended in 100 µl of F(ab’)2 MK104 in 1 % BSA/PBS (4 µg ml−1) for 45 min at room temperature followed by centrifugation and washing 1 × in 1 % BSA/PBS. Bacteria were then stained with goat anti-mouse AF-488 (1 : 1000; Invitrogen) or goat anti-mouse PE (1;1000; Abcam) secondary antibody using the above conditions. Bacteria incubated in the absence of serum was used as a negative control.
Bound C4BP was assessed using FACS CANTO (BD) flow cytometer using wavelength 488 and 633 nm. Stained and unstained bacteria were employed for accurate gating of bacteria; a minimum of 20 000 events were used in all experiments. Data were analysed by FlowJo v10 software (BD Life Sciences).
Complement C9 deposition analysis
S. aureus and S. pyogenes were prepared as above for C4BP interaction. Bacteria were incubated with 8 % NHS and increasing concentrations of IgG-free C4BP diluted in GVB++ for 1 h at 37 °C. Bacteria were centrifuged and washed once in 1 % BSA/PBS and resuspended in 100 µl FcR blocking reagent (1 : 5, miltenyibiotec). Plates were incubated at 4 °C for 30 min. After washing with 1 % BSA/PBS, bacteria were stained with goat anti-human C9 (Complement Technologies) at 4 °C for 30 min and washing once with 1 % BSA/PBS. Bacteria were when stained with rabbit-anti-goat AF-488 secondary antibody (1 : 1000, Invitrogen) at 4 °C for 30 min. Bacteria incubated in HI-S were used as negative control. C9 deposition was examined as described above for C4BP detection using flow cytometry. The fluorescent intensity for each sample was normalized using NHS without added C4BP as the maximum value and HI-S as the minimum value.
Statistical analysis
A one-way or two-way ANOVA (GraphPad Prism 9.4.1) was used to examine differences between experimental results where a P value<0. 05 was considered to be significant.
Results
Interaction of plasma purified C4BP with S. aureus and L. lactis expressing SdrE
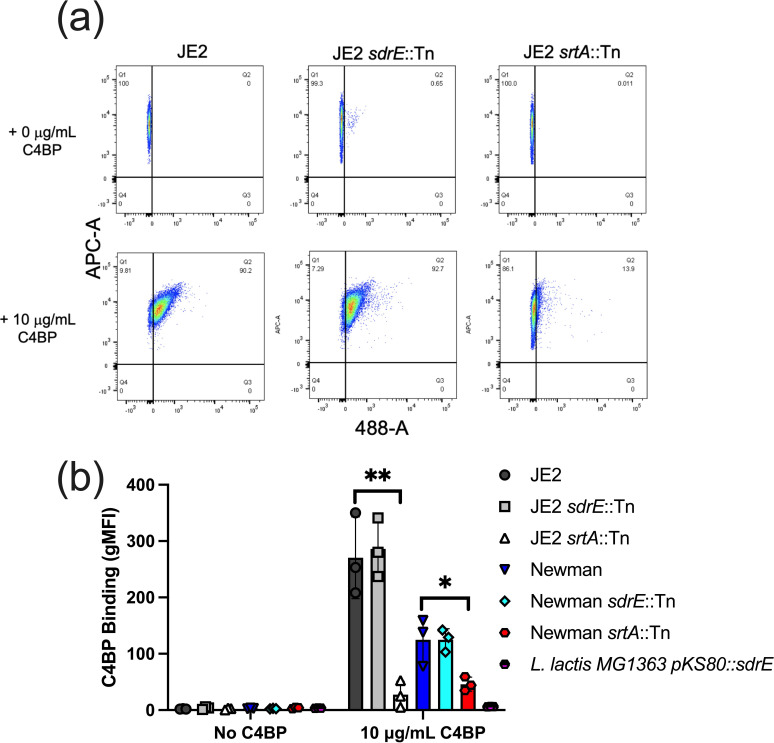
Our original aim was to test whether SdrE was the sole staphylococcal protein that mediated C4BP recruitment to the bacterial surface. To test this, we developed a simple S. aureus – C4BP binding assay using DyLight 488-fluorescently labelled plasma purified C4BP (Fig. S1, available in the online version of this article), Cell-trace far red labelled S. aureus and flow cytometry. We tested binding of plasma purified C4BP using two commonly used S. aureus strains, JE2 and Newman, both of which showed binding of C4BP (10 µg ml−1) (Fig. 1a-b). Next, we tested whether inactivation of SdrE, the only described S. aureus C4BP binding protein [11], impacted C4BP recruitment. Surprisingly we saw no difference in binding of C4BP between WT and sdrE::Tn mutants. Interestingly, when we overexpressed the gene coding for SdrE in the heterologous host, Lactococcus lactis , we did not observe any C4BP binding (Fig. 1b). We confirmed that L. lactis was expressing SdrE following extraction of cell-wall proteins and Western blotting using anti-SdrE serum [16] (Fig. S2A). Lastly, we tested the importance of a functional sortase A gene in the recruitment of C4BP. Sortase A is a central enzyme in the covalent linkage of staphylococcal cell-wall-anchored (CWA) proteins to peptidoglycan [20]. Here we observed significantly less C4BP binding in both JE2 and Newman generated srtA mutants (Fig. 1b), indicating the importance of CWA proteins for plasma purified C4BP interaction.
Fig. 1.
Interaction of plasma purified C4BP with S. aureus and L. lactis expressing SdrE. Dy488-labelled plasma purified C4BP (10 µg ml−1) was incubated with S. aureus strains and L. lactis expressing SdrE. Bacterial strains were labelled using cell trace Far Red (APC-A). C4BP binding was analysed by measuring the geometric mean fluorescence intensity (gMFI) using a BD FACS Canto flow cytometer. Bars indicate the mean; data points represent three biological replicates and error bars inform the standard deviation. Statistical differences were calculated using a one-way ANOVA analysis using Dunnett’s multiple comparisons test comparing mutants to respective isogenic WT control. *P<0.05, **P<0.01,.
Protein A mediates interaction with plasma purified C4BP
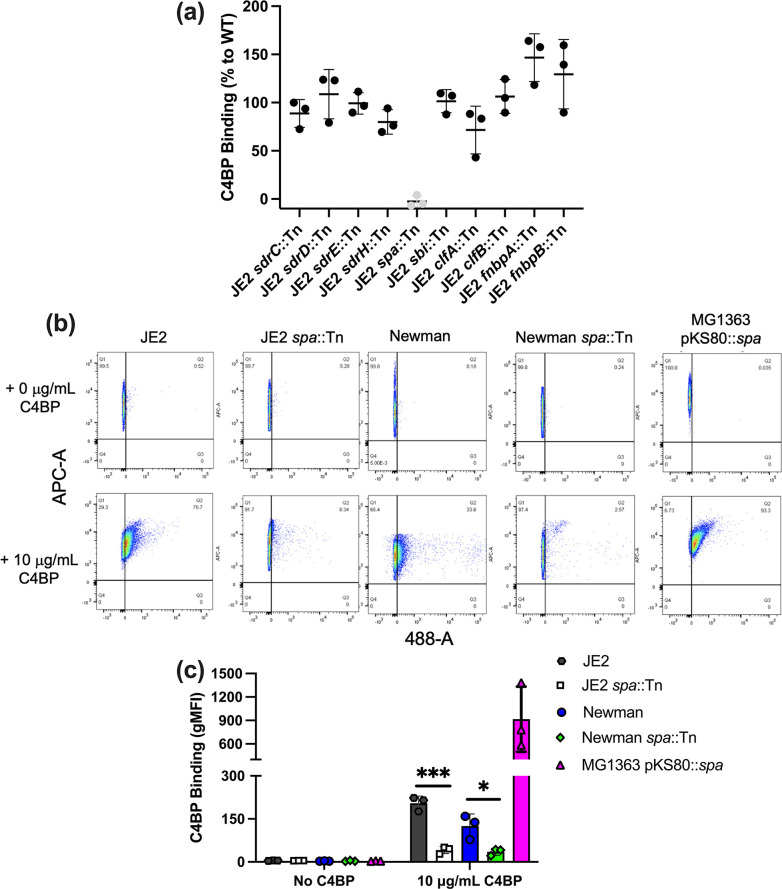
To assess the importance of other CWA proteins in binding C4BP, we screened defined surface proteins [SdrC, SdrD, SdrH, protein A, Staphylococcus aureus binder of immunoglobulin protein (Sbi), clumping factor A (ClfA), ClfB, fibronectin binding protein A (FnBPA) and FnBPB] known to interact with serum and/or plasma proteins [21] (Fig. 2a). Interestingly, only the protein A mutant (spa::Tn) showed a significant decrease in C4BP binding in both JE2 and Newman backgrounds and similar to the no C4BP control (Fig. 2b, c). Overexpression of the gene encoding protein A (spa) in L. lactis resulted in binding of C4BP. The presence of Spa in the cell wall was confirmed using Western blotting (Fig. S2B). These results indicated that protein A played a major role in the recruitment of plasma-purified C4BP to the staphylococcal surface.
Fig. 2.
Protein A mediates interaction with plasma purified C4BP. (a) Illustrates the C4BP binding capacity of multiple cell wall-anchored protein transposon mutants (SdrC, SdrD, SdrE, protein A, ClfA, ClfB, FnbpA, and FnbpB) and Sbi, a non-covalently associated protein that is related to protein A. (b), (c) C4BP binding to S. aureus WT and transposon mutants of protein A and L. lactis expressing protein A. C4BP binding was analysed by measuring the geometric mean fluorescence intensity (gMFI) using a BD FACS Canto flow cytometer. Bars indicate the mean; data points represent three biological replicates and error bars inform the standard deviation. Statistical differences were calculated using a one-way ANOVA using Dunnett’s multiple comparisons test analysis comparing mutants to respective isogenic WT control. *P<0.05, ***P<0.001,.
C4BP is not recruited to S. aureus when incubated in human serum
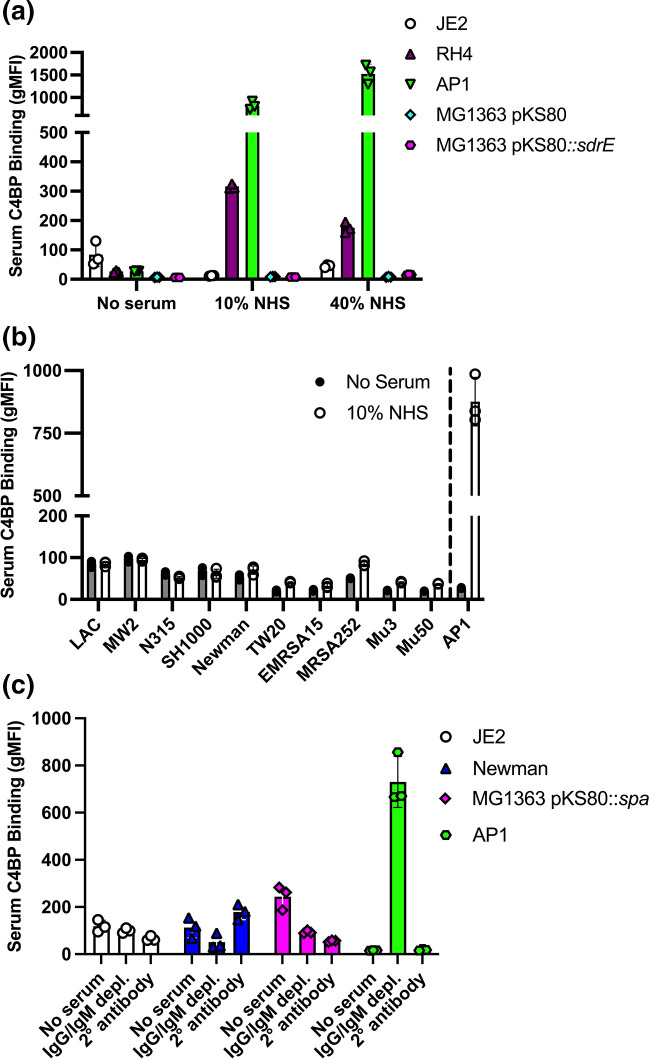
Our results indicated that protein A was responsible for mediating the recruitment of C4BP to the staphylococcal surface. We next investigated C4BP binding from human serum using WT S. aureus strain JE2 and L. lactis expressing protein A (Fig. 3a) . To limit protein A mediated non-specific binding, we generated F(ab)’2 antibodies of the murine anti-human C4BP antibody MK104. Importantly, we could not observe C4BP binding from human serum using JE2 or L. lactis expressing protein A (Fig. 3a) . As additional controls we used previously characterised C4BP binders Streptococcus pyogenes strain AP1 and Moraxella catarrhalis strain RH4, which showed significant binding in line with previous data [22, 23] (Fig. 3a). In addition, we screened ten different S. aureus reference strains representing six different clonal complex types for serum C4BP binding. Again, we did not observe significant C4BP binding between serum and non-serum control (Fig. 3b). To investigate further we examined C4BP binding from IgM/IgG depleted serum as antibodies in serum may have prevented C4BP interaction with protein A, however we observed no C4BP interaction using depleted serum but did observe significant C4BP binding using S. pyogenes strain AP1 (Fig. 3c).
Fig. 3.
S. aureus does not recruit C4BP when incubated in human serum. (a) C4BP recruitment to the bacterial surface was analysed following incubation of bacteria [ S. aureus (JE2), M. catarrhalis (RH4), S. pyogenes (AP1) and L. lactis MG1363 with pKS80 empty control or pKS80 expressing SdrE] in either 10 % or 40% normal human serum (NHS). (b) Ten different S. aureus reference strains [LAC (clonal complex, CC))8; MW2 (CC1); N315 (CC5); SH1000 (CC8); Newman (CC8); TW20 (CC239); EMRSA15 (CC22); MRSA252 (CC30); Mu3 (CC5); Mu50 (CC5)] were examined for serum C4BP binding in 10 % NHS. All strains tested except SH1000 carry the sdrE gene. S. pyogenes strain AP1 is shown as a reference for a known C4BP binder. (c) C4BP recruitment was assessed using IgM/IgG depleted serum and compared to no serum and secondary antibody only controls. (a–c) Bound C4BP was detected using F(ab’)2 murine anti-human C4BP MK104 and goat anti-mouse AF-488 secondary antibody. The geometric mean fluorescence intensity (gMFI) was measured using a BD FACS Canto flow cytometer. Bars indicate the mean; data points represent three biological replicates and error bars inform the standard deviation.
Plasma purified C4BP interaction with S. aureus occurred due to IgG co-purified with C4BP
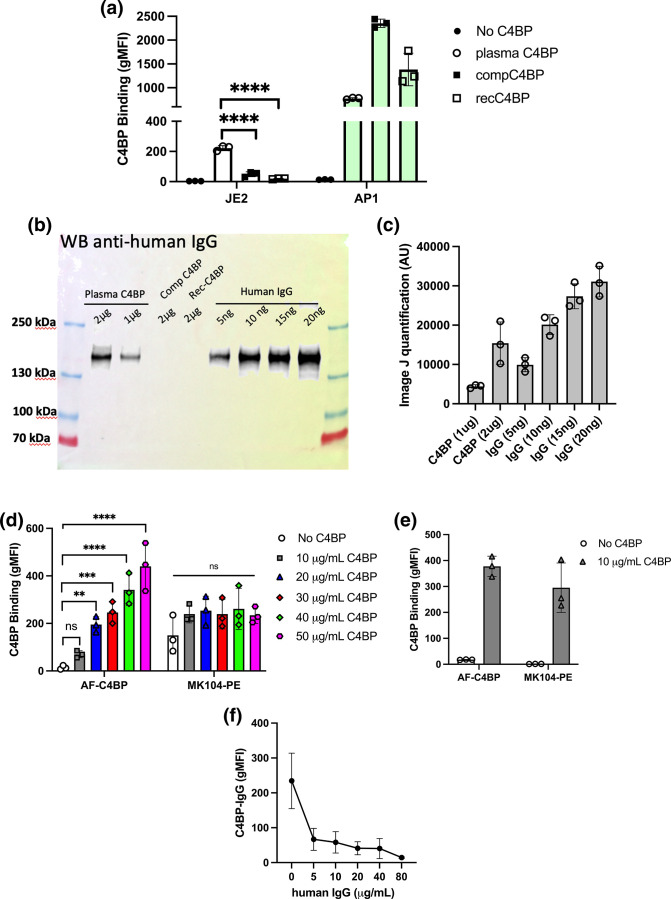
To understand why S. aureus could bind plasma purified C4BP but not C4BP present in human serum, we tested two other sources of C4BP; we used recombinantly produced C4BP (recC4BP) expressed and purified from human kidney 293 cells and C4BP obtained commercially (compC4BP). Both proteins were Dy-488 fluorescently labelled and the ability of S. aureus to bind to them was monitored using flow cytometry. Interestingly, S. aureus had significantly reduced binding to both recC4BP and compC4BP (Fig. 4a) compared to plasma C4BP preparation. As protein A was identified as the main staphylococcal C4BP binding protein, we tested if during preparation C4BP was in complex or co-purified with another protein A binding molecule, which led us to investigate IgG contamination. Using Western blotting and antibodies against human IgG we determined that our plasma purified C4BP was contaminated with human IgG whereas recombinantly produced and commercially obtained C4BP contained no traces of IgG (Figs 4b-c and S3). Image J quantification of anti-human IgG bands detected from our plasma purified C4BP compared to purified human IgG indicates 1 µg C4BP contains between 2.5–5 ng human IgG. To determine if IgG could form a complex with C4BP and mediate the recruitment of C4BP to the bacterial surface we determined the presence of labelled plasma purified C4BP on S. aureus using F(ab’)2 MK104 antibodies and a secondary antibody labelled with a non-overlapping phycoerythrin fluorophore (Fig. 4d). Increasing concentrations of Dy-488 labelled C4BP resulted in increased 488 fluorescence in a dose-dependent manner, however no increase in C4BP binding using the MK104-PE combination was observed and no statistically significant difference was observed between no C4BP control and 10–50 μg ml−1 C4BP. These results confirm that S. aureus was interacting solely with fluorophore labelled IgG that was co-purified with C4BP during plasma C4BP purification. As a control we tested S. pyogenes AP1 binding to 10 µg ml−1 Dy-488 labelled C4BP and observed binding both via Dy-488 and PE gating (Fig. 4e), indicating the presence of C4BP on the streptococcal surface in line with previous results.
Fig. 4.
Plasma purified C4BP interaction with S. aureus occurred via IgG co-purified with C4BP. (a) Dy488-labelled plasma purified C4BP (plasma C4BP), commercially obtained C4BP (compC4BP) and recombinantly produced C4BP (recC4BP) were incubated with S. aureus strain JE2 or S. pyogenes strain AP1. Binding was analysed by measuring the geometric mean fluorescence intensity (gMFI) using a BD FACS Canto flow cytometer. (b) Plasma C4BP (1 and 2 µg), compC4BP (2 µg) and recC4BP (2 µg) were probed for human IgG contamination using goat anti-human IgG-HRP. Purified human IgG at concentrations between 5–20 ng was used as a control C) Image J quantification of western blot analysis (n=3; Fig. S2). (d) Dy488-labelled plasma C4BP was incubated with S. aureus strain JE2 at concentrations between 0–50 µg ml−1. Fluorescence was either measured at 488 or following incubation of bacteria with F(ab’)2 MK104 mouse anti-C4BP antibodies and secondary goat anti-mouse – PE antibodies. (e) Dy488-labelled plasma C4BP (10 µg ml−1) was incubated with S. pyogenes strain AP1 and binding analysed as described in (d). (f) S. aureus strain JE2 was pre-incubated with increasing concentrations of human IgG. Dy488-labelled plasma purified C4BP (10 µg ml−1) binding was analysed by measuring the gMFI using a BD FACS Canto flow cytometer. Bars indicate the mean; data points represent three biological replicates and error bars inform the standard deviation. (a–d) Statistical differences were calculated using a one-way ANOVA analysis using Dunnett’s multiple comparisons test. **P<0.01, ***P<0.001 ****P<0.0001,.
Lastly, pre-incubation of S. aureus strain JE2 with human IgG prevented binding of IgG contaminated C4BP (C4BP-IgG) indicating that protein A mediated binding of C4BP-IgG was via protein A domains that recruit human IgG (Fig. 4f).
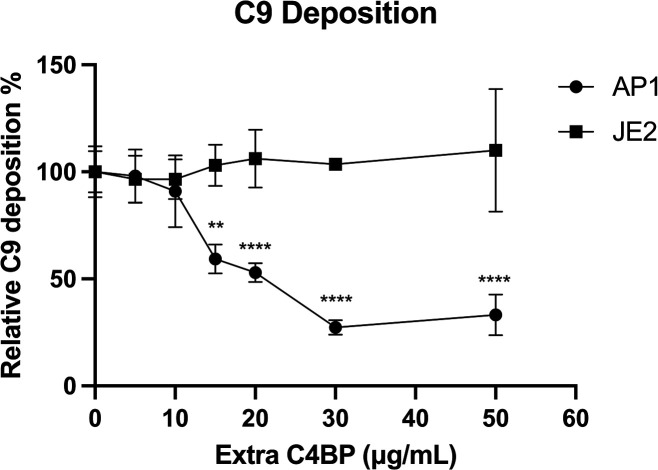
Pre-incubation of S. aureus with C4BP had no effect in limiting complement deposition
We tested the outcome of pre-incubating S. aureus or S. pyogenes with increasing concentrations of C4BP (containing no bound IgG) on complement deposition. S. pyogenes incubated with exogenously added C4BP concentrations above 15 µg ml−1 in human serum resulted in a statistically significant reduction of C9 deposition compared to S. aureus JE2 (Fig. 5). This data shows that C4BP was functional in reducing complement deposition on S. pyogenes when the serum was supplied. Noteworthy, under the same conditions we observed no difference in the level of C9 deposition on S. aureus when pre-incubated with concentrations of up to 50 µg ml−1 C4BP (Fig. 5).
Fig. 5.
Pre-incubation of S. aureus with C4BP had no effect in limiting complement deposition. S. aureus strain JE2 or S. pyogenes strain AP1 were pre-incubated with increasing concentrations of C4BP prior to incubation in 8 % NHS. C9 deposition was analysed using goat anti-human C9 antibody and rabbit-anti-goat AF-488 secondary antibody with the geometric mean fluorescence intensity (gMFI) using a BD FACS Canto flow cytometer. Symbols represent the mean of three biological replicates and error bars inform the standard deviation. Statistical differences were calculated using a two-way ANOVA using Sidak’s multiple comparison test. **P<0.01, ****P<0.0001,.
Discussion
S. aureus is a master of immune evasion, expressing multiple proteins that interfere with complement activity, antibody recognition, and immune cell recruitment, activation and killing mechanisms [24]. In this study we investigated whether S. aureus , like other human pathogens, utilizes the recruitment of the soluble complement regulator C4BP as an immune evasive defence mechanism. Initially our results indicated that S. aureus bound C4BP via protein A and not through the previously identified SdrE protein. However, we could not observe any C4BP binding from serum using a panel of diverse S. aureus strains but did observe significant binding of C4BP by pathogenic bacteria, S. pyogenes and M. catarrhalis , that have been previously identified as C4BP binders [22, 23], validating our flow cytometric, bacteria-protein binding experiments.
Protein A is a highly abundant cell-wall-anchored protein expressed by the vast majority of S. aureus isolates [25]. Protein A can capture IgG molecules via the Fc region, interrupting antibody-mediated phagocytosis [26] and preventing IgG hexamerization required for classical complement pathway activation [27]. We tested the ability of S. aureus to bind to recombinantly produced (rec-C4BP) and commercially purchased (compC4BP) C4BP, however we observed no interaction with S. aureus . This indicated that plasma purified C4BP may contain contaminants that have been fluorescently labelled along with the purified C4BP and are recognized by protein A. We tested the three sources of C4BP and found that plasma purified C4BP contained minute quantities of human IgG whereas recC4BP and compC4BP contained no detectable IgG. Previous work has shown that C4BP has several different binding partners including plasma, ECM and amyloid proteins [6], therefore we hypothesized that C4BP may form complexes with IgG in serum and S. aureus could recruit C4BP to the surface via interaction with bound IgG. Using antibodies that recognize labelled plasma purified C4BP we were able to determine that C4BP was not localized to the staphylococcal surface, and S. aureus was interacting solely with IgG that was co purified during C4BP purification and Dy-488 labelled along with C4BP. C4BP interacts with C4b which can be bound to immune complexes, therefore during C4BP purification very minor contamination of plasma purified C4BP with IgG/IgM and C4b may occur as observed in this study where IgG was detected at 2.5–5 ng per μg of C4BP. Combined, our results indicate that S. aureus does not recruit C4BP to the bacterial surface. Importantly, analysis of the cell surface proteome of S. aureus and its interaction with human serum proteins did not identify C4BP on the cell surface of S. aureus strain LAC or Newman [28].
Previous experiments to determine C4BP recruitment to the staphylococcal surface incubated S. aureus in human serum and then employed 2 % SDS to strip bacterial cells of surface proteins, determining the presence of C4BP through dot blot using monoclonal anti-C4BP antibodies [10, 11]. Using this approach, it was shown that significant C4BP binding was observed on the non-pathogenic L. lactis empty vector control, with binding of greater than 1 µg ml−1 C4BP reported [10]. This is surprising as to date, all confirmed C4BP binders are pathogenic microorganisms [8]; experiments that investigated the ability of endogenous microflora to recruit complement inhibitors showed no interaction [29], however, a thorough investigation using diverse microflora has not been reported. In addition, experiments using purified C4BP were not tested for IgG contamination and dot blot analysis was not carried out using F(ab)’2 generated antibodies and therefore the results may be distorted by the presence of protein A in the stripped cell extracts. Furthermore, human serum proteins have a high propensity for binding polymers such as polypropylene and polystyrene [28, 30], which form the basis of standard microcentrifuge tubes and microplates. Adsorption of proteins onto polymers is predominantly mediated via hydrophobic interactions [31]. Therefore, it is feasible that during the incubation of S. aureus with NHS, human serum proteins including C4BP can bind to the reaction vessels and remain following the washing stages. Treatment with SDS during the cell-stripping stage would release proteins including C4BP from the reaction vessel producing erroneous dot blot results informing that S. aureus binds C4BP. In contrast, our analysis described here uses highly sensitive flow cytometry of intact bacterial cells to determine C4BP recruitment, which would limit any binding artefacts that may arise following treatment with SDS and only report on bound protein on the bacterial surface.
Gaining a complete understanding of the intricate methods used by pathogens to manipulate immune responses is crucial for the design of future immunotherapeutics. Confirming whether microbial pathogens recruit soluble complement inhibitors is central for the development of chimeric fusion proteins designed to simultaneously disrupt the recruitment of complement inhibitors while activating complement on microbial surfaces, as we and others have recently developed as novel anti-infective immunotherapeutics [32–34]. Work presented in this study offers new insights into complement evasion strategies employed by S. aureus and may help design future intervention strategies to combat this human pathogen.
Supplementary Data
Funding information
This work was supported by an Academy of Medical Sciences Springboard Grant (ML; SBF006\1023), ESCMID research grant (ML) and a Swedish Research Council (AB).
Acknowledgements
We would like to thank Professor Foster (Trinity College Dublin) for providing anti-SdrE serum and Professor Riesbeck (Lund University) for providing the Moraxella catarrhalis strain.
Author contributions
M.L. conceived the study and wrote the original manuscript, S.L., F.M. and S.B. performed experiments and analysed data, A.M.B. and J.A.G. participated in writing the manuscript and supported the project
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Healthy volunteers provided written informed consent according to the recommendations of the University of Bath, Research Ethics Approval Committee for Health (reference: EP 18/19 108).
Footnotes
Abbreviations: C4BP, C4b-binding protein; FH, factor H; FI, Factor I; gMFI, geometric mean fluorescence intensity; MAC, membrane attack complex; NHS, normal human serum; Sdr, serine-aspartate repeat.
References
- 1.Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. 2019;17:203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foster TJ. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol Rev. 2017;41:430–449. doi: 10.1093/femsre/fux007. [DOI] [PubMed] [Google Scholar]
- 3.WHO Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. World Health Organization. 2017 [Google Scholar]
- 4.Merle NS, Church SE, Fremeaux-Bacchi V, Roumenina LT. Complement system part I - molecular mechanisms of activation and regulation. Front Immunol. 2015;6:262. doi: 10.3389/fimmu.2015.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noris M, Remuzzi G. Overview of complement activation and regulation. Semin Nephrol. 2013;33:479–492. doi: 10.1016/j.semnephrol.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ermert D, Blom AM. C4B-binding protein: the good, the bad and the deadly. Novel functions of an old friend. Immunol Lett. 2016;169:82–92. doi: 10.1016/j.imlet.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Ferreira VP, Pangburn MK, Cortés C. Complement control protein factor H: the good, the bad, and the inadequate. Mol Immunol. 2010;47:2187–2197. doi: 10.1016/j.molimm.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ermert D, Ram S, Laabei M. The hijackers guide to escaping complement: lessons learned from pathogens. Mol Immunol. 2019;114:49–61. doi: 10.1016/j.molimm.2019.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Tung H s, Guss B, Hellman U, Persson L, Rubin K, et al. A bone sialoprotein-binding protein from Staphylococcus aureus: a member of the staphylococcal Sdr family. Biochem J. 2000;345:611–619. [PMC free article] [PubMed] [Google Scholar]
- 10.Hair PS, Foley CK, Krishna NK, Nyalwidhe JO, Geoghegan JA, et al. Complement regulator C4BP binds to Staphylococcus aureus surface proteins SdrE and Bbp inhibiting bacterial opsonization and killing. Results Immunol. 2013;3:114–121. doi: 10.1016/j.rinim.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hair PS, Wagner SM, Friederich PT, Drake RR, Nyalwidhe JO, et al. Complement regulator C4BP binds to Staphylococcus aureus and decreases opsonization. Mol Immunol. 2012;50:253–261. doi: 10.1016/j.molimm.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Sabat A, Melles DC, Martirosian G, Grundmann H, van Belkum A, et al. Distribution of the serine-aspartate repeat protein-encoding sdr genes among nasal-carriage and invasive Staphylococcus aureus strains. J Clin Microbiol. 2006;44:1135–1138. doi: 10.1128/JCM.44.3.1135-1138.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Lv J, Qi X, Ding Y, Li D, et al. The carriage of the serine-aspartate repeat protein-encoding sdr genes among Staphylococcus aureus lineages. Braz J Infect Dis. 2015;19:498–502. doi: 10.1016/j.bjid.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy AJ, Lindsay JA. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol. 2010;10:173. doi: 10.1186/1471-2180-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio. 2013;4:e00537-00512. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien L, Kerrigan SW, Kaw G, Hogan M, Penadés J, et al. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Mol Microbiol. 2002;44:1033–1044. doi: 10.1046/j.1365-2958.2002.02935.x. [DOI] [PubMed] [Google Scholar]
- 17.Mohlin FC, Blom AM. Purification and functional characterization of C4b-binding protein (C4BP) Methods Mol Biol. 2014;1100:169–176. doi: 10.1007/978-1-62703-724-2_14. [DOI] [PubMed] [Google Scholar]
- 18.Blom AM, Webb J, Villoutreix BO, Dahlbäck B. A cluster of positively charged amino acids in the C4BP alpha-chain is crucial for C4b binding and factor I cofactor function. J Biol Chem. 1999;274:19237–19245. doi: 10.1074/jbc.274.27.19237. [DOI] [PubMed] [Google Scholar]
- 19.Härdig Y, Hillarp A, Dahlbäck B. The amino-terminal module of the C4b-binding protein alpha-chain is crucial for C4b binding and factor I-cofactor function. Biochem J. 1997;323:469–475. doi: 10.1042/bj3230469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 21.Geoghegan JA, Foster TJ. Cell wall-anchored surface proteins of Staphylococcus aureus: many proteins, multiple functions. Curr Top Microbiol Immunol. 2017;409:95–120. doi: 10.1007/82_2015_5002. [DOI] [PubMed] [Google Scholar]
- 22.Laabei M, Liu G, Ermert D, Lambris JD, Riesbeck K, et al. Short Leucine-rich proteoglycans modulate complement activity and increase killing of the respiratory pathogen Moraxella catarrhalis . J Immunol. 2018;201:2721–2730. doi: 10.4049/jimmunol.1800734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ermert D, Laabei M, Weckel A, Mörgelin M, Lundqvist M, et al. The molecular basis of human IgG-mediated enhancement of C4b-binding protein recruitment to group A Streptococcus . Front Immunol. 2019;10:1230. doi: 10.3389/fimmu.2019.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Jong NWM, van Kessel KPM, van Strijp JAG. Immune evasion by Staphylococcus aureus . Microbiol Spectr. 2019;7 doi: 10.1128/microbiolspec.gpp3-0061-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatlin CL, Pieper R, Huang S-T, Mongodin E, Gebregeorgis E, et al. Proteomic profiling of cell envelope-associated proteins from Staphylococcus aureus . Proteomics. 2006;6:1530–1549. doi: 10.1002/pmic.200500253. [DOI] [PubMed] [Google Scholar]
- 26.Peterson PK, Verhoef J, Sabath LD, Quie PG. Effect of protein A on staphylococcal opsonization. Infect Immun. 1977;15:760–764. doi: 10.1128/iai.15.3.760-764.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cruz AR, Boer M den, Strasser J, Zwarthoff SA, Beurskens FJ, et al. Staphylococcal protein A inhibits complement activation by interfering with IgG hexamer formation. Proc Natl Acad Sci U S A. 2021;118:e2016772118. doi: 10.1073/pnas.2016772118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dreisbach A, van der Kooi-Pol MM, Otto A, Gronau K, Bonarius HPJ, et al. Surface shaving as a versatile tool to profile global interactions between human serum proteins and the Staphylococcus aureus cell surface. Proteomics. 2011;11:2921–2930. doi: 10.1002/pmic.201100134. [DOI] [PubMed] [Google Scholar]
- 29.Nissilä E, Douillard FP, Ritari J, Paulin L, Järvinen HM, et al. Genotypic and phenotypic diversity of Lactobacillus rhamnosus clinical isolates, their comparison with strain GG and their recognition by complement system. PLoS One. 2017;12:e0181292. doi: 10.1371/journal.pone.0181292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Husby S, Holmskov-Nielsen U, Jensenius JC, Erb K. Increased non-specific binding of heat-treated proteins to plastic surfaces analyzed by ELISA and HPLC-fractionation. J Immunoassay. 1985;6:95–110. doi: 10.1080/01971528508063023. [DOI] [PubMed] [Google Scholar]
- 31.Weikart CM, Breeland AP, Taha AH, Maurer BR. Enhanced recovery of low concentration protein and peptide solutions on ultra-low binding microplates. Future Sci OA. 2019;5:FSO367. doi: 10.4155/fsoa-2018-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bettoni S, Shaughnessy J, Maziarz K, Ermert D, Gulati S, et al. C4BP-IgM protein as a therapeutic approach to treat Neisseria gonorrhoeae infections. JCI Insight. 2019;4:e131886. doi: 10.1172/jci.insight.131886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laabei M, Colineau L, Bettoni S, Maziarz K, Ermert D, et al. Antibacterial fusion proteins enhance Moraxella catarrhalis killing. Front Immunol. 2020;11:2122. doi: 10.3389/fimmu.2020.02122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaughnessy J, Chabeda A, Tran Y, Zheng B, Nowak N, et al. An optimized Factor H-Fc fusion protein against multidrug-resistant Neisseria gonorrhoeae . Front Immunol. 2022;13:975676. doi: 10.3389/fimmu.2022.975676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.